Abstract
Mapping-by-sequencing (or SHOREmapping) has revitalized the powerful concept of forward genetic screens in plants. However, as in conventional genetic mapping approaches, mapping-by-sequencing requires phenotyping of mapping populations established from crosses between two diverged accessions. In addition to the segregation of the focal phenotype, this introduces natural phenotypic variation, which can interfere with the recognition of quantitative phenotypes. Here, we demonstrate how mapping-by-sequencing and candidate gene identification can be performed within the same genetic background using only mutagen-induced changes as segregating markers. Using a previously unknown suppressor of mutants of like heterochromatin protein1 (lhp1), which in its functional form is involved in chromatin-mediated gene repression, we identified three closely linked ethyl methanesulfonate-induced changes as putative candidates. In order to assess allele frequency differences between such closely linked mutations, we introduced deep candidate resequencing using the new Ion Torrent Personal Genome Machine sequencing platform to our mutant identification pipeline and thereby reduced the number of causal candidate mutations to only one. Genetic analysis of two independent additional alleles confirmed that this mutation was causal for the suppression of lhp1.
In Arabidopsis (Arabidopsis thaliana) research, ethyl methanesulfonate (EMS) mutagenesis is a powerful tool that has been widely explored to uncover the functionality of many genes in a broad spectrum of pathways (Page and Grossniklaus, 2002). Recent advances in sequencing technology have greatly reduced the time required to pinpoint induced mutations. In a proof-of-principle experiment, mapping-by-sequencing (SHOREmapping) was first demonstrated on a mutant in the background of the Arabidopsis reference accession Columbia (Col-0) crossed to the diverged accession Landsberg erecta. A pool of DNA isolated from bulked segregants was sequenced and used for the simultaneous mapping and mutant identification (Schneeberger et al., 2009b). This first application was followed by other studies successfully applying similar methods (Cuperus et al., 2010; Austin et al., 2011).
Although all described approaches are straightforward and extremely fast, their application is hindered by the requirement for interaccession crosses that impedes the success rate of screens based on quantitative traits, such as screens for genetic modifiers. The major obstacle is that the considerable phenotypic variation in F2 populations from crosses between diverged accessions impairs the recognition of mutants with subtle phenotypic alterations. In addition, if genetic screens involve modifiers of a preexisting mutant, the mapping depends on the availability of the primary mutant in another suitable accession, the introgression of the mutation in such a background, or the laborious additional genotyping for the presence of the first-site mutation.
Avoiding these disadvantages, Ashelford et al. (2011) have demonstrated that the isolation of a causative EMS-induced change is possible by direct resequencing of a complete mutant genome. However, their approach initially resulted in 103 putative causal mutations that had the potential to change the amino acid sequences of 48 putative proteins. In addition, the mutations were clustered in two separate regions of the genome, even though the mutant had been backcrossed four times to the parental line.
Recently, Abe et al. (2012) reduced the large number of candidate mutations by backcrossing mutant genomes to their nonmutagenized progenitor, followed by sequencing bulk segregants from these crosses. This drastically reduced the number of causal candidates, although it was not possible to pinpoint the causal change from the sequencing data alone. The main problem remains the short-read coverage at each of the candidate mutations, which is typically lower than the number of individuals combined within the bulked DNA. This hinders accurate allele frequency estimations based on the whole-genome sequencing data alone and thus makes it impossible to distinguish between causal and closely linked mutations.
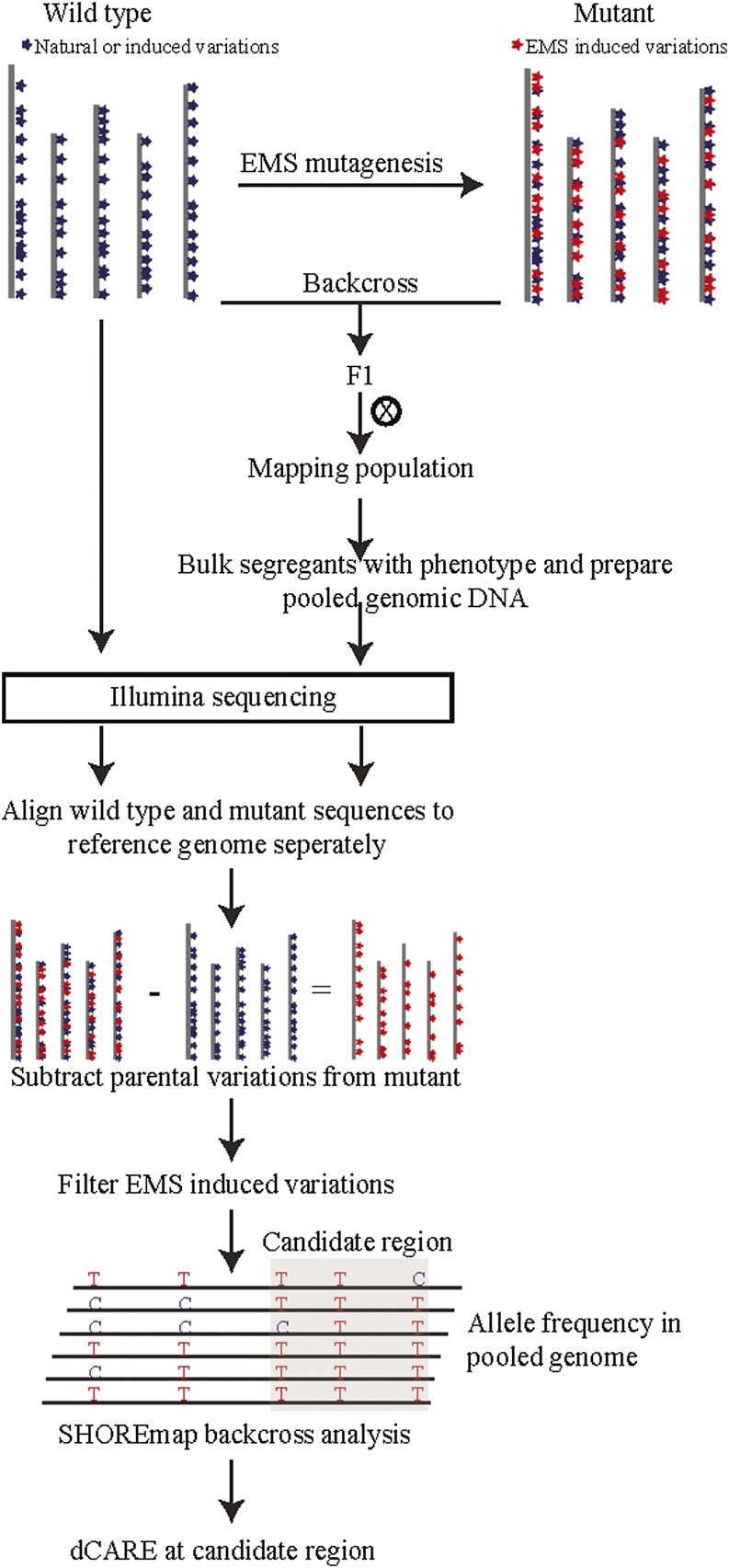
In this study, we combined isogenic bulk segregant analysis with deep candidate resequencing (dCARE) to facilitate the mutation identification of genetic modifiers based on bulked DNA and sequencing data alone. Our approach relies on the assumption that in pools of bulked segregants, the causative change occurs with the highest frequency among all EMS-induced changes (Fig. 1). Using resequencing data alone, it is not possible to distinguish between the subtle allele frequencies of EMS changes that are closely linked. However, dCARE of all candidate mutations using the new Ion Torrent sequencing technology enables quick and cost-effective detection of subtle allele frequency differences between closely linked mutations and thus allows the identification of causal candidates.
