Abstract
A comparative proteomic approach was performed to identify differentially expressed proteins in plastids at three stages of tomato (Solanum lycopersicum) fruit ripening (mature-green, breaker, red). Stringent curation and processing of the data from three independent replicates identified 1,932 proteins among which 1,529 were quantified by spectral counting. The quantification procedures have been subsequently validated by immunoblot analysis of six proteins representative of distinct metabolic or regulatory pathways. Among the main features of the chloroplast-to-chromoplast transition revealed by the study, chromoplastogenesis appears to be associated with major metabolic shifts: (1) strong decrease in abundance of proteins of light reactions (photosynthesis, Calvin cycle, photorespiration) and carbohydrate metabolism (starch synthesis/degradation), mostly between breaker and red stages and (2) increase in terpenoid biosynthesis (including carotenoids) and stress-response proteins (ascorbate-glutathione cycle, abiotic stress, redox, heat shock). These metabolic shifts are preceded by the accumulation of plastid-encoded acetyl Coenzyme A carboxylase D proteins accounting for the generation of a storage matrix that will accumulate carotenoids. Of particular note is the high abundance of proteins involved in providing energy and in metabolites import. Structural differentiation of the chromoplast is characterized by a sharp and continuous decrease of thylakoid proteins whereas envelope and stroma proteins remain remarkably stable. This is coincident with the disruption of the machinery for thylakoids and photosystem biogenesis (vesicular trafficking, provision of material for thylakoid biosynthesis, photosystems assembly) and the loss of the plastid division machinery. Altogether, the data provide new insights on the chromoplast differentiation process while enriching our knowledge of the plant plastid proteome.
One of the most visible events occurring during fruit ripening is the loss of chlorophyll and the synthesis of colored compounds. In many fruit, such as the tomato (Solanum lycopersicum), the change in color from green to red is due to the differentiation of chloroplasts into chromoplasts and is accompanied by the accumulation of carotenoids.
Numerous studies have been devoted to ultrastructural events underlying the conversion of chloroplast to chromoplast. The events investigated include, in particular, the formation of protein-accumulating bodies and the remodeling of the internal membrane system. At the structural level, the differentiation of chromoplasts consists mainly of the lysis of the grana and thylakoids (Spurr and Harris, 1968) but also includes new synthesis of membranes derived from the inner plastid membrane and that become the site for the formation of carotenoids (Simkin et al., 2007).
At the biochemical and molecular level chromoplast differentiation studies have been largely dedicated to the synthesis of carotenoids (Camara et al., 1995; Bramley, 2002). However, although highly specialized, chromoplasts carry out a variety of functions, many of them persisting from the chloroplast (Bouvier and Camara, 2007; Egea et al., 2010). The biochemical and structural events during chromoplast differentiation have been reviewed in a number of articles over the last decades (Thomson and Whatley, 1980; Ljubesić et al., 1991; Marano et al., 1993; Camara et al., 1995; Waters and Pyke, 2004; Lopez-Juez, 2007; Egea et al., 2010; Bian et al., 2011).
In the recent years high-throughput technologies have provided novel and extensive information on the plastid proteome of higher plants with strong emphasis on the chloroplast (for review, see van Wijk and Baginsky, 2011). Less information is available on the chromoplast proteome to the studies focusing on pepper (Capsicum annuum; Siddique et al., 2006), tomato (Barsan et al., 2010), and sweet orange (Citrus sinensis; Zeng et al., 2011). A comparison of the chromoplast proteome of sweet orange and tomato shows a high level of conservation although some specificities have been encountered (Zeng et al., 2011). Nevertheless, the global changes occurring during the differentiation of chromoplasts from chloroplasts have not been evaluated so far. In this work, a quantitative proteomic analysis was carried out to understand the regulation of the metabolic and structural changes occurring in tomato fruit plastids during the transformation of chloroplasts into chromoplasts.
RESULTS AND DISCUSSION
Inventory of Proteins Present in Tomato Fruit Plastids during the Chloroplast-to-Chromoplast Transition
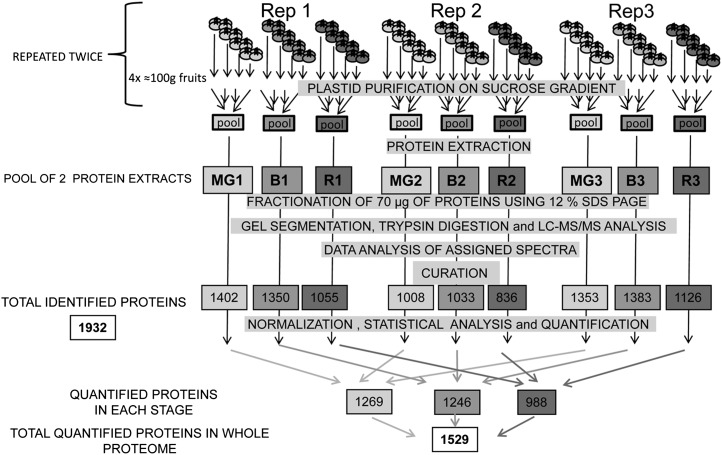
The experimental design used for fruit sampling and biological replications is presented in Figure 1. Plastids from four sets of about 100 g of pericarp (corresponding to 25–30 fruits) were isolated separately in three independent replicates (Rep 1–3) for each developmental stage, mature green (MG; 1–3), breaker (B; 1–3), and red (R; 1–3). This procedure was repeated twice. Two protein extracts from four individual plastid fractions were pooled for each replicate. The raw data coming from the analysis of the replicates of the tomato plastid proteins were curated by comparing the set of proteins with five databases (AT-CHLORO, Plprot, PPDB, SUBA, and Uniprot) and three predictors for subcellular localization (TargetP, Predotar, iPSORT). Only proteins present in at least two databases or predicted to be plastid localized by one of the predictors were retained for analysis. In addition manual curation was performed on the basis of the information available in the literature. By combining the curated list of proteins encountered in the three replicates of the tomato plastids at three stages of development, an inventory of 1,932 proteins (including 47 proteins encoded by the plastid genome) was drawn up and cataloged in Supplemental Table S1. Previous work with fruit chromoplasts identified 988 proteins in tomato (Barsan et al., 2010), 151 in pepper (Siddique et al., 2006), and 493 in sweet orange fruit (Zeng et al., 2011). The inventory reported here is in the same range of magnitude as other plastids database: 1,345 in AT-CHLORO, 2,043 in plprot, 1,367 in PPDB, and 2,112 in SUBA. Our work brings 362 proteins that had not yet been referenced in plastid databases and that are predicted to be plastid localized by at least one predictor. Among these, 38 had already been cataloged in the tomato chromoplast proteome (Barsan et al., 2010). The size of the plant plastid proteome has been evaluated by applying a combination of chloroplast transit peptides predictors programs to the screening of the nucleus-encoded amino acid sequences available in databases. In Arabidopsis (Arabidopsis thaliana) predictions vary between 2,100 (Richly and Leister, 2004) and 2,700 (Millar et al., 2006). Armbruster et al. (2011) come to similar predictions of between 2,000 and 3,000 proteins. In rice (Oryza sativa), predictions estimate at 4,800 the number of chloroplast proteins (Richly and Leister, 2004). Therefore it seems that marked differences exist between plant species. However discrepancies are observed in the estimation of the size of the plastid proteome probably due to the use of different evaluation methods and to the fact that many plastid proteins are not predicted by prediction tools (Kleffmann et al., 2006). By combining three different approaches for protein localization (experimental determination by cell biology methods, homology-based identification, and predictions by targeting programs), Pierleoni et al. (2007) estimated the Arabidopsis plastid proteome size at 4,875 proteins. With the availability of the tomato sequence genome, we have been able to calculate that the number of proteins predicted to be plastidial in the tomato genome is of 2,696, 4,651, and 4,142 using Predotar, iPSORT, and TargetP, respectively. Therefore it can be concluded that the present inventory of plastid proteins in databases is probably not exhaustive.
Figure 1.
Summarized experimental design used in this proteomic study. Plastids from four sets of four fruits were isolated separately in three independent replicates (Rep1, Rep2, and Rep3) for each developmental stage (MG 1–3; B 1–3; and R 1–3). This procedure was repeated twice and plastids corresponding to each stage were pooled for each experiment before protein extraction. The different steps from purification of plastids to curation of data are indicated and described in detail in “Material and Methods.” The number of proteins encountered in each individual analysis is given under the denomination of total resources, resulting in a nonredundant total list of 1,932 proteins that are reported in Supplemental Table S1. After normalization and statistical analysis, the number of proteins that could be quantified is indicated under the denomination of total quantified proteins. The list and quantitative information of the 1,529 quantified proteins is given in Supplemental Table S2.
