Abstract
Proteins decorated with arabinogalactan (AG) have important roles in cell wall structure and plant development, yet the structure and biosynthesis of this polysaccharide are poorly understood. To facilitate the analysis of biosynthetic mutants, water-extractable arabinogalactan proteins (AGPs) were isolated from the leaves of Arabidopsis (Arabidopsis thaliana) plants and the structure of the AG carbohydrate component was studied. Enzymes able to hydrolyze specifically AG were utilized to release AG oligosaccharides. The released oligosaccharides were characterized by high-energy matrix-assisted laser desorption ionization-collision-induced dissociation mass spectrometry and polysaccharide analysis by carbohydrate gel electrophoresis. The Arabidopsis AG is composed of a β-(1→3)-galactan backbone with β-(1→6)-d-galactan side chains. The β-(1→6)-galactan side chains vary in length from one to over 20 galactosyl residues, and they are partly substituted with single α-(1→3)-l-arabinofuranosyl residues. Additionally, a substantial proportion of the β-(1→6)-galactan side chain oligosaccharides are substituted at the nonreducing termini with single 4-O-methyl-glucuronosyl residues via β-(1→6)-linkages. The β-(1→6)-galactan side chains are occasionally substituted with α-l-fucosyl. In the fucose-deficient murus1 mutant, AGPs lack these fucose modifications. This work demonstrates that Arabidopsis mutants in AGP structure can be identified and characterized. The detailed structural elucidation of the AG polysaccharides from the leaves of Arabidopsis is essential for insights into the structure-function relationships of these molecules and will assist studies on their biosynthesis.
Arabinogalactans (AGs) are structurally complex large-branched polysaccharides attached to Hyp residues of many plant cell wall polypeptides. Most proteins glycosylated with AGs (AGPs) have both AG glycosylated domains (glycomodules) and structural or enzymatic domains. However, typical AGPs commonly contain less than 10% protein, suggesting that the AG is the functional part of the molecule (Clarke et al., 1979; Fincher et al., 1983; Kieliszewski and Lamport, 1994; Borner et al., 2003; Xu et al., 2008). Hyp is the most characteristic amino acid present at the glycosylated domain of the AGP, but other amino acids such as Ser, Ala, and Thr are also very common. Type II AG polysaccharides share common structural features based on a β-(1→3)-galactan backbone with β-(1→6)-linked galactan side chains and can be found both on AGPs and rhamnogalacturonan-I (RG-I) pectin (Renard et al., 1991). The galactopyranosyl (Galp) residues can be further substituted with l-arabinofuranosyl (l-Araf) and occasionally also l-rhamnosyl (l-Rha), l-fucosyl (l-Fuc), and glucuronosyl (GlcA; with or without 4-O-methylation) residues (Tsumuraya et al., 1988; Tan et al., 2004; Tryfona et al., 2010). (Sugars mentioned in this work belong to the D-series unless otherwise stated.)
The structure of AGs is poorly characterized, and this is mainly due to the great heterogeneity of glycan structures, not only between different AGPs but also even on the same peptide sequence in the same tissue (Estévez et al., 2006). The glycan structure can also be different depending on the developmental stage and tissue type (Tsumuraya et al., 1988), adding to the great heterogeneity of these molecules and therefore limiting their detailed characterization. Molecular and biochemical evidence has indicated that AGPs have specific functions during root formation, promotion of somatic embryogenesis (van Hengel et al., 2002), and attraction of pollen tubes to the style (Cheung et al., 1995). In addition, enhanced secretion efficiency or stability in the cell wall are properties that the AG may confer on the glycosylated protein (Borner et al., 2003). However, it has been difficult to differentiate one species of AGP from another in plant tissues and to assign specific roles to individual AGPs.
l-Fuc is present in AGPs in Arabidopsis (Arabidopsis thaliana; van Hengel et al., 2002), radish (Raphanus sativus; Nakamura et al., 1984; Tsumuraya et al., 1984a, 1984b, 1988), and several other dicot plants such as thyme (Thymus vulgaris; Chun et al., 2001) and celery (Apium graveolens; Lin et al., 2011). Reduction in l-Fuc by 40% in roots of murus1 (mur1) plants resulted in a decrease of 50% in root cell elongation, and eel lectin binding assays suggested that the phenotype was the result of alterations in the composition of root AGPs (van Hengel and Roberts, 2002). An α-(1→2)-fucosyltransferase (FUT) activity for radish primary root AGPs has been described, where an α-l-Araf-(1→3)-β-Galp-(1→6)-Galp trisaccharide was used as exogenous substrate acceptor to mimic an AG polysaccharide in the enzymatic assay (Misawa et al., 1996). Linkage analysis, reactivity with eel lectin, and digestion with α-(1→2)-fucosidase indicated that the l-Fuc residues added are terminal and attached via an α-linkage to the C-2 position of an adjacent l-Araf residue (Nakamura et al., 1984; Tsumuraya et al., 1984a, 1984b, 1988). Recently, Wu et al. (2010) identified AtFUT4 and AtFUT6 genes encoding FUT proteins specific to AGPs, but the structures of the fucosylated AG generated have not been fully characterized.
To gain insights into the synthesis and function of plant AGPs, it would be useful to have mutants altered in their carbohydrate moieties. However, no AG-specific biosynthetic mutants have been characterized, and this, among other reasons, is due to the very limited knowledge of the structure of Arabidopsis AGs (Qu et al., 2008). Moreover, characterization of AG in candidate mutants remains challenging. Even though the structures of some AGs have been proposed using NMR and sugar linkage analyses, the complete structural elucidation of a native AG still remains a formidable task, because NMR spectroscopy and methylation analysis have been largely used to provide information regarding the amount and type of linkages between adjacent glycosyl residues, and AG heterogeneity can confound attempts to build complete structural models. Recently, a modular structure was proposed for AGs on heterologously expressed proteins in tobacco (Nicotiana tabacum; Tan et al., 2010). Tan et al. (2010) proposed that approximately 15-residue repeating blocks of decorated β-(1→3)-trigalactosyl subunits connected by β-(1→6)-linkages were the building blocks of type II AG polysaccharides and concluded that these molecules are far less complex than commonly supposed. Most characterized β-(1→6)-galactan side chains in AGs are reported to be short, of one or two residues (Neukom and Markwalder, 1975; Gane et al., 1995; Gaspar et al., 2001). On the contrary, there are reports of long β-(1→6)-galactan side chains in radish root AGPs (Haque et al., 2005). Similarly, we recently found evidence that wheat (Triticum aestivum) flour endosperm AGP extracts contained long β-(1→6)-galactan side chains heavily substituted with l-Araf at C-3 (Tryfona et al., 2010). This partial structure of the carbohydrate component of wheat flour AGP isolated from water extracts of wheat endosperm was elucidated utilizing a combination of analytical approaches, such as the use of enzymes able to release oligosaccharides specifically from AGs, high-energy matrix-assisted laser desorption ionization (MALDI)-collision-induced dissociation (CID) mass spectrometry (MS), and polysaccharide analysis by carbohydrate gel electrophoresis (PACE; Tryfona et al., 2010). In this work, we applied these techniques to study the carbohydrate component of Arabidopsis leaf AGPs. AG-specific enzyme digestion products were analyzed by PACE and MS, allowing a partial structure to be proposed. We show that endogenous Arabidopsis leaf AG is composed of a β-(1→3)-galactan backbone with β-(1→6)-galactan side chains. These side chains are substituted with l-Araf residues via α-(1→3)-linkages and can vary in length from one up to at least 20 Galp residues. We also found that the β-(1→6)-galactan side chains are substituted mainly with 4-O-methyl-glucuronosyl (4-O-Me-GlcA) at their nonreducing termini, while occasional l-Fuc substitutions were also present via α-(1→2)-linkages on l-Araf residues. In addition, AG oligosaccharides from leaves of the mur1 mutant were identified, and their structures were compared with those isolated from wild-type plants.
RESULTS
Arabidopsis Leaf AGPs Have a β-(1→3)-Galactan Backbone with β-(1→6)-Galactan Side Chains Decorated by l-Araf, 4-O-Me-GlcA, and Deoxyhexose Residues
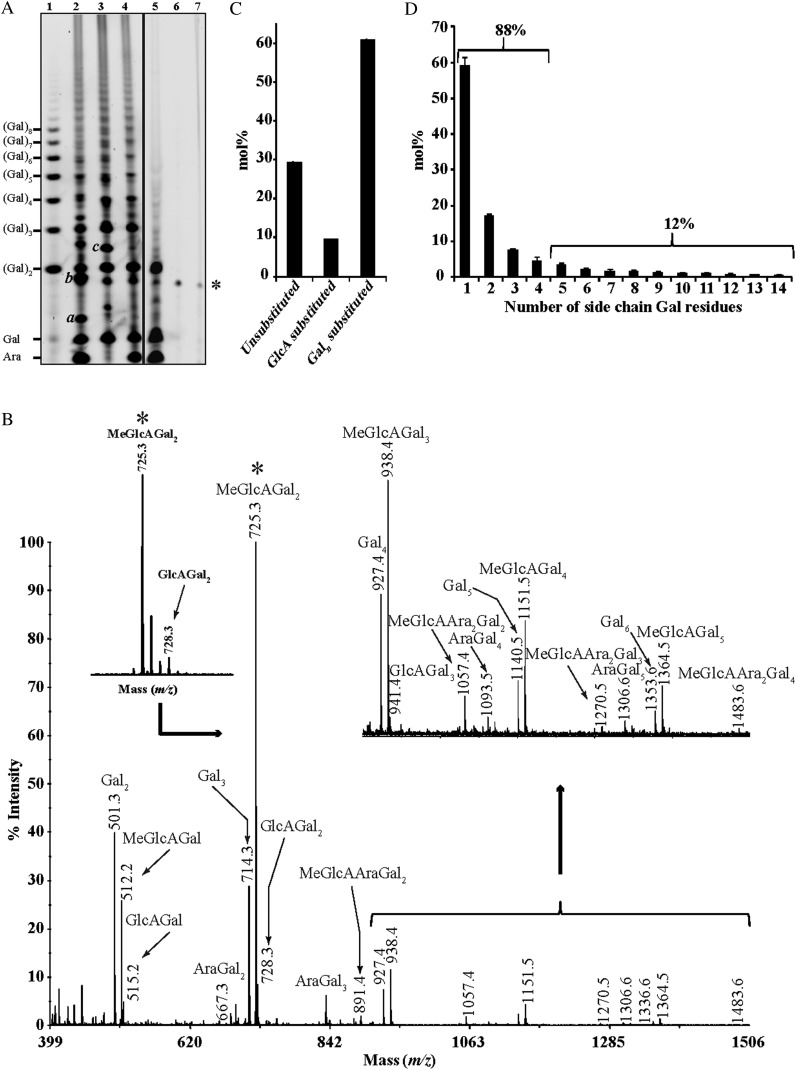
The exo-β-(1→3)-galactanase specifically cleaves terminal β-(1→3)-galactosidic linkages and can bypass branching points, liberating any β-(1→6)-linked galactosyl side chains in the AG as oligomers (Tsumuraya et al., 1990). Our hypothesis was that the exo-β-(1→3)-galactanase might hydrolyze Arabidopsis leaf AGPs, releasing oligosaccharides of different degrees of polymerization (DP). Use of this AG-specific enzyme avoids the need to obtain pure AGP for analysis. Released oligosaccharides can be resolved with PACE. We have recently shown that PACE can resolve AG oligosaccharides with various DP, glycosidic linkage, or composition, and it has also been demonstrated that the band intensity is proportional to the quantity of the oligosaccharides (Goubet et al., 2002; Tryfona et al., 2010). Therefore, we subjected Arabidopsis leaf AGP extracts to digestion with exo-β-(1→3)-galactanase and characterized the oligosaccharide products by PACE. From Figure 1, it is clear that the Arabidopsis leaf AGs are susceptible to the exo-β-(1→3)-galactanase. Treatment with exo-β-(1→3)-galactanase alone released diverse oligosaccharides, but the observation of high-Mr saccharides was hampered due to the smearing on the PACE gel, suggesting great heterogeneity of the hydrolysis products (data not shown). α-l-Arabinofuranosidase has been shown to hydrolyze α-(1→3)- and α-(1→5)-l-Araf residues from AGPs (Takata et al., 2010). Following the α-l-arabinofuranosidase hydrolysis to remove accessible l-Araf residues, clear ladders of oligosaccharides were visible (Fig. 1A). Three of these released oligosaccharides comigrated with Gal, β-(1→6)-galactobiose, and β-(1→6)-galactotriose, suggesting that the exo-β-(1→3)-galactanase releases side chains of substituted β-(1→6)-galactan.