Figure 1.
Schematic illustration of the fast isogenic mapping approach. Chemical mutagens typically introduce hundreds of novel mutations. Within the M2 generation, mutants are screened for phenotypes. Selected plants are backcrossed to the nonmutagenized progenitor. The F2 offspring of such a cross forms an isogenic mapping population, as only novel mutations are segregating. Backcrossed individuals that display the mutant phenotype are selected, bulked, and their DNA is prepared as a pool and whole-genome sequenced. If the parental line is genetically different from the reference line Col-0, it needs to be resequenced in order to control for naturally occurring differences that need to be differentiated from novel mutations. Thus, all novel EMS-induced mutations can be selected for SHOREmap analysis by filtering for mutations that do not reside in the parental line. Candidate mutations (gray box) that show high mutant allele frequencies and linkage are selected for dCARE to pinpoint the causal mutation.
The mutant identified by this fast isogenic mapping approach was isolated as a suppressor of developmental aberrations caused by defects in LIKE HETEROCHROMATIN PROTEIN1 (LHP1), which participates in the Polycomb Group (PcG) gene regulatory pathway in Arabidopsis. Enhancer/suppressor screens have been successfully used to identify genes that play a role in chromatin-mediated gene repression and activation in Drosophila melanogaster. For example, many components of the repressive PcG pathway were isolated as genetic enhancers or suppressors of homeotic mutations, whereas components of the Trithorax Group protein pathway were originally identified as suppressors of PcG-related mutations (Landecker et al., 1994; Gildea et al., 2000; Alonso et al., 2007).
RESULTS
Mutant Selection for Fast Isogenic Mapping-by-Sequencing
We selected an EMS-induced mutant that was isolated as a suppressor of the lhp1 mutant phenotype to validate the isogenic mapping approach. LHP1 is part of one or several distinct POLYCOMB REPRESSIVE COMPLEXES1 (PRC1s) in Arabidopsis (Xu and Shen, 2008; Bratzel et al., 2010). PRC1s are targeted to nucleosomes that carry Lys-27 trimethylated H3 (H3K27me3), and the chromodomain of LHP1 directly binds H3K27me3 (Turck et al., 2007; Zhang et al., 2007; Exner et al., 2009).
Mutated lhp1 plants display a pleiotropic phenotype. They are shorter, have smaller, downwardly curled leaves, flower earlier than the wild type independent of daylength, and form terminal flowers (Kotake et al., 2003). However, the phenotype is relatively mild if compared with that of mutants that have globally reduced H3K27me3 levels or are severely impaired in their PRC1 function. In these mutant plants, developmental structures are not maintained, resulting in a callus-like growth phenotype (Makarevich et al., 2006; Bratzel et al., 2010).
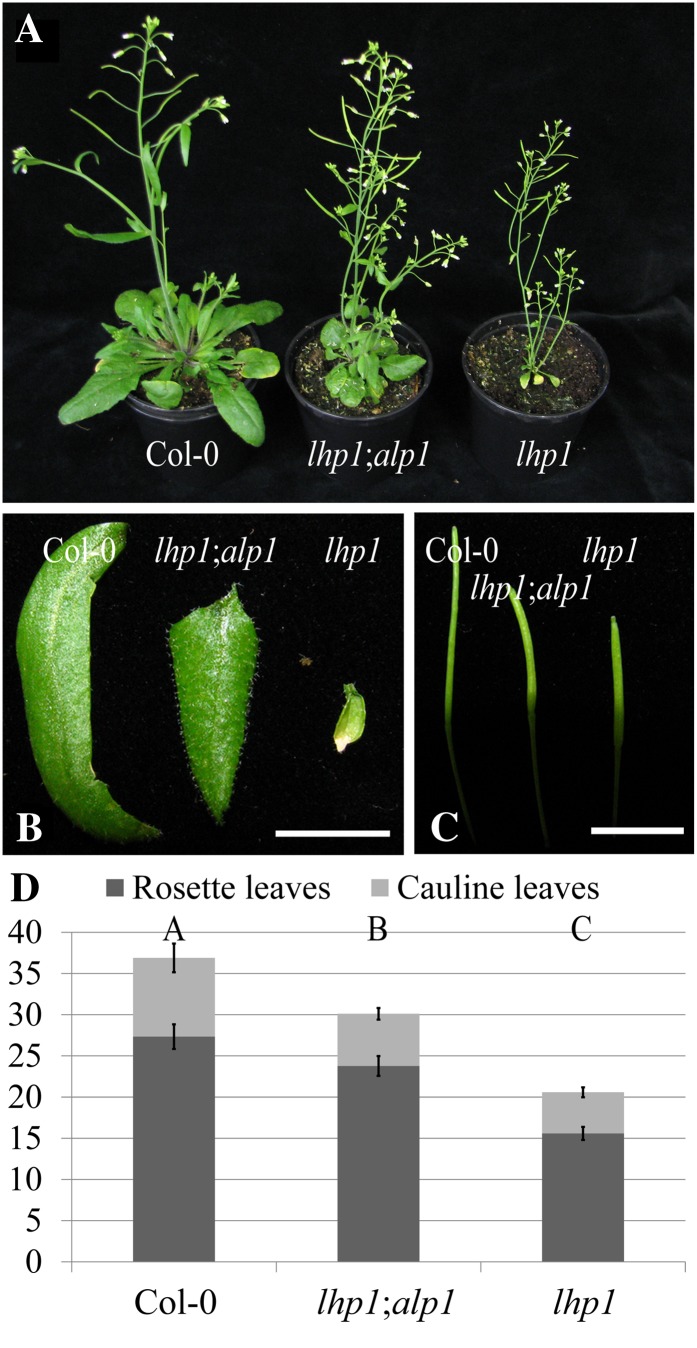
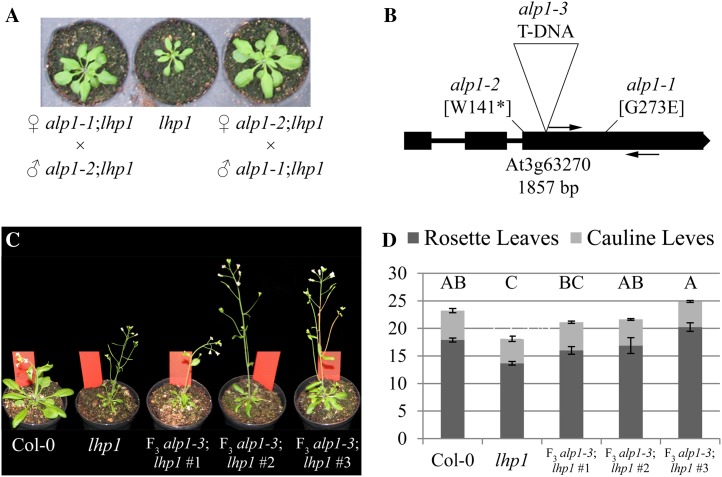
We performed an EMS mutagenesis of the lhp1-3 (alternative name, terminal flower2-2; Larsson et al., 1998) mutant to perform a large forward genetic screen for genetic suppressors and enhancers of lhp1. Among other mutants, the screen led to the isolation of the EMS-induced antagonist of lhp1-1 (alp1);lhp1 double mutant as a suppressor of the lhp1 phenotype. Height, cauline and rosette leaf size, and silique length were increased in alp1;lhp1 compared with the lhp1 single mutant, resulting in an intermediate phenotype between the wild type Col-0 reference and the lhp1 mutant (Fig. 2, A–C).