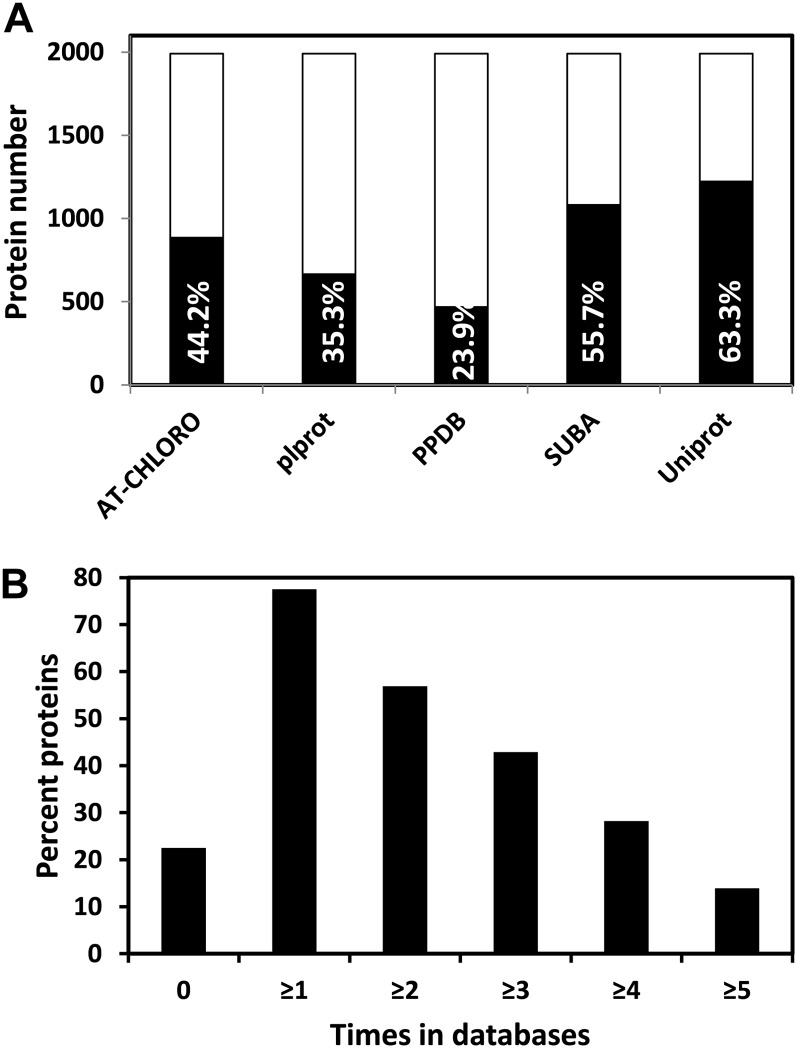
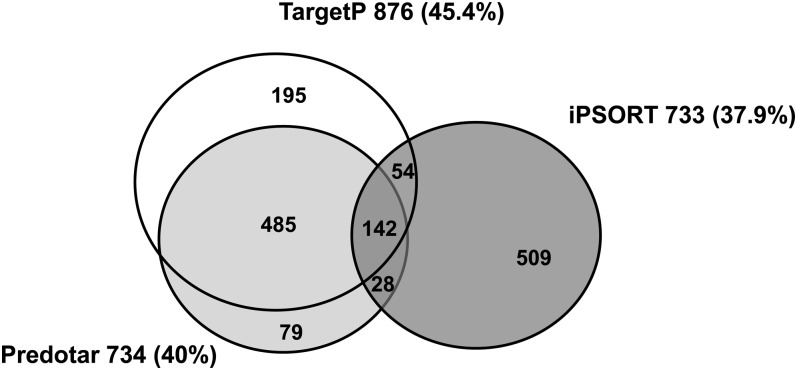
The number of proteins of the tomato plastid proteome referenced in each of the five databases is presented in Figure 2A. It indicates that the overlap is higher than 50% for SUBA (55.7%) and Uniprot (63.3%), more than 35% for Plprot (35.3%) and AT-CHLORO (44.2%). PPDB has the lowest overlap with 23.9%. The percentage of tomato proteins that are not referenced in any of the five databases is 22.5% only while the percentage referenced at least one, two, three, four, and five times is 77.5%, 56.9%, 44.9%, 28.3%, and 13.9%, respectively (Fig. 2B). A large majority of proteins (1,492 = 77.2%) were predicted to be plastid localized by at least one prediction software. The overlap of tomato plastid proteins predicted by the three softwares is presented in Figure 3. TargetP, iPSORT, and Predotar forecast a plastid localization of 45.4% (876 proteins), 37.9% (733), and 40% (734) proteins, respectively. The percentage of proteins predicted by all three programs is only 7.3% (142). Previous reports have indicated similar proportions of mitochondrial proteins predicted in common by four prediction programs (Heazlewood et al., 2004). Predotar and TargetP were the closest in terms of predictions with 485 proteins in common. It is noticeable that predictions specific to iPSORT were the highest with 509 proteins (Fig. 3). Predictions made by the three programs within the existing chloroplast databases are extremely variable from 83% with TargetP in PPDB to 43% with iPSORT in SUBA (Table I).
Figure 2.
Proteins of the tomato plastid proteome referenced in five plastid databases. A, Number and percentage of proteins (in black bars) referenced in each of the five databases, calculated on the basis of the 1,932 proteins listed in Supplemental Table S1. B, Percentage of the tomato plastid proteins not referenced (0) or referenced at least one, two, three, four, and five times in the different databases. The five databases are the following: AT-CHLORO, Plprot, PPDB, Uniprot, and SUBA.
Figure 3.
Venn diagram of the number of proteins of the tomato plastid proteome predicted to be plastid localized by three predictors. A total of 1,492 proteins are predicted by at least one predictor, TargetP, Predotar, or iPSORT. The 47 plastid-encoded proteins are not concerned.
Table I. Number of proteins predicted by three predictors in the tomato plastid proteome and in four plastid databases.
Predictions are made only on nuclear-encoded proteins. For plprot and PPDB, only the subset of Arabidopsis plastidial proteins was considered (indicated by an asterisk).
| Database | No. of Nuclear-Encoded Proteins | TargetP | iPSORT | Predotar |
|---|---|---|---|---|
| Tomato plastid | 1,885 | 876 (46.5%) | 733 (38.9%) | 730 (38.7%) |
| AT-CHLORO | 1,345 | 928 (68.9%) | 660 (49%) | 764 (56.8%) |
| plprot* | 1,006 | 493 (49.0%) | 489 (48.6%) | 546 (54.3%) |
| PPDB* | 1,178 | 980 (83.2%) | 771 (65.4%) | 908 (77.1%) |
| SUBA | 2,112 | 1,208 (57.1%) | 913 (43.2%) | 1,039 (49.2%) |
The inventory of tomato fruit plastid proteins present at different stages of plastid differentiation enriches the knowledge of the plant plastid proteome. Most of the data available were related to chloroplasts (Armbruster et al., 2011; van Wijk and Baginsky, 2011), although some information was also available for nonphotosynthetic plastid structures such as amyloplasts (Andon et al., 2002; Balmer et al., 2006), etioplasts (von Zychlinski et al., 2005), proplastids (Baginsky et al., 2004), and embryoplasts (Demartini et al., 2011). The present inventory also provides information that complements previous articles published on the chromoplast proteome (bell pepper: Siddique et al., 2006; tomato: Barsan et al., 2010; and sweet orange: Zeng et al., 2011).
The plastid proteome of tomato fruit at the MG stage was compared with the proteome of Arabidopsis chloroplasts reported in the AT-CHLORO database (Ferro et al., 2010) and in Zybailov et al. (2008). It appears that the percentage of proteins classified according to the MapMan functional classes is very similar in the two proteomes, particularly for the class corresponding to photosynthesis (Supplemental Fig. S1). This indicates that MG plastids have the general characteristics of chloroplasts although fruit photosynthesis is very low compared with leaf photosynthesis (Blanke and Lenz, 1989; Hetherington et al., 1998) and plays an unimportant role in fruit metabolism (Lytovchenko et al., 2011).
Proteomic Specificities of the Three Stages of Plastid Differentiation in Terms of Protein Abundance
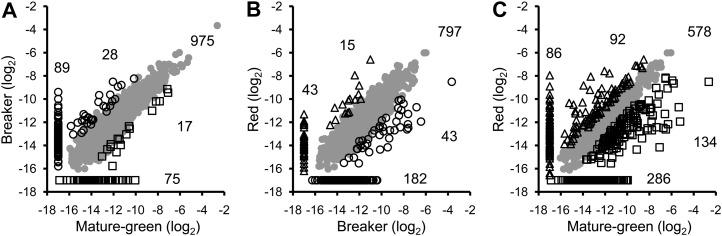
After normalization and statistical analysis, 1,529 proteins out of the inventory of 1,932 were quantified and their pattern of abundance determined (Fig. 1; Supplemental Table S2). A comparison of the protein pattern at different stages two by two is presented as scatter plots in Figure 4. Between the MG and B stages (Fig. 4A) a large number of proteins underwent no significant change in abundance (975 = 63.7%). Only 17 proteins were significantly more abundant at the green stage and 28 at the B stage, while 75 proteins are specific to the MG stage and 89 to the B stage. Between the B and R stages 797 (52.1%) proteins underwent no quantitative changes. Among proteins showing differences in abundance, 43 are more abundant at the B stage and 15 at the R stage while 182 and 43 are specific to the B and R stages, respectively (Fig. 4B). Comparing the two most distant stages (MG and R), the number of proteins undergoing no changes between the two stages was found to be only 578, representing 37.8% of the proteome. Therefore around 60% of the proteins change in abundance during the whole differentiation process (Fig. 4C) with 134 proteins overexpressed in the MG and 92 in the R plastids. The number of proteins encountered at one stage only was 286 in the MG and 86 in the R plastids (Fig. 4C).
Figure 4.
Scatter plots comparing log2 protein abundance between two differentiation stages of tomato fruit plastids. A, MG versus B. B, B versus R. C, MG versus R. Shades of gray represent proteins that are equally abundant in both stages; open squares correspond to proteins overexpressed at the MG stage; open circles are proteins overexpressed at the B stage, and open triangles represent proteins overabundant at the R stage. Data along the axis of the scatter plots indicate proteins that have been quantified at one of the two stages only and totally absent at the other stage. Numbers correspond to the number of proteins in each category. To draw the graphs, a log2 value of −17 was arbitrarily affected to proteins for which no abundance value was available in Supplemental Table S2.
Proteomic data on the whole tomato fruit are available in which quantitative data on plastidial proteins can be encountered (Rocco et al., 2006; Faurobert et al., 2007). Forty-seven proteins have been identified both in the plastid proteome described in this work and in the whole fruit proteome of Rocco et al. (2006) and Faurobert et al. (2007). Although the tomato varieties and the quantification methods were very different, the ratios of abundance between the three stages of plastid development (MG, B, and R) were identical or similar, thus confirming the accuracy of the quantification methods.