Figure 1.
Characterization of oligosaccharides released by AG-specific enzymes from Arabidopsis leaf AGP extracts. A, PACE. Oligosaccharide products of exo-β-(1→3)-galactanase followed by α-l-arabinofuranosidase (lane 2), exo-β-(1→3)-galactanase followed by β-glucuronidase (lane 3), exo-β-(1→3)-galactanase, α-l-arabinofuranosidase, and β-glucuronidase (lane 4), exo-β-(1→3)-galactanase, α-l-arabinofuranosidase, β-glucuronidase, and endo-β-(1→6)-galactanase (lane 5), enzyme control (lane 6), and AGP extract control (lane 7) were reductively aminated with 2-aminonaphthaline trisulfonic acid and separated by electrophoresis on acrylamide gels. An oligosaccharide ladder prepared from β-(1→6)-galactan was used as a migration marker (lane 1): a, MeGlcAGal; b, MeGlcAGal2; c, MeGlcAAraGal2. The asterisk indicates a background band. B, MALDI-ToF-MS spectra of per-deuteromethylated oligosaccharides released by exo-β-(1→3)-galactanase followed by α-l-arabinofuranosidase. Peaks marked with asterisks were selected for MALDI-CID structural analysis. C, Quantification of substitution of the β-(1→3)-galactan backbone: none, MeGlcA, or galactan. Arabidopsis leaf AGP extracts were digested with AG-specific α-l-arabinofuranosidase and exo-β-(1→3)-galactanase, and the amount of the hydrolysis products was calculated by PACE [Galn corresponds to β-(1→6)-galactan chains with two to 15 Gal residues]. Values are means ± sd of one representative biological replicate analyzed six times. D, Quantification by PACE of the different-length β-(1→6)-galactan side chains (e.g. chain 1 corresponds to Gal2, chain 2 corresponds to Gal3) released by sequential digestion with α-l-arabinofuranosidase, exo-β-(1→3)-galactanase, and β-glucuronidase of Arabidopsis leaf AGP extracts. Values are means ± sd of two biological replicates analyzed six times.
Type II AGs with β-(1→3)- and β-(1→6)-linked-galactosyl residues may occur attached to the RG-I (Renard et al., 1991) in addition to their attachment to protein in AGPs. We next determined whether the type II AG oligosaccharides detected by PACE may be derived in part from cell wall pectic polysaccharides. The leaf AGP extracts and alcohol-insoluble residue (AIR) from Arabidopsis leaf cell walls as a control were subjected to RG-I lyase treatment, and the hydrolysis products were analyzed by PACE to detect any RG-I backbone (Supplemental Fig. S1A). Following the RG-I lyase treatment, a number of abundant oligosaccharides were released from Arabidopsis leaf AIR, but a negligible amount were released from the Arabidopsis leaf AGP extracts, suggesting that RG-I was not substantially extracted with the AGPs. In addition, a very small amount of Rha was detected in monosaccharide analysis of neutral sugars of crude Arabidopsis leaf AGP extracts by high-pH anion-exchange chromatography (HPAEC) with amperometric detection (PAD; Supplemental Fig. S1, B and C). Taken together, these findings suggest that the AGP extract contained little RG-I and that the majority of type II AG polysaccharides characterized here modify AGPs. Recent work characterizing purified wheat flour AGP (Tryfona et al., 2010) showed similar decorated long-chain β-(1→6)-linked-galactan, supporting the view that these oligosaccharides are derived from AGPs.
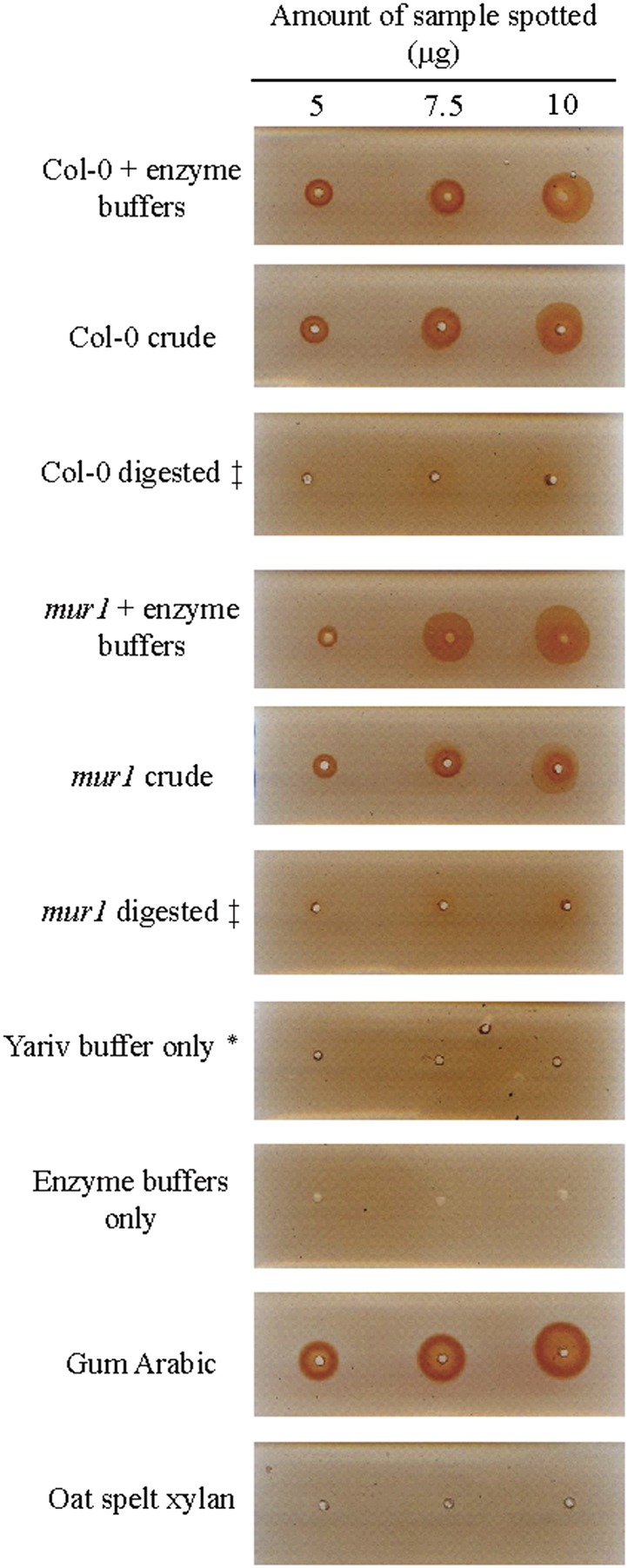
A Yariv diffusion assay showed that the strong Arabidopsis leaf AGP reactivity with β-galactosyl Yariv is completely lost after treatment with AGP hydrolytic enzymes. This indicates that the bulk of the AGPs are sensitive to enzymatic digestion (Fig. 2).
Figure 2.
The bulk of Arabidopsis leaf AGPs are sensitive to AG-specific enzymes as shown by a Yariv diffusion assay. Leaf AGP extracts from wild-type and mur1 mutant plants were subjected to sequential hydrolysis by AG-specific enzymes: exo-β-(1→3)-galactanase, 2× α-l-arabinofuranosidase, β-glucuronidase, and endo-β-(1→6)-galactanase. The AGP extracts or the enzyme hydrolysis products were dissolved in NaCl (0.15 m) and sodium azide (0.02%) solution and were spotted on agarose gels (1%) containing the β-galactosyl Yariv reagent (0.002%). The controls included reagents without the hydrolysis products: gum Arabic (positive) and oat spelt xylan (negative). ‡exo-β-(1→3)-galactanase, 2× α-l-arabinofuranosidase, β-glucuronidase, and endo-β-(1→6)-galactanase sequential digestion; *NaCl (0.15 m) and sodium azide (0.02%) buffer solution. Col-0, Ecotype Columbia.