Figure 2.
Phenotype comparisons. A to C, Col-0 wild-type, alp1;lhp1 double mutant, and lhp1 single mutant plants 36 d after germination and growth in climate chamber conditions (12 h of light/16°C, 12 h of dark/14°C). One representative example for each genotype is shown as whole plant (A), the third oldest cauline leaf of the main shoot (B), and the seventh youngest silique of the main shoot (C). Bars = 1 cm. D, Flowering-time analysis. Plants grown as in A to C were scored when the main shoot had bolted to about 1-cm height. The leaf number is indicated on the y axis. Error bars represent se (n = 9). Statistical significance was evaluated by single-factor ANOVA followed by an honestly significant difference Tukey test. Letters above the bars indicate significantly different groups (P < 0.05). [See online article for color version of this figure.]
Double-mutant plants flowered earlier than Col-0 wild-type plants but later than lhp1 mutants in our screening conditions (Fig. 2D). Early flowering of lhp1 mutant plants is caused by the up-regulation of FLOWERING LOCUS T (FT) expression (Kotake et al., 2003). Obvious candidates for suppressors of the early-flowering phenotype of lhp1 are, apart from mutations in FT, mutations in the autonomous pathway. Autonomous pathway mutations result in a strong up-regulation of the floral repressor FLOWERING LOCUS C (FLC) that directly targets and represses FT (Simpson, 2004; Searle et al., 2006; Jiang et al., 2008). FLC levels are also moderately increased in the PcG pathway mutants curly leaf (clf) and lhp1 (Mylne et al., 2006; Jiang et al., 2008). However, this moderate increase is not able to suppress FT up-regulation caused by the loss of PcG-mediated repression (Kotake et al., 2003; Farrona et al., 2011). FLC levels were not dramatically increased and FT levels were not significantly altered between alp1;lhp1 and lhp1 compared with other suppressor mutations that were likely to be affected in the autonomous pathway (Supplemental Fig. S1). In contrast to alp1;lhp1 plants, the leaf size is not increased in ft;lhp1 double mutants, which made it also unlikely that the suppression was caused by a mutation in FT. No differences in flower morphology could be observed when flowers of lhp1 and the double mutant were compared with each other (Supplemental Fig. S2).
The pleiotropic phenotype of lhp1 mutant plants differs quantitatively between accessions such as Col-0 and Wassilewskija-2, making it difficult to create a robust mapping population for subtle modifiers (Supplemental Fig. S3). Progeny of an alp1;lhp1 cross to the original lhp1 allele segregated with a 3:1 ratio for the suppressor phenotype in the F2 generation, indicating that a single mutation was responsible for the suppression. One of the F2 plants with a suppressor phenotype was randomly picked and backcrossed a second time to lhp1 and gave rise to another F2 generation (BC2F2).
Fast Isogenic Mapping-by-Sequencing Reveals Candidate Mutations
Leaf samples of 270 BC2F2 alp1;lhp1 plants were pooled. DNA prepared from the pooled material was sequenced on a single lane of an Illumina Genome Analyzer IIx. In parallel, DNA of 48 pooled lhp1 single mutant leaves from the parental line was sequenced as a reference on a separate sequencing reaction. The parental line had been generated from an EMS mutagenesis in the background of Col-0 and had been backcrossed to the parental Col-0 for an unknown number of times (Larsson et al., 1998).
Out of 43.4 and 42.2 million high-quality reads, 93% and 94% aligned to the reference sequence and yielded an average nucleic genome coverage of 41- and 49-fold for lhp1 and alp1;lhp1, respectively (Supplemental Table S1). Differences between the reference sequence and both sequence sets were independently identified with SHORE (Ossowski et al., 2008; see “Materials and Methods”). Within the resequencing data of the BC2F2 alp1;lhp1, short-read analysis was performed to identify all mutations with an allele frequency higher than 20%, in order to identify fixed as well as nonfixed EMS mutations segregating in the pool (see “Materials and Methods”). By removing all sequence differences that had their origin in the lhp1 genome from the alp1;lhp1 sequence, we defined a set of 852 novel EMS changes (G/C:A/T) that were specific for the BC2F2 alp1;lhp1 pool.
