Abstract
Monoacylglycerol acyltransferase (MGAT) catalyzes the synthesis of diacylglycerol, the precursor of triacylglycerol biosynthesis and an important signaling molecule. Here, we describe the isolation and characterization of the peanut (Arachis hypogaea) MGAT gene. The soluble enzyme utilizes invariant histidine-62 and aspartate-67 residues of the acyltransferase motif for its MGAT activity. A sequence analysis revealed the presence of a hydrolase (GXSXG) motif, and enzyme assays revealed the presence of monoacylglycerol (MAG) and lysophosphatidylcholine (LPC) hydrolytic activities, indicating the bifunctional nature of the enzyme. The overexpression of the MGAT gene in yeast (Saccharomyces cerevisiae) caused an increase in triacylglycerol accumulation. Similar to the peanut MGAT, the Arabidopsis (Arabidopsis thaliana) homolog (At1g52760) also exhibited both acyltransferase and hydrolase activities. Interestingly, the yeast homolog lacks the conserved HX4D motif, and it is deficient in the acyltransferase function but exhibits MAG and LPC hydrolase activities. This study demonstrates the presence of a soluble MGAT/hydrolase in plants. The predicted three-dimensional homology modeling and substrate docking suggested the presence of two separate substrate (MAG and LPC)-binding sites in a single polypeptide. Our study describes a soluble bifunctional enzyme that has both MGAT and hydrolase functions.
Plant vegetative cells contain 5% to 10% lipid by dry weight, and almost this entire lipid is found in the membranes. Although lipids, proteins, and carbohydrate are the major forms of carbon in oilseeds that are used for germination and metabolism, lipids constitute up to 60% of the oilseed dry weight (Ohlrogge and Browse, 1995). Diacylglycerol (DAG) consists of a glycerol backbone linked to two fatty acids via ester bonds. It is a component of biological membranes and functions as an intermediate for lipid biosynthesis. In animal systems, DAG plays a role in cell signaling (Ron and Kazanietz, 1999), intestinal fat absorption (Bell and Coleman, 1980; Lehner and Kuksis, 1996; Phan and Tso, 2001), energy storage in muscle and adipose tissues (Swanton and Saggerson, 1997), and lactation (Bell and Coleman, 1980; Smith et al., 2000). In addition to its role in metabolism and signaling, DAG also increases the oil (triacylglycerol [TAG]) content in plants.
DAG is an important branch point between the neutral lipid and membrane phospholipid biosynthetic pathways. De novo biosynthesis of DAG mainly takes place through two pathways in higher plants. One is the prokaryotic pathway of the chloroplast inner envelope, and the second is the eukaryotic pathway that occurs in the endoplasmic reticulum (Ohlrogge and Browse, 1995). Both the pathways start with acylation of glycerol-3-phosphate to generate lysophosphatidic acid (LPA) in plant systems (Ohlrogge and Browse, 1995; Shen et al., 2010). The eukaryotic pathway uses acyl-CoAs and the prokaryotic pathway uses acyl-ACPs, and intermediates such as DAG could be exchanged between the two systems. In animal (Hajra et al., 2000) and yeast (Saccharomyces cerevisiae; Athenstaedt et al., 1999; Rajakumari et al., 2008) systems, LPA is also synthesized by the acylation and subsequent reduction of dihydroxyacetone phosphate (DHAP) catalyzed by DHAP acyltransferase and the NADPH-dependent acyl-DHAP reductase. In contrast, most prokaryotes utilize the PlsX/Y pathway, which produces LPA using acyl-phosphate (Zhang and Rock, 2008). The LPA is either acylated to form phosphatidic acid (PA) or dephosphorylated to form monoacylglycerol (MAG; a PA-independent pathway). In the PA-dependent pathway, DAG is produced by dephosphorylation of PA by PA phosphatase. Alternatively, a nucleotide-activated form of DAG (i.e. CDP-DAG) is produced from the reaction of PA with CTP. Conversely, CDP-DAG reacts with myoinositol, Ser, and glycerol-3-phosphate and results in the formation of phosphatidylinositol, phosphatidylserine, and phosphatidylglycerol, respectively. Plants are also able to synthesize phosphatidylserine through exchange reactions with phosphatidylcholine and phosphatidylethanolamine (Vincent et al., 2001).
In the PA-independent pathway, LPA is hydrolyzed by a specific phosphatase and thus produces MAG. LPA phosphatase is well characterized in animals (Xie and Low, 1994; Hiroyama and Takenawa, 1999), and a soluble magnesium-dependent LPA phosphatase from Arabidopsis (Arabidopsis thaliana; Reddy et al., 2010) and yeast (Reddy et al., 2008) have also been reported. The synthesized MAG is then converted to DAG by monoacylglycerol acyltransferase (MGAT), and this step may be one of the major reactions for DAG synthesis in eukaryotes (Coleman and Haynes, 1986; Yen et al., 2002; Cao et al., 2003). In mammals, the role of acyl-CoA:MGAT has been clearly established (Coleman and Haynes, 1986), where the enzyme plays a predominant role in dietary fat absorption in the small intestine (Cao et al., 2003). Recently, peanut (Arachis hypogaea) oleosin was shown to have both MGAT and phospholipase A2 activities (Parthibane et al., 2012b), and the bifunctional activities were shown to be regulated by Ser/Thr/Tyr protein kinase (Parthibane et al., 2012a).
We have identified earlier an acyl-CoA-dependent MGAT in the soluble fraction of immature peanut seeds (Tumaney et al., 2001). However, the amino acid sequence of the enzyme was not known. This study describes the molecular cloning and functional characterization of MGAT of immature peanut seeds (AhMGAT). AhMGAT exhibits both MGAT and hydrolase activities. At1g52760 and YKL094W are the homologs of AhMGAT in Arabidopsis and yeast, respectively, and both exhibit MAG and lysophosphatidylcholine (LPC) hydrolase activities. In silico data clearly indicate the existence of two binding sites on AhMGAT, one on the surface that is needed for the interaction with MAG and a second site that is located in the groove for LPC binding. Overall, these studies support the experimental results showing AhMGAT with multiple functions.
RESULTS
Identification of the Gene Encoding AhMGAT
A full-length complementary DNA (cDNA) clone encoding a putative MGAT protein was isolated by screening a peanut seed-specific λ phage library with the oligonucleotide primer designed based on the N-terminal sequence of the peanut MGAT (Tumaney et al., 2001). A number of positive clones were isolated, and the clones with the longest inserts (approximately 1.0–1.5 kb) were sequenced from both ends. The analysis of nucleotide sequence revealed an open reading frame of 321 amino acid residues. The 3′-untranslated region contained a noncanonical polyadenylation signal (AATAT) near the poly(A) tail and 5′-end starts with a stretch of untranslated region containing the start codon in the middle of it. This indicates that the isolated clone possibly represents a full-length cDNA. Following sequencing of the positive clone, only the open reading frame starting with an N-terminal Met (5′-end) and the stop codon at the 3′-end was cloned. A Kyte-Doolittle hydropathy plot (Kyte and Doolittle, 1982) indicated the likely absence of transmembrane domains.
Domain Organization and Phylogenetic Analysis of AhMGAT
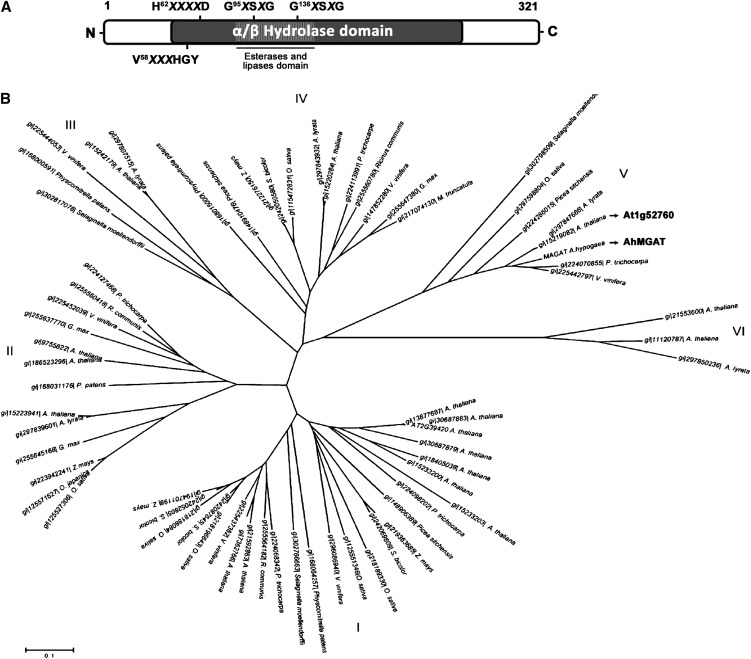
AhMGAT (GenBank accession no. JF340215) is a member of α/β-hydrolase family. In addition, it possesses an esterase (COG1647), a hydrolase/acyltransferase domain (COG0596/COG1073), and a lysophospholipase domain (COG2267). The protein sequence was analyzed for the presence of structural motifs, and we found that it has putative phosphorylation/glycosylation sites. The GXSXG motifs form a highly conserved stretch of amino acids in the majority of known lipases, phospholipases, lysophospholipases, esterases, and Ser proteases (Wei et al., 1999). AhMGAT also possesses a lipid-binding motif (58VX3HGF64). An interesting feature of this protein is that it has a distinct acyltransferase motif, 62HX4D67, and it also has two lipase/esterase motifs, G95XSXG and G136XSXG (Fig. 1A). Using AhMGAT as a query sequence, 67 AhMGAT-related sequences from 25 different plant species were identified that belong to the α/β-hydrolase family. Most of them are unannotated, and some are predicted as lipases, lysophospholipases, and esterases. For 67 AhMGAT-related amino acid sequences, the phylogenetic tree was constructed with a neighbor-joining algorithm using the MEGA software package. All of the sequences belong to α/β-hydrolase broad family. In this, six distinct subgroups were identified, and AhMGAT comes under subgroup V (Fig. 1B). The orthologous protein of AhMGAT in yeast, YKL094W, belongs to clade VI.
Figure 1.
The domain structure and phylogenetic analysis of peanut MGAT. A, Schematic representation of domains retrieved from the conserved domain database at the NCBI: α/β-hydrolase fold (COG0596 and COG1073); PldB, lysophospholipase (COG2267); esterase (COG1647). The presence of an acyltransferase motif (H62X4D) and two lipase motifs (G95XSXG and G136XSXG) are indicated. B, Phylogenetic tree of AhMGAT and its homologs in plants. The evolutionary history was inferred using the neighbor-joining method with 500 replicates. The length of the branches is proportional to the degree of divergence and, thus, corresponds to the statistical significance of the phylogeny between the protein sequences. The evolutionary distances were computed using the Poisson correction method; units used were number of amino acid substitutions per site. All positions with gaps and missing data were eliminated from the data set (complete deletion option). A phylogenetic analysis was performed using MEGA4.