Changes in Abundance of Proteins Encoded by the Plastid Genome
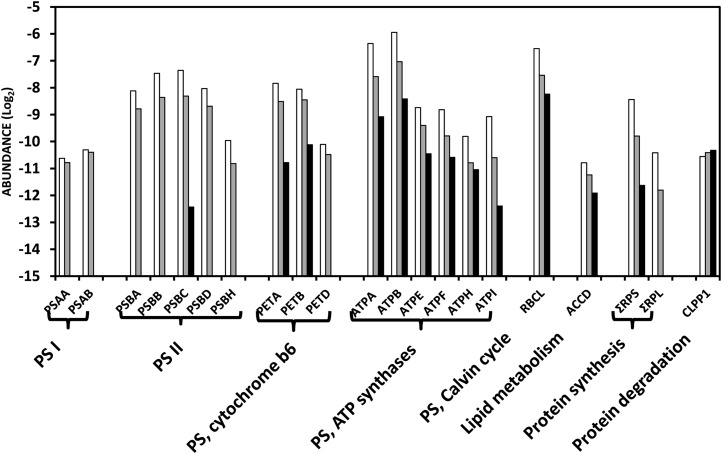
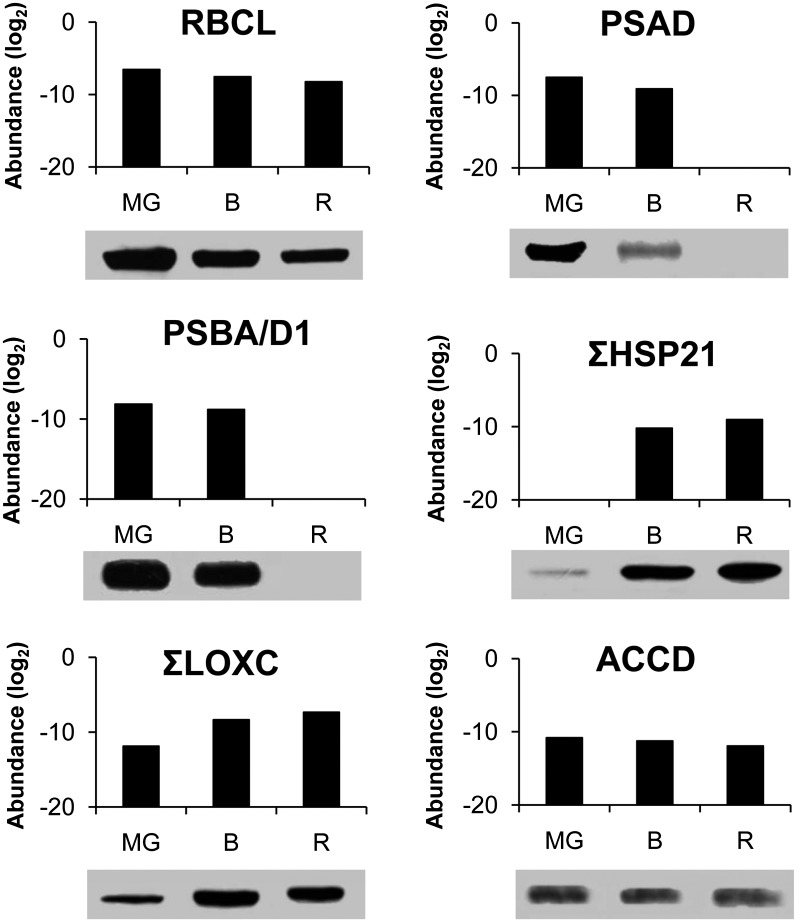
Among the 87 proteins predicted to be encoded by the plastid genome, 47 were encountered in our study (Supplemental Table S1) and the abundance of 32 of them is presented in Figure 5 for all three stages of plastid development (Supplemental Table S2). These comprise several proteins participating in photosynthesis whose abundance remains essentially stable between the MG and B stages but disappear or strongly decrease at the R stage when the photosynthetic system is dismantled: two proteins of PSI, five of PSII, three of PS cytochrome b6, and five of PS ATP synthase. Only one protein of PSII (PSBC), and two proteins of cytochrome b6 (PETA and PETB) are present at the R stage at substantial amounts. Immunoblots were performed for the PSAD protein of PSI and PSBA/D of PSII (Fig. 6). They are decreasing in abundance between the MG and B stages and are totally undetectable at the R stage, which is globally in agreement with the proteomic analysis. Surprisingly all the six ATP synthase subunits encoded by the plastid genome were found. They all undergo a continuous decrease in abundance, but are still present at significant amount at the R stage, indicating that the ATP synthesis machinery is maintained at a good level throughout chromoplast development. The large subunit of Rubisco (RBCL) continuously decreases in abundance but is present at significant levels in red plastids (Fig. 5). The pattern of changes of RBCL is identical when evaluated by western blot (Fig. 6), thus providing another confirmation of the reliability of the quantification procedure. Another interesting information given in Figure 5 is that the sum of ribosomal proteins of the small 30S subunit (RPS) and of the large 50S subunit (RPL) strongly decline in abundance, with the RPL proteins absent at the R stage. This is in agreement with the gradual down-regulation of plastid translation observed by Kahlau and Bock (2008) during chromoplast differentiation in tomato. In contrast, the only caseinolytic protease encoded by the plastid genome, CaseinoLytic Plastidial Protease1 (CLPP1) is not changing in abundance, indicating a high level of protein processing throughout the differentiation process. This is in line with the sustained abundance of elements of the protein import machinery discussed below. In our study special attention has been given to the acetyl CoA carboxylase (ACCD) protein involved in fatty acid biosynthesis because previous work has shown that chromoplast gene expression largely serves the production of ACCD (Kahlau and Bock, 2008). In our proteomic analysis it is decreasing in abundance, although at a significance of P = 0.06 only (Supplemental Table S2). Western blot of ACCD also shows a decrease in abundance between the MG and R stages (Fig. 6). This is in apparent contrast with the western-blot data of Kahlau and Bock (2008) showing an increase of the ACCD protein between the green and the turning stages. To explain the discrepancy, we performed another series of immunoblots blots by including an earlier stage of fruit development. The tendency observed in Figure 6 of a decline of ACCD protein between the MG and R stages was confirmed, but, most importantly the abundance of the protein was much lower in immature green fruit (Supplemental Fig. S2). These data indicate that the green fruit of Kahlau and Bock (2008) were probably sampled well before the MG stage. In our conditions, the MG stage was selected because the fruit has gained the capacity to ripen and to respond to the plant hormone ethylene (Pech et al., 2012) but plastids still have a chloroplastic structure with high chlorophyll, low carotenoid content, and absence of lycopene. Western blots of the ACCD protein indicate that important changes occur in plastids between the green and MG stages of development (Fig. 6). Whether these changes are part of the chromoplast differentiation process may be a matter of discussion. However, a unified view could be that plastid differentiation is a continuous process during fruit development in which the final steps of the differentiation corresponding to the chromatogenesis process per se are triggered by the plant hormone ethylene. In such a scheme, the early accumulation of the ACCD protein that is involved in fatty acid biosynthesis can be considered as a prerequisite for chromoplast differentiation by providing a storage matrix for the accumulation of carotenoids.
Figure 5.
Abundance of proteins encoded by the plastid genome. Proteins present at all three stages of plastid development (MG: white bars; B: gray bars; and R: black bars) were classified according the MapMan functional classes. Protein abundance is expressed as a log2. The graphs were generated using the data and symbols of Supplemental Table S2. PSA, PSB, PET, and ATP correspond to proteins of the subunits of PSI, PSII, cytochrome b6, and ATP synthase complexes, respectively. RBCL corresponds to the large subunit of Rubisco, ACCD to the D subunit of acetyl CoA carboxylase, and CLPP1 to the caseinolytic protease P1. ΣRPS and ΣRPL represent the sum of proteins of the small 30S and the large 50S subunits, respectively.
Figure 6.
Comparison of protein abundance determined by proteomic analysis and immunoblotting. The abundance of proteins determined by proteomic analysis is expressed as log2. RBCL corresponds to the large subunit of Rubisco (GI89241679), PSAD to the D subunit of PSI (Solyc06g054260), PSBA/D1 to the A/D1 subunit of PSII (GI89241651), HSP21 to Heat Shock Proteins21 (Solyc03g082420 and Solyc05g014280), LOXC to lipoxygenase C (Solyc01g006540, Solyc01g006560, and Solyc12g011040), and ACCD to the D subunit of acetyl CoA carboxylase (GI89241680). For western blots, proteins were extracted from partially purified plastids as indicated in “Material and Methods.”
Changes in Subplastidial Compartmentation
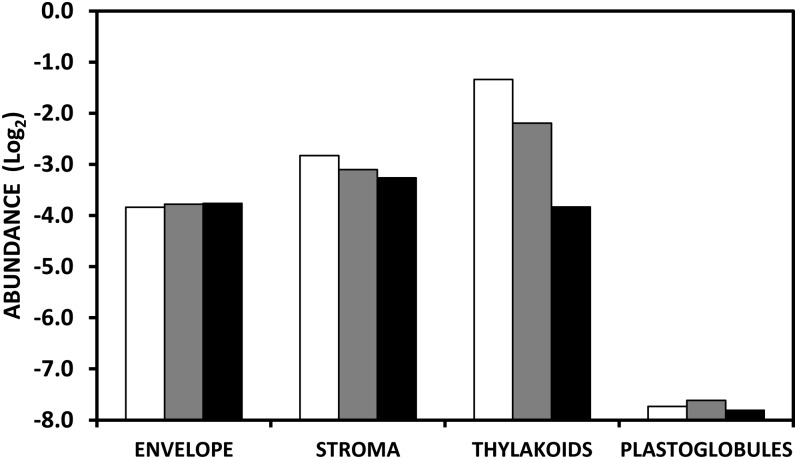
The tomato plastid proteome referenced in Supplemental Table S2 has been screened with the AT_Chloro database (Ferro et al., 2010) to isolate proteins present in the stroma, thylakoids, and envelope membrane (Supplemental Table S3) and with the list of proteins of the plastoglobules established by Lundquist et al. (2012). In agreement with the structural remodeling of the internal membrane system (Spurr and Harris, 1968), this study clearly shows that the abundance of thylakoid proteins fell mostly during the transition from B to R stages while the abundance of proteins of the envelope and of the plastoglobules remained essentially unchanged and proteins of the stroma underwent a slight decrease in abundance (Fig. 7). The observation that the plastoglobule proteins underwent no changes during the transition from chloroplasts to chromoplasts is in line with the fact that the bulk of the carotenoids of tomato fruit are stored predominantly in the form of lycopene crystalloids in membrane-shaped structures (Harris and Spurr, 1969).
Figure 7.
Abundance of proteins in the subplastidial compartments of tomato fruit plastids. Plastids were isolated from MG (white bars), B (gray bars), and R (black bars) fruit. Protein abundance is expressed as a log2. The present graph corresponds to Supplemental Table S3 generated by screening the tomato plastid proteome on the base of AT homologs with the AT-CHLORO subplastidial database (Ferro et al., 2010) for stroma, thylakoids, and envelope proteins and in Lundquist et al. (2012) for plastoglobule proteins.