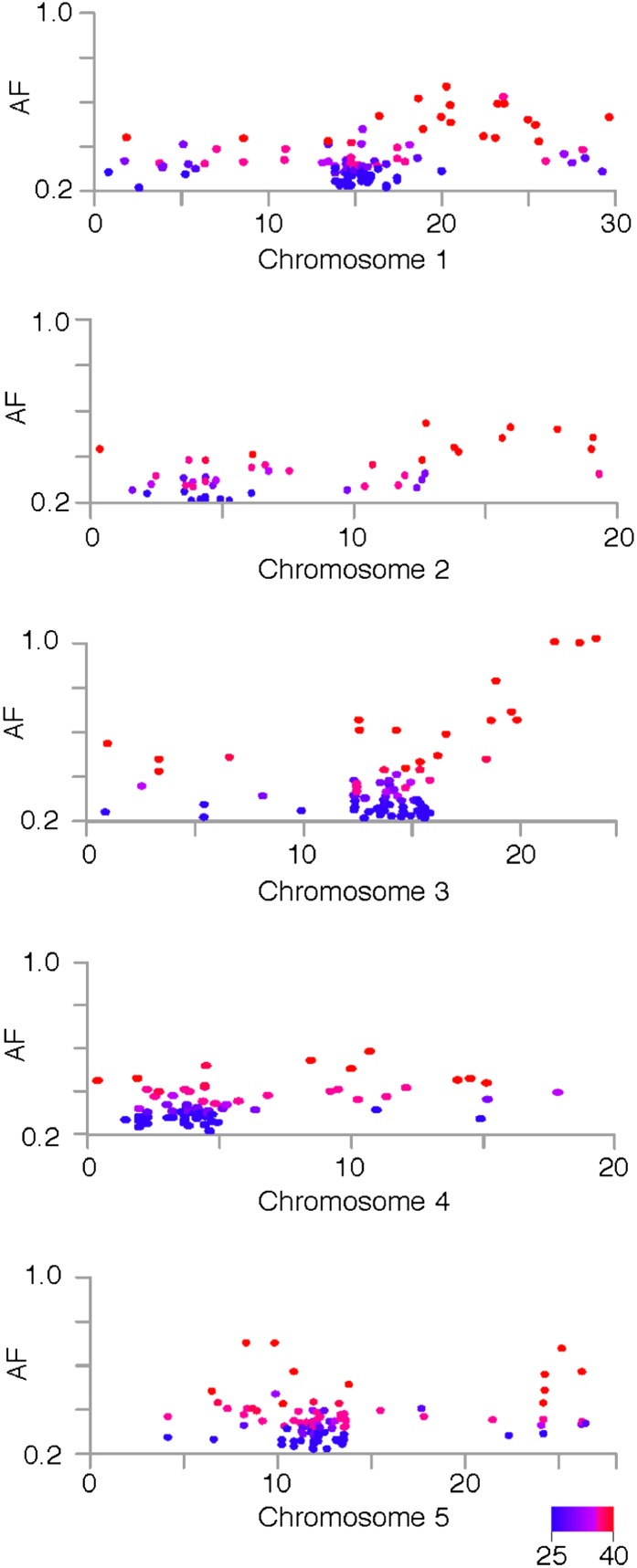
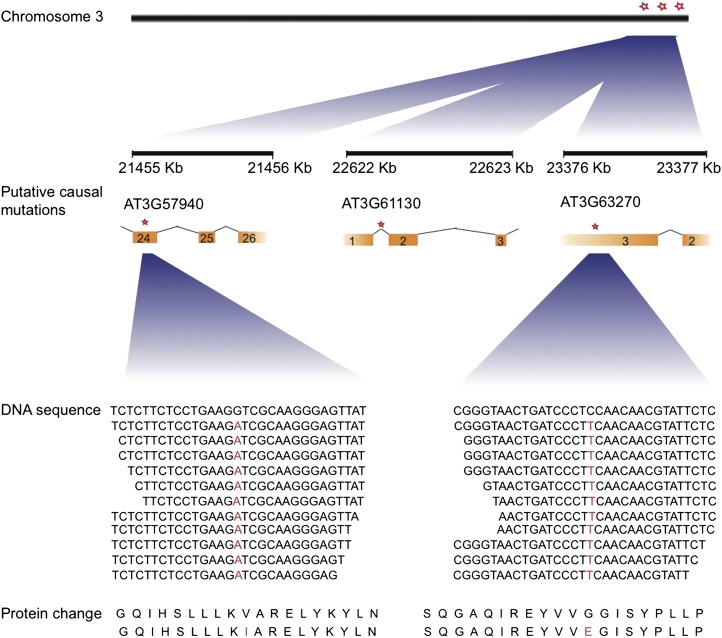
Using SHOREmap to visualize the allele frequency estimations at the mutant loci, selection for the lower arm of chromosome 3 became apparent through an allele frequency distortion in this region (Fig. 3; see “Materials and Methods”). Out of three EMS mutations that had a mutant allele frequency higher than 80%, two were found to be located in exons of At3g57940 and At3g63270 and one in an intron of At3g61130. The first two mutations caused missense mutations leading to amino acid changes of Val→Ile and Gly→Glu, respectively (Fig. 4; Supplemental Table S2).
Figure 3.
Allele frequency estimations at EMS changes. Allele frequency estimations at EMS-induced mutations of alp1;lhp1 across all five chromosomes are shown (Mb; x axis). Allele frequencies (AF; y axis) were estimated as fractions of short reads supporting the mutant allele divided by the number of all reads aligning to a given marker. The color indicates the resequencing consensus (SHORE) score, and only base calls with a quality score of more than 25 have been considered. The long arm of chromosome 3 was found to be under selection, as local allele frequencies appeared higher as compared with other regions in the genome.
Figure 4.
Annotation of putative causal mutations. The genomic regions of candidate EMS mutations (red asterisks) along with gene annotations are shown. Orange boxes indicate exons. Locations of EMS mutations that have putative effects on amino acid sequences are shown in red letters; for clarity, the DNA sequences in the graph do not reflect the actual number of reads at these locations (coverage was 50-fold).
dCARE Identifies Causal Change
Nearly complete linkage between the three candidate mutations was apparent in the pooled DNA, even though the mutations were spaced over 2 Mb apart. Based on Arabidopsis genetic maps, this physical distance corresponds to approximately 7 to 8 centimorgan, suggesting that several recombination events between these mutations are expected in a pool of 270 recombinants (Giraut et al., 2011). Our analysis of the raw reads covering the three mutations revealed two Col-0 wild-type reads for the mutation in At3g57940 as well as for the intronic change in At3G61130 but only one wild-type read for the mutation in At3g63270 (Supplemental Table S3). Although the mutation in At3g63270 could therefore act as main candidate, the disparity was too minor to reliably exclude the other mutations. This is a sampling problem, as usually the number of individuals pooled in bulk segregant analyses is considerably larger than the read coverage, which, therefore, is not powerful enough to resolve the real allele frequency accurately. However, an increased number of short-read alignments at the mutations would help to resolve the real allele frequency of the mutant allele in the bulked DNA much more precisely (Supplemental Fig. S4). In order to generate more sequencing data for the mutated regions, we amplified regions across the mutations by PCR using the pooled DNA from bulked segregants as template and sequenced the amplicons with the Ion Torrent Personal Genome Machine (PGM; Rothberg et al., 2011). This dCARE analysis generated 20,111, 4,390, and 19,203 reads across the changes affecting At3g57940, At3g61130, and At3g63270, respectively. For the changes in At3g57940 and At3g61130, we found 5.7% and 2.1% reads not supporting the mutant allele, whereas only 0.45% of the reads at At3g63270 supported the wild-type allele.
The presence of Col-0 wild-type reads at all candidate mutations can be explained by contamination of the segregant bulk, possibly due to misscoring of mutants or by sequencing errors that occur at a low rate. Both types of error affect mutations independently of their linkage to the causative change and represent background noise. In fact, the rate of nonmutant alleles at At3g63270 is even slightly lower than the rate of sequencing errors reported for Ion Torrent PGM sequencing (Rothberg et al., 2011). As a consequence, we could not reliably identify any wild-type alleles for the mutation affecting At3g63270, whereas the wild-type allele was clearly apparent for both linked mutations (Supplemental Table S3). Thus, dCARE reduced the list of candidates to At3g63270.