AhMGAT Encodes MGAT
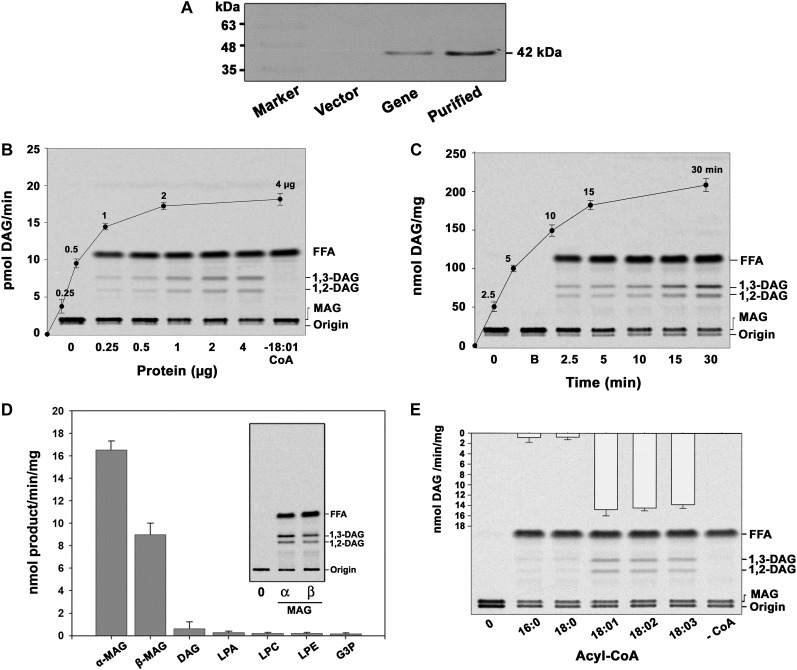
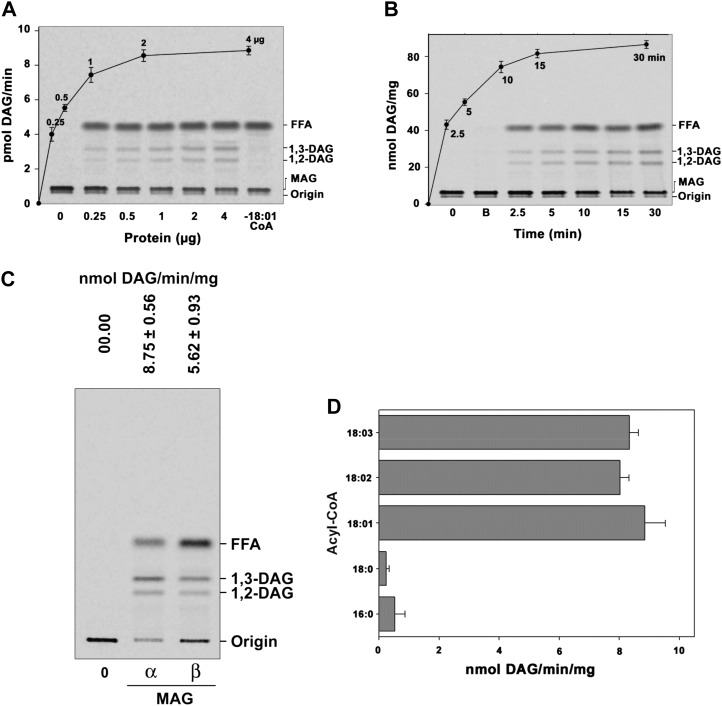
The AhMGAT protein possesses a conserved acyltransferase domain, and we studied the possible acyltransferase activity with dialyzed yeast recombinant AhMGAT using [14C]MAG and oleoyl-CoA. Prior to assays, the expression of AhMGAT in yeast cells was confirmed by immunoblot with anti-His6 monoclonal antibody (Fig. 2A). Yeast (BY4741) has no intrinsic MGAT activity. The protein-dependent (Fig. 2B) and time-dependent (Fig. 2C) formation of DAG was observed in the presence of AhMGAT when compared with no-enzyme and boiled-enzyme controls. To assess the in vitro acyl acceptor specificity of AhMGAT, we performed the assay with DAG, LPA, LPC, lysophosphatidylethanolamine, and glycerol-3-phosphate. The enzyme exhibited an insignificant acyltransferase activity toward these substrates; the highest activity was observed with MAG when compared with other substrates (Fig. 2D). These data suggest that the enzyme has a preference toward MAG. To investigate the acyl donor specificity, the MGAT assay was performed using different acyl-CoAs, and we found that the enzyme exhibited preference toward unsaturated fatty acyl-CoAs (Fig. 2E). Our results demonstrate that AhMGAT preferred to acylate MAG, specifically with unsaturated fatty acid.
Figure 2.
The isolated cDNA clone encodes MGAT. AhMGAT and its corresponding vector were transformed into wild-type yeast. A, Yeast cells overexpressing AhMGAT was confirmed by western-blot analysis using anti-His monoclonal antibody. B, Recombinant AhMGAT was purified by Ni2+-NTA column chromatography, and the dialyzed protein was used as the enzyme source. MGAT activity was determined with increasing amounts of protein (0–4 µg) for 15 min with [14C]MAG and 20 µm oleoyl-CoA. Lane 0, Enzyme was added after stopping the reaction. FFA, Free fatty acid. C, The time-dependent acylation of MAG; the reaction was initiated by the addition of 2 µg of protein. The reaction was stopped by extracting lipids, and the lipids were separated on a silica-TLC plate using petroleum ether:diethyl ether:acetic acid (70:30:1, v/v) as the solvent system. Lane 0, Enzyme was added after stopping the reaction; lane B, enzyme fraction was boiled for 5 min, and assay was performed (boiled enzyme control). D, The preference for various acyl acceptors was determined using 2 µg of protein, 50 μm acyl acceptor, and 20 μm [14C]oleoyl-CoA. The inset represents a typical phosphor image of MGAT assay with [14C]oleoyl-CoA and α,β-MAG as substrates. Lane 0, Enzyme was added after stopping the reaction. G3P, Glycerol-3-phosphate; LPE, lysophosphatidylethanolamine. Values are means ± sd of three independent experiments. E, Preference of acyl-CoAs. The assay was performed with 50 μm [14C]MAG with 20 μm of different acyl-CoAs. Lane 0, Enzyme was added after stopping the reaction. Values are means ± sd of three independent determinations.
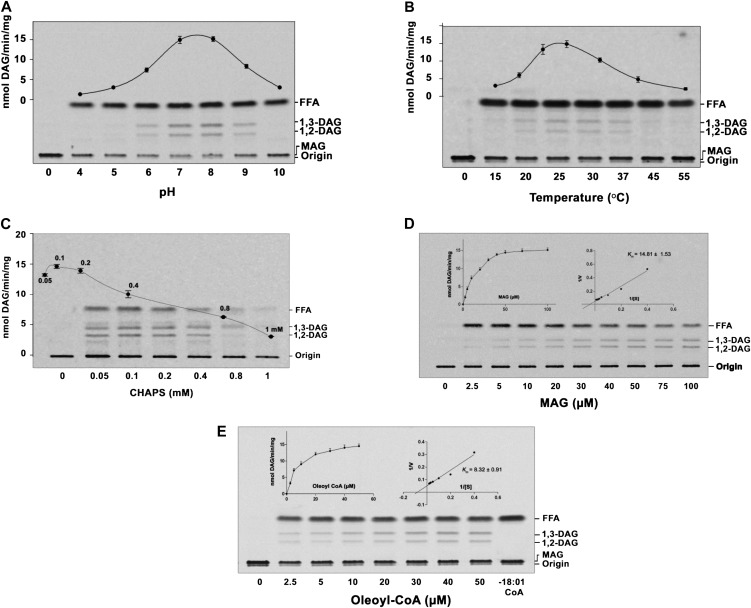
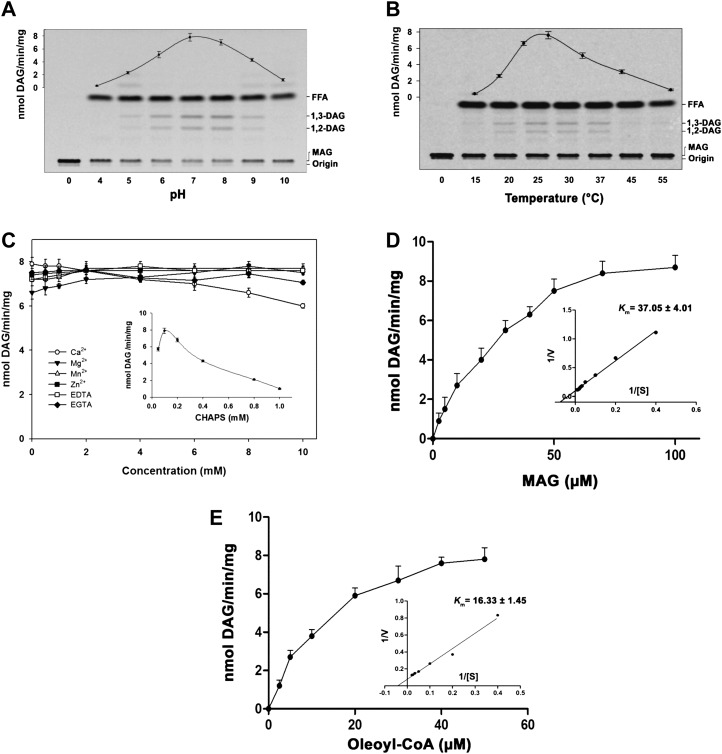
MGAT activity was characterized using [14C]MAG and oleoyl-CoA. Acyltransferase showed a maximum activity at pH 7.0 to 8.0 (Fig. 3A) and an optimum temperature of 30°C (Fig. 3B). CHAPS at low concentration showed an increased activity, but high concentrations had an inhibitory effect (Fig. 3C). However, divalent cations like Ca2+, Mg2+, Mn2+, Zn2+, EDTA, and EGTA (0–10 mm) had no effect on the enzyme activity. The increase in substrate concentrations of [14C]MAG or oleoyl-CoA with a fixed concentration of oleoyl-CoA or [14C]MAG obeyed the saturation kinetics, and the maximum velocity was approximately 15 nmol DAG formed min−1 mg−1. Apparent Km values for α-MAG and oleoyl-CoA were calculated to be 14.81 µm (Fig. 3D) and 8.32 µm (Fig. 3E), respectively.
Figure 3.
Characterization of AhMGAT. The enzyme activity was performed under different assay conditions with the purified AhMGAT protein for 10 min. A, MGAT assay was performed at different pH values with 50 µm [14C]MAG (prepared with CHAPS) and 20 µm oleoyl-CoA at 30°C. FFA, Free fatty acid. B, Temperature dependence of MGAT activity. C, MGAT activity was monitored in the presence of various concentrations of CHAPS (0.05–1 mm). Values are means ± sd of three independent experiments. D, Lineweaver-Burk plot of AhMGAT toward MAG. Activity was measured as a function of MAG concentration, and 20 µm [14C]oleoyl-CoA was kept constant. Values are averages of two independent determinations. E, Lineweaver-Burk plot of AhMGAT toward oleoyl-CoA. Activity was measured with 50 µm [14C]MAG and increasing concentrations of oleoyl-CoA. Lane 0, Enzyme was added after stopping the reaction. Values are averages of two independent determinations.