Kinetics of Changes in the Functional Classes during the Chloroplast-to-Chromoplast Transition
The kinetics of changes in protein abundance occurring during the three stages of chromoplast development have been classified into seven categories: stable, decreasing early, decreasing late, decreasing continuously, increasing early, increasing late, and increasing continuously (Table II). Among the seven categories, proteins whose abundance remained statistically constant slightly outweighed (569) proteins undergoing an increase in abundance (104 + 289 + 186 = 547). Those of the increasing category are the less abundant (72 + 82 + 100 = 254). Proteins showing a late decrease are the second-most numerous, indicating that important changes occur during the last step between B and R stages (Table II).
Table II. Abundance pattern of the tomato plastid proteins classified into seven categories.
Some proteins (127) for which we were unable to establish a consistent and logical pattern of abundance have been omitted. Data are from Supplemental Table S2.
Among proteins showing the same abundance during the whole differentiation process, some functional classes were seen to be more stable than others (Fig. 8A). Sulfur assimilation, although represented by few proteins, is 100% stable, followed by tricarboxylic acid (TCA)/organic acid (85.3%), metal handling (66.7%), electron transport/ATP synthesis (62.5%), and glycolysis (56.5%). Representatives of classes comprising around one-third of stable proteins only are: amino acid metabolism, protein synthesis, lipid metabolism, secondary metabolism, Calvin cycle, and oxidative pentose phosphate pathway (OPP). Some classes have a very low percentage of stable proteins such as major carbohydrate (CHO; 10.7%), hormone metabolism (6.3%), and photosynthesis (2.4%). This picture allows the identification of a basal background of functions and structures that are roughly maintained during the differentiation of chromoplasts as well as profound changes in some functions that contribute to redirecting the plastid metabolism.
Figure 8.
Number and percentage of proteins in the MapMan functional classes for seven patterns of abundance. A, Stable. B, Decreasing early. C, Decreasing late. D, Decreasing continuously. E, Increasing early. F, Increasing late. G, Increasing continuously. The abundance patterns are described in Table II. Numbers in % represent the percentage of proteins within each functional class.
Photosynthesis is the most representative of the functional classes showing decreasing abundance during the chromoplast differentiation process with a much higher percentage of proteins (60.7%) decreasing late between the B and R stages (Fig. 8C) than decreasing early (17.9%; Fig. 8B) or continuously (15.5%; Fig. 8D). Tetrapyrrole biosynthesis (which comprises the biosynthesis of chlorophylls), OPP, and posttranslational event classes follows the same trend with a larger proportion of proteins undergoing a decline in the later stages (Fig. 8C). The situation where a high percentage of proteins decrease early is represented by the cofactors/vitamin metabolism, nitrogen metabolism, and biodegradation of xenobiotics classes with a decrease between the MG and B stages ranging from 25% to 36.4% of the proteins (Fig. 8B). The major CHO metabolism class comprises proteins decreasing in abundance in about the same proportions in the early (25.0%) and late (25.0%) stages of differentiation (Fig. 8, B and C).
The proportion of proteins showing increasing abundance (Fig. 8, E–G) in the different classes is generally low as compared with the decreasing categories (Fig. 8, B–D). One noticeable exception is proteins involved in fermentation of which 66.7% increase continuously (Fig. 8G). Interestingly stress-related proteins are those showing the highest percentage of increase with 15.7% (Fig. 8G), 15.7% (Fig. 8E), and 9.8% (Fig. 8F) in the increasing continuously, early or late categories, respectively. The significant proportion of cell division/organization proteins (15.1%) in the increasing continuously category (Fig. 8G) may account for the structural changes occurring in the formation of chromoplasts. Another remarkable observation is the high percentage (>30%) of hormone-related and DNA-related proteins showing an early (Fig. 8E) and a late (Fig. 8F) increase in abundance, respectively. The changes in hormone-related proteins are correlated to the increase in the synthesis of some hormones, mainly between B and R stages, such as abscisic acid (Zhang et al., 2009) and jasmonates (Fan et al., 1998). A small proportion of proteins of the major metabolisms, such as lipid, major and minor CHO, and OPP still underwent an increase in abundance. Noteworthy is the absence of proteins of the photosynthesis and Calvin cycle classes in the increasing categories (Fig. 8, E–G), a very low proportion of them remaining at constant amounts throughout chromoplast differentiation (Fig. 8A). Interestingly, two carbonic anhydrases of the TCA/organic acid class (Supplemental Table S2) were present at increasing levels (Solyc02g067750 and Solyc05g005490). Carbonic anhydrases have been found associated with the Rubisco complex (Jebanathirajah and Coleman, 1998). Rubisco being still present at late stages of chromoplast formation, carbonic anhydrases may contribute to the provision of CO2 to Rubisco by catalyzing the dehydration of HCO3− in the alkaline stroma in close proximity to Rubisco. Some of the proteins increasing in abundance participate in the development of the sensory quality of the fruit, such as lipoxygenase C (LOXC; Solyc01g006540, Solyc01g006560, and Solyc12g011040), which is responsible for the biosynthesis of fatty-acid-derived aroma volatiles (Chen et al., 2004). The protein undergoes continuous increase from the MG to the R stages in proteomic analysis (Supplemental Table S2) as well as in western blots (Fig. 6). The increase in LOXC is coincident with the increase in the production of aroma volatiles (Birtić et al., 2009).
Overview of Metabolic and Regulatory Changes Occurring during Chromoplastogenesis
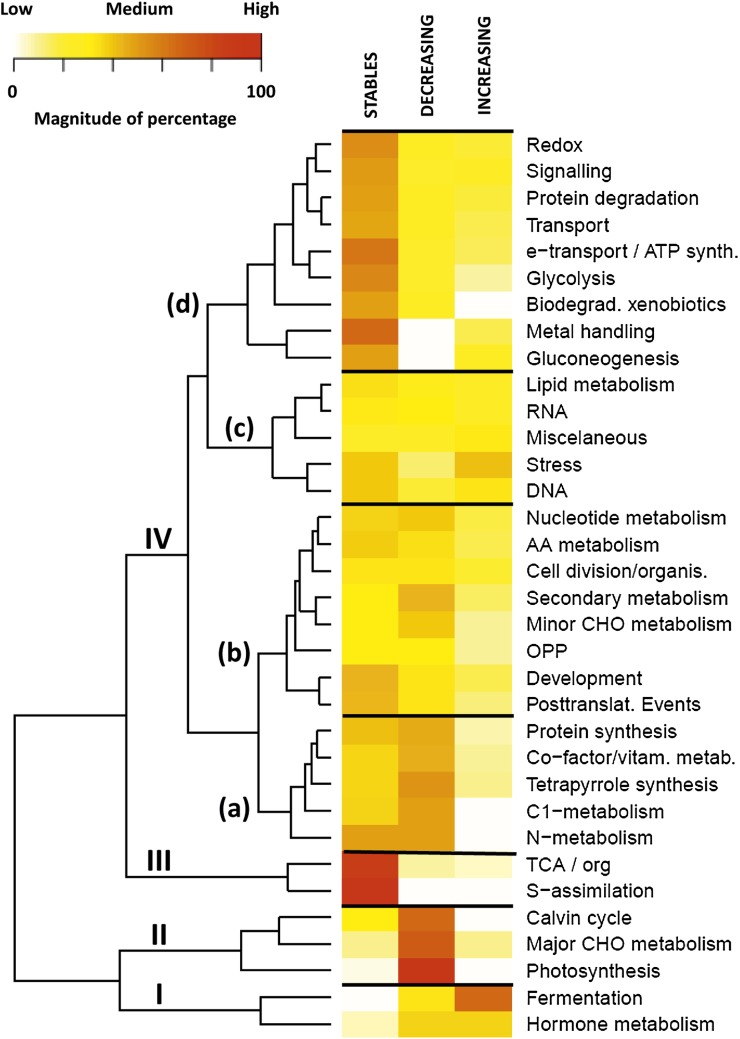
A heatmap showing the percentage of proteins in each functional class according to their abundance patterns allows various clusters to be distinguished (Fig. 9) Two clusters comprise a high percentage of stable proteins: III (sulfur assimilation and TCA/organic acid classes), IVd (redox, signaling, protein degradation, transport, electron transport/ATP synthesis, glycolysis, biodegradation/xenobiotics, metal handling, and gluconeogenesis). Two clusters include a majority of decreasing proteins corresponding to cluster II (Calvin cycle, major CHO metabolism and photosynthesis) and to cluster IVa (protein synthesis, cofactor/vitamin metabolism, tetrapyrrole synthesis, C1 and nitrogen metabolism). Cluster I is the only one comprising proteins with a strong increase in abundance, essentially fermentation. Clusters IVb and IVc have an almost equal percentage of proteins with stable, decreasing, and increasing patterns, indicating profound redirections in the corresponding functional classes, for instance OPP, metabolism of nucleotides, amino acids, and lipids as well as RNA synthesis.
Figure 9.
Heatmap showing the percentage of proteins of each functional class according to the abundance pattern. The three abundance patterns considered during the differentiation of chromoplasts are: stable, decreasing, and increasing. The magnitude of the percentage is represented by a color scale (top left) going from low (white), to medium (yellow), and high (red).