To characterize further the released oligosaccharides, enzyme-digested samples were per-deuteromethylated and analyzed by MALDI-time of flight (ToF)-MS (Fig. 1B). The exo-β-(1→3)-galactanase and α-l-arabinofuranosidase sequential treatment released oligosaccharides consisting predominantly of hexosyl (Hex) and hexuronosyl (HexA) residues. A major series of ions corresponding to Hex2-5 and HexAHex1-5 were seen, and some pentose-containing oligosaccharides still remained after α-l-arabinofuranosidase digestion (e.g. PentHex3). The pentose and hexosyl are l-Ara and Gal, since the oligosaccharides arise from AG-specific enzyme digestion, and these oligosaccharides were sensitive to further α-l-arabinofuranosidase or exo-β-(1→3)-galactanase hydrolysis (see below) and some comigrated in PACE with standards of known structure. We were also able to detect minor ions with mass-to-charge ratio (m/z) values that correspond to short oligosaccharides possibly carrying deoxyhexose (l-Rha or l-Fuc) modifications (data not shown). This finding is discussed below.
To determine the nature of the hexuronosyl substitutions, the oligosaccharides released by the sequential exo-β-(1→3)-galactanase/α-l-arabinofuranosidase digestion were treated with AG-specific β-glucuronidase. PACE showed a substantial alteration of the fingerprint after β-glucuronidase digestion (Fig. 1A), indicating that many oligosaccharides are substituted with terminal β-GlcA residues. Since both GlcA (Tryfona et al., 2010) and 4-O-Me-GlcA (Haque et al., 2005) have been reported in AGPs, we performed per-deuteromethylation to distinguish by MS between the methylated and nonmethylated forms (Fig. 1B). Signals corresponding to MeGlcAGal1-5, MeGlcAAraGal2, and MeGlcAAra2Gal2-4 ([M + Na]+ m/z values are indicated in Fig. 1B) were detected, but also minor signals corresponding to GlcAGal1-3 (m/z 515.2, 728.3, and 941.4, respectively) were present in the spectra. The relative amounts of the oligosaccharides carrying the 4-O-Me-GlcA modifications were much higher than their nonmethylated counterparts (Fig. 1B). Interestingly, an ion with m/z 512.2, which corresponds to a MeGlcAGal disaccharide (or m/z 515.2, which corresponds to the nonmethylated counterpart) was detected. This oligosaccharide is also seen by PACE (Fig. 1A). Given that exo-β-(1→3)-galactanase liberates the side chains attached to a Gal residue from the backbone, these data indicate that 4-O-Me-GlcA residues are directly linked to the β-(1→3)-galactan backbone. Similar findings have been reported for radish AGPs (Tsumuraya et al., 1990; Ichinose et al., 2006).
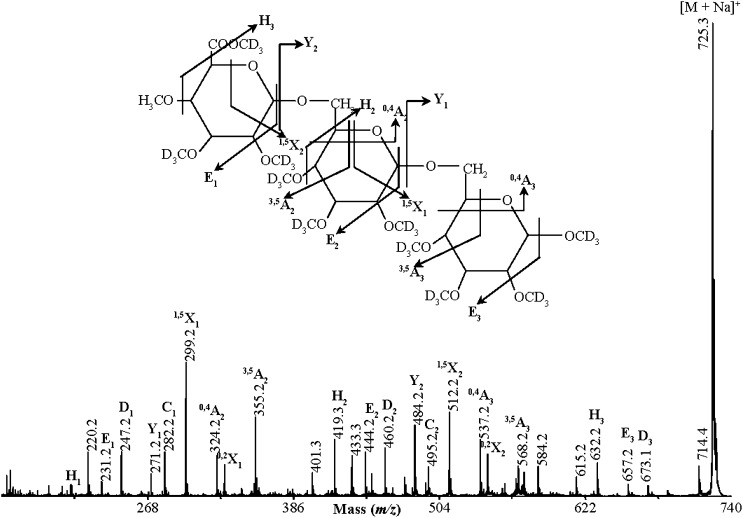
To investigate the structure of the AG side chain oligosaccharides, we carried out high-energy MALDI-CID. The tandem mass spectrometry (MS/MS) spectrum of a MeGlcAGal2 per-deuteromethylated (m/z 725.3) oligosaccharide is shown in Figure 3. The series of 1,5X and Y ions (Domon and Costello, 1988) show the distribution of Gal residues, while the nonreducing end 0,4A3 and 0,4A2 ions confirm the sequence and also indicate the presence of (1→6)-glycosidic linkages not only between the two Gal residues but also between the GlcA and Gal residues. The methyl group on GlcA is likely to be attached to C-4, as 4-O-Me-GlcA has been found in radish AGP (Haque et al., 2005). This is supported by the presence of the H3 “elimination” ion (Maslen et al., 2007). Thus, the oligosaccharide was identified as 4-O-Me-β-GlcA-(1→6)-β-Galp-(1→6)-Galp. β-glucuronidase was able to hydrolyze several oligosaccharides including MeGlcAGal and MeGlcAGal2 but not MeGlcAAraGal2 (Fig. 1A), suggesting that this enzyme is inhibited by adjacent l-Araf. However, after sequential hydrolysis with α-l-arabinofuranosidase and finally β-glucuronidase, the oligosaccharide pattern was much simplified and dominated by a ladder comigrating with the β-(1→6)-galactan ladder. The ladder extended at least to DP 20 (Fig. 1A, lane 4). The susceptibility of the side chain decorations to these two enzymes suggests that most are α-l-Araf or 4-O-β-Me-GlcA. Taken together, the PACE and MS results indicate that the side chains of Arabidopsis leaf AGPs are decorated with α-l-Araf. Some terminate in β-4-O-Me-GlcA through (1→6)-linkages or, less often, GlcA residues.
Figure 3.
Structural characterization of an acidic AG oligosaccharide by MALDI-CID. Arabidopsis leaf AGP extracts were sequentially hydrolyzed with exo-β-(1→3)-galactanase and α-l-arabinofuranosidase, and the resulting oligosaccharides were analyzed by MALDI-ToF-MS as shown in Figure 1B. The per-deuteromethylated oligosaccharide with m/z 725.3 was selected for MALDI-CID analysis and is identified as 4-O-Me-β-GlcA-(1→6)-β-Galp-(1→6)-Galp. Glycosidic and cross-ring fragments are identified according to the nomenclature of Domon and Costello (1988).
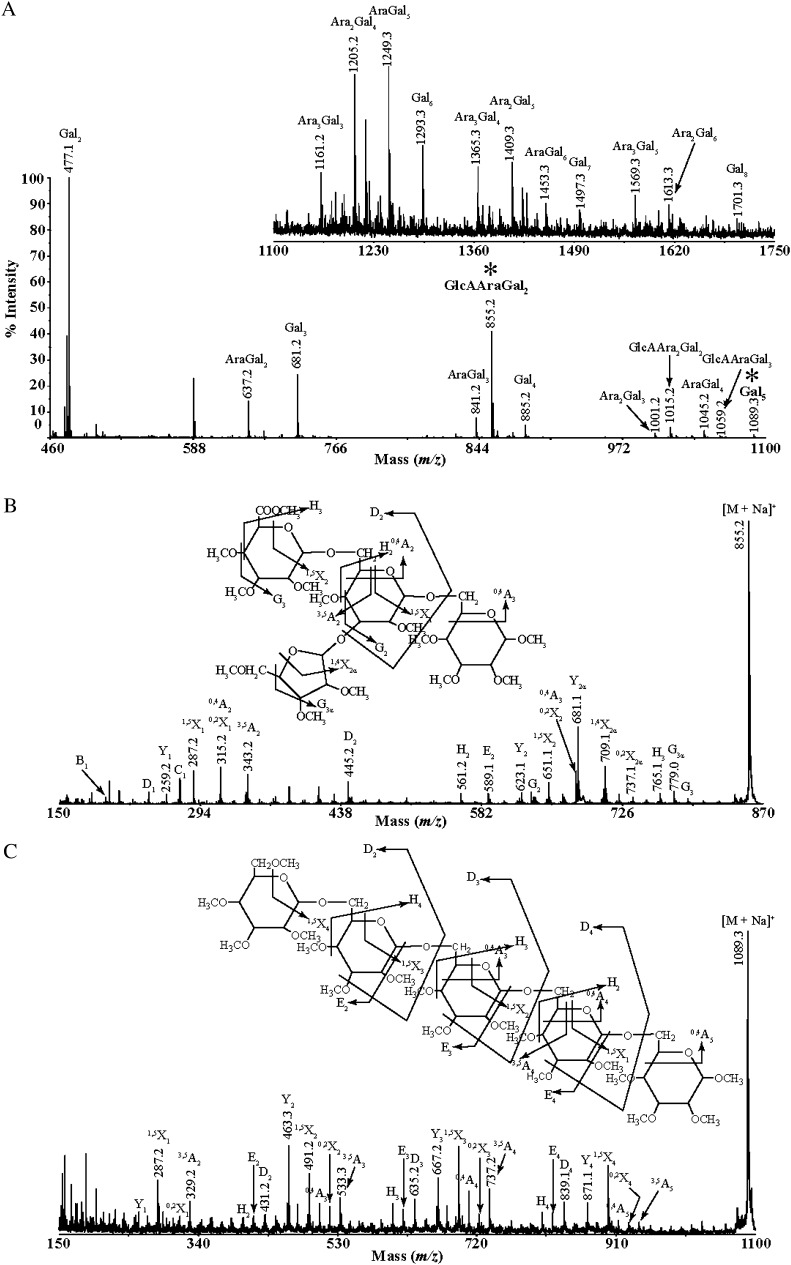
To study the position of the l-Araf decorations on the AG side chains, we performed MALDI-ToF-MS and MALDI-CID. Figure 4A shows the MALDI-ToF-MS spectrum for per-methylated products (thus combining the signals for 4-O-Me-GlcA and GlcA substituted oligosaccharides) after sequential AG digestion with exo-β-(1→3)-galactanase and β-glucuronidase. Diverse oligosaccharides containing Galp and l-Araf were observed. In many oligosaccharides, fewer l-Ara residues were present than Galp, indicating that not all Galp residues are substituted by l-Ara. The Ara3Gal3 and GlcAAra2Gal2 oligosaccharides likely have a side chain of at least two l-Ara residues, since the reducing end Galp released by exo-β-(1→3)-galactanase cannot be substituted. As discussed above, the MS data further suggested that GlcA residues are not removed by β-glucuronidase if they are adjacent to Araf, since oligosaccharides carrying GlcA plus l-Araf were detected (m/z 855.2, 1,015.2, and 1,059.2). To determine their structure, we performed MALDI-CID of the per-methylated GlcAAraGal2 (m/z 855.2) oligosaccharide (Fig. 4B). As seen for MeGlcAGal2 in Figure 3, the spectrum suggests that GlcA is linked on the C-6 of the nonreducing end Gal and also indicates the presence of (1→6)-linkages between the two Gal residues. The cross-ring fragments (0,4A2 ion, m/z 324.2; 3,5A2 ion, m/z 355.2) and elimination ions (H2 elimination ion, m/z 419.3; D2 elimination ion, m/z 460.2; and E2 elimination ion, m/z 444.2; Chai et al., 2001; Spina et al., 2004) indicate that Ara is linked on the C-3 of the nonreducing end Gal. Another elimination ion the G3α at m/z 779.0 suggests that the l-Ara residue may be present in its furanose form (Maslen et al., 2007). Thus, the oligosaccharide was identified as 4-O-Me-β-GlcA-(1→6)-[α-Araf-(1→3)]-β-Galp-(1→6)-Galp. After removal of their l-Araf and (4-O-Me)GlcA decorations, the AGP side chains comigrated on PACE with the β-(1→6)-galactan ladder. To confirm the structure of these side chains, an oligosaccharide product of the exo-β-(1→3)-galactanase, α-l-arabinofuranosidase, and β-glucuronidase sequential digestion (m/z 1,089.3 mass of Gal5) was analyzed by MALDI-CID (Fig. 4C). The presence of a complete series of 1,5X and Y ions differing by m/z 204 (a Gal residue) in the MALDI-CID spectrum suggested that the chain is linear. The presence of the series of 0,4A and 3,5A nonreducing end ions shows the presence of (1→6)-linkages between the Gal residues. Therefore, the oligosaccharide was identified as β-(1→6)-galactopentaose. Finally, the ladder of galactan side chains was shown using PACE to be sensitive to endo-β-(1→6)-galactanase (Fig. 1A, lane 5).