AhMGAT Is a Multifunctional Enzyme
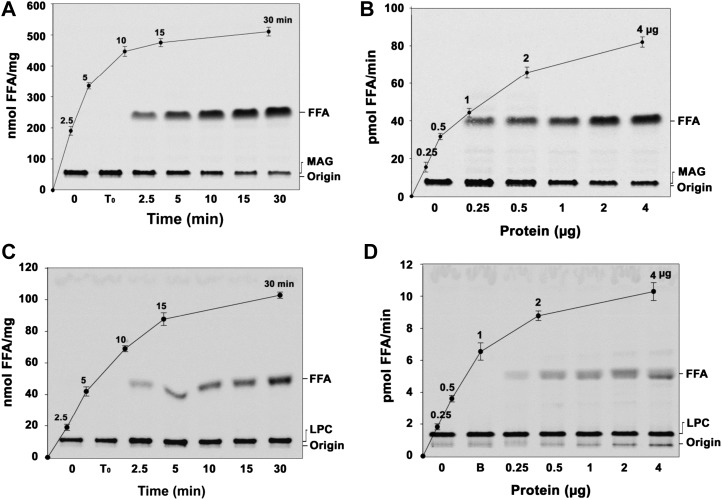
Figure 2, B and C, depict the formation of DAG accompanied with the reduced MAG in a protein- and time-dependent manner. In addition, we also observed the increased amount of free fatty acid. Based on the sequence analysis of AhMGAT, the protein was predicted to have phospholipase-like activity. We hypothesized that AhMGAT is a multifunctional enzyme with hydrolytic activities. Hence, we tested the hydrolase and phospholipase assays using radiolabeled substrates. The enzyme effectively hydrolyzed [14C]MAG in a time-dependent (Fig. 4A) and protein-dependent (Fig. 4B) manner. Apart from MAG, the enzyme also hydrolyzed [14C]LPC in a time-dependent (Fig. 4C) and protein-dependent (Fig. 4D) manner. The MAG hydrolase activity was 6.6-fold higher than LPC hydrolysis. Acyltransferase activity was approximately 2-fold higher than the LPC hydrolase activity and 3-fold lower than the MAG hydrolase activity. There was no hydrolytic activity observed with [14C]TAG, [14C]DAG, or [14C]phosphatidylcholine. Overall, these data suggest that peanut MGAT also exhibits MAG and LPC hydrolase activities.
Figure 4.
The peanut MGAT exhibits both MAG and LPC hydrolase activities. A, Time-dependent hydrolysis of MAG at 30°C with 2 µg of recombinant protein. Lane 0, Enzyme was added after stopping the reaction; lane T0, zero time point (reaction was stopped immediately after adding the enzyme). FFA, Free fatty acid. B, Protein-dependent MAG hydrolase assay was conducted with 50 µm [14C]MAG for 10 min at 30°C. Lane 0, Enzyme was added after stopping the reaction. The reaction was stopped by extracting lipids, and the lipids were separated on a silica-TLC plate using petroleum ether:diethyl ether:acetic acid (70:30:1, v/v) as the solvent system. Values are means ± sd of three independent experiments. C, The time-dependent LPC hydrolase assay was performed using 50 µm [14C]LPC with 2 µg of recombinant AhMGAT. Lane 0, Enzyme was added after stopping the reaction; lane T0, zero time point (reaction was stopped immediately after adding the enzyme). D, The LPC hydrolase assay was performed for 10 min at 30°C with increasing amounts of purified AhMGAT. Lane 0, Enzyme was added after stopping the reaction; lane B, enzyme fraction was boiled for 5 min, and assay was performed. The reaction was stopped and lipids were resolved on a TLC plate using chloroform:methanol:28% ammonia (65:25:5, v/v) as the solvent system. Values are means ± sd of three independent experiments.
AhMGAT Overexpression Enhances TAG Accumulation in Yeast
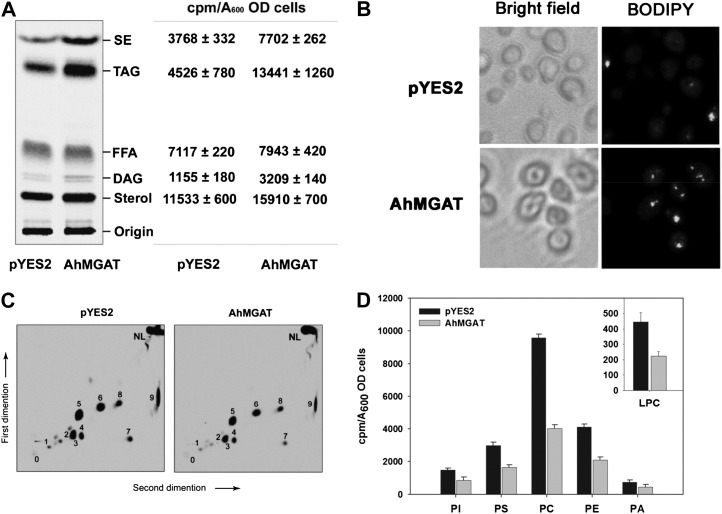
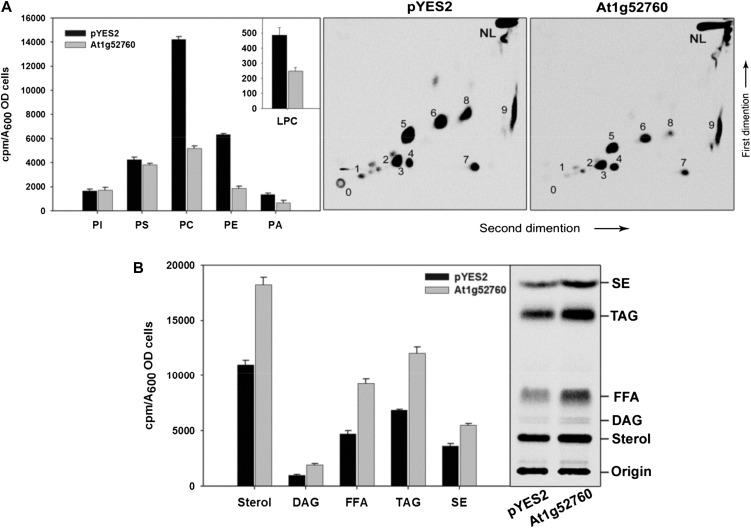
To assess the effect of AhMGAT overexpression on cellular neutral lipids, yeast was transformed with pYES2 vector and pYES2-AhMGAT. Next, we performed the incorporation of [14C]acetate into neutral lipids in recombinant yeast cells. In AhMGAT-expressing yeast, there was a higher accumulation of TAG (3-fold) than in vector control (Fig. 5A). In addition, there was a 2-fold increase in steryl ester level. These data suggest that the DAG synthesized by AhMGAT may subsequently be converted into TAG by the endogenous acyltransferases. Accumulation of TAG was further confirmed by staining of neutral lipids (Gocze and Freeman, 1994) with BODIPY493/503, a fluorescent neutral lipid probe (Fig. 5B). Apart from the TAG accumulation, there was a decrease in the overall phospholipid content observed in AhMGAT-expressing yeast cells (Fig. 5C), particularly phosphatidylcholine and LPC (approximately 50%) followed by phosphatidylethanolamine, when compared with vector control (Fig. 5D). Even though we observed a reduction in phosphatidylcholine, there was no LPC formation, which could be due to LPC hydrolysis by AhMGAT.
Figure 5.
Overexpression of AhMGAT increases TAG formation. MGAT was overexpressed in wild-type yeast in the presence of [14C]acetate (0.5 μCi mL−1). The yeast cells were grown in SM-U containing 2% Gal for 24 h, and equal absorbance (A600 = 20) of cells was harvested and lipids extracted using chloroform:methanol (1:2, v/v). The neutral lipids and phospholipids were separated by silica-TLC, and the radioactivity was quantified by a liquid scintillation counter. A, Phosphor image of TLC showing the [14C]acetate labeling of yeast neutral lipids. Lipids were identified by comparison with known standards. The table represents the incorporation of radiolabels as cpm quantified by liquid scintillation counter using toluene-based scintillation fluid. sd is for three independent experiments. FFA, Free fatty acid. B, Lipid droplet formation in wild-type cells was confirmed by BODIPY493/503 (green) staining. Top panels, wild-type yeast cells expressing pYES2 vector alone; bottom panels, yeast cells overexpressing AhMGAT. C, Incorporation of [14C]acetate into yeast phospholipids, separated by two-dimensional TLC using chloroform:methanol:ammonia (65:25:5, v/v) in the first-dimension solvent system followed by chloroform:methanol:acetone:acetic acid:water (50:10:20:15:5, v/v) as the second-dimension solvent systems. 0, Origin; 1, LPC; 2, PI, phosphatidylinositol; 3, PS, phosphatidylserine; 5, PC, phosphatidylcholine; 6, PE, phosphatidylethanolamine; 4, 7, 8, and 9, unknown; NL, neutral lipid. D, Quantification of phospholipids. Values are means ± sd of three independent experiments. OD, Optical density.
At1g52760 Is Homologous to Peanut MGAT
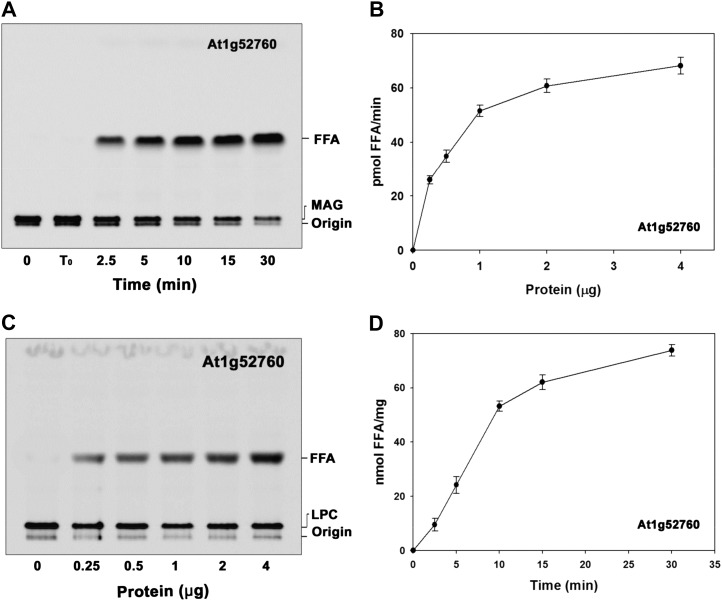
The BLASTx search analysis of AhMGAT in the Arabidopsis database identified At1g52760, which corresponds to gi:15219082 as the closest homolog. At1g52760 protein shares 86.9% similarity and 76.4% identity with peanut MGAT protein. Considering this homology, At1g52760 was cloned into pYES2/NT B and overexpressed in yeast. The recombinant protein was purified and assayed for MGAT activity. We observed an increased formation of DAG in protein (Fig. 6A) and time-dependent (Fig. 6B) experiments. The AtMGAT activity was stable only for 2 d at 4°C. Hence, the assays were performed with fresh protein preparation. At1g52760 preferred to acylate MAG (Fig. 6C) over other acyl acceptors like DAG, LPA, LPC, and glycerol-3-phosphate (data not shown); the acyl-CoA preference was with unsaturated fatty acids (Fig. 6D). Arabidopsis MGAT showed a maximum activity at pH 7.0 to 8.0 (Fig. 7A) and temperatures of 25°C to 30°C (Fig. 7B). Low concentration (0.1 mm) of CHAPS showed an increased activity, and the subsequent high concentrations had an inhibitory effect (Fig. 7C). Moreover, Ca2+, Mg2+, Mn2+, Zn2+, and chelating agents had no significant effect on the enzyme activity (Fig. 7C). Apparent Km values for α-MAG and oleoyl-CoA were calculated to be 37.05 µm (Fig. 7D) and 16.33 µm (Fig. 7E).