Validation of the Mutation Causing the Phenotype
A second suppressor mutant of lhp1, also identified in our forward genetic screen, displayed a phenotype similar to the alp1;lhp1 double mutant. Reciprocal crosses between the two suppressor mutants showed that they were likely allelic to each other, since all F1 individuals of reciprocal crosses looked like alp1;lhp1 (Fig. 5A). The three candidate loci analyzed by dCARE were sequenced in the second suppressor and in a single alp1;lhp1 M3 plant. We could confirm all mutations in alp1;lhp1, but in the second suppressor, only At3g63270 was disrupted by a G-to-A change leading to a premature stop codon (Fig. 5B). We designated the allele underlying the original suppressor mutation as alp1-1 and the allele with the internal stop codon as alp1-2. A third allele, alp1-3, was caused by a transferred DNA (T-DNA) insertion from an enhancer trap line that disrupted the third exon of ALP1 (ET1398; http://genetrap.cshl.edu). The alp1-3 allele was in the Landsberg erecta background, and the F2 generation from a cross between alp1-3 and lhp1 showed a range of suppression phenotypes of lhp1. Therefore, we scored three F3 families that were homozygous for alp1-3;lhp1 and compared their flowering time with that of lhp1 and wild-type Col-0 (Fig. 5, C and D). The data confirmed that alp1-3 suppressed the early flowering of lhp1 and increased the leaf size to a value that was intermediate between lhp1 and wild-type Col-0.
Figure 5.
Validation of the causal mutation. A, Complementation test of two EMS-induced alleles of alp1. F1 progeny were from reciprocal crosses of two suppressors of lhp1 with similar phenotypes (alp1-1;lhp1 and alp1-2;lhp1). Single lhp1 mutants are shown as references as indicated. B, EMS-induced changes at ALP1 and point T-DNA insertion of alp1-3. In addition to the mutations identified in alp1-1;lhp1 by whole-genome sequencing, a point mutation leading to a premature stop codon was found in alp1-2;lhp1. Arrows indicate the positions of PCR primers used for dCARE. C and D, Analysis of the T-DNA insertion allele alp1-3. C, Representatives of three double homozygous F3 families derived from the cross of alp1-3 with lhp1. D, Flowering-time measurements in three double homozygous alp1-3;lhp1 F3 families as in C. Plants were grown in long days in Percival cabinet conditions (16 h of light/22°C, 8 h of dark/20°C). The y axis shows leaf number until bolting for rosette and cauline leaves. Error bars represent se. Statistical significance was evaluated by single-factor ANOVA followed by an honestly significant difference Tukey test. Letters above the bars indicate significantly different groups (P < 0.05). [See online article for color version of this figure.]
ALP1 Is Related to Harbinger-Like Transposases
ALP1 encodes a gene related to Harbinger-like transposases. Harbinger transposases belong to the P Instability Factor superfamily of transposases (Walker et al., 1997) and code for a transposase as well as an accessory protein with a potential DNA-binding Myb/SANT domain. In particular, ALP1 encodes the transposase component, which features an endonuclease domain of the DDE-4 superfamily (position-specific scoring matrix id cl15789, e-value 3.54e-19 in a National Center for Biotechnology Information [NCBI] Conserved Domain Database [CDD] search; Marchler-Bauer et al., 2011). These endonucleases contain a catalytic triad of three acidic amino acid residues (DDE) that coordinate metal ions needed for catalysis (Yuan and Wessler, 2011). An N-terminal helix-turn-helix domain between amino acids 110 and 141 of ALP1 is supported by the NCBI CDD (position-specific scoring matrix id cl00088, e-value 5.05e-3).
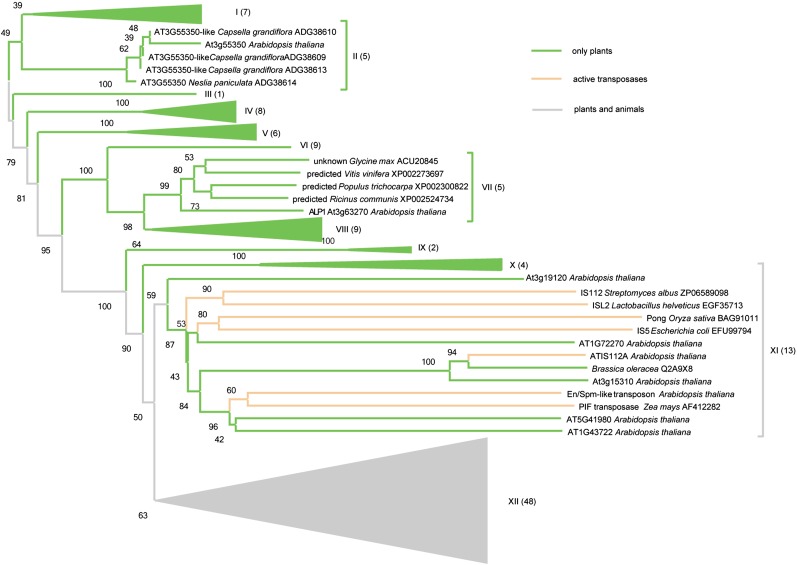
To evaluate if ALP1 was an active transposon showing expansion in the Arabidopsis genome, we compared ALP1 with its closest homologs available from a GenBank BLAST search using unique protein sequences from all species. ClustalW sequence alignment and calculation of a neighbor-joining tree showed that ALP1 amino acid sequence did not cluster together with the seven other Harbinger-like genes from Arabidopsis or with an outgroup of functional Harbinger-related transposases such as IS5 from bacteria (Fig. 6). The ALP1 clade included four additional plant proteins of unknown function from soybean (Glycine max), poplar (Populus spp.), grapevine (Vitis vinifera), and castor bean (Ricinus communis), whereas the closest Arabidopsis homolog, At3g55350, was present in a distinct branch of the tree. The data show that ALP1 is encoded by a single-copy gene in Arabidopsis and is found in different plant families. Notably, the helix-turn-helix domain, which could represent a DNA-binding motif, was shared within the ALP1 clade but was not detected in any of the other Arabidopsis homologs of ALP1 by a CCD search.
Figure 6.
Phlyogenetic analysis of ALP1. Nine of the 12 subclades of the neighbor-joining tree have been collapsed to emphasize the ALP1 (VII), At3g55350 (II), and IS5 (XI) subclades. The number of proteins within collapsed clades is indicated in brackets. The tree is drawn with branch lengths linear to the evolutionary distances used to infer the phylogenetic tree. Four unknown proteins from different plant species cluster with ALP1 in subclade VII. ALP1 and its closest homolog At3g55350 were assigned to two distinct clades. The XI outgroup contains active transposons from bacteria and plants, four of the seven ALP1 Arabidopsis homologs, and an additional Arabidopsis protein that is most closely related to the transposase encoded by ATIS112A, an annotated Harbinger transposon from Arabidopsis. Clade XII contains 48 proteins from plants and animals, including the human HARB1 protein. [See online article for color version of this figure.]