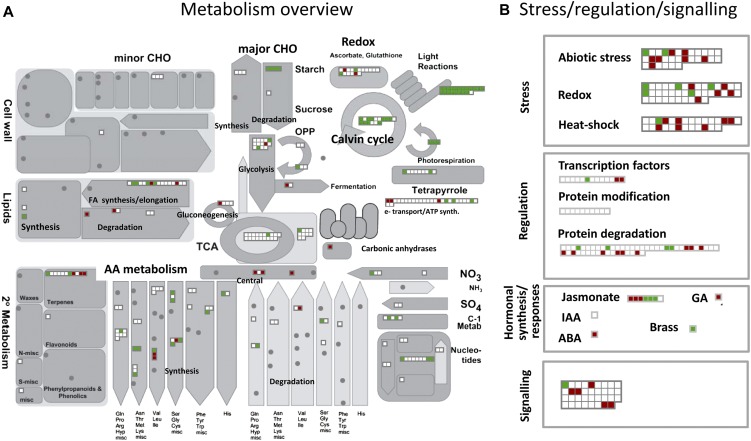
Changes in the abundance of individual proteins participating in the central metabolism are represented in a MapMan metabolic display (Fig. 10A). This illustrates the major shifts in metabolism occurring during the differentiation of chromoplasts. A large number of proteins involved in light reactions (including photosynthesis, Calvin cycle, and photorespiration) and major CHO metabolism (starch metabolism) are more abundant in the MG plastid and therefore decrease in abundance during chromoplastogenesis. Many of the proteins involved in the provision of energy to the plastid remain unchanged (TCA cycle, glycolysis, and electron transport/ATP synthesis). Some proteins increase in abundance such as carbonic anhydrases and some proteins involved in the metabolism of terpenoids (including carotenoids), lipids, amino acids, and ascorbate glutathione cycle. The increase in activity of enzymes of the ascorbate glutathione cycle has already been demonstrated during fruit ripening (Jimenez et al., 2002). The abundance of carbonic anhydrases in red chromoplasts has been discussed previously as possibly participating in providing CO2 to Rubisco that is still present at high levels. Another feature of the chloroplast-to-chromoplast transition is related to changes in stress-related, regulatory, and signaling proteins (Fig. 10B). Abiotic stress, redox, and heat shock classes comprise a majority of proteins not changing in abundance but also a number of proteins exhibiting either an increase or a decrease in abundance. The simultaneous changes in redox and abiotic stress proteins are consistent with the role of redox signaling in the response to abiotic stress in plants (Suzuki et al., 2012). The role of some heat shock proteins in promoting color changes during tomato fruit ripening has been reported (Neta-Sharir et al., 2005). Moderate redistribution can also be observed in transcription factors, signaling, and proteins involved in protein modification and degradation. Of particular interest are the signaling proteins undergoing an increase in levels. Quantitative changes also occur in proteins involved in the response to hormones and in hormones synthesis. The jasmonate biosynthetic pathway is located in the plastid and several proteins increase in abundance during chromoplastogenesis. This is consistent with the role of jasmonate in the biosynthesis of carotenoid biosynthesis in interaction with ethylene (Fan et al., 1998). Proteins annotated as ethylene related are absent from the MapMan mapping. This could be considered as surprising considering that ethylene plays a major role in the ripening process of climacteric fruit such as the tomato (Pirrello et al., 2009; Pech et al., 2012) and in the synthesis of carotenoids (Bramley, 2002). However, the ethylene biosynthesis and signaling pathways are not located in the plastid and it is expected that the regulation of ethylene-responsive genes occurs mainly at the nucleus. In a microarray analysis of tomato fruit treated by the ethylene antagonist 1-methylcyclopropene, Tiwari and Paliyath (2011) found that at least nine nuclear genes encoding plastidial proteins were up-regulated by ethylene and three down-regulated. Genes involved in the biosynthesis of carotenoids, fatty acids, and jasmonic acid and in gluconeogenesis were up-regulated, while genes involved in starch degradation were down-regulated. Nevertheless, a full inventory of ethylene-responsive genes encoding plastid-localized proteins remains to be performed. In addition, the cross talk between hormones in ripening fruit is not yet well understood at the molecular level.
Figure 10.
Metabolic overview comparing the protein abundance in MG and R tomato plastids. Red squares represent proteins with decreasing levels while blue squares correspond to proteins with increasing levels. A, Metabolic overview. B, Stress, regulation, and signaling proteins. MapMan software (Thimm et al., 2004; http://gabi.rzpd.de/projects/MapMan/).
Loss of the Machinery for the Build Up of Thylakoids and Photosystems
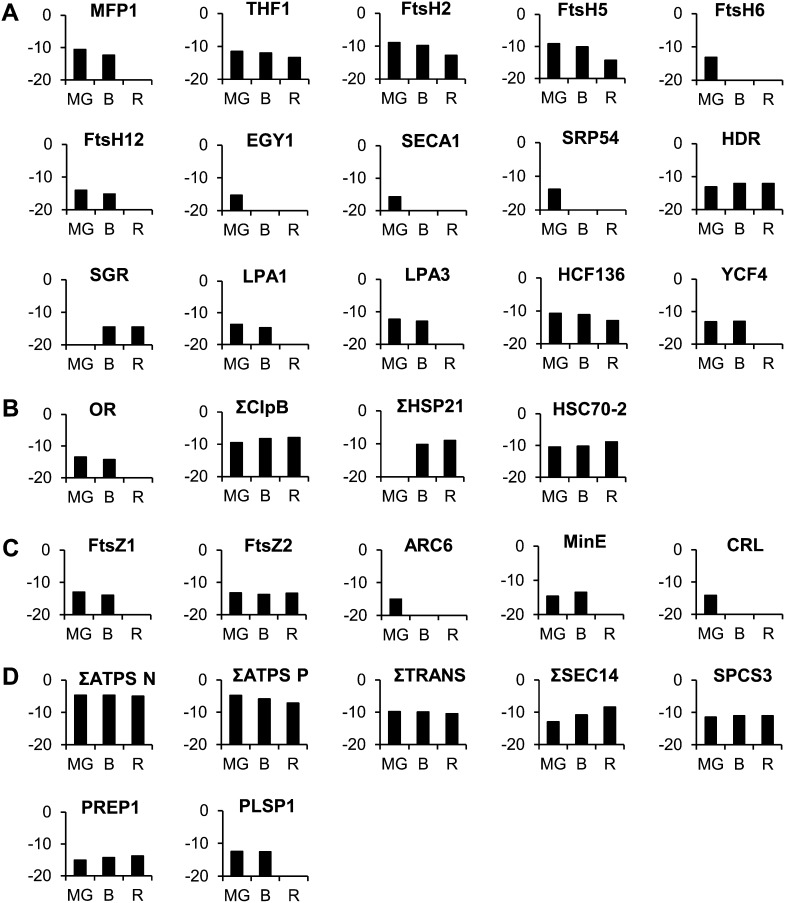
A number of proteins participating in the build up of thylakoids have been encountered that decrease in abundance during the chloroplast-to-chromoplast transition (Fig. 11A; Supplemental Table S4). A nucleoid-binding protein classified in the RNA functional class, MATRIX ATTACHMENT REGION FILAMENT BINDING PROTEIN1 (Solyc03g120230), which is tightly associated with the accumulation of thylakoid membranes (Jeong et al., 2003) decreases in abundance between the green and B stages and is not detectable at the R stage. In addition, the THYLAKOID FORMATION1 (Solyc07g054820) protein, which is involved in thylakoid formation through vesicular trafficking, continuously decreases in abundance. It is thought to interact with a plasma membrane G protein to provide a sugar-signaling mechanism necessary for thylakoid formation (Huang et al., 2006). The tomato plastid proteome comprises a number of membrane-bound ATP-dependent metalloproteases of the filamentation temperature-sensitive metalloprotease class (FtsH2: Solyc07g055320; FtsH5: Solyc04g082250; FtsH6: Solyc02g081550; and FtsH12: Solyc02g079000). Members of these proteases are involved in the repair of PSII (Liu et al., 2010a) as well as in the maintenance of the thylakoid structure (Kato et al., 2012). Similarly, another type of ATP-independent metalloprotease, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW MUTANT1 (EGY1; Solyc10g081470), present in the tomato plastid proteome has been described as required for chloroplast development via its role in thylakoid membrane biogenesis (Chen et al., 2005). The build up of the thylakoids requires the import of proteins within the plastid. A thylakoid Sec translocase subunit (SECA1, Solyc01g080840) is essential for chloroplast biogenesis (Liu et al., 2010b). Interestingly, the FtsH and EGY1 proteins decrease in abundance, mostly between the B and R stages, which correlates well with the dismantling of the thylakoid membranes, while SECA was present only at the MG stage, indicating an early end to the provision of material for thylakoid biogenesis. Recently the lutescent2 mutant of tomato has been identified as mutated for a homolog of EGY1 of Arabidopsis (Barry et al., 2012). The mutation is responsible for an altered chloroplast development and delay in fruit ripening but this not precluded chromoplast differentiation, indicating that chromoplast development does not depend on functional chromoplasts (Barry et al., 2012). A signal recognition particle subunit (SRP54, Solyc09g009940) involved in the integration of the light-harvesting chlorophyll a/b protein into the thylakoid membrane (Li et al., 1995; Rutschow et al., 2008) is present at the MG stage and then absent, giving another indication of the disappearance of photosynthetic protein import during the chloroplast-to-chromoplast transition.
Figure 11.
Abundance of proteins (y axis in log2) involved in structural modifications of plastids, provision of energy, and translocation of precursors during the chloroplast-to-chromoplast transition. A, Proteins involved in the biogenesis of thylakoids and photosystems. B, Proteins involved in plastid differentiation. C, Proteins involved in plastid division. D, Proteins involved in energy and translocation. Protein abundance is expressed as a log2. The full name of the proteins is indicated in the text and in Supplemental Table S4. Abbreviations preceded by Σ correspond to the sum of several proteins harboring the same function. Individual values are given in Supplemental Table S4.
Consistent with the decrease in the chlorophyll biosynthetic branch, an increase is observed of the STAY-GREEN (Solyc08g080090) protein that regulates chlorophyll degradation by modulating pheophorbide a oxygenase activity (Ren et al., 2007). Another protein, HYDROXYMETHYLBUTENYL DIPHOSPHATE REDUCTASE (Solyc01g109300) produces MEP-derived precursors for plastid carotene biosynthesis. It increases slightly during the three stages that is consistent with the accumulation of carotenoids and in agreement with the up-regulation of the HYDROXYMETHYLBUTENYL DIPHOSPHATE REDUCTASE genes during tomato fruit ripening (Botella-Pavía et al., 2004). In addition, there is also an abrupt decrease, between the B and R stages, of two low PSII accumulation proteins (LPA1: Solyc09g074880 and LPA3: Solyc06g068480) involved in the assembly of PSII (Peng et al., 2006; Cai et al., 2010). The two proteins have been clearly identified by Peng et al. (2006) and Cai et al. (2010) but still annotated as unknown gene in The Arabidopsis Information Resource and Solyc databases. Another protein necessary for the assembly of PSII high chlorophyll fluorescence (HCF136, Solyc02g014150; Plücken et al., 2002) decreases continuously. A plastid genome-encoded protein, YCF4 (Solyc01g007360) belonging to the hypothetical chloroplast reading frame family (YCF) decreases strongly between the B and R stages in parallel with the late disassembly of PSI. The Ycf4 protein participates in the assembly of PSI in the alga Chlamydomonas reinhardtii (Onishi and Takahashi, 2009) and in higher plants (Krech et al., 2012). However, unlike its role in algae, in higher plants it is not essential. The knockdown of the YCF4 gene in tobacco (Nicotiana tabacum) results in an only partial reduction of the amount of PSI complex, indicating that other proteins can replace YCF4 (Krech et al., 2012). Similarly to the decline in the YCF4 protein during the transition from chloroplast to chromoplast observed here in tomato fruit, YCF4 declines continuously in old leaves of tobacco concomitantly with the decrease of photosynthetic activity (Krech et al., 2012).