Figure 6.
Characterization of At1g52760, a homolog of AhMGAT. A, MGAT activity was determined under standard assay conditions with an increasing amount of protein for 15 min with [14C]MAG and 20 µm oleoyl-CoA. FFA, Free fatty acid. B, The time-dependent acylation of MAG with 2 µg of protein. Lane 0, Enzyme was added after stopping the reaction; lane B, enzyme fraction was boiled for 5 min, and assay was performed. C, A typical phosphor image of MGAT assay with [14C]oleoyl-CoA and α,β-MAG as substrates. D, Preference of acyl-CoAs. The assay was performed with 50 μm [14C]MAG with 20 μm of different acyl-CoAs. The values are means ± sd of three independent determinations.
Figure 7.
Characterization of At1g52760. MGAT activity was determined under different assay conditions with purified At1g52760 protein for 10 min. A, MGAT assay was performed at different pH values with [14C]MAG (prepared in CHAPS) and 20 µm oleoyl-CoA at 30°C. The experiment was repeated a minimum of three times. Representative TLC is shown. FFA, Free fatty acid. B, Temperature dependence of MGAT activity. Lane 0, Enzyme was added after stopping the reaction. C, MGAT activity was monitored in the presence of CHAPS (0.05–1 mm), various divalent cations, and chelating agents (concentration used was up to 10 mm). The experiment was repeated three times. D, Lineweaver-Burk plot of Arabidopsis MGAT toward MAG. Activity was measured as a function of MAG concentration, and 20 µm [14C]oleoyl-CoA was kept constant. Values are averages of two independent determinations. E, Lineweaver-Burk plot of AhMGAT toward oleoyl-CoA. Activity was measured with 50 µm [14C]MAG and increasing concentrations of oleoyl-CoA. Values are averages of two independent determinations.
We performed the MAG hydrolase assay and observed time-dependent (Fig. 8A) and protein-dependent (Fig. 8B) hydrolysis of MAG by At1g52760. It was previously reported that At1g52760 functions as a LPC hydrolase (Gao et al., 2010). To study the multiple functions of this protein, protein- and time-dependent [14C]LPC hydrolysis were performed (Fig. 8, C and D), and the LPC activity of At1g52760 was approximately 6-fold higher than AhMGAT. These in vitro assays revealed that At1g52760 functions as both MAG and LPC lipases.
Figure 8.
The MAG and LPC hydrolase activities of At1g52760. A, The time-dependent hydrolysis of [14C]MAG in the presence of 2 µg of protein. Lane 0, Enzyme was added after stopping the reaction; lane T0, zero time point (reaction was stopped immediately after adding the enzyme). FFA, Free fatty acid. B, Hydrolysis of [14C]MAG with increasing amounts of purified At1g52760 protein for 10 min. The reaction was stopped by extracting lipids, and the lipids were separated on a silica-TLC plate using petroleum ether:diethyl ether:acetic acid (70:30:1, v/v) as the solvent system. Lane 0, No-enzyme control; lane B, enzyme fraction was boiled for 5 min, and assay was performed. Values are means ± sd of three independent experiments. C, The LPC hydrolase assay was performed with increasing amounts of purified At1g52760 for 10 min at 30°C. Lane 0, Enzyme was added after stopping the reaction. D, Time-dependent LPC acylhydrolase assay using [14C]LPC with 2 µg of purified At1g52760 protein. The reaction was stopped, and lipids were resolved on a TLC plate using chloroform:methanol:28% ammonia (65:25:5, v/v) as the solvent system. Lane 0, No-enzyme control. Values are means ± sd of three independent experiments.
The overexpression of At1g52760 in wild-type yeast caused a 2-fold increase in the formation of TAG as compared with the vector control (Fig. 9A). In addition, AtMGAT overexpression caused a significant decrease in overall phospholipids, particularly phosphatidylethanolamine followed by phosphatidylcholine (Fig. 9B). LPC was also decreased in the At1g52760-overexpressed yeast.
Figure 9.
[14C]Acetate labeling of phospholipids and neutral lipids in At1g52760-overexpressing yeast cells. The peanut MGAT homolog of Arabidopsis At1g52760 was overexpressed in wild-type yeast in the presence of [14C]acetate (0.5 μCi mL−1). The yeast cells were grown in SM-U containing 2% Gal for 24 h, and equal absorbance (A600 = 20) of cells was harvested and lipids were extracted. A, Incorporation of [14C]acetate into neutral lipids. Lipids were separated on TLC using petroleum ether:diethyl ether:acetic acid (70:30:1, v/v). 0, Origin; 1, LPC; 2, PI, phosphatidylinositol; 3, PS, phosphatidylserine; 5, PC, phosphatidylcholine; 6, PE, phosphatidylethanolamine; 4, 7, 8, and 9, unknown; NL, neutral lipid. B, [14C]Acetate incorporation into yeast phospholipids. Two-dimensional TLC using chloroform:methanol:ammonia (65:25:5, v/v) was used in the first dimension followed by chloroform:methanol:acetone:acetic acid:water (50:10:20:15:5, v/v) in the second dimension. Values are means ± sd of three independent experiments. FFA, Free fatty acid; OD, optical density.
YKL094W Is Homologous to Peanut MGAT in Yeast
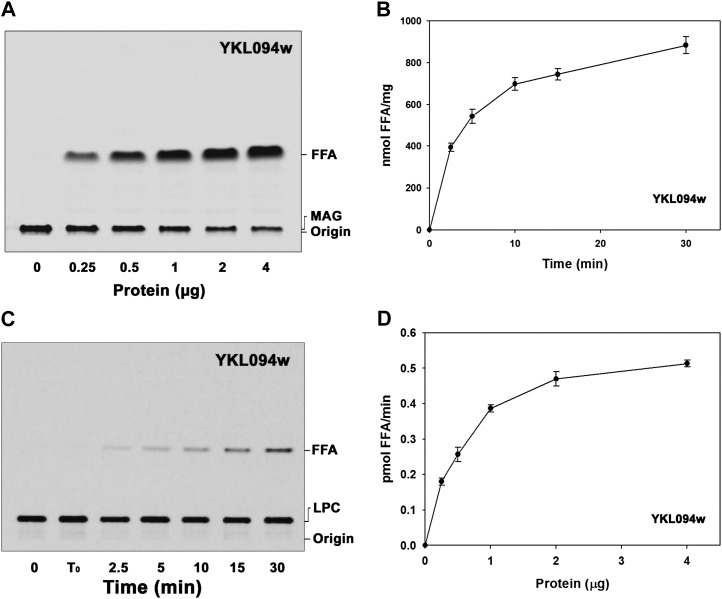
YKL094W is the closest homolog of AhMGAT in yeast that was characterized as a MAG hydrolase (Heier et al., 2010). It shows 45% similarity with AhMGAT. The YKL094W gene was cloned in pYES2, and enzymatic assays were performed with the dialyzed protein. We observed protein-dependent (Fig. 10A) and time-dependent (Fig. 10B) hydrolysis of MAG with recombinant YKL094W. The MGAT assay was also conducted with YKL094W protein, but it did not show acyltransferase activity, in contrast to its orthologs (AhMGAT and At1g52760). Interestingly, the recombinant enzyme showed protein- and time-dependent hydrolysis of LPC (Fig. 10, C and D). These results suggested that the homologous gene has a similar function across the phylogeny.
Figure 10.
Characterization of YKL094W, a homolog of AhMGAT. YKL094W was purified by Ni2+-NTA column chromatography, and the enzyme assays were performed with dialyzed protein. A, Protein-dependent hydrolysis of [14C]MAG with increasing amounts of protein for 10 min. Lane 0, Enzyme was added after stopping the reaction. FFA, Free fatty acid. B, Time-dependent MAG hydrolysis assay was performed at different time intervals at 30°C. The reaction was stopped by the addition of chloroform:methanol:2% orthophosphoric acid (1:2:1, v/v). Lipids were extracted and separated on a silica-TLC plate using petroleum ether:diethyl ether:acetic acid (70:30:1, v/v) as the solvent system. C, The time-dependent LPC hydrolase assay was conducted using [14C]LPC with 2 µg of purified protein. Lane 0, Enzyme was added after stopping the reaction; lane T0, zero time point (reaction was stopped immediately after adding the enzyme). D, The LPC hydrolase assay was performed for 10 min at 30°C in the presence of increasing amounts of purified YKL094W. The reaction was stopped, and lipids were resolved on a TLC plate using chloroform:methanol:28% ammonia (65:25:5, v/v) as the solvent system. Values are means ± sd of at least three independent experiments.