Figure 4.
Structural characterization of an acidic AG oligosaccharide and of a long galacto-oligosaccharide by MALDI-CID. A, Oligosaccharide products of exo-β-(1→3)-galactanase followed by β-glucuronidase were per-methylated and analyzed by MALDI-ToF-MS. Peaks marked with asterisks were selected for MALDI-CID structural analysis. B and C, MALDI-CID of GlcAAraGal2 and Gal5 oligosaccharide, respectively.
Taken together, the PACE and MS characterization of the enzyme products suggests that the Arabidopsis leaf AG contains a β-(1→3)-galactan backbone. Oligosaccharide quantification by PACE revealed that approximately 30% of Gal backbone residues are unsubstituted, 10% are decorated with β-(1→6)-linked MeGlcA/GlcA, and 60% are branched with β-(1→6)-Gal side chains (Fig. 1C). The distribution of side chain lengths shows that single Gal substitutions are most frequent, with a smooth decay in the frequency of longer side chains. Approximately 90% of these galactan side chains are shorter than four residues (Fig. 1D), but chains up to DP 14 were quantifiable and longer chains were easily detectable. The side chains are substituted with α-(1→3)-linked l-Araf, and many chains have terminal β-(1→6)-linked 4-O-Me-GlcA or GlcA.
Deoxyhexose Modifications on Arabidopsis Leaf AGPs Are l-Fuc Residues
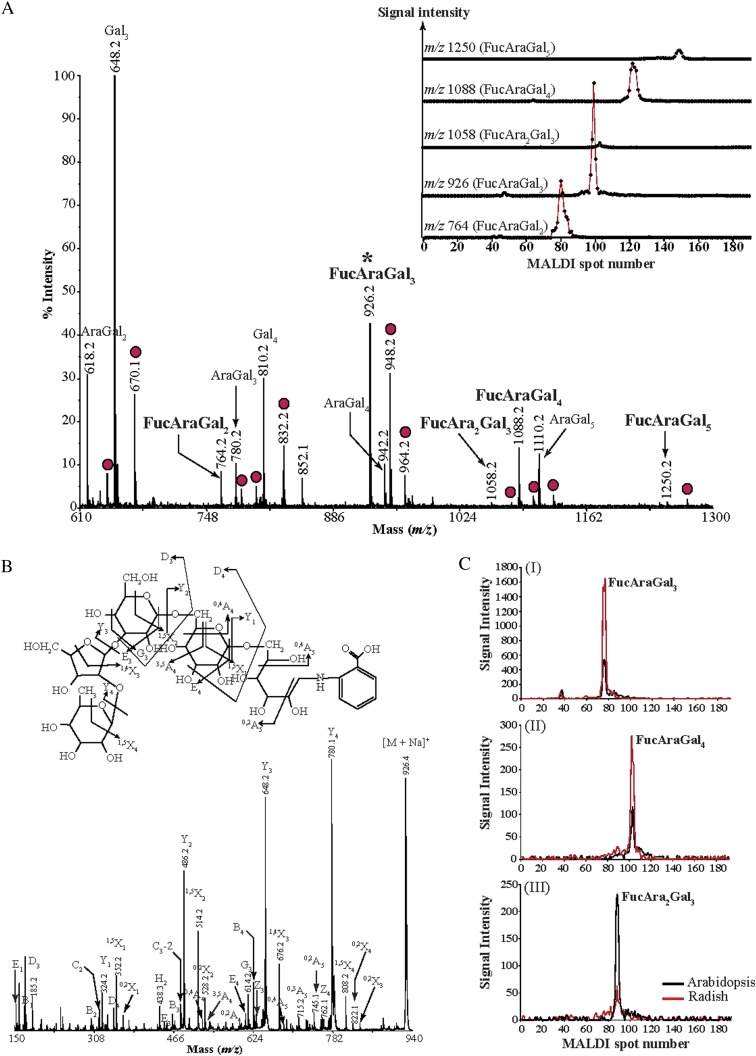
We investigated whether the occasional deoxyhexose modifications on Arabidopsis leaf AGPs correspond to l-Fuc or l-Rha, both of which have been found in AGPs from other species (Tsumuraya et al., 1988; Tan et al., 2004). Although the HPAEC-PAD monosaccharide analysis revealed that no Rha was present in the Arabidopsis AGP preparation, this sugar may not have been detectable due to the low frequency of these decorations. Pure radish leaf AGPs are known to contain solely l-Fuc as the deoxyhexose modification, but in unknown structures (Tsumuraya et al., 1988). Therefore, we took a comparative approach to determine whether the same oligosaccharide structures are present in AGPs from Arabidopsis and radish leaves. Since fucosylation is a minor decoration, radish leaf AGPs were subjected to α-arabinofuranosidase, exo-β-(1→3)-galactanase, β-glucuronidase, and endo-β-(1→6)-galactanase sequential digestion to enrich for the enzyme-resistant fucosylated oligosaccharides. Initial experiments involved the direct analysis of 2-aminobenzoic acid (2-AA)-labeled digestion products by MALDI-ToF-MS (Fig. 5A), where a series of adduct ions ([M + Na]+ and [M + 2Na – H]+) were observed for each oligosaccharide. The oligosaccharides released consisted predominantly of Gal and l-Ara, but short oligosaccharides carrying l-Fuc residues corresponding to FucAraGal2-5 were detected (m/z 764.2, 926.2, 1,088.2, and 1,250.2, respectively). A low-intensity signal (m/z 1,058.2) corresponding to FucAra2Gal3 was also present in the spectrum. To separate possible isomeric isobaric structures in the oligosaccharide mixture, we performed hydrophilic interaction liquid chromatography (HILIC) coupled offline to MALDI-ToF-MS. The extracted ion chromatograms showed the presence of single structural isomers for each m/z (Fig. 5A, inset). The structure of each of these low-abundance oligosaccharides was determined by CID. The resulting spectrum of FucAraGal3 (m/z 926 [M + Na]+) is shown in Figure 5B. The complete series of Y and 1,5X ions shows that FucAraGal3 is linear, while the D3 and G3 elimination ions show that the Fuc-Ara disaccharide is linked on the third Gal from the reducing end via a (1→3)-linkage between the l-Ara and Gal residues. The nonreducing end cross-ring 0,4A4 and 0,4A5 ions show that the Gal residues are linked at C-6. Moreover, the 0,2X3 cross-ring ion indicates that the terminal l-Fuc residue is (1→2)-linked at l-Ara. Misawa et al. (1996) have reported the attachment of l-Fuc to an l-Araf residue at O-2 by an α-glycosidic linkage; thus, the oligosaccharide here was identified as α-Fuc-(1→2)-α-Araf-(1→3)-β-Galp-(1→6)-β-Galp-(1→6)-Galp. Similarly, the FucAraGal2 oligosaccharide was identified as α-Fuc-(1→2)-α-Araf-(1→3)-β-Galp-(1→6)-Gal. The MALDI-CID spectrum of the low-abundance FucAra2Gal3 did not resolve the linkage between the two l-Araf residues but indicated that it has the structure of α-l-Fuc-(1→2)-α-l-Araf-(1→?)-α-l-Araf-(1→3)-β-Galp-(1→6)-β-Galp-(1→6)-Galp (data not shown).
Figure 5.
Identification of l-Fuc-modified oligosaccharides from radish leaf AGP extracts. Purified radish and Arabidopsis leaf AGPs were subjected to a sequential digestion of α-l-arabinofuranosidase, exo-β-(1→3)-galactanase, β-glucuronidase, and endo-β-(1→6)-galactanase. The hydrolysis products were reductively aminated with 2-AA (A and B) or [12C6]aniline (Arabidopsis oligosaccharides) and [13C6]aniline (radish oligosaccharides; C). The labeled glycans were either directly analyzed by MALDI-ToF-MS or were separated by HILIC and subsequently analyzed by MALDI-ToF-MS (inset in A). Peaks marked with asterisks were selected for MALDI-CID structural analysis, and the corresponding spectrum is shown in B. A, AGP extracts from radish leaves contain five different types of l-Fuc-modified oligosaccharides. For every oligosaccharide, sodiated molecular ions ([M + Na]+; peak assignment in this figure) and corresponding doubly sodiated molecular ions ([M + 2Na − H]+; 22 D larger, red circles) are observed. Separation of the structural isomers by HILIC (inset) shows that a single structural isomer is present for each oligosaccharide. B, MALDI-CID of the radish leaf 2-AA-labeled FucAraGal3 AG oligosaccharide. C, Extracted ion chromatograms for l-Fuc-modified oligosaccharides originating from radish leaf (red lines) or Arabidopsis leaf (black lines) AGPs hydrolyzed by AG-specific enzymes. Arabidopsis leaf AGP extracts contain three different l-Fuc-modified oligosaccharides.