Several Elements of Plastid Differentiation Correlate with Chromoplast Formation
Interestingly, the tomato fruit plastid proteome contains a number of proteins known to participate in the differentiation of plastids (Fig. 11B; Supplemental Table S4). One of them is a dnaj-like chaperone (Solyc03g093830) and corresponds to the product of the Or gene that controls chromoplast differentiation and carotenoid accumulation (Li and Van Eck, 2007). The Or protein is more abundant at the B than at the green stage and was not detected at the R stage when carotenoids are at their maximum level.
Some ATP-dependent casein lytic proteinases (Clp) located in the stroma are described as participating in chloroplast development (Lee et al., 2007). Six of them classified in the stress-related proteins (ClpB: Solyc06g082560, Solyc03g115230, Solyc02g088610, Solyc06g011400, Solyc06g011370, and Solyc06g011380) are encountered in the tomato plastids. Summing the abundance of the six proteins remains essentially constant, suggesting a sustained function during chromoplast differentiation. Also interesting is the absence at the G stage and the sharp accumulation at the B and R stages of two HEAT SHOCK PROTEIN21 (HSP21) heat shock proteins (Solyc03g082420 and Solyc05g014280) that have been reported to be involved in the promotion of color changes during tomato fruit maturation (Neta-Sharir et al., 2005). Western blots show the presence of HSP21 proteins at very low level at the MG stage and a sudden and large increase at the B stage (Fig. 6). Except for the presence of small amounts at the MG stage in western blots, these data are in agreement with the proteomic analysis. Another heat shock protein, HSC70-2 (Solyc11g020040), highly expressed in the tomato plastids, increases continuously. It has been described as essential for plant development in Arabidopsis (Su and Li, 2008). Its role in chromoplast differentiation would deserve elucidation.
Loss of the Plastid Division Machinery
Many proteins involved in the division of plastids have been encountered in the tomato plastid proteome (Fig. 11C; Supplemental Table S4). Two nuclear-encoded forms of filamenting temperature-sensitive Z mutants (FtsZ1, Solyc07g065050 and FtsZ2, Solyc09g009430) play a major role in the initiation and progression of plastid division in plant cells (McAndrew et al., 2008). They are both present at MG and B stages, but FtsZ1 was absent at the R stage while FtsZ2 remained stable at all three stages. An accumulation and replication of chloroplasts protein (ARC6, Solyc04g081070) has been characterized in a mutant that has only one or two chloroplast per cell instead of >100 in wild-type cells (Vitha et al., 2003). The mutant exhibits abnormal localization of the two key plastid division proteins FtsZ1 and FtsZ2, indicating that ARC6 promotes FtsZ filament formation in the chloroplast (Aldridge et al., 2005). ARC6 was encountered at the MG stage only. The MinE protein (Solyc05g012710), which supports and maintains FtsZ filament formation (Maple and Møller, 2007) and is required for correct plastid division in Arabidopsis (Maple et al., 2002), disappears between the MG and B stages. Similarly, the crumpled leaf protein (CRL, Solyc06g068760), also described as important for the division of plastids, was found only at the MG stage. Asano et al. (2004) showed that cells of the CRL mutant contained a reduced number of plastids. The disappearance or strong decrease of the majority of proteins involved in plastid division mentioned above during the differentiation of chromoplasts indicates that plastid division ceases. This is in agreement with our previous observation that no plastid division occurred in ripening tomatoes since all preexisting chloroplasts were differentiating into chromoplasts (Egea et al., 2011). These data provide target proteins potentially responsible for the cessation of plastid division.
Proteins Involved in Energy Provision and Translocation Activities
ATP synthase is an important enzyme that provides energy for the cell to use through the synthesis of ATP. Thirteen nuclear-encoded ATP synthase units were quantified in the tomato plastid proteome (Supplemental Table S4). The summation of the abundance of all these ATP synthase units was stable during chromoplast differentiation (Fig. 11D). On the other hand, six ATP synthase units encoded by the plastid genome were quantified (Supplemental Table S4) and slightly decreased during tomato fruit ripening (Fig. 11D). These data indicate that the machinery for the provision of energy stays very present throughout the differentiation process. Plastids can import cytosolic Glc-6-P via the Glc-6-P/phosphate translocator (GPT, Solyc07g064270). Glc-6-P is used either for the synthesis of starch and fatty acids or is fed into the plastidic OPP (Emes and Neuhaus, 1997). Niewiadomski et al. (2005) have shown that the loss of GPT1 function results in a disruption of the oxidative pentose phosphate cycle and affects fatty acid biosynthesis. Another translocator, phosphoenolpyruvate phosphate translocator (Solyc03g112870), has been identified that delivers the energy-rich glycolytic intermediate phosphoenolpyruvate into the plastids (Fischer et al., 1997). In addition, a triose phosphate/phosphate translocator (Solyc10g008980) has also been encountered that participates in Suc biosynthesis (Cho et al., 2012). The sum of the three translocators decreases very lightly during plastid transition (Fig. 11), indicating that the machinery for provision of energy and precursors remained in place to allow the synthesis of fatty acids and Suc within the plastid.
Elements for the direct import of lipids into the plastid are also present (Fig. 11D; Supplemental Table S4). Two proteins of the SEC translocase system are highly expressed and increase continuously in abundance (SEC14, Solyc11g051160, Solyc06g064940). They are the homologs of the yeast (Saccharomyces cerevisiae) SFH5 phosphatidylinositol transfer protein (Yakir-Tamang and Gerst, 2009) and could be involved in Golgi vesicle transport of phosphoinositides in plant plastids. The transport of lipids to the plastids via vesicles derived from the endoplasmic reticulum membrane and fused with Golgi membranes is more than a hypothesis (Andersson and Dörmann, 2009; Benning, 2009). Elements of the protein import machinery were also encountered. One signal peptidase complex subunit (SPCS3, Solyc01g098780) and one presequence protease (PREP1, Solyc01g108600) remained roughly stable, indicating that elements of the machinery for import of proteins remain present during chromoplast differentiation, while the synthesis of protein by the plastid translational machinery strongly declines, as mentioned above. Another signal peptidase, plastidic signal peptidase (PLSP1, Solyc12g007120) involved in thylakoid development through its involvement in processing the Toc75 envelope protein and 0E33 thylakoid luminal protein (Shipman-Roston et al., 2010) was absent at the R stage, which is consistent with thylakoid dismantling.
CONCLUSION
High-throughput technologies have been used in the recent years for elucidating the mechanisms of fruit ripening, such as transcriptomics (Alba et al., 2004), metabolomics (Schauer et al., 2006; Deborde et al., 2009; Moing et al., 2011), proteomics (Rocco et al., 2006; Faurobert et al., 2007; Palma et al., 2011), and more recently, a combination of all of them (Osorio et al., 2011). However these technologies have been scarcely used in the study of subcellular organelles such as chromoplasts. In this article, we used high-throughput proteomics to make an inventory of the proteins present at different stages of plastid differentiation in ripening tomato fruit. This inventory enriches the knowledge of the plant plastid proteome as reported in plastid databases. In addition, it provides an in-depth description of the changes occurring in the plastidial proteome during chloroplast-to-chromopast differentiation. The mechanisms governing the differentiation of plastids such as the conversion of proplastids to chloroplasts or chloroplasts to chromoplasts have received little attention and are barely mentioned in recent reviews on plastid proteomics (Armbruster et al., 2011; van Wijk and Baginsky, 2011). The pattern of changes in protein abundance reported here is in agreement with proteomic data generated in whole fruit by others (Rocco et al., 2006; Faurobert et al., 2007). The western blots of six proteins representative of several metabolic or regulatory pathways give a pattern of changes that is similar to proteomic analysis. Therefore, these data confirm the reliability of the tandem mass spectrometry (MS/MS) analysis and the spectral counting quantification procedure carried out in this study. In addition, the evolutions of protein abundance are in agreement with the most important metabolic changes described in the literature such as chlorophyll degradation, loss of photosynthetic activity, dismantling of thylakoid membranes, and increase in the synthesis of lycopene (Egea et al., 2010). Previous studies had sequentially described individual metabolisms or even specific steps of metabolism. In this work a global and extensive quantitative picture is given of the major shifts occurring during the differentiation of chromoplasts that integrate both the strong decrease in proteins of the light reactions and major CHO metabolism and the increase in abundance of proteins involved in carotenoid biosynthesis. A remarkable finding not described so far is the stability of proteins of the TCA cycle, glycolysis, and the high level of electron transport/ATP synthesis, indicating that the machinery providing energy to the plastid is largely conserved. Particularly noticeable is also the increase in a number of proteins of the ascorbate glutathione cycle, abiotic stress, redox, and heat shock classes, indicating that the transition seems to be governed at least partly by a redox-signaling pathway causing a stress-related response. Strong redistribution can also be observed in transcription factors, signaling, proteins involved in protein-modification/degradation, and in proteins involved in the response to and synthesis of hormones. This indicates that chromoplastogenesis involves profound changes in signaling events whose details and interactions remain to be elucidated. The demonstration that chromoplasts can develop from altered chloroplasts in lutescent2 tomato fruit mutants (Barry et al., 2012) indicates that the differentiation of chromoplasts and the associated signaling events do not depend on functional chloroplasts. However, although with some delay, chromoplast formation always accompanies fruit ripening.