Homology Modeling and Docking Studies of Peanut MGAT
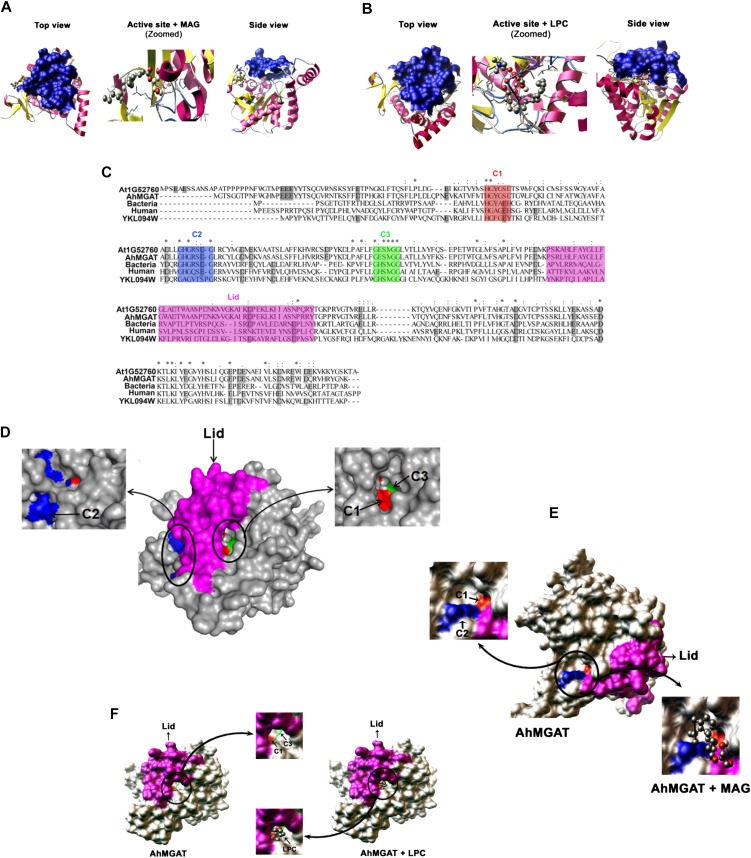
A predicted homology model was developed for AhMGAT to understand the spatial arrangement of conserved motifs. The docking of various lipid substrates was performed, and the stability of docked complexes was evaluated using binding free energies. It was found that MAG (Gibbs free energy, −3.2 kcal mol−1) and LPC (Gibbs free energy, −4.81 kcal mol−1) stably interacted with AhMGAT in two different orientations (Fig. 11, A and B). Multiple sequence analysis of AhMGAT with bacteria, yeast, Arabidopsis, and human homologs revealed three conserved amino acid regions termed C1 (HXYXXD), C2 (GXSXG), and C3 (GXSXG) motifs that are highlighted in red, blue, and green in Figure 11C. These motifs are essential for protein function. The C1 motif contains conserved residues for acyltransferase function, and the other two motifs (GXSXG) are highly conserved in lipases. The amino acids Tyr-64 and Asp-67 of the C1 motif, responsible for acyltransferase function, are found only in AhMGAT and Arabidopsis sequences but not in other homologs from bacteria, yeast, and human. The binding sites for lipid substrates on AhMGAT were identified with a semiflexible autodocking approach. The molecular surface analysis of AhMGAT revealed that C1 motif residues are shared by both C2 and C3 motifs (Fig. 11D). The conserved C1 and C2 motifs (comprising His-62, Tyr-64, Asp-67, Gly-95, Ser-97, and Gly-99) are located on the surface of AhMGAT, where MAG forms a stable interaction (Fig. 11E). The LPC binds near the C3 motif (comprising Gly-135, Ser-138, and Gly-140), which is accessible for the substrate through a tunnel-like entry (Fig. 11F). All these residues are located under a lid-like structure. The MAG- and LPC-binding orientations are completely different from each other.
Figure 11.
Homology modeling and molecular docking studies on AhMGAT protein. A and B, The three-dimensional model for AhMGAT protein was built using a MAG lipase (Protein Data Bank code 3PE6) structure, and the quality of the homology model was assessed using a Ramachandran plot. The docking of lipid substrates demonstrated the binding orientations of MAG (1-oleoyl; A) and LPC (B) on AhMGAT protein. The snapshots of docking complexes (AhMGAT protein with MAG or LPC) are shown as top and side views. MAG and LPC substrates are shown in ball-and-stick representation. The lid structure that covers the LPC binding region is shown as a blue-colored surface. C, Multiple sequence alignment of AhMGAT protein and its homologs. The conserved regions are denoted as C1 (HXYXXD), C2 (GHGXSXG), and C3 (GXSXG) motifs. The color codes are as follows: C1, red; C2, blue; C3, green; lid, magenta. D, A surface view of AhMGAT illustrates the spatial organization of the conserved regions. C2 and C1 are located on the AhMGAT protein surface, and MAG forms a stable interaction with the binding free energy of −3.23 kcal mol−1. E, A surface view of AhMGAT protein structure revealed that C1 and C3 are closely arranged under the lid structure where the LPC binds (−4.81 kcal mol−1). F, The C1 region is located at an equal distance from the C2 and C3 regions.
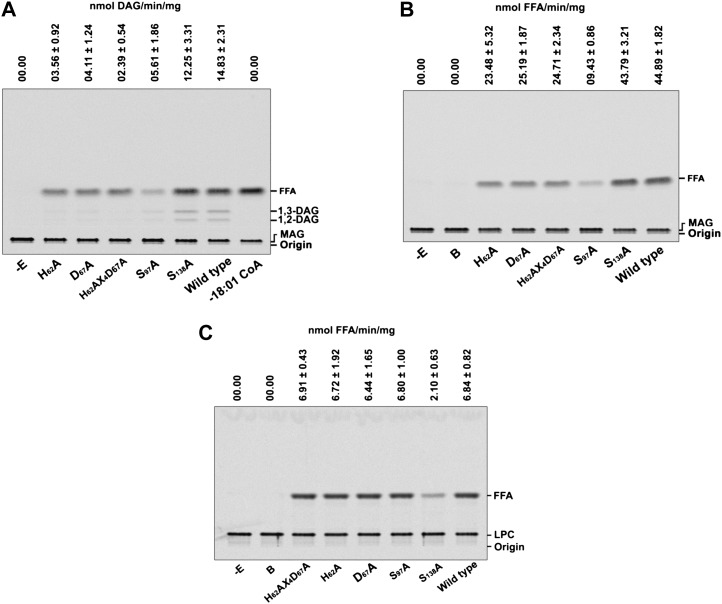
Site-directed mutagenesis was conducted to assess the role of H(X)4D and GXSXG motifs in the acyltransferase and hydrolase functions, respectively. The predicted amino acid residues, His-62, Asp-67, Ser-97, and Ser-138, were replaced with Ala. The recombinant AhMGAT mutant proteins were purified similarly to wild-type AhMGAT protein, and enzyme assays were performed. The MGAT activity was drastically reduced (83%) in the H62A/D67A double mutant, and there was approximately 72% reduction in the single mutants H62A and D67A as compared with the wild type (Fig. 12A). Interestingly, one of the lipase motif mutants (S97A) also showed a drastic reduction in both acyltransferase and MAG hydrolase functions (Fig. 12, A and B), but S138A was able to hydrolyze MAG with approximately 50% efficiency. The S138A mutant showed a 3.25-fold lower LPC hydrolase activity as compared with the wild type and other mutants (Fig. 12C). These results suggest that the H(X)4D motif could be important for the acyltransferase function and the 95GXSXG motif could be important for lipase function.
Figure 12.
Site-directed mutagenesis of acyltransferase and lipase motifs. A, The MGAT assay was performed for 15 min with [14C]MAG and 20 µm oleoyl-CoA using wild-type and HX4D mutant recombinant AhMGAT proteins: H62A, His-62 was replaced with Ala; D67A, Asp-67 was replaced with Ala; H62AX4D67A, double mutant, His-62 and Asp-67 were replaced with Ala; S97A, Ser-97 was replaced with Ala; S138A, Ser-138 was replaced with Ala; wild-type AhMGAT. FFA, Free fatty acid. B, The MAG hydrolase assay was performed with 50 μm [14C]MAG using wild-type and mutant recombinant AhMGAT proteins. The reaction was stopped by extracting lipids, and the lipids were separated on a silica-TLC plate using petroleum ether:diethyl ether:acetic acid (70:30:1, v/v) as the solvent system. Lane −E, No-enzyme control; lane B, enzyme fraction was boiled for 5 min, and assay was performed. Values are means ± sd of three independent experiments. C, The LPC hydrolase assay was performed with 50 μm [14C]LPC using wild-type and mutant recombinant AhMGAT proteins. The reaction was stopped, and lipids were resolved on a TLC plate using chloroform:methanol:28% ammonia (65:25:5, v/v) as the solvent system. Results are represented as percentage means ± sd of three independent experiments.
DISCUSSION
The biosynthesis of TAG has been shown to occur in the microsomal membranes of biological systems (Cases et al., 2001; Nykiforuk et al., 2002; Kalscheuer and Steinbüchel, 2003), although TAG biosynthetic enzymes have been well documented in the soluble fractions of plants. Soluble LPA phosphatase (Shekar et al., 2002) and soluble MGAT (Tumaney et al., 2001) have been observed in developing peanut cotyledons. A soluble DGAT has also been observed in peanut (Saha et al., 2006) and Arabidopsis (Rani et al., 2010). Here, we report, to our knowledge for the first time, the isolation of soluble MGAT genes from peanut and Arabidopsis. Two independent pieces of evidence in this study indicate that the acyltransferase in peanut MGAT is soluble in nature: (1) the activity was associated with the 150,000g supernatant (Tumaney et al., 2001); and (2) the membrane-spanning region was absent in the hydropathy analysis.
Our results show that the cDNA obtained from the seed-specific library encodes MGAT, as indicated by the following: (1) the expressed recombinant protein showed a time- and protein-dependent increase in the formation of DAG; (2) the MGAT reaction produced DAG, which in turn it could be used as a substrate for TAG biosynthesis by the endogenous DGATs; (3) the overexpression of AhMGAT in yeast caused an increased incorporation of labeled acetate into TAG; (4) the acyltransferase reaction was found to prefer MAG; and (5) the predicted homology modeling and substrate docking supported the isolated gene showing MGAT activity.
In an animal system, it was reported that the role of hepatic MGAT is to preserve essential fatty acids in specific tissues through reesterification cycles (Xia et al., 1993). In plants, it is understood that MGAT might have a similar role in preserving unsaturated fatty acids. It has been observed that significant acyl remodeling occurs during TAG synthesis in plants such as Ricinus communis, Brassica napus, and maize (Zea mays; Baud and Lepiniec, 2010). Hence, we have investigated other acyl-CoAs such as 18:2 and 18:3 that also exhibited significant MGAT activity. Based on the above observations, it is clear that peanut MGAT is a soluble MGAT that has broad unsaturated acyl donor substrate preferences and plays a possible role in DAG biosynthesis. The fatty acid profile of peanut is in agreement with the acyl preference of AhMGAT. The enzyme is also involved in the deacylation of MAG and LPC. The enzyme uses MAG for both acylation and deacylation to give DAG and free fatty acid, respectively. We hypothesize that the availability of the acyl-CoA in the system could regulate the dual function of the enzyme.
A BLASTx analysis of peanut MGAT in Arabidopsis revealed that At1g52760 is one of the closest homologs. Arabidopsis Lysophospholipase2 (At1g52760) was identified as a protein interactor of Acyl-CoA-Binding Protein2 (ACBP2), which was shown to be involved in tolerance to cadmium-induced oxidative stress (Gao et al., 2010). The biochemical characterization of At1g52760 showed that it has an acyl-CoA-dependent MGAT activity that, to our knowledge, was not hitherto reported. It is possible that ACBP2 interaction with MGAT promotes MAG hydrolysis instead of acyltransferase function by sequestering the acyl-CoA and making them unavailable for MGAT.