The alignment of the ALP1 clade with At3g55350 and HARBI1 from human (Homo sapiens) and zebra fish (Danio rerio) provided evidence that the acidic triad with the conserved amino acid residues DDE is disrupted in all members of the ALP1 clade (Supplemental Fig. S5). As the DDE triad is required for catalysis, it is likely that members of the ALP1 clade have lost their endonuclease activity.
ALP1 is an expressed gene that is not directly regulated by LHP1 and the PcG pathway (Supplemental Fig. S6, A and B). Expression levels were not altered in lhp1 seedlings compared with the wild type, and the epigenetic landscape of ALP1 was free of H3K27me3 and LHP1 (Zhang et al., 2007; Farrona et al., 2011).
Thus, we hypothesize that ALP1 is derived from an ancient Harbinger transposon but seems to have acquired a plant-specific function over time.
DISCUSSION
Conventional genetic mapping requires outcrossing to a diverged accession for the establishment of a mapping population. However, differences in phenotypes that segregate between Arabidopsis accessions are likely to mask subtle phenotypes caused by mutations. We have bypassed this problem by backcrossing an EMS-induced double mutant plant to its single mutant parent, generating an isogenic mapping population. Consequently, conventional markers are absent in the population and cannot be used to distinguish parental alleles. However, as we performed whole-genome sequencing, it was possible to identify mutagen-induced changes and to use them as markers, as these are only specific to the mutant genome and are absent in the original genome. This allowed scoring of a mapping population for the mutant phenotype only using the original genetic background. This method opens possibilities for the identification of subtle phenotypes that were previously inaccessible.
In addition, fast isogenic mapping-by-sequencing saves a large amount of time and labor in comparison with classical mapping approaches (Abe et al., 2012). After sequencing, data analysis to produce putative candidate genes will take only 1 d using automated pipelines, like the one provided for download with this report (http://shoremap.org).
Whole-genome sequencing of pooled DNA from bulked segregants usually does not allow for a unique identification of the causal change but results in a list of linked candidate changes. Mutations that are closely linked with the causal mutation are only influenced by a minor number of recombinations, and the coverage of whole-genome resequencing does not allow distinguishing between homozygous and nearly homozygous changes. If the complement of pooled segregants is likely to introduce a low rate of recombination between closely linked candidate mutations, noncausative mutations can be excluded by a quantitative detection of rare wild-type alleles. Introducing dCARE to the mapping pipeline allowed us to drastically increase the coverage for linked changes, which reduced the list of candidates to one (causal) change, with comparably little additional effort.
The number of segregants required to unambiguously identify a single mutation as the main candidate depends on various factors, including EMS load, recombination frequency, and the error rate in scoring the phenotype. The detected load of EMS mutation in alp1;lhp1 was low and helped in reducing the number of candidate genes to three, but the dCARE could have been easily extended to more sequence differences at very low additional cost. A low mutation rate harbors other drawbacks, as it reduces the number of mutants identified in an EMS screen. In particular, the availability of a second allele with the same phenotype from the screen proved to be a big advantage in confirming the resequencing results (Fig. 5).
ALP1 is related to type II double-stranded DNA d (dsDNA) transposases, which are the most abundant and possibly most essential elements for evolution in viral, bacterial, and eukaryotic genomes (Aziz et al., 2010). They can fulfill essential functions for an organism, such as DNA processing (Nowacki et al., 2009). The catalytic acidic triad DDE that was found to be disrupted in ALP1 is characteristic of the transposase/integrase supergroup and is essential in coordinating metal ions involved in the “cut-and-paste” mechanism of double-stranded DNA transposases (Craig, 2002; Casola et al., 2007; Yuan and Wessler, 2011). The fact that functionally relevant amino acid residues were not conserved in the ALP1 protein supports our hypothesis that the protein does not function as an active transposase. Of seven homologs of ALP1 in Arabidopsis, four clustered together with active transposases in bacteria in a neighbor-joining analysis, whereas the others were present in distinct clades of the tree. These clades contained only plant proteins from other species, suggesting coexistence between active and inactive Harbingers in Arabidopsis (Fig. 6). ALP1 might be able to bind DNA through one or two helix-turn-helix motifs toward the N-terminal end of the protein (Iwahara et al., 1998). This function could be exclusive for the ALP1 clade, since a CDD search did not detect the same domain structure in the other Arabidopsis homologs.
In conclusion, ALP1 is an actively transcribed gene that is related to Harbinger transposases but is likely to have lost its ability to transpose. To reveal the function of ALP1 in detail, and in particular to elucidate its interaction with the lhp1 mutation, remains a challenging task for the future.
MATERIALS AND METHODS
Treatment of Seeds
Germination rates of lhp1 and wild-type seeds were scored on 1/2 Murashige and Skoog growth medium plates after 10 long days at 22°C in a Percival plant growth chamber (CLF Plant Climatics). For EMS treatment, 200 mg of seeds was wrapped into Miracloth and imbibed on a shaker at 4°C in 0.1% KCl solution for 14 h. Seeds were then washed with distilled water and treated with 100 mL of 30 mm EMS diluted in distilled water on a magnetic stirrer for 12 h.
Two washing steps with 100 mL of 100 mm sodium thiosulfate for 15 min and three washing steps with 500 mL of deionized water for 30 min followed. After washing, seeds were equally divided into five bottles containing 500 mL of 0.1% Universal Agarose (Bio-Budget Technologies). Seeds were sown in 7.5-mL aliquots onto 9- × 9-cm pots using plastic pipettes.
Selection of Potential Mutants
M1 plants were bagged in small bulks averaging three plants. For each M1 plant, 10 M2 seeds were sown onto 9- × 9-cm pots in four rows of five plants each. Potential mutants were primarily screened in short-day conditions and grown together with lhp1 (Col-0), also termed terminal flower2-2 (Larsson et al., 1998), lhp1;clf (Salk 006658; Col-0), and lhp1 (Col-0);emf2-10 (Wassilewskija-2; Chanvivattana et al., 2004) mutants and Col-0 wild-type plants as controls.
Bulks with potential mutants were rescreened in a Percival chamber at 60% humidity, 12 h of light, and 16°C day and 14°C night temperatures. At least 10 plants of each potential mutant were grown in the M3 at the same conditions to confirm the stability of the previously recorded phenotype. Randomly selected M3 plants scored as confirmed mutants were backcrossed to Col-0 and lhp1 to generate BC1F1 seeds. For the Col-0 backcross, the following BC1F2 generation was also scored for the absence or presence of additional segregating phenotypes. The stability and segregation rate of the mutant phenotype were scored in the lhp1 backcross. One randomly selected BC1F2 alp1-1;lhp1 plant was again backcrossed to lhp1 to generate a BC2F2 population.