Among the most important events of chromoplast differentiation the dismantling of thylakoids have been well described in the literature by electron microscopy. This study brings novel information on the target proteins participating in the loss of the machinery for the build up of thylakoids and for the assembly of photosystems. For instance, proteins involved in thylakoid formation through vesicular trafficking, in the provision of material for thylakoid biosynthesis, and in the assembly of photosystems undergo a strong decrease in abundance to be essentially absent in red chromoplasts. In agreement with the loss of chlorophylls, the stay-green protein that stimulates chlorophyll degradation increases in abundance. Proteins of the plastid division machinery have been identified that disappear during the differentiation of chromoplasts. Their absence in fully differentiated chromoplasts can therefore be considered as responsible for the cessation of plastid division. An unexpected finding is that chromoplast differentiation is accompanied with the maintenance of the major elements providing energy and metabolites to the plastid such as ATP synthase, hexose, and triose phosphate translocators, as well as lipid import elements. The translation machinery of the plastid probably undergoes a great loss of efficiency with the strong decrease in abundance of ribosomal proteins of both the 30S and 50S complexes. However, several elements of the protein import machinery remain at a high level, suggesting that nuclear-encoded proteins continue to be transferred at a high rate till the last stage of chromoplast differentiation.
Our results complement recent studies on chloroplast-to-chromoplast differentiation, showing that chromoplast gene expression largely serves the production of a single protein, ACCD (Kahlau and Bock, 2008). We now demonstrate that this occurs early during fruit development while the fruit is unable to ripen autonomously and well before any visible transition from chloroplast to chromoplast. We conclude the early accumulation of the ACCD protein that is involved in fatty acid biosynthesis is a prerequisite for chromoplast differentiation by providing a storage matrix for the accumulation of carotenoids. The final steps of differentiation during which the metabolic shifts occur are associated with the initiation of the ripening process by the plant hormone ethylene.
These data demonstrate that the shifts in metabolism are preceded by the increase of the ACCD protein that could account for the generation of a cartenoids storage matrix. They also show that the metabolic changes are associated with a high abundance of proteins involved in providing energy and import of metabolites. In addition, regulatory proteins have been identified that are potentially responsible for the overall differentiation process as well as for individual differentiation events such as the dismantling of thylakoids and photosystems and cessation of division.
MATERIALS AND METHODS
Plant Material
Tomato (Solanum lycopersicum ‘MicroTom’) plants were cultivated under standard greenhouse conditions. Fruit were collected at three stages of ripening after careful selection: (1) MG fruit corresponds to fruit having reached full size and able to ripen autonomously. They produce very low amounts of ethylene but respond to ethylene in terms of ripening. MG fruit were selected by the presence of gel in the locules and an a*/b* chromatic ratio between −0.32 and −0.38 measured by the Minolta chromameter; (2) B + 2 corresponds to fruit harvested 2 d after the B stage. B is characterized by a change in color from green to pale orange over about 30% of the surface. At B + 2, the fruit reach peak ethylene production; (3) R fruit has the whole surface colored red and were harvested 10 d after B.
Plastid Isolation from Fruit at Various Stages of Ripening and Fractionation of Proteins
Plastids were isolated from the fruit pericarp at three stages of ripening according to the procedure described in Egea et al. (2011). After separation on Suc gradient, plastid fractions were analyzed by fluorescence confocal microscopy associated with spectrophotometric analysis. MG plastids were characterized by the almost exclusive presence of chlorophylls, characteristic of chloroplasts; B plastids corresponding to intermediate forms of plastids were selected in a band of gradient containing reduced amounts of chlorophylls and substantial amounts of carotenoids; R plastids contained almost exclusively carotenoids, typical of chromoplasts. The plastid bands collected were washed twice with extraction buffer (250 mm HEPES, 330 mm sorbitol, 0.1 mm EDTA, 5 mm β-mercaptoethanol, pH 7.6). For the proteomic analysis plastids were resuspended in 1 m HEPES buffer (pH 7.6), 2 mm dithiothreitol (DTT), and kept at −20°C until protein precipitation. After storage at −20°C plastids were broken by osmotic shock by resuspension in 1 m HEPES buffer (pH 7.6) complemented with 2 mm DTT, followed by freeze/thawing and homogenization in a Potter-Elvehjem tissue grinder. Proteins of each fraction were precipitated with methanol/chloroform (3:8 v/v) according to Seigneurin-Berny et al. (1999) with some modifications. The resulting protein pellet was dissolved in 2% SDS, 62.5 mm Tris-HCl buffer (pH 6.8). Protein concentrations were determined using the bicinchoninic acid method (Smith et al., 1985). Finally the proteins were reduced with DTT (20 mm) and alkylated with chloroacetamide (60 mm) before solubilization in Laemmli buffer and SDS-PAGE (12% acrylamide) separation. Each lane of the one-dimensional SDS-PAGE gel was loaded with 70 μg of proteins originating from the three types of plastids. After electrophoresis, proteins were stained with PageBlue protein staining solution (Fermentas). Three independent biological replicates of SDS-PAGE gel were carried out for each plastid development stage (Fig. 1).
Western-Blot Analysis
Western-blot analysis was performed using polyclonal antibodies at appropriate dilution against chloroplastic RBCL (53 kD, at 1:50,000 dilution), PSI PSAD (23 kD, at 1:25,000 dilution), PSII PSBA/D1 protein (39 kD, at 1:50,000 dilution), HSP21 (26 kD, at 1:30,000 dilution), and LOXC (102 kD, at 1:50,000 dilution) from Agrisera. ACCD (58 kD) antibodies were purchased from the Plant Antibody facility of the Ohio State University and used at 1:25,000 and 1:15,000 dilution in Figure 6 and Supplemental Figure S2, respectively. Extraction of proteins, separation by SDS-PAGE, and detection by antibodies were performed as described in Barsan et al. (2010). Proteins were extracted from partially purified plastids recovered at the step before separation on Suc gradient as described in Egea et al. (2011).
Trypsin Digestion and Liquid Chromatography MS/MS Analyses of Gel Segments
The protocol was the same as in Barsan et al. (2010) except that each lane of the gel was cut into 10 slices.
Protein Identification and Quantification by Spectral Counting
Data were analyzed using Xcalibur software (version 2.1.0, Thermo Fisher Scientific) and MS/MS centroid peak lists were generated using the extract_msn.exe executable (Thermo Fisher Scientific) integrated into the Mascot Daemon software (Mascot version 2.3.2, Matrix Sciences). The following parameters were set to create peak lists: parent ions in the mass range 400 to 4,500, no grouping of MS/MS scans, and threshold at 1,000. A peaklist was created for each fraction (i.e. each gel slice) analyzed and individual Mascot searches were performed for each fraction. Data were searched against iTAG2.3 proteome sequence (ftp://ftp.solgenomics.net/tomato_genome/annotation/ITAG2.3_release/ITAG2.3_proteins.fasta) predicted from the tomato genome sequence by the International Tomato Genome Annotation (iTAG) team and against the proteome sequences predicted from the chloroplastic genome (accession no. AM087200). Mass tolerances in mass spectrometry and MS/MS were set to 5 ppm and 0.8 D, respectively, and the instrument setting was specified as ESI Trap. Trypsin was designated as the protease (specificity set for cleavage after Lys or Arg) and one missing cleavage was allowed. Carbamido methylation of Cys was fixed and oxidation of Met was searched as variable modification. Mascot results were parsed with the homemade and developed software MFPaQ version 4.0 (Mascot File Parsing and Quantification; Bouyssié et al., 2007). To evaluate false positive rates, all the initial database searches were performed using the decoy option of Mascot, i.e. the data were searched against a combined database containing the real specified protein sequences (iTAG2.3 tomato proteome sequence) and the corresponding reversed protein sequences (decoy database). MFPaQ used the same criteria to validate decoy and target hits, calculated the false discovery rate (FDR; FDR = number of validated decoy hits/[number of validated target hits + number of validated decoy hits] × 100) for each gel slice analyzed, and made the average of FDR for all slices belonging to the same gel lane (i.e.to the same sample). FDRs were below 1.6%. From all the validated result files corresponding to the fractions of a one-dimensional gel lane, MFPaQ was used to generate a unique nonredundant list of proteins that were identified and characterized by homology-based comparisons with iTA1G2.3 tomato proteome sequence. Output files from Mascot searches were uploaded with MFPaQ for spectral counting. The total number of spectra corresponding to each identified protein was extracted from each lane of analyses. The filtering parameters were P values > 0.05, more than seven amino acids in the peptides and more than one peptide encountered. Among all strategies used for quantitative proteomics (Thelen and Peck, 2007; van Wijk and Baginsky, 2011) we followed a label-free option of spectral counting (Liu et al., 2004; Booth et al., 2011) described in Bouyssié et al. (2007). The spectral-count-based label free has been reported as allowing quantification of protein abundance with similar efficiency to isotope labeling (Zhu et al., 2009). This methodology had been applied in a number of other studies related to the plastid proteome (Bräutigam et al., 2008; Ferro et al., 2010; Demartini et al., 2011).
Comparison with Existing Databases, Targeting Predictions, Functional Classification, and Curation
Protein descriptions were performed using annotations associated with each protein entry and through homology-based comparisons with the TAIR9 protein database (http://www.arabidopsis.org/) using BasicLocal Alignment Search Tool BLASTX (Altschul et al., 1990) with an e-value cutoff of 1e-5 to avoid false positives, and linked. MapMan Bins were used for functional assignments (http://gabi.rzpd.de/projects/MapMan/). The protein list was compared with five plastidial databases either specific to plastids (AT-CHLORO: Ferro et al., 2010; http://www.grenoble.prabi.fr/proteome/grenoble-plant-proteomics/; plprot: Kleffmann et al., 2006; http://www.plprot.ethz.ch) or general databases comprising subcellular subsets (PPDB: Sun et al., 2009; http://ppdb.tc.cornell.edu; SUBA: Heazlewood et al., 2007; http://www.suba.bcs.uwa.edu.au; Uniprot: The Uniprot Consortium, 2010: http://www.uniprot.org). Predictions of subcellular localization were undertaken using three predictors (TargetP: Emanuelsson et al., 2000; http://www.cbs.dtu.dk/services/TargetP/; iPSORT: Bannai et al., 2002; http://hypothesiscreator.net/iPSORT/; Predotar: Small et al., 2004; http://genoplante-info.infobiogen.fr/predotar/). Predictions were made on the basis of tomato proteins when harboring an N-terminal sequence. A curation procedure was used to select, in the protein data set, those having confirmed plastid location. Proteins were retained in the final dataset if they meet with at least one of the two following criteria: (1) presence in at least two of the five databases mentioned above (AT-CHLORO, plprot, PPDB, SUBA, and Uniprot) and (2) predicted by at least one of the three predictors (TargetP, iPSORT, and Predotar). Proteins predicted to be encoded by the plastid genome were all retained. This resulted in the cataloging of 1,932 proteins in Supplemental Table S1.
Normalization and Differential Abundance Analysis
The raw data arising from the LTQ-Orbitrap mass spectrometer analysis (1,932 proteins listed in Supplemental Table S1 were submitted to a normalization process that consisted in standardizing the MS/MS counts by the length of the corresponding protein, and by an integral normalization, as described by Paoletti et al., 2006). The sum of abundances in all samples was set to 1 so as to eliminate the effect of the size of the total MS/MS counts in the different samples. Then, the Probabilistic Quotient Normalization method (Dieterle et al., 2006) was applied by taking a reference sample at random among the replicates of the MG stage. This allowed the calculation of the quotients of protein abundances (as log2 ratios) in each sample and hence the median of the quotients. By dividing the abundance of proteins by the corresponding median, the effect of the difference in proteome size between samples was eliminated. The normalization process is now widespread and considered as a standard way to preprocess data in metabolomics and proteomics (Torgrip et al., 2008; Issaq et al., 2009; Kato et al., 2011).
The differential abundance analysis was carried out with the Anapuce R package in the R environment (http://www.R-project.org/) and calculated using a paired t test on normalized data. An estimation of the variance for each protein observation was performed using the mixed model of Delmar et al. (2005a, 2005b) implemented in the anapuce R package and consisting in the estimation of several groups of proteins with the same variance. Finally, a classical FDR procedure was applied to correct for multiple comparison tests according to the procedure of Benjamini and Hachberg (1995) implemented in the anapuce R package. Normalization and statistical analysis led to the cataloging of 1,529 proteins considered as quantifiable out of the initial 1,932 proteins (Supplemental Table S2). Protein abundance values (expressed as log2) were retained only when a given protein had been detected at least twice in a given stage. Supplemental Table S2 comprises the log2-abundance values, the log2 ratios, and the P value of the t test for each protein. The trends of changes in abundance between stages were calculated with a 5% significance level and referred to as 0 for no change, +1 for increasing, and −1 for decreasing.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparison of the percentage of proteins classified into MapMan functional classes between the tomato fruit chloroplast proteome described in this article (white bars) and the Arabidopsis chloroplast proteome described in the AT-CHLORO database (Ferro et al., 2010; gray bars) and in Zybailov et al. (2008; black bars).
Supplemental Figure S2. Western-blot analysis of the ACCD protein in plastids isolated at four stages of fruit development: IG (Immature-Green).
Supplemental Table S1. Inventory of the 1,932 proteins representing the compilation of the curated list of proteins encountered in the three replicates of the tomato plastids at three stages of development.
Supplemental Table S2. Inventory of 1,529 proteins quantified from the curated list.
Supplemental Table S3. List of proteins in the subplastidial compartments: envelope, stroma, thylakoids, and plastoglobules.
Supplemental Table S4. List of proteins involved in the build up of thylakoids and photosystems, plastid differentiation, plastid division, and energy and translocation.
Acknowledgments
We are grateful to Dr. Yasushi Yoshioka (Nagoya University, Japan) for generously providing antibodies against CRL protein and Dr. Basil Nikolau (Iowa State University) for his help in getting the antibodies against the ACCD protein.
Glossary
- RBCL
large subunit of Rubisco
- OPP
oxidative pentose phosphate pathway
- DTT
dithiothreitol
- FDR
false discovery rate
- MG
mature green
- B
breaker
- R
red
- CHO
carbohydrate
- MS/MS
tandem mass spectrometry
References
- Alba R, Fei Z, Payton P, Liu Y, Moore SL, Debbie P, Cohn J, D’Ascenzo M, Gordon JS, Rose JK, et al. (2004) ESTs, cDNA microarrays, and gene expression profiling: tools for dissecting plant physiology and development. Plant J 39: 697–714 [DOI] [PubMed] [Google Scholar]
- Aldridge C, Maple J, Møller SG. (2005) The molecular biology of plastid division in higher plants. J Exp Bot 56: 1061–1077 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Andersson MX, Dörmann P. (2009) Chloroplast membrane lipid biosynthesis and transport. In AS Sandelius, H Aronson, eds, The Chloroplast; Interactions with the Environment. Springer-Verlag, Berlin, pp 125–158
- Andon NL, Hollingworth S, Koller A, Greenland AJ, Yates JR, III, Haynes PA. (2002) Proteomic characterization of wheat amyloplasts using identification of proteins by tandem mass spectrometry. Proteomics 2: 1156–1168 [DOI] [PubMed] [Google Scholar]
- Armbruster U, Pesaresi P, Pribil M, Hertle A, Leister D. (2011) Update on chloroplast research: new tools, new topics, and new trends. Mol Plant 4: 1–16 [DOI] [PubMed] [Google Scholar]
- Asano T, Yoshioka Y, Kurei S, Sakamoto W, Machida Y, Sodmergen (2004) A mutation of the CRUMPLED LEAF gene that encodes a protein localized in the outer envelope membrane of plastids affects the pattern of cell division, cell differentiation, and plastid division in Arabidopsis. Plant J 38: 448–459 [DOI] [PubMed] [Google Scholar]
- Baginsky S, Siddique A, Gruissem W. (2004) Proteome analysis of tobacco bright yellow-2 (BY-2) cell culture plastids as a model for undifferentiated heterotrophic plastids. J Proteome Res 3: 1128–1137 [DOI] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, DuPont FM, Buchanan BB, Hurkman WJ. (2006) Proteome of amyloplasts isolated from developing wheat endosperm presents evidence of broad metabolic capability. J Exp Bot 57: 1591–1602 [DOI] [PubMed] [Google Scholar]
- Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S. (2002) Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18: 298–305 [DOI] [PubMed] [Google Scholar]
- Barry CS, Aldridge GM, Herzog G, Ma Q, McQuinn RP, Hirschberg J, Giovannoni JJ. (2012) Altered chlorolast development and delayed fruit ripening caused by mutations in a zinc metalloprotease at the lutescent2 locus of tomato. Plant Physiol 159: 1086–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsan C, Sanchez-Bel P, Rombaldi C, Egea I, Rossignol M, Kuntz M, Zouine M, Latché A, Bouzayen M, Pech JC. (2010) Characteristics of the tomato chromoplast revealed by proteomic analysis. J Exp Bot 61: 2413–2431 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hachberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Benning C. (2009) Mechanisms of lipid transport involved in organelle biogenesis in plant cells. Annu Rev Cell Dev Biol 25: 71–91 [DOI] [PubMed] [Google Scholar]
- Bian W, Barsan C, Egea I, Purgatto E, Chervin C, Zouine M, Latché A, Bouzayen M, Pech JC. (June 3, 2011) Metabolic and molecular events occurring during chromoplast biogenesis: review article. J Bot http://dx.doi.org/10.1155/2011/289859 [Google Scholar]
- Birtić S, Ginies C, Causse M, Renard CM, Page D. (2009) Changes in volatiles and glycosides during fruit maturation of two contrasted tomato (Solanum lycopersicum) lines. J Agric Food Chem 57: 591–598 [DOI] [PubMed] [Google Scholar]
- Blanke MM, Lenz F. (1989) Fruit photosynthesis. Plant Cell Environ 12: 31–46 [Google Scholar]
- Booth JG, Eilertson KE, Olinares PD, Yu H. (2011) A bayesian mixture model for comparative spectral count data in shotgun proteomics. Mol Cell Proteomics 10: M110. 007203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella-Pavía P, Besumbes O, Phillips MA, Carretero-Paulet L, Boronat A, Rodríguez-Concepción M. (2004) Regulation of carotenoid biosynthesis in plants: evidence for a key role of hydroxymethylbutenyl diphosphate reductase in controlling the supply of plastidial isoprenoid precursors. Plant J 40: 188–199 [DOI] [PubMed] [Google Scholar]
- Bouvier F, Camara B. (2007) The role of plastids in ripening fruits. In RR Wise, JK Hoober, eds, The Structure and Functions of Plastids. Springer, Dordrecht, The Netherlands, pp 419–432
- Bouyssié D, Gonzalez de Peredo A, Mouton E, Albigot R, Roussel L, Ortega N, Cayrol C, Burlet-Schiltz O, Girard JP, Monsarrat B. (2007) Mascot file parsing and quantification (MFPaQ), a new software to parse, validate, and quantify proteomics data generated by ICAT and SILAC mass spectrometric analyses: application to the proteomics study of membrane proteins from primary human endothelial cells. Mol Cell Proteomics 6: 1621–1637 [DOI] [PubMed] [Google Scholar]
- Bramley PM. (2002) Regulation of carotenoid formation during tomato fruit ripening and development. J Exp Bot 53: 2107–2113 [DOI] [PubMed] [Google Scholar]
- Bräutigam A, Hoffmann-Benning S, Weber APM. (2008) Comparative proteomics of chloroplast envelopes from C3 and C4 plants reveals specific adaptations of the plastid envelope to C4 photosynthesis and candidate proteins required for maintaining C4 metabolite fluxes. Plant Physiol 148: 568–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W, Ma J, Chi W, Zou M, Guo J, Lu C, Zhang L. (2010) Cooperation of LPA3 and LPA2 is essential for photosystem II assembly in Arabidopsis. Plant Physiol 154: 109–120 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Camara B, Hugueney P, Bouvier F, Kuntz M, Monéger R. (1995) Biochemistry and molecular biology of chromoplast development. Int Rev Cytol 163: 175–247 [DOI] [PubMed] [Google Scholar]
- Chen G, Bi YR, Li N. (2005) EGY1 encodes a membrane-associated and ATP-independent metalloprotease that is required for chloroplast development. Plant J 41: 364–375 [DOI] [PubMed] [Google Scholar]
- Chen G, Hackett R, Walker D, Taylor A, Lin Z, Grierson D. (2004) Identification of a specific isoform of tomato lipoxygenase (TomloxC) involved in the generation of fatty acid-derived flavor compounds. Plant Physiol 136: 2641–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MH, Jang A, Bhoo SH, Jeon