To attempt to gain further understanding of the role of peanut MGAT, we explored the in silico microarray data of the homolog At1g52760 using the electronic Fluorescent Pictograph Browser, available at http://www.bar.utoronto.ca/. The developmental map for At1g52760 suggested that the gene is maximally expressed during seed stage 7 without silique. However, expression is also observed during seed stages 4, 5, and 6, suggesting the importance of this gene during seed development, particularly in those stages when the TAG accumulation is shown to be high. The expression analysis following exposure to abiotic stresses indicated a maximum expression of the gene under heat, osmotic, and salt stresses, suggesting a possible role of this protein under certain cellular stress conditions and signal transduction. It appears that TAG synthesis increases in plants in response to stress (Moellering et al., 2010). Alternatively, MGAT might serve a function besides or in addition to TAG biosynthesis.
DAG is a signaling molecule and an intermediate for the synthesis of both neutral and membrane lipids. The MGAT pathway may operate for a storage purpose, whereas the DAG molecule formed by the phospholipase C reaction could be used for signaling. With this study, the relative importance between the MGAT pathway and the phospholipase reaction for the accumulation of DAG is difficult to understand. It needs further detailed study to know the relative importance. Many mechanisms have evolved in the cell to maintain the correct levels of DAG (Milligan et al., 1997; Katagiri and Shinozaki, 1998; Lu et al., 2001; Litvak et al., 2005; Mousley et al., 2006). In an alternative mechanism, cells may produce a permanent reservoir of DAG wherein DAG is stored in the form of TAG, thereby reducing the total DAG concentration in the cell. It was reported earlier that during cutin biosynthesis in Arabidopsis, a distinct glycerol-3-phosphate acyltransferase, which also functions as a phosphatase, results in the production of MAG (Yang et al., 2010). The MAG formed can also serve as a precursor for cutin biosynthesis. Subsequently, a DAG acyltransferase was identified and shown to be involved in TAG biosynthesis (Rani et al., 2010). TAG that is formed by a defective cuticular ridges gene (At5g23940) may further be involved in cutin biosynthesis. However, the enzyme that converts MAG to DAG has not been reported to date in cutin biosynthesis. We hypothesize that the soluble MGAT identified in this study could possibly participate in cutin biosynthesis by forming DAG.
In our study, a possible catalytic function of the AhMGAT protein was also studied by molecular docking analysis. The uncharacterized AhMGAT protein belongs to the α/β-hydrolase superfamily, and its sequence showed 47% similarity with hMGLL, for which an experimental structure is available (http://www.rcsb.org/pdb/). To build a homology model, a minimum of 30% global similarity is enough to get better three-dimensional folding. The global sequence identity of AhMGAT with hMGLL is 29%. Using MODELER, we have generated 100 AhMGAT models, and the quality of the each model was assessed by Ramachandran plot. The bad contacts of side chains were removed using model refinement. After the quality assessment of the homology model of AhMGAT, the best model was chosen for molecular docking. The knowledge-based model for AhMGAT was generated with the help of hMGLL. The AhMGAT homology model, like hMGLL, has the characteristic regulatory lid-type structure. Most of the lipid hydrolytic enzymes have a regulatory hood-like structure (Jennens and Lowe, 1994). Possibly, this lid controls the entry and exit of suitable substrates (Supplemental Video S1). However, the precise mechanism of lid opening and closing is still unclear. Analysis of the AhMGAT predicted homology model suggested that the spatial organization of these essential residues could have resulted in two different binding sites on AhMGAT. This confirms that it plays multiple roles in lipid metabolism. Earlier studies reported that mutation of His and Asp residues resulted in drastic reduction of acyltransferase activity (Brumlik and Buckley, 1996; Heath and Rock, 1998). The sequence-structure analysis of AhMGAT revealed conserved Tyr-64 and Asp-67 present in the C1 motif. But these residues are not conserved across bacteria, yeast, and human. The critical role of these residues has been experimentally shown to be involved in acyltransferase activity (Robertson et al., 1994; Brumlik and Buckley, 1996). The molecular docking results are consistent with our in vitro characterization of the AhMGAT protein. The LPC reaches the binding site through a deep tunnel-like opening and is accommodated near the C3 motif. On the basis of this study, we propose that two independent binding sites are required for different substrate catalysis.
It was shown in the animal systems that the MAG pathway may play an important role in the regulation of lipid metabolism by controlling the chain length of fatty acids (Knudsen et al., 1975), by controlling the intracellular concentrations of acyl-CoA (Gross, 1983), or by facilitating the selective retention of essential fatty acids during hepatic oxidation (Pelech and Vance, 1989). From our data, in vitro MGAT activity is more pronounced with unsaturated fatty acids than saturated. It could be understood that MGAT might have a possible role in preserving unsaturated fatty acids in plants. Another possibility is that the MAG pathway contributes to a separate intracellular pool of DAG to be used for a different set of metabolic reactions. Therefore, the identification of soluble MGAT operating in a PA-independent pathway (Parthibane et al., 2012a, 2012b) has a significant implication for our understanding of the regulation of DAG and the biosynthesis of TAG in plants. However, the physiological relevance of the enzyme in plants needs to be addressed further.
MATERIALS AND METHODS
Materials
[1-14C]Oleoyl-CoA (54 mCi mmol−1), [1-14C]palmitoyl-CoA (54 mCi mmol−1), [1-14C]stearoyl-CoA (54 mCi mmol−1), and [14C]acetate (51 mCi mmol−1) were purchased from Perkin-Elmer. [1-14C]2-MAG (55 mCi mmol−1) and [oleoyl-1-14C]LPC (55 mCi mmol−1) were obtained from American Radiolabeled Chemicals. The silica-thin-layer chromatography (TLC) plates were obtained from Merck. The acyl donors and acyl acceptors were obtained from Avanti Polar Lipids. The Arabidopsis (Arabidopsis thaliana) open reading frame clone was from the Arabidopsis Stock Center. Yeast clone YKL094W was obtained from Open Biosystems-Saf Labs. Yeast expression vectors pYES2/CT and pYES2/NT were purchased from Invitrogen BioServices and are referred as pYES2 in “Results.” Nickel-nitrilotriacetic acid agarose (Ni2+-NTA) was purchased from Qiagen. The monoclonal anti-poly(His) antibody produced in mouse, phenylmethanesulfonyl fluoride, and all the other reagents were obtained from Sigma unless specified otherwise.
Strains and Culture Conditions
Luria-Bertani broth (1% [w/v] tryptone, 0.5% [w/v] yeast extract, and 1% [w/v] NaCl) was used for bacterial growth, and the bacteria were cultivated at 37°C. The wild-type yeast (Saccharomyces cerevisiae) strain (BY4741: MATα; HIS3Δ1; LEU2Δ0; MET15Δ0; URA3Δ0) was used as a heterologous host to study the expression of the MGAT genes. The transformed yeast cells were cultivated at 30°C in a minimal medium (synthetic dropout medium lacking uracil [SM-U]), and the medium was supplemented with either 2% (w/v) Glc or Gal as the carbon source.
Sequence Retrieval, Alignment, and Comparison
The cDNA, EST, and protein sequences were identified by searching the public domain databases available at the National Center for Biotechnology Information (NCBI) and The Arabidopsis Information Resource with the BLAST algorithms. The sequences were aligned using the ClustalX program. The nonredundant protein sequence database was searched using default parameters, and the sequences with E values of 10−5 and a score of 85 were retrieved and subjected to sequence alignment.
The conserved protein domains were examined using the conserved domain database at the NCBI (http://www.ncbi.nih.gov/Structure/cdd/cdd.shtml) and the pfam database (http://pfam.sanger.ac.uk/). The multiple sequence alignment was performed using the ClustalX program with the bootstrap resampling method. The bootstrap replicate alignments were then used to construct a phylogenetic tree using the neighbor-joining method. The phylogenetic analyses were conducted using MEGA4 software. For the construction of the phylogenetic tree, approximately 67 sequences were used from 25 different species.
cDNA Phage Library Screening
A seed-specific cDNA library of immature peanut (Arachis hypogaea) was constructed in λ ZAP (Stratagene). The expression library was screened using a 3′-end radiolabeled oligonucleotide primer (5′-GGAACGAGCGGAGGCACCCCGAATTTTTGGGGGCACATGCCC-3′), which was designed based on the N-terminal sequence (GTSGGTPNFWGHMP) of the peanut MGAT protein (Tumaney et al., 2001). Several probes were designed and then screened the cDNA library. The MGAT sequence was obtained by using the oligonucleotide primer given above. A total of 4.5 × 106 plaques were screened, and the positive plaques were purified by three additional screenings. Plasmid DNA was sequenced on both strands.
Cloning and Expression of the Peanut AhMGAT
The coding sequence of the AhMGAT gene was cloned into the pYES2/CT shuttle vector using a forward primer (5′-AAAAGCTTAATGGGCACGAGCGGAG-3′) with a HindIII restriction site and a reverse primer (5′-GCCTCGAGTTTATTTCCATACCTATGAACC-3′) with a XhoI restriction site. The clone was confirmed by both double digestion and nucleotide sequencing. The construct and pYES2/CT were transformed into the yeast cells using the lithium acetate method (Schiestl and Gietz, 1989). The resulting transformants were confirmed by colony PCR using AhMGAT gene-specific primers and grown to the late log phase in SM-U containing 2% (w/v) Glc. Protein expression was induced by Gal; hence, the galactokinase (GAL1) promoter is present in the pYES2/CT vector, which is induced by the presence of Gal in the medium. Cells were harvested and washed with cold water by centrifugation and inoculated at a concentration of absorbance at 600 nm (A600) = 0.4 in an induction medium (SC-U containing 2% [w/v] Gal) for 24 h. The expressed protein was purified using a Ni2+-NTA matrix, and it was confirmed by immunoblot analysis. Briefly, cells were resuspended in yeast lysis buffer (50 mm Tris-HCl, pH 7.5, 1 mm phenylmethanesulfonyl fluoride, 5 mm MgCl2, and 1% [v/v] glycerol) and then lysed using glass beads. The recombinant AhMGAT was purified by Ni2+-NTA column chromatography. The purified proteins were resolved on a 12% (w/v) SDS-PAGE gel and transferred onto a nitrocellulose membrane. The expression of AhMGAT was confirmed using an anti-His6 tag monoclonal antibody (1:10,000 dilution [v/v]).
Cloning and Expression of the Recombinant At1g52760
The open reading frame of At1g52760 in the pUNI vector (from the Arabidopsis Stock Center) was used as a template for amplification of the gene. The coding sequence of At1g52760 was amplified under specific thermal cycling conditions (1 min of denaturation at 94°C, 1 min of annealing at 58°C, and 1 min of elongation at 72°C, repeated for 32 cycles with a final elongation of 10 min) using a forward primer (5′-AGGTACCATGCCGTCGGAAGCGG-3′) with a KpnI restriction site and a reverse primer (5′-CCCTCGAGAGCGGTTTTAGATCCATACTTCTT-3′) with a XhoI restriction site. The amplified At1g52760 and pYES2/NT B shuttle vector were double digested and ligated directionally. The positive clone was confirmed by both double digestion and nucleotide sequencing. The construct and pYES2/NT B were transformed into yeast cells using the lithium acetate method (Schiestl and Gietz, 1989). The resulting transformants were confirmed by colony PCR using At1g52760 gene-specific primers. Protein expression was induced by growing the cells (A600 = 0.4) in an induction medium (SM-U containing 2% [w/v] Gal) for 24 h. The expressed protein was purified using a Ni2+-NTA matrix, and it was confirmed by immunoblot analysis.