Recently, we demonstrated that the chromatographic elution positions of an oligosaccharide labeled with [13C6]- and [12C6]aniline are identical, a property that can be used for comparison of the abundance of oligosaccharides between samples (Ridlova et al., 2008). Our hypothesis was that if Arabidopsis leaf AGP enzyme digests contain any oligosaccharides with l-Fuc modifications identical to the ones derived from radish leaf AGPs, then the chromatographic elution positions for these oligosaccharides derivatized with the 13C6- and 12C6-labeled isotope tags should be identical. Indeed, we identified three deoxyhex-decorated oligosaccharides from Arabidopsis leaf AGPs that coeluted with their isobaric l-Fuc-modified counterparts from radish leaf AGPs (Fig. 5C). This indicates that the deoxyhex-modified oligosaccharides detected in Arabidopsis leaf AGPs are l-Fuc modified. Taken together with the structural characterization data for radish leaf l-Fuc-modified AG oligosaccharides (Fig. 5, A and B), we can identify the l-Fuc-modified oligosaccharides present in AGP extracts from Arabidopsis leaves as α-l-Fuc-(1→2)-α-l-Araf-(1→3)-β-Galp-(1→6)-Galp, α-l-Fuc-(1→2)-α-l-Araf-(1→3)-β-Galp-(1→6)-β-Galp-(1→6)-Galp, and α-l-Fuc-(1→2)-α-l-Araf-(1→?)-α-l-Araf-(1→3)-β-Galp-(1→6)-β-Galp-(1→6)-Galp.
Arabidopsis mur1 Mutants Lack l-Fuc Modifications on Leaf AGPs
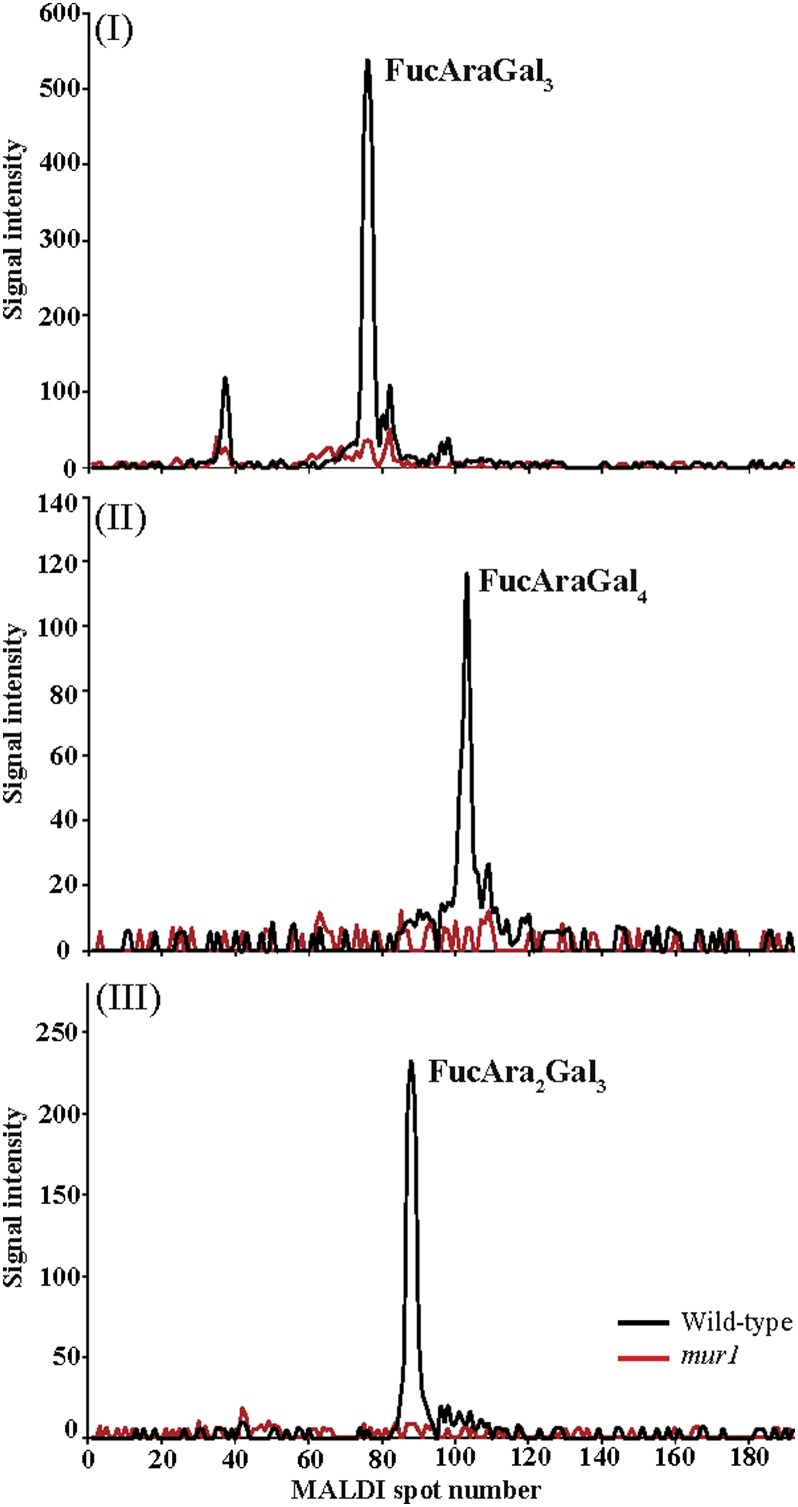
Arabidopsis mur1 mutants are affected in the l-Fuc biosynthetic pathway (Bonin et al., 1997). Alterations in the composition of root AGPs caused by the limited availability of l-Fuc have been reported, but the structure of the fucosylated AGP is unknown (Reiter et al., 1997). To obtain further evidence that leaf AGPs contain l-Fuc decorations, and to investigate whether this mutation alters AG structure, we investigated the structure of leaf AGPs in mur1 mutants. AGPs from mur1 Arabidopsis leaves were extracted and subjected to α-arabinofuranosidase, exo-β-(1→3)-galactanase, β-glucuronidase, and endo-β-(1→6)-galactanase sequential digestion to release the enzyme-resistant fucosylated oligosaccharides. The resulting oligosaccharides were compared with those prepared in the same way from leaf AGP extracts from wild-type plants using the [13C6]aniline/[12C6]aniline labeling approach. The HILIC-MALDI-ToF-MS extracted ion chromatograms for FucAraGal3, FucAraGal4, and FucAra2Gal3 oligosaccharides from the wild type (m/z 882, 1,086, and 1,014, respectively, labeled with the light isotope [12C6]) and the mur1 mutant (m/z 888, 1,092, and 1,020, respectively, labeled with the heavy isotope [13C6]) are shown in Figure 6. One main structural isomer is observed for each of the three oligosaccharides in the wild-type sample, while all three oligosaccharides are clearly absent from the extracted ion chromatograms of the 13C6-tagged oligosaccharides from mur1 samples. These results indicate that mur1 mutant leaf AGP extracts lack oligosaccharides with l-Fuc modifications.
Figure 6.
Capillary HILIC-MALDI-ToF-MS using stable isotopic labeling of l-Fuc-modified oligosaccharides from Arabidopsis leaf AGP extracts of wild-type (black line) and mur1 (red line) plants. Purified Arabidopsis leaf AGPs from wild-type and mur1 plants were subjected to a sequential digestion of α-l-arabinofuranosidase, exo-β-(1→3)-galactanase, β-glucuronidase, and endo-β-(1→6)-galactanase. The hydrolysis products were purified on a C18 cartridge (elution with 5% acetic acid) to remove the enzymes, followed by cation-exchange cleanup (Dowex resin; elution with 5% acetic acid) to remove salt, and were reductively aminated with [12C6]aniline (wild-type oligosaccharides; black lines) and [13C6]aniline (mur1 oligosaccharides; red lines). The labeled glycans were purified from the reduction buffers on a normal-phase cartridge (Glyko Clean S), and the purified oligosaccharides were separated by HILIC and analyzed by MALDI-ToF-MS. Although three l-Fuc-modified oligosaccharides were separated from the wild-type leaf AGP extracts of AG-specific hydrolyzed samples, no corresponding signals were detected from leaf AGP extracts from mur1 plants.
DISCUSSION
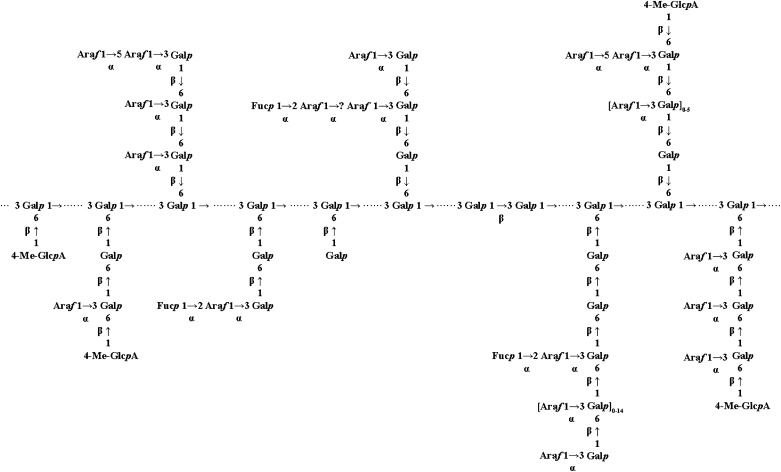
Even though Arabidopsis plants express substantial numbers of AGPs, our knowledge of the carbohydrate structure and biosynthesis of these molecules is very limited. In this work, we describe the structural characterization of the carbohydrate component of Arabidopsis leaf AGPs. Combining data from AG-specific enzyme digestion, PACE, and MALDI-ToF-MS (for a detailed list of some of the saccharide fragments identified, see Supplemental Table S1), we propose a model showing the structures present in Arabidopsis leaf AGs (Fig. 7). The model is similar to that proposed for radish root (Haque et al., 2005) and wheat flour AGPs (Tryfona et al., 2010) but very different from the modular structure proposed for AG on heterologously expressed proteins in tobacco and Arabidopsis (Tan et al., 2004, 2010). The AG structures identified here and shown in our model come from many different AGPs in the leaf extracts, and it will be interesting to determine if some of these structures are found specifically on particular AGP protein backbones. Although our extracts did not contain substantial RG-I, it is possible that some of the identified structures are found on free type II AG.
Figure 7.