Flowering-Time Measurements
Seeds were stratified for 3 d at 4°C on soil in the dark and transferred either to Percival plant growth chambers (CLF Plant Climatics) set to long-day conditions (16 h of light/22°C, 8 h of dark/20°C) or set to screening long-day conditions (12 h of light/16°C, 12 h dark/14°C). Rosette and cauline leaves were counted as measures for flowering time. Statistical significance was evaluated by single-factor ANOVA followed by an honestly significant difference Tukey test. Letters above bars in the figures indicate significantly different groups (P < 0.05).
Library Preparation and Sequencing
Approximately 1,000 BC2F2 plants were sown, and leaf samples of equal size were collected from 270 plants scored as alp1-1;lhp1. In parallel, leaf samples were collected from 48 lhp1 plants. The leaves were bulked prior to DNA extraction with the DNeasy Plant Maxi Kit (Qiagen). DNA was eluted with 500 µL of water in four steps. DNA concentration and quality were determined with a Nanodrop 1000 (Peqlab) on a 1% agarose gel. DNA samples were concentrated to more than 50 ng µL−1 with a Speed-Vac when necessary.
Samples of more than 3 µg of total high-quality DNA extract (260:280 ratio > 1,8) were sequenced by the Cologne Center for Genomics. There, a quality check of the samples was performed with an Agilent 2100 bioanalyzer. Libraries were generated using the Illumina Genomic DNA sample kit according to the manufacturer’s instruction. DNA concentration of the amplified libraries was measured with the DNA 1000 kit as well as the DNA high-sensitivity kit for diluted libraries (both Agilent). The samples were sequenced on an Illumina Genome Analyzer GAIIx in a 96-bp paired end run.
Resequencing Analysis
We applied SHORE to independently align the read sets of the lhp1 mutant and the alp1-1;lhp1 double mutant to the Col-0 reference genome using GenomeMapper as an alignment tool (Ossowski et al., 2008; Schneeberger et al., 2009a, 2009b; Arabidopsis Genome Consortium; The Arabidopsis Information Resource 10). Using the function SHORE import, raw reads were trimmed or discarded based on quality values with a cutoff Phred score of +38. After correcting the paired-end alignments with an expected insert size of 300 bp, we applied SHORE consensus to identify variations between the mutants and reference. We removed the background of alp1;lhp1 by filtering out all the difference between lhp1 and the reference sequence from alp1;lhp1. alp1;lhp1-specific canonical EMS changes with high quality (SHORE score > 24) and supported by more than seven reads were used in SHOREmap backcross for allele frequency analysis. Allele frequency estimates were calculated as the ratio of the reads of mutant alleles divided by all reads at a particular locus. Sequence changes in the region that featured evidence for selection were annotated for their effect on gene identity using The Arabidopsis Information Resource 10 gene annotation. See Supplemental Table S4 for command line calls for the resequencing and mapping-by-sequencing.
dCARE
Primers for dCARE were designed with the help of Primer3 (version 0.4.0) to amplify 80- to 150-bp amplicons that contained the candidate mutations at a distance from +1 to +50 from the 3′ end of the primer that contained the A-type extension required for Ion Torrent PGM sequencing (primers are listed in Supplemental Table S5). DNA was amplified from the same pool of DNA as used for whole-genome resequencing. Amplicons were purified using Agencourt AMPure beads (Beckmann Coulter) according to the manufacturer’s instruction. Amplicons were quantified by optical density at 260 nm, pooled with samples from other customers, and sequenced in an Ion Torrent PGM (Life Technologies) using a 316K chip to a depth of 5,000 to 20,000 reads per amplicon.
Allele frequencies of both the wild type and mutant were estimated from raw reads. Using a 21-mer around the mutation site, an ad-hoc script was used to count the allele occurrence with perfect match or one mismatch. Coverage at each locus was calculated by the sum of satisfying reads from the above criteria.
Quantification of mRNA Abundance
After the removal of roots, total RNA was extracted from the aerial part of 10-d-old seedlings grown on soil or on 1/2 Murashige and Skoog growth medium plates with an RNeasy Plant Mini kit (Qiagen) according to the manufacturer’s instructions. One microgram of RNA of each sample was loaded onto a 1% agarose gel to control for RNA degradation by visualizing the two distinct rRNA bands.
When quality control was passed, 5 µg of RNA was treated with DNase I using a DNA-free kit (Ambion, Life Technologies). After DNase treatment, complementary DNA was synthesized with a dT(18) primer and a SuperScript II reverse transcriptase kit (Invitrogen, Life Technologies) according to the manufacturer’s instructions. Samples were diluted to 150 µL with deionized water, and 1 to 3 µL of complementary DNA was used for quantitative reverse transcription-PCR. All quantitative reverse transcription-PCRs were performed in a Bio-Rad iCycler iQ5 with EvaGreen (Biotium) as a chelating fluorescent dye to quantify the real-time signal. Primers used for the quantification of mRNA are listed in Supplemental Table S5.
Neighbor-Joining Analysis
Amino acid sequences were aligned using ClustalW implemented in MEGA5 (Tamura et al., 2011). The analysis involved 105 amino acid sequences that were identified by BLAST against nonredundant proteins in the NCBI database. The evolutionary history was inferred using the neighbor-joining method (Saitou and Nei, 1987). The bootstrap consensus tree inferred from 10,000 replicates was taken to represent the evolutionary history of the taxa analyzed (Felsenstein, 1985). Branches showing partitions reproduced in less than 50% bootstrap replicates are collapsed. The evolutionary distances were computed using the number of differences method (Nei and Kumar, 2000) and are in units of the number of base differences per sequence. There were a total of 3,082 positions in the final data set. Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2011).
Short read sequence data from this article can be downloaded from (http://shoremap.org).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression of FLC and FT in suppressors of lhp1.
Supplemental Figure S2. Flower morphology of lhp1 and lhp1;alp1 flowers.
Supplemental Figure S3. Comparison of lhp1 loss-of-function alleles between two Arabidopsis accessions.
Supplemental Figure S4. Effect of random sampling on mutant allele frequency estimations.
Supplemental Figure S5. Alignment of ALP1 clade, At3g63270 clade, and HARBI1 proteins from human and zebra fish.
Supplemental Figure S6. Chromatin landscape of ALP1 and expression.
Supplemental Table S1. Summary of genome resequencing of lhp1 and alp1;lhp1.
Supplemental Table S2. Annotation of high-scoring mutations from fast isogenic mapping.
Supplemental Table S3. Raw data for allele frequency calculations at three high-scoring mutations.
Supplemental Table S4. Exemplary command line calls for SHOREmap and preceding resequencing using SHORE.
Supplemental Table S5. List of primers used in this study.