MGAT Assay
Acyltransferase activity was measured by either incorporation of [1-14C]oleoyl-CoA into MAG or acylation of oleoyl-CoA into [1-14C]MAG (oleoyl) to form DAG. All assays were performed with dialyzed protein samples. The assay mixture consisted of 50 mm Tris-HCl (pH 8.0), 1 mm MgCl2, 10 mm NaCl, 50 μm MAG, 20 μm [1-14C]oleoyl-CoA (0.025 µCi) or 50 μm [14C]MAG (0.025 µCi), 20 μm oleoyl-CoA, and the enzyme source (0–4 μg) in a total volume of 100 μL. Substrate was prepared in CHAPS detergent (0.01 mm final concentration). MGAT assays were also performed in various experimental conditions like different pH and temperature, the presence of CHAPS (0–1 mm), and various divalent cations and chelating agents (0–10 mm). The reaction was conducted at 30°C for different time intervals (0–30 min) and stopped by the addition of 2:1 (v/v) CHCl3:CH3OH. The lipids were separated on a silica-TLC plate using petroleum ether:diethyl ether:acetic acid (70:30:1, v/v) as the solvent system. The individual lipid moieties were identified by migration with respect to the standards. Enzymatic products were monitored by phosphor imager, corresponding spots were scraped from the TLC plate, and the radioactivity was quantified with a liquid scintillation counter.
Site-Directed Mutagenesis
The mutations of acyltransferase (H62X4D67A) motif and lipase (GXSXG) motifs (S97A and S136A) in AhMGAT were introduced by amplification of whole plasmid using the following primers: H62A, 5′-CGTCTTCATGACCGCCGGCTACGGCTCCGAC-3′; D67A, 5-′CCACGGCTACGGCTCCGCTACCGGTTGGCTCT-3′; H62AX4D67A, 5′-CGTCTTCATGACCGCCGGCTACGGCTCCGCTACCGGTTGGCTCT-3′; S97A, 5′-TGGCCTCCGCTGGCCCCTCGGCGACAT-3′; and S138A, 5′-GGGTGGTCTCGCGCCCCTGCTTATGTA-3′. An oligonucleotide sequence with the reverse complementary sequence of the forward primer was used as the reverse primer. The underlined bases represent the sites of the mutations. The amplified whole plasmid was first treated with 10 units of DpnI at 37°C for 1 h to digest the methylated parent template. The newly amplified unmethylated plasmid was then transformed into Escherichia coli DH5α competent cells. The colonies were screened for the presence of mutation in the gene. The mutants were confirmed by sequencing the plasmid DNA by using gene-specific primers. The enzymatic assays were performed with purified recombinant AhMGAT proteins from all mutants and the wild type.
MAG and LPC Hydrolysis Assays
Hydrolase activity was measured by monitoring the release of 14C-fatty acid from [1-14C]MAG or [14C]LPC. The assay mixture consisted of 50 mm Tris-HCl (pH 8), 1 mm dithiothreitol, 50 μm [14C]MAG (0.025 µCi) or 50 μm [14C]LPC (0.025 µCi), and the enzyme source (0–4 μg) in a total volume of 100 μL. The reaction was conducted at 30°C for different time interval (0–30 min) and stopped by the addition of 2:1 (v/v) CHCl3:CH3OH. The lipids were extracted and then resolved on a silica-TLC plate using petroleum ether:diethyl ether:acetic acid (70:30:1, v/v) for the MAG hydrolysis and chloroform:methanol:28% ammonia (65:25:5, v/v) or chloroform:acetone:methanol:acetic acid:water (50:20:10:10:5, v/v) for the LPC hydrolase activity.
Incorporation of [14C]Acetate into Yeast Lipids
The transformants (pYES2/CT, pYES2/CT+AhMGAT or pYES2/NT, pYES2/NT+At1g52760) in wild-type yeast were grown until late log phase in 5 mL of SM-U with 2% (w/v) Glc. For labeling, a cell culture with A600 = 0.4 was inoculated in fresh medium containing 2% (w/v) Gal and 0.25 μCi of [14C]acetate mL−1 and grown for 24 h. Cells (A600 = 20) were harvested and lipids were extracted. Neutral lipids were separated by using petroleum ether:diethyl ether:glacial acetic acid (70:30:1, v/v) as a solvent system, and for phospholipids, chloroform:methanol:ammonia (65:25:5, v/v) was used in the first dimension followed by chloroform:methanol:acetone:acetic acid:water (50:10:20:15:5, v/v) as the second-dimension solvent systems. The TLC plate was exposed to a phosphor imaging screen, and individual lipid spots were scraped off from the plate to determine their radioactivity in a liquid scintillation counter.
Cloning and Expression of the Recombinant YKL094W
The gene YKL094W was obtained from Open Biosystems (Saf Labs-cDNA Clone Center). The open reading frame of YKL094W was cloned into yeast expression vector pYES2/NT B shuttle vector using a forward primer (5′-ATATGGATCCATGGCTCCGATCCATACAAA-3′) with a BamHI restriction site and a reverse primer (5′-ATATGAATTCTTATGGTTTACTTCGGTCGT-3′) with an EcoRI restriction site. The construct was transformed into yeast cells using the lithium acetate method (Schiestl and Gietz, 1989). The cells were grown in dextrose medium and later transferred to Gal-containing SM-U for protein expression. The expressed protein was purified using a Ni2+-NTA matrix, and it was confirmed by immunoblot analysis.
Homology Modeling
The multiple sequence alignment of AhMGAT and its homologs from bacteria, yeast, and human was generated using the ClustalX program (Thompson et al., 1997). The homology modeling was performed using MODELER 6v4 (Sali et al., 1995). The experimental structure of human MAG lipase (hMGLL) was used as a template for model construction, which shared 47% homology with AhMGAT. The x-ray structure coordinates of hMGLL were obtained from the Protein Data Bank (http://www.rcsb.org/pdb/). In brief, the template (hMGLL) and target (AhMGAT) sequences were aligned and saved as alignment format. Information about the alignment file and three-dimensional coordinate file of hMGLL was fed into model_default python script. The program was executed using a mod6v4 model_default.py command. There were 100 models generated after successful run of the program, and the quality of each model was validated using PROCHECK (Laskowski et al., 1993) and Ramachandran plot (Ramachandran and Venkatachalam, 1968). The best-quality model was chosen for docking studies.
Docking of Lipid Substrates
The two-dimensional structures of various lipid substrates were created with JME molecular editor (Ertl and Jacob, 1997), and atomic force fields were assigned using ProDrg (Schüttelkopf and van Aalten, 2004). The resultant ligands were used directly for docking studies. The ligand flexibilities and bond rotational properties were generated using the autotor program. The electrostatic charges, solvation parameters, and missing hydrogen were added to the protein. The grid box where substrate molecules find favorable interaction was set to 47 × 41 × 41 (x, y, and z dimensions) for LPC and 46 × 40 × 40 for MAG. The possible conformations of ligands were generated using a genetic algorithm (Morris et al., 1998), and binding energy for each conformation was generated. The total energy evaluation was achieved by calculating potentials of each atom of the substrates (Goodford, 1985), which was set to 27,000,000 evaluations. The best-fit population size was set to 250 dockings. The docking of lipid substrates on the AhMGAT homology model was carried out using AutoDock version 3.05 (Morris et al., 1996). The docked complex images were visualized and analyzed by MGL and PYMOL softwares.
The nucleotide sequence of peanut MGAT reported in this study has been submitted to GenBank with accession number JF340215.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Video S1. Molecular docking studies on AhMGAT protein with MAG and LPC.
Acknowledgments
P.V. acknowledges a research associateship from the Department of Biotechnology, New Delhi.