A proposed model of the average structure of the carbohydrate component of Arabidopsis leaf AGPs. Structures determined in this work are shown in Supplemental Table S1. The dotted line indicates the β-(1→3)-linked backbone, but the arrangement or presence of the different side chains on specific AGPs is unknown. Some of these structures may also be present on pectic type II AG. We cannot exclude the presence of kinks of β-(1→6)-linked residues in the β-(1→3)-galactan backbone, as proposed by Tan et al. (2004).
The Arabidopsis AG has a backbone of β-(1→3)-galactan that is susceptible to exo-β-(1→3)-galactanase digestion. This enzyme releases some unsubstituted Gal, indicating that not all backbone residues are substituted. Nevertheless, most of the β-(1→3)-linked Gal residues are branched with β-(1→6)-galactan side chains, and many of these are longer than DP 5 and can reach DP 20 or more. Many β-(1→6)-galactan side chains in AGs characterized by NMR or linkage analysis are short, of only one or two residues (Gane et al., 1995; Gaspar et al., 2001; Tan et al., 2004). However, using AG-specific enzymes, long β-(1→6)-galactans have been reported in radish root AGPs (Haque et al., 2005) and more recently also in wheat flour AGPs (Tryfona et al., 2010). It is possible that the structures vary between species, but the use of the enzymatic profiling approach may facilitate the detection of these longer chains. We are unable to determine whether the backbone contains “kinks” of β-(1→6)-linked Gal residues, as proposed for certain AGPs (Tan et al., 2010). We did not observe any such mixed linkage galacto-oligosaccharides, but they might not be generated by exo-β-(1→3)-galactanase digestion. It is also possible that the structures of AG on heterologously expressed proteins (Tan et al., 2004) may be different from those found on endogenous AGPs.
We have also shown that GlcA residues are located on the nonreducing end of some β-(1→6)-galactan chains. These were easily detectable on most of the short side chains but may also be present on the longer branches. This is in accordance with reports on type II AG side chain substitutions (Gane et al., 1995; Redgwell et al., 2002; Tryfona et al., 2010). Predominantly, these GlcA residues are modified at O-4 by a methyl group, and only a small proportion of the GlcA residues present on Arabidopsis leaf AGPs are not methylated. We also found single 4-O-Me-GlcA and GlcA residues directly attached to the β-(1→3)-galactan backbone via a β-(1→6)-linkage.
Many, but not all, Gal residues in the β-(1→6)-galactan side chains are substituted with l-Araf residues at C-3, consistent with previous reports for type II AGs (Redgwell et al., 2002; Tan et al., 2004; Haque et al., 2005; Tryfona et al., 2010). We observed some oligosaccharides that likely contain side chains of two contiguous l-Araf residues, but we have not determined the linkage between them. Tan et al. (2004) and Hata et al. (1992) have also shown that AG polysaccharides in other species contain terminal l-Araf residues linked through α-(1→3)- or α-(1→5)-linkages, so the oligosaccharides we observed may contain α-l-Araf-(1→3 or 5)-α-l-Araf-(1→3) sequence linked to Galp.
Arabidopsis leaf AGP extracts are occasionally modified with l-Fuc, supporting previous reports of fucosylated type II AGPs in radish leaves (Nakamura et al., 1984; Tsumuraya et al., 1984a, 1984b) and Arabidopsis roots (van Hengel and Roberts, 2002). Using a strategy that combined sequential enzyme digestions with AG-specific enzymes and MALDI-ToF-MS, we identified three types of fucosylated oligosaccharides from Arabidopsis leaf AGP extracts. By comparison with radish leaf AG samples, we identified the radish and Arabidopsis leaf fucosylated AG oligosaccharides as α-l-Fuc-(1→2)-α-l-Araf-(1→3)-β-Galp-(1→6)-Galp, α-l-Fuc-(1→2)-α-l-Araf-(1→3)-β-Galp-(1→6)-β-Galp-(1→6)-Galp, and α-l-Fuc-(1→2)-α-l-Araf-(1→?)-α-l-Araf-(1→3)-β-Galp-(1→6)-β-Galp-(1→6)-Galp. The terminal α-l-Fuc-(1→2) linkage is consistent with the results of Misawa et al. (1996), who used a trisaccharide with terminal l-Araf residues α-(1→3)-linked to Gal as an acceptor for a radish AGP fucosyltransferase activity and showed that the enzyme fucosylated AGPs via an α-(1→2)-linkage onto l-Araf residues. The presence of differently structured fucosylated products is consistent with the view that AtFUT4.1 and AtFUT6 transfer l-Fuc residues onto different l-Araf acceptor residues in AGP molecules (Wu et al., 2010). We have not yet determined whether the fucosyl residues are solely at the nonreducing end of the β-(1→6)-galactan side chains or are also internal. We found them solely at the nonreducing end of the enzyme-released galacto-oligosaccharides, but they could arise by cleavage by the endo-β-(1→6)-galactanase adjacent to a fucosylated branch.
The mur1 Arabidopsis mutant, which cannot synthesize l-Fuc (Reiter et al., 1993, 1997), exhibits alterations in the composition of root AGPs (van Hengel and Roberts, 2002). In this work, we showed that the l-Fuc-modified oligosaccharides we identified on leaf AGP extracts from wild-type plants are not present in leaf AGP extracts from mur1 mutants, revealing an altered AG structure. This demonstrates that this AG characterization approach can reveal subtle changes in AG structure and opens the possibility of discovering and characterizing further mutants in the biosynthesis of AG in Arabidopsis.
The abundance of AGPs at the cell surface and their involvement in various processes of growth and development are clear (Seifert and Roberts, 2007), yet the AGP function at molecular level is relatively unexplored. Our structural elucidation of the carbohydrate component of Arabidopsis leaf AGPs can provide useful insights into the structure-function relationships of these molecules or even support existing hypotheses on their roles. For example, it has been suggested that the close proximity of GlcA residues on the AG polysaccharides may favor Ca+ chelation, which taken together with the signaling functions attributed to AGPs may suggest an involvement of those molecules in calcium signaling (Tan et al., 2010). Moreover, it has been suggested that type II AGPs may be associated sometimes with pectin, presumably contributing this way to the rigidification of the cell wall by oxidative cross-linking (Seifert and Roberts, 2007). The structural elucidation of endogenous Arabidopsis leaf AGPs described here may help to test these hypotheses. In addition, major bottlenecks in identifying the roles of candidate glycosyltransferases of AGP biosynthesis are the difficulty of making appropriate acceptor substrates for these enzymes and in characterizing changes in type II AG structure. The structures reported in this work will help to improve our knowledge of the enzymology of AGP biosynthesis.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) seeds were surface sterilized and sown on solid medium containing 0.5× Murashige and Skoog salts (Murashige and Skoog, 1962) including vitamins (Sigma) and Suc (1%, w/v). Following stratification for 48 h at 4°C in the dark, plates were transferred to a growth room at 20°C under white light with a 16-h-light/8-h-dark cycle and 60% humidity. After 2 to 3 weeks, these plants were transferred to soil (John Innes Compost No. 1) in pots of 8 cm × 6.5 cm (one plant per pot) under the same growth conditions. Alternatively, mur1 plants were grown on the above medium supplemented with 1 mm boric acid solution (O’Neill et al., 2001). During the initial growth stage, plastic domes were placed over these growing plants for 1 week to increase humidity. The age of plants in this work was defined as the number of days post stratification. Four to 6 weeks later, healthy green leaves from the rosettes were collected and frozen immediately in liquid nitrogen and then stored at −80°C before any further processing.
AIR Preparation
Arabidopsis leaves were harvested, submerged in 96% (v/v) ethanol, and boiled at 70°C for 30 min to inactivate enzymes. Following homogenization using a ball mixer mill (Glen Creston), the pellet was collected by centrifugation (4,000g for 15 min). The pellet was then washed with 100% (v/v) ethanol, twice with chloroform:methanol (2:1), followed by successive washes with 65% (v/v), 80% (v/v), and 100% (v/v) ethanol. The remaining pellet of AIR was air dried at 70°C overnight. Aqueous suspensions of AIR were prepared using a glass homogenizer.
AGP-Specific Enzymes, β-Galactosyl Yariv Reagent Synthesis, and Preparation of AG Proteins
α-l-Arabinofuranosidase (EC 3.2.1.55), exo-β-(1→3)-galactanase (EC 3.2.1.145), β-glucuronidase (EC 3.2.1.31), and endo-β-(1→6)-galactanase (EC 3.2.1.164) were prepared by methods described previously (Tsumuraya et al., 1990; Kotake et al., 2004; Konishi et al., 2008; Takata et al., 2010). Arabidopsis leaf AGPs were prepared using a protocol adapted from Tsumuraya et al. (1984a, 1988). Arabidopsis leaves were submerged in three times their weight of 1× phosphate-buffered saline with 0.05% sodium azide and homogenized in a Mill MM200 ball miller (Glen Creston) for 30 min. The resultant homogenate were heated in a boiling-water bath with occasional shaking for 30 min, cooled, and filtered through 5.5-cm Whatman No. 1 filter paper on a Buchner funnel to remove insoluble residues. Then, 2.5 volumes of ethanol was added to the clean filtrate. The mixture was shaken vigorously and left standing overnight at 4°C for the sediments to settle. These sediments were then collected by centrifugation at 12,000g for 20 min and suspended in 10 volumes of distilled water. After extraction at 4°C for 10 h, the insoluble materials were removed by centrifugation at 12,000g for 20 min. The resultant supernatant was dialyzed with SnakeSkin Dialysis Tubing in a molecular mass of 10 kD (Thermo Scientific) against several changes of distilled water and lyophilized to give a white powder. Radish (Raphanus sativus) leaf AGPs were prepared as described (Tsumuraya et al., 1984a). Crude Arabidopsis and radish leaf AGP preparations were freeze dried and kept at −20°C in 1-mg aliquots. β-Galactosyl Yariv reagent was prepared by the method of Yariv et al. (1967).
Enzymatic Hydrolysis of AGPs and PACE Analysis
AG peptide preparations (1 mg) were digested with recombinant α-l-arabinofuranosidase (16 milliunits) expressed in Pichia pastoris in 50 mm ammonium acetate buffer (pH 5.0, 100 μL) at 37°C for 24 h. The enzyme was inactivated for 5 min at 100°C, and the sample was dried in a rotary evaporator. For consecutive digestions with different AGP-specific enzymes, samples were dissolved in the respective digestion buffer and were incubated at 37°C for 24 h. The buffer composition was as follows: 20 mm ammonium acetate, pH 4.6, 10 mm ammonium acetate, pH 4.3, containing 50 mm KCl, and 20 mm ammonium acetate, pH 3.5, containing 150 mm NaCl for exo-β-(1→3)-galactanase, β-glucuronidase, and endo-β-(1→6)-galactanase, respectively. For the RG-I lyase (Jensen et al., 2010) treatment experiments, Arabidopsis leaf AIR and leaf AGP extracts were dissolved in 500 μL of 0.05 m Tris-HCl solution, pH 8.0, containing 2 mm CaCl2 and were incubated at 21°C for 24 h. The derivatization of carbohydrates was performed according to previously developed protocols (Goubet et al., 2002). Carbohydrate electrophoresis and PACE gel scanning and quantification were performed as described by Goubet et al. (2002, 2009). Control experiments without substrates or enzymes were performed under the same conditions to identify any nonspecific compounds in the enzymes, polysaccharides/cell walls, or labeling reagents. Standards for quantification (Gal, Man2, and Man3) were separated alongside samples on each gel to obtain a standard curve of pmol quantity of fluorophore-labeled oligosaccharide. The quantity of hydrolysis products (DP 1–16) in 1 μL of sample was calculated using this standard curve.
AGP Oligosaccharide Sample Desalting and Cleanup
Following the enzyme digestions and prior to per-deuteromethylation or per-methylation, released peptides and enzymes were removed using reverse-phase Sep-Pak C18 cartridges (Waters) as described previously (Tryfona et al., 2010).
per-Methylation and per-Deuteromethylation of AG Polysaccharides
The per-methylation of glycans was performed using the method described by Ciucanu and Kerek (1984). Glycans were lyophilized in a glass tube, and in order to avoid undermethylation, samples were kept in the lyophilizer until the time of per-methylation. In a dry mortar, a few NaOH pellets were placed and about 3 mL of dry dimethyl sulfoxide (Romil) was added. The NaOH pellets were ground until a slurry was formed, and 0.5 to 1 mL of the slurry was added to each sample tube. Accordingly, 1 mL of methyl iodide (Fluka; or deuteromethyl iodide in the case of per-deuteromethylation) was added, and the glass tubes were sealed with a screw cap. The samples were mixed vigorously for 10 min, and the reaction was quenched with the addition of 1 mL of water. Consequently, 2 mL of chloroform was added, and the samples were mixed well and allowed to settle into two layers. The upper aqueous phase was removed by a Pasteur pipette and discarded, while the lower phase was washed several times with water. Samples were then dried under a gentle N2 stream, and dry samples were resuspended in 100 μL of methanol and kept at room temperature for MALDI-ToF-MS analysis.
MALDI-ToF/ToF-MS/MS
Methylated methanol-dissolved samples (5 μL) were mixed with 5 μL of 2,5-dihydroxybenzoic acid matrix (10 mg mL−1 dissolved in 50% methanol), and 1 μL of the mixture was spotted on a MALDI target plate and rapidly dried in a vacuum desiccator in order to produce small crystals for easy spectral acquisition. Samples were analyzed by MALDI-ToF/ToF-MS/MS (4700 Proteomics Analyzer; Applied Biosystems) as described previously (Maslen et al., 2007). High-energy MALDI-CID spectra were acquired with an average 10,000 laser shots per spectrum using a high collision energy (1 kV). The oligosaccharide ions were allowed to collide in the CID cell with argon at a pressure of 2 × 10−6 Torr.
Reductive Amination of AG Oligosaccharides and Purification
The AG oligosaccharides were reductively aminated with 2-AA (Sigma) or [12C6]- and [13C6]aniline (Sigma) using optimized labeling conditions described previously (Ridlova et al., 2008; Tryfona and Stephens, 2010). The saccharides were then purified from the reductive amination reagents using a Glyko Clean S cartridge (Prozyme) as described previously (Tryfona and Stephens, 2010). When isotopic labeling was performed, samples labeled with the two aniline isotopes were mixed equally prior to purification from the reductive amination reagents.
HILIC-MALDI-ToF/ToF-MS/MS
Capillary HILIC was carried out using an LC-Packings Ultimate system (Dionex), which was used to generate the gradient that flowed at 3 μL min−1. Solvent A was 50 mm ammonium formate adjusted to pH 4.4 with formic acid. Solvent B was 5% solvent A in acetonitrile. The labeled oligosaccharides dissolved in 95% acetonitrile were loaded onto an amide-80 column (300 μm × 25 cm, 3-μm particle size; Dionex) and eluted with increasing aqueous concentrations. The following gradient conditions were applied: 0 min, 5% solvent A, 95% solvent B; 6 min, 25% solvent A, 75% solvent B; 86 min, 45% solvent A, 55% solvent B. The column eluent passed through a capillary UV detector (set at 254 nm) to the MALDI sample spotter. For HILIC-MALDI-ToF/ToF-MS/MS, a Probot sample fraction system (Dionex) was employed for automated spotting of the HPLC eluent onto a MALDI target at 20-s intervals. After air drying, the sample spots were overlaid with 2,5-dihydroxybenzoic acid matrix and analyzed by MALDI-ToF/ToF-MS/MS as described above.
Trifluoroacetic Acid Hydrolysis and HPAEC-PAD Monosaccharide Analysis
Samples were hydrolyzed in 2 m trifluoroacetic acid for 1 h at 120°C. Trifluoroacetic acid was removed by evaporation under vacuum. Following resuspension in 200 µL of water, the monosaccharide sugars were separated using protocols adapted from Currie and Perry (2006) on a Dionex ICS3000 system equipped with a PA20 column, a PA20 guard column, and a borate trap (Dionex). In particular, for the separation of neutral sugars, the initial isocratic wash with 12 mm KOH from 0 to 5 min followed a linear gradient from 5 to 20 min, down to 1 mm KOH. Finally, an isocratic wash step in 100 mm KOH was maintained for 5 min (from 20 to 25 min). Postcolumn addition of 100 mm KOH was performed to increase PAD sensitivity. Acidic sugars were separated using a linear gradient from 20 to 200 mm ammonium acetate in 100 mm NaOH over 10 min followed by an isocratic step in 200 mm ammonium acetate in 100 mm NaOH for 10 min.
β-Galactosyl Yariv Diffusion Assay
A Yariv diffusion assay for the AGPs was performed using a protocol adapted from van Holst and Clarke (1985). Arabidopsis leaf AGP samples (0.5 mg) were dissolved in 0.15 m NaCl buffer solution containing 0.02% sodium azide. Agarose gels (1%) containing the β-galactosyl Yariv reagent (0.002%), 0.15 m NaCl, and 0.02% sodium azide were poured onto microscope slides to give a layer of agarose approximately 3 mm thick. Wells were made in the gel, and 5, 7.5, and 10 μg of the polysaccharide sample were pipetted into the wells. The reagents without the polysaccharides were used as blanks, and gum arabic (Sigma) and oat (Avena sativa) spelt xylan (Sigma) were used as positive and negative controls, respectively. The microscope slides were incubated in a sealed moist, dark chamber at ambient temperature for 2 d to allow the halo to develop.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. PACE, HPAEC-PAD monosaccharide analysis of neutral sugars and monosaccharide composition of AGP extracts from Arabidopsis leaves.
Supplemental Table S1. Saccharide fragments characterized in this study.
Acknowledgments
We thank Mrs. Zhinong Zhang and Dr. Tina Theys for technical support and Dr. Jenny Mortimer for her assistance with the monosaccharide analysis. We thank Novozymes for the gift of RG-I lyase (EC 4.2.2.23).
Glossary
- AG
arabinogalactan
- AGP
arabinogalactan protein
- RG-I
rhamnogalacturonan-I
- Galp
galactopyranosyl
- GlcA
glucuronosyl
- l-Araf
l-arabinofuranosyl
- l-Rha
l-rhamnosyl
- l-Fuc
l-fucosyl
- MALDI
matrix-assisted laser desorption ionization
- CID
collision-induced dissociation
- MS
mass spectrometry
- PACE
polysaccharide analysis by carbohydrate gel electrophoresis
- 4-O-Me-GlcA
4-O-methyl-glucuronosyl
- DP
degrees of polymerization
- AIR
alcohol-insoluble residue
- HPAEC
high-pH anion-exchange chromatography
- PAD
amperometric detection
- ToF
time of flight
- m/z
mass-to-charge ratio
- MS/MS
tandem mass spectrometry
- 2-AA
2-aminobenzoic acid
- HILIC
hydrophilic interaction liquid chromatography
References
- Bonin CP, Potter I, Vanzin GF, Reiter W-D. (1997) The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-D-mannose-4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-L-fucose. Proc Natl Acad Sci USA 94: 2085–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GHH, Lilley KS, Stevens TJ, Dupree P. (2003) Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis: a proteomic and genomic analysis. Plant Physiol 132: 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai W, Piskarev V, Lawson AM. (2001) Negative-ion electrospray mass spectrometry of neutral underivatized oligosaccharides. Anal Chem 73: 651–657 [DOI] [PubMed] [Google Scholar]
- Cheung AY, Wang H, Wu HM. (1995) A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell 82: 383–393 [DOI] [PubMed] [Google Scholar]
- Chun H, Shin DH, Hong BS, Cho HY, Yang HC. (2001) Purification and biological activity of acidic polysaccharide from leaves of Thymus vulgaris L. Biol Pharm Bull 24: 941–946 [DOI] [PubMed] [Google Scholar]
- Ciucanu I, Kerek F. (1984) A simple and rapid method for the permethylation of carbohydrates. Carbohydr Res 131: 209–217 [Google Scholar]
- Clarke AE, Anderson RL, Stone BA. (1979) Form and function of arabinogalactans and arabinogalactan-proteins. Phytochemistry 18: 521–540 [Google Scholar]
- Currie HA, Perry CC. (2006) Resolution of complex monosaccharide mixtures from plant cell wall isolates by high pH anion exchange chromatography. J Chromatogr A 1128: 90–96 [DOI] [PubMed] [Google Scholar]
- Domon B, Costello CE. (1988) A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj J 5: 397–409 [Google Scholar]
- Estévez JM, Kieliszewski MJ, Khitrov N, Somerville C. (2006) Characterization of synthetic hydroxyproline-rich proteoglycans with arabinogalactan protein and extensin motifs in Arabidopsis. Plant Physiol 142: 458–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher GB, Stone BA, Clarke AE. (1983) Arabinogalactan-proteins: structure, biosynthesis, and function. Annu Rev Plant Physiol 34: 47–70 [Google Scholar]
- Gane AM, Craik D, Munro SLA, Howlett GJ, Clarke AE, Bacic A. (1995) Structural analysis of the carbohydrate moiety of arabinogalactan-proteins from stigmas and styles of Nicotiana alata. Carbohydr Res 277: 67–85 [DOI] [PubMed] [Google Scholar]
- Gaspar Y, Johnson KL, McKenna JA, Bacic A, Schultz CJ. (2001) The complex structures of arabinogalactan-proteins and the journey towards understanding function. Plant Mol Biol 47: 161–176 [PubMed] [Google Scholar]
- Goubet F, Barton CJ, Mortimer JC, Yu X, Zhang Z, Miles GP, Richens J, Liepman AH, Seffen K, Dupree P. (2009) Cell wall glucomannan in Arabidopsis is synthesised by CSLA glycosyltransferases, and influences the progression of embryogenesis. Plant J 60: 527–538 [DOI] [PubMed] [Google Scholar]
- Goubet F, Jackson P, Deery MJ, Dupree P. (2002) Polysaccharide analysis using carbohydrate gel electrophoresis: a method to study plant cell wall polysaccharides and polysaccharide hydrolases. Anal Biochem 300: 53–68 [DOI] [PubMed] [Google Scholar]
- Haque MA, Kotake T, Tsumuraya Y. (2005) Mode of action of β-glucuronidase from Aspergillus niger on the sugar chains of arabinogalactan-protein. Biosci Biotechnol Biochem 69: 2170–2177 [DOI] [PubMed] [Google Scholar]
- Hata K, Tanaka M, Tsumuraya Y, Hashimoto Y. (1992) α-L-Arabinofuranosidase from radish (Raphanus sativus L.) seeds. Plant Physiol 100: 388–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose H, Kuno A, Kotake T, Yoshida M, Sakka K, Hirabayashi J, Tsumuraya Y, Kaneko S. (2006) Characterization of an exo-β-1,3-galactanase from Clostridium thermocellum. Appl Environ Microbiol 72: 3515–3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MH, Otten H, Christensen U, Borchert TV, Christensen LLH, Larsen S, Leggio LL. (2010) Structural and biochemical studies elucidate the mechanism of rhamnogalacturonan lyase from Aspergillus aculeatus. J Mol Biol 404: 100–111 [DOI] [PubMed] [Google Scholar]
- Kieliszewski MJ, Lamport DTA. (1994) Extensin: repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant J 5: 157–172 [DOI] [PubMed] [Google Scholar]
- Konishi T, Kotake T, Soraya D, Matsuoka K, Koyama T, Kaneko S, Igarashi K, Samejima M, Tsumuraya Y. (2008) Properties of family 79 β-glucuronidases that hydrolyze β-glucuronosyl and 4-O-methyl-β-glucuronosyl residues of arabinogalactan-protein. Carbohydr Res 343: 1191–1201 [DOI] [PubMed] [Google Scholar]
- Kotake T, Kaneko S, Kubomoto A, Haque MA, Kobayashi H, Tsumuraya Y. (2004) Molecular cloning and expression in Escherichia coli of a Trichoderma viride endo-β-(1→6)-galactanase gene. Biochem J 377: 749–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LY, Ker YB, Chang CH, Chen KC, Peng RY. (2011) Arabinogalactan present in the mountain celery seed extract potentiated hypolipidemic bioactivity of coexisting polyphenols in hamsters. Pharm Biol 49: 319–326 [DOI] [PubMed] [Google Scholar]
- Maslen SL, Goubet F, Adam A, Dupree P, Stephens E. (2007) Structure elucidation of arabinoxylan isomers by normal phase HPLC-MALDI-TOF/TOF-MS/MS. Carbohydr Res 342: 724–735 [DOI] [PubMed] [Google Scholar]
- Misawa H, Tsumuraya Y, Kaneko Y, Hashimoto Y. (1996) α-l-Fucosyltransferases from radish primary roots. Plant Physiol 110: 665–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nakamura K., Tsumuraya Y, Hashimoto Y, Yamamoto S. (1984) Arabinogalactan-proteins reacting with Eel anti-H agglutinin from leaves of cruciferous plants. Agric Biol Chem 48: 753–760 [Google Scholar]
- Neukom H, Markwalder H. (1975) Isolation and characterization of an arabinogalactan from wheat flour. Carbohydr Res 39: 387–389 [Google Scholar]
- O’Neill MA, Eberhard S, Albersheim P, Darvill AG. (2001) Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science 294: 846–849 [DOI] [PubMed] [Google Scholar]
- Qu Y, Egelund J, Gilson PR, Houghton F, Gleeson PA, Schultz CJ, Bacic A. (2008) Identification of a novel group of putative Arabidopsis thaliana β-(1,3)-galactosyltransferases. Plant Mol Biol 68: 43–59 [DOI] [PubMed] [Google Scholar]
- Redgwell RJ, Curti D, Fischer M, Nicolas P, Fay LB. (2002) Coffee bean arabinogalactans: acidic polymers covalently linked to protein. Carbohydr Res 337: 239–253 [DOI] [PubMed] [Google Scholar]
- Reiter W-D, Chapple C, Somerville CR. (1997) Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J 12: 335–345 [DOI] [PubMed] [Google Scholar]
- Reiter W-D, Chapple CCS, Somerville CR. (1993) Altered growth and cell walls in a fucose-deficient mutant of Arabidopsis. Science 261: 1032–1035 [DOI] [PubMed] [Google Scholar]
- Renard CMGC, Voragen AGJ, Thibault JF, Pilnik W. (1991) Studies on apple protopectin. V. Structural studies on enzymatically extracted pectins. Carbohydr Polym 16: 137–154 [Google Scholar]
- Ridlova G, Mortimer JC, Maslen SL, Dupree P, Stephens E. (2008) Oligosaccharide relative quantitation using isotope tagging and normal-phase liquid chromatography/mass spectrometry. Rapid Commun Mass Spectrom 22: 2723–2730 [DOI] [PubMed] [Google Scholar]
- Seifert GJ, Roberts K. (2007) The biology of arabinogalactan proteins. Annu Rev Plant Biol 58: 137–161 [DOI] [PubMed] [Google Scholar]
- Spina E, Sturiale L, Romeo D, Impallomeni G, Garozzo D, Waidelich D, Glueckmann M. (2004) New fragmentation mechanisms in matrix-assisted laser desorption/ionization time-of-flight/time-of-flight tandem mass spectrometry of carbohydrates. Rapid Commun Mass Spectrom 18: 392–398 [DOI] [PubMed] [Google Scholar]
- Takata R, Tokita K, Mori S, Shimoda R, Harada N, Ichinose H, Kaneko S, Igarashi K, Samejima M, Tsumuraya Y, et al. (2010) Degradation of carbohydrate moieties of arabinogalactan-proteins by glycoside hydrolases from Neurospora crassa. Carbohydr Res 345: 2516–2522 [DOI] [PubMed] [Google Scholar]
- Tan L, Qiu F, Lamport DTA, Kieliszewski MJ. (2004) Structure of a hydroxyproline (Hyp)-arabinogalactan polysaccharide from repetitive Ala-Hyp expressed in transgenic Nicotiana tabacum. J Biol Chem 279: 13156–13165 [DOI] [PubMed] [Google Scholar]
- Tan L, Varnai P, Lamport DTA, Yuan C, Xu J, Qiu F, Kieliszewski MJ. (2010) Plant O-hydroxyproline arabinogalactans are composed of repeating trigalactosyl subunits with short bifurcated side chains. J Biol Chem 285: 24575–24583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryfona T, Liang H-C, Kotake T, Kaneko S, Marsh J, Ichinose H, Lovegrove A, Tsumuraya Y, Shewry PR, Stephens E, et al. (2010) Carbohydrate structural analysis of wheat flour arabinogalactan protein. Carbohydr Res 345: 2648–2656 [DOI] [PubMed] [Google Scholar]
- Tryfona T, Stephens E. (2010) Analysis of carbohydrates on proteins by offline normal-phase liquid chromatography MALDI-TOF/TOF-MS/MS. Methods Mol Biol 658: 137–151 [DOI] [PubMed] [Google Scholar]
- Tsumuraya Y, Hashimoto Y, Yamamoto S, Shibuya N. (1984a) Structure of l-arabino-d-galactan-containing glycoproteins from radish leaves. Carbohydr Res 134: 215–228 [Google Scholar]
- Tsumuraya Y, Mochizuki N, Hashimoto Y, Kovác P. (1990) Purification of an exo-beta-(1→3)-D-galactanase of Irpex lacteus (Polyporus tulipiferae) and its action on arabinogalactan-proteins. J Biol Chem 265: 7207–7215 [PubMed] [Google Scholar]
- Tsumuraya Y, Nakamura K, Hashimoto Y, Yamamoto S. (1984b) Immunological properties of arabinogalactan proteins from leaves of cruciferous plants. Agric Biol Chem 48: 2915–2917 [Google Scholar]
- Tsumuraya Y, Ogura K, Hashimoto Y, Mukoyama H, Yamamoto S. (1988) Arabinogalactan-proteins from primary and mature roots of radish (Raphanus sativus L.). Plant Physiol 86: 155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hengel AJ, Roberts K. (2002) Fucosylated arabinogalactan-proteins are required for full root cell elongation in Arabidopsis. Plant J 32: 105–113 [DOI] [PubMed] [Google Scholar]
- van Hengel AJ, Van Kammen A, De Vries SC. (2002) A relationship between seed development, arabinogalactan-proteins (AGPs) and the AGP mediated promotion of somatic embryogenesis. Physiol Plant 114: 637–644 [DOI] [PubMed] [Google Scholar]
- van Holst G-J, Clarke AE. (1985) Quantification of arabinogalactan-protein in plant extracts by single radial gel diffusion. Anal Biochem 148: 446–450 [DOI] [PubMed] [Google Scholar]
- Wu Y, Williams M, Bernard S, Driouich A, Showalter AM, Faik A. (2010) Functional identification of two nonredundant Arabidopsis alpha(1,2)fucosyltransferases specific to arabinogalactan proteins. J Biol Chem 285: 13638–13645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Tan L, Lamport DTA, Showalter AM, Kieliszewski MJ. (2008) The O-Hyp glycosylation code in tobacco and Arabidopsis and a proposed role of Hyp-glycans in secretion. Phytochemistry 69: 1631–1640 [DOI] [PubMed] [Google Scholar]
- Yariv J, Lis H, Katchalski E. (1967) Precipitation of arabic acid and some seed polysaccharides by glycosylphenylazo dyes. Biochem J 105: 1C–2C [DOI] [PMC free article] [PubMed] [Google Scholar]