Acknowledgments
We thank Mitzi Villajuana-Bonequi for helpful discussions on EMS mutagenesis and Dr. Mbaye Tine for help with Ion Torrent sequencing.
Glossary
- EMS
ethyl methanesulfonate
- Col-0
Columbia
- dCARE
deep candidate resequencing
- PcG
Polycomb Group
- H3K27me3
Lys-27 trimethylated H3
- PGM
Personal Genome Machine
- NCBI
National Center for Biotechnology Information
- CDD
Conserved Domain Database
- T-DNA
transferred DNA
References
- Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M, et al. (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 30: 174–178 [DOI] [PubMed] [Google Scholar]
- Alonso AG, Gutiérrez L, Fritsch C, Papp B, Beuchle D, Müller J. (2007) A genetic screen identifies novel Polycomb group genes in Drosophila. Genetics 176: 2099–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashelford K, Eriksson ME, Allen CM, D’Amore R, Johansson M, Gould P, Kay S, Millar AJ, Hall N, Hall A. (2011) Full genome re-sequencing reveals a novel circadian clock mutation in Arabidopsis. Genome Biol 12: R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin RS, Vidaurre D, Stamatiou G, Breit R, Provart NJ, Bonetta D, Zhang JF, Fung P, Gong YC, Wang PW, et al. (2011) Next-generation mapping of Arabidopsis genes. Plant J 67: 715–725 [DOI] [PubMed] [Google Scholar]
- Aziz RK, Breitbart M, Edwards RA. (2010) Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res 38: 4207–4217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratzel F, López-Torrejón G, Koch M, Del Pozo JC, Calonje M. (2010) Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr Biol 20: 1853–1859 [DOI] [PubMed] [Google Scholar]
- Casola C, Lawing AM, Betrán E, Feschotte C. (2007) PIF-like transposons are common in Drosophila and have been repeatedly domesticated to generate new host genes. Mol Biol Evol 24: 1872–1888 [DOI] [PubMed] [Google Scholar]
- Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J. (2004) Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131: 5263–5276 [DOI] [PubMed] [Google Scholar]
- Craig NL. (2002) Mobile DNA II. ASM Press, Washington, DC
- Cuperus JT, Montgomery TA, Fahlgren N, Burke RT, Townsend T, Sullivan CM, Carrington JC. (2010) Identification of MIR390a precursor processing-defective mutants in Arabidopsis by direct genome sequencing. Proc Natl Acad Sci USA 107: 466–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner V, Aichinger E, Shu H, Wildhaber T, Alfarano P, Caflisch A, Gruissem W, Köhler C, Hennig L. (2009) The chromodomain of LIKE HETEROCHROMATIN PROTEIN 1 is essential for H3K27me3 binding and function during Arabidopsis development. PLoS ONE 4: e5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrona S, Thorpe FL, Engelhorn J, Adrian J, Dong X, Sarid-Krebs L, Goodrich J, Turck F. (2011) Tissue-specific expression of FLOWERING LOCUS T in Arabidopsis is maintained independently of Polycomb group protein repression. Plant Cell 23: 3204–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1985) Confidence-limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Gildea JJ, Lopez R, Shearn A. (2000) A screen for new trithorax group genes identified little imaginal discs, the Drosophila melanogaster homologue of human retinoblastoma binding protein 2. Genetics 156: 645–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraut L, Falque M, Drouaud J, Pereira L, Martin OC, Mézard C. (2011) Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet 7: e1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahara J, Kigawa T, Kitagawa K, Masumoto H, Okazaki T, Yokoyama S. (1998) A helix-turn-helix structure unit in human centromere protein B (CENP-B). EMBO J 17: 827–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Wang Y, Wang Y, He Y. (2008) Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS ONE 3: e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake T, Takada S, Nakahigashi K, Ohto M, Goto K. (2003) Arabidopsis TERMINAL FLOWER 2 gene encodes a heterochromatin protein 1 homolog and represses both FLOWERING LOCUS T to regulate flowering time and several floral homeotic genes. Plant Cell Physiol 44: 555–564 [DOI] [PubMed] [Google Scholar]
- Landecker HL, Sinclair DAR, Brock HW. (1994) Screen for enhancers of Polycomb and Polycomblike in Drosophila melanogaster. Dev Genet 15: 425–434 [DOI] [PubMed] [Google Scholar]
- Larsson AS, Landberg K, Meeks-Wagner DR. (1998) The TERMINAL FLOWER2 (TFL2) gene controls the reproductive transition and meristem identity in Arabidopsis thaliana. Genetics 149: 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevich G, Leroy O, Akinci U, Schubert D, Clarenz O, Goodrich J, Grossniklaus U, Köhler C. (2006) Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep 7: 947–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu SN, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39: D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylne JS, Barrett L, Tessadori F, Mesnage S, Johnson L, Bernatavichute YV, Jacobsen SE, Fransz P, Dean C. (2006) LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc Natl Acad Sci USA 103: 5012–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S. (2000) Molecular Evolution and Phylogenetics. Oxford University Press, Oxford
- Nowacki M, Higgins BP, Maquilan GM, Swart EC, Doak TG, Landweber LF. (2009) A functional role for transposases in a large eukaryotic genome. Science 324: 935–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Clark RM, Lanz C, Warthmann N, Weigel D. (2008) Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res 18: 2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page DR, Grossniklaus U. (2002) The art and design of genetic screens: Arabidopsis thaliana. Nat Rev Genet 3: 124–136 [DOI] [PubMed] [Google Scholar]
- Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, Leamon JH, Johnson K, Milgrew MJ, Edwards M, et al. (2011) An integrated semiconductor device enabling non-optical genome sequencing. Nature 475: 348–352 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Schneeberger K, Hagmann J, Ossowski S, Warthmann N, Gesing S, Kohlbacher O, Weigel D. (2009a) Simultaneous alignment of short reads against multiple genomes. Genome Biol 10: R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jørgensen JE, Weigel D, Andersen SU. (2009b) SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods 6: 550–551 [DOI] [PubMed] [Google Scholar]
- Searle I, He YH, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G. (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG. (2004) The autonomous pathway: epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr Opin Plant Biol 7: 570–574 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, Buisine N, Gagnot S, Martienssen RA, Coupland G, Colot V. (2007) Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet 3: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker EL, Eggleston WB, Demopoulos D, Kermicle J, Dellaporta SL. (1997) Insertions of a novel class of transposable elements with a strong target site preference at the r locus of maize. Genetics 146: 681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Shen WH. (2008) Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr Biol 18: 1966–1971 [DOI] [PubMed] [Google Scholar]
- Yuan YW, Wessler SR. (2011) The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc Natl Acad Sci USA 108: 7884–7889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Germann S, Blus BJ, Khorasanizadeh S, Gaudin V, Jacobsen SE. (2007) The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat Struct Mol Biol 14: 869–871 [DOI] [PubMed] [Google Scholar]