Glossary
- DAG
diacylglycerol
- MGAT
monoacylglycerol acyltransferase
- TAG
triacylglycerol
- LPA
lysophosphatidic acid
- DHAP
dihydroxyacetone phosphate
- PA
phosphatidic acid
- MAG
monoacylglycerol
- LPC
lysophosphatidylcholine
- Ni2+-NTA
nickel-nitrilotriacetic acid agarose
- TLC
thin-layer chromatography
- SM-U
synthetic dropout medium lacking uracil
- NCBI
National Center for Biotechnology Information
- cDNA
complementary DNA
- A600:
absorbance at 600 nm
References
- Athenstaedt K, Weys S, Paltauf F, Daum G. (1999) Redundant systems of phosphatidic acid biosynthesis via acylation of glycerol-3-phosphate or dihydroxyacetone phosphate in the yeast Saccharomyces cerevisiae. J Bacteriol 181: 1458–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Lepiniec L. (2010) Physiological and developmental regulation of seed oil production. Prog Lipid Res 49: 235–249 [DOI] [PubMed] [Google Scholar]
- Bell RM, Coleman RA. (1980) Enzymes of glycerolipid synthesis in eukaryotes. Annu Rev Biochem 49: 459–487 [DOI] [PubMed] [Google Scholar]
- Brumlik MJ, Buckley JT. (1996) Identification of the catalytic triad of the lipase/acyltransferase from Aeromonas hydrophila. J Bacteriol 178: 2060–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Burn P, Shi Y. (2003) Properties of the mouse intestinal acyl-CoA:monoacylglycerol acyltransferase, MGAT2. J Biol Chem 278: 25657–25663 [DOI] [PubMed] [Google Scholar]
- Cases S, Stone SJ, Zhou P, Yen E, Tow B, Lardizabal KD, Voelker T, Farese RV., Jr (2001) Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. J Biol Chem 276: 38870–38876 [DOI] [PubMed] [Google Scholar]
- Coleman RA, Haynes EB. (1986) Monoacylglycerol acyltransferase: evidence that the activities from rat intestine and suckling liver are tissue-specific isoenzymes. J Biol Chem 261: 224–228 [PubMed] [Google Scholar]
- Ertl P, Jacob O. (1997) WWW-based chemical information system. Theochem 419: 113–120 [Google Scholar]
- Gao W, Li HY, Xiao S, Chye ML. (2010) Acyl-CoA-binding protein 2 binds lysophospholipase 2 and lysoPC to promote tolerance to cadmium-induced oxidative stress in transgenic Arabidopsis. Plant J 62: 989–1003 [DOI] [PubMed] [Google Scholar]
- Gocze PM, Freeman DA. (1994) Factors underlying the variability of lipid droplet fluorescence in MA-10 Leydig tumor cells. Cytometry 17: 151–158 [DOI] [PubMed] [Google Scholar]
- Goodford PJ. (1985) A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J Med Chem 28: 849–857 [DOI] [PubMed] [Google Scholar]
- Gross RW. (1983) Purification of rabbit myocardial cytosolic acyl-CoA hydrolase, identity with lysophospholipase, and modulation of enzymic activity by endogenous cardiac amphiphiles. Biochemistry 22: 5641–5646 [DOI] [PubMed] [Google Scholar]
- Hajra AK, Larkins LK, Das AK, Hemati N, Erickson RL, MacDougald OA. (2000) Induction of the peroxisomal glycerolipid-synthesizing enzymes during differentiation of 3T3-L1 adipocytes: role in triacylglycerol synthesis. J Biol Chem 275: 9441–9446 [DOI] [PubMed] [Google Scholar]
- Heath RJ, Rock CO. (1998) A conserved histidine is essential for glycerolipid acyltransferase catalysis. J Bacteriol 180: 1425–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heier C, Taschler U, Rengachari S, Oberer M, Wolinski H, Natter K, Kohlwein SD, Leber R, Zimmermann R. (2010) Identification of Yju3p as functional orthologue of mammalian monoglyceride lipase in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 1801: 1063–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroyama M, Takenawa T. (1999) Isolation of a cDNA encoding human lysophosphatidic acid phosphatase that is involved in the regulation of mitochondrial lipid biosynthesis. J Biol Chem 274: 29172–29180 [DOI] [PubMed] [Google Scholar]
- Jennens ML, Lowe ME. (1994) A surface loop covering the active site of human pancreatic lipase influences interfacial activation and lipid binding. J Biol Chem 269: 25470–25474 [PubMed] [Google Scholar]
- Kalscheuer R, Steinbüchel A. (2003) A novel bifunctional wax ester synthase/acyl-CoA:diacylglycerol acyltransferase mediates wax ester and triacylglycerol biosynthesis in Acinetobacter calcoaceticus ADP1. J Biol Chem 278: 8075–8082 [DOI] [PubMed] [Google Scholar]
- Katagiri T, Shinozaki K. (1998) Disruption of a gene encoding phosphatidic acid phosphatase causes abnormal phenotypes in cell growth and abnormal cytokinesis in Saccharomyces cerevisiae. Biochem Biophys Res Commun 248: 87–92 [DOI] [PubMed] [Google Scholar]
- Knudsen J, Clark S, Dils R. (1975) Acyl-CoA hydrolase(s) in rabbit mammary gland which control the chain length of fatty acids synthesised. Biochem Biophys Res Commun 65: 921–926 [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Macarthur MW, Moss DS, Thornton JM. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26: 283–291 [Google Scholar]
- Lehner R, Kuksis A. (1996) Biosynthesis of triacylglycerols. Prog Lipid Res 35: 169–201 [DOI] [PubMed] [Google Scholar]
- Litvak V, Dahan N, Ramachandran S, Sabanay H, Lev S. (2005) Maintenance of the diacylglycerol level in the Golgi apparatus by the Nir2 protein is critical for Golgi secretory function. Nat Cell Biol 7: 225–234 [DOI] [PubMed] [Google Scholar]
- Lu C, Peng YW, Shang J, Pawlyk BS, Yu F, Li T. (2001) The mammalian retinal degeneration B2 gene is not required for photoreceptor function and survival. Neuroscience 107: 35–41 [DOI] [PubMed] [Google Scholar]
- Milligan SC, Alb JG, Jr, Elagina RB, Bankaitis VA, Hyde DR. (1997) The phosphatidylinositol transfer protein domain of Drosophila retinal degeneration B protein is essential for photoreceptor cell survival and recovery from light stimulation. J Cell Biol 139: 351–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering ER, Muthan B, Benning C. (2010) Freezing tolerance in plants requires lipid remodeling at the outer chloroplast membrane. Science 330: 226–228 [DOI] [PubMed] [Google Scholar]
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK. (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19: 1639–1662 [Google Scholar]
- Morris GM, Goodsell DS, Huey R, Olson AJ. (1996) Distributed automated docking of flexible ligands to proteins: parallel applications of AutoDock 2.4. J Comput Aided Mol Des 10: 293–304 [DOI] [PubMed] [Google Scholar]
- Mousley CJ, Tyeryar KR, Ryan MM, Bankaitis VA. (2006) Sec14p-like proteins regulate phosphoinositide homoeostasis and intracellular protein and lipid trafficking in yeast. Biochem Soc Trans 34: 346–350 [DOI] [PubMed] [Google Scholar]
- Nykiforuk CL, Furukawa-Stoffer TL, Huff PW, Sarna M, Laroche A, Moloney MM, Weselake RJ. (2002) Characterization of cDNAs encoding diacylglycerol acyltransferase from cultures of Brassica napus and sucrose-mediated induction of enzyme biosynthesis. Biochim Biophys Acta 1580: 95–109 [DOI] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J. (1995) Lipid biosynthesis. Plant Cell 7: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthibane V, Iyappan R, Vijayakumar A, Venkateshwari V, Rajasekharan R. (2012a) Serine/threonine/tyrosine protein kinase phosphorylates oleosin, a regulator of lipid metabolic functions. Plant Physiol 159: 95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthibane V, Rajakumari S, Venkateshwari V, Iyappan R, Rajasekharan R. (2012b) Oleosin is bifunctional enzyme that has both monoacylglycerol acyltransferase and phospholipase activities. J Biol Chem 287: 1946–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelech SL, Vance DE. (1989) Signal transduction via phosphatidylcholine cycles. Trends Biochem Sci 14: 28–30 [Google Scholar]
- Phan CT, Tso P. (2001) Intestinal lipid absorption and transport. Front Biosci 6: D299–D319 [DOI] [PubMed] [Google Scholar]
- Rajakumari S, Grillitsch K, Daum G. (2008) Synthesis and turnover of non-polar lipids in yeast. Prog Lipid Res 47: 157–171 [DOI] [PubMed] [Google Scholar]
- Ramachandran GN, Venkatachalam CM. (1968) Stereochemical criteria for polypeptides and proteins. IV. Standard dimensions for the cis-peptide unit and conformation of cis-polypeptides. Biopolymers 6: 1255–1262 [DOI] [PubMed] [Google Scholar]
- Rani SH, Krishna TH, Saha S, Negi AS, Rajasekharan R. (2010) Defective in cuticular ridges (DCR) of Arabidopsis thaliana, a gene associated with surface cutin formation, encodes a soluble diacylglycerol acyltransferase. J Biol Chem 285: 38337–38347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VS, Rao DK, Rajasekharan R. (2010) Functional characterization of lysophosphatidic acid phosphatase from Arabidopsis thaliana. Biochim Biophys Acta 1801: 455–461 [DOI] [PubMed] [Google Scholar]
- Reddy VS, Singh AK, Rajasekharan R. (2008) The Saccharomyces cerevisiae PHM8 gene encodes a soluble magnesium-dependent lysophosphatidic acid phosphatase. J Biol Chem 283: 8846–8854 [DOI] [PubMed] [Google Scholar]
- Robertson DL, Hilton S, Wong KR, Koepke A, Buckley JT. (1994) Influence of active site and tyrosine modification on the secretion and activity of the Aeromonas hydrophila lipase/acyltransferase. J Biol Chem 269: 2146–2150 [PubMed] [Google Scholar]
- Ron D, Kazanietz MG. (1999) New insights into the regulation of protein kinase C and novel phorbol ester receptors. FASEB J 13: 1658–1676 [PubMed] [Google Scholar]
- Saha S, Enugutti B, Rajakumari S, Rajasekharan R. (2006) Cytosolic triacylglycerol biosynthetic pathway in oilseeds: molecular cloning and expression of peanut cytosolic diacylglycerol acyltransferase. Plant Physiol 141: 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. (1995) Evaluation of comparative protein modeling by MODELLER. Proteins 23: 318–326 [DOI] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD. (1989) High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet 16: 339–346 [DOI] [PubMed] [Google Scholar]
- Schüttelkopf AW, van Aalten DMF. (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 60: 1355–1363 [DOI] [PubMed] [Google Scholar]
- Shekar S, Tumaney AW, Rao TJ, Rajasekharan R. (2002) Isolation of lysophosphatidic acid phosphatase from developing peanut cotyledons. Plant Physiol 128: 988–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Li JQ, Dauk M, Huang Y, Periappuram C, Wei Y, Zou J. (2010) Metabolic and transcriptional responses of glycerolipid pathways to a perturbation of glycerol 3-phosphate metabolism in Arabidopsis. J Biol Chem 285: 22957–22965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SJ, Cases S, Jensen DR, Chen HC, Sande E, Tow B, Sanan DA, Raber J, Eckel RH, Farese RV., Jr (2000) Obesity resistance and multiple mechanisms of triglyceride synthesis in mice lacking Dgat. Nat Genet 25: 87–90 [DOI] [PubMed] [Google Scholar]
- Swanton EM, Saggerson ED. (1997) Glycerolipid metabolizing enzymes in rat ventricle and in cardiac myocytes. Biochim Biophys Acta 1346: 93–102 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. (1997) The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumaney AW, Shekar S, Rajasekharan R. (2001) Identification, purification, and characterization of monoacylglycerol acyltransferase from developing peanut cotyledons. J Biol Chem 276: 10847–10852 [DOI] [PubMed] [Google Scholar]
- Vincent P, Maneta-Peyret L, Cassagne C, Moreau P. (2001) Phosphatidylserine delivery to endoplasmic reticulum-derived vesicles of plant cells depends on two biosynthetic pathways. FEBS Lett 498: 32–36 [DOI] [PubMed] [Google Scholar]
- Wei Y, Contreras JA, Sheffield P, Osterlund T, Derewenda U, Kneusel RE, Matern U, Holm C, Derewenda ZS. (1999) Crystal structure of brefeldin A esterase, a bacterial homolog of the mammalian hormone-sensitive lipase. Nat Struct Biol 6: 340–345 [DOI] [PubMed] [Google Scholar]
- Xia T, Mostafa N, Bhat BG, Florant GL, Coleman RA. (1993) Selective retention of essential fatty acids: the role of hepatic monoacylglycerol acyltransferase. Am J Physiol 265: R414–R419 [DOI] [PubMed] [Google Scholar]
- Xie M, Low MG. (1994) Identification and characterization of an ecto-(lyso)phosphatidic acid phosphatase in PAM212 keratinocytes. Arch Biochem Biophys 312: 254–259 [DOI] [PubMed] [Google Scholar]
- Yang W, Pollard M, Li-Beisson Y, Beisson F, Feig M, Ohlrogge J. (2010) A distinct type of glycerol-3-phosphate acyltransferase with sn-2 preference and phosphatase activity producing 2-monoacylglycerol. Proc Natl Acad Sci USA 107: 12040–12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen CL, Stone SJ, Cases S, Zhou P, Farese RV., Jr (2002) Identification of a gene encoding MGAT1, a monoacylglycerol acyltransferase. Proc Natl Acad Sci USA 99: 8512–8517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YM, Rock CO. (2008) Thematic review series: glycerolipids. Acyltransferases in bacterial glycerophospholipid synthesis. J Lipid Res 49: 1867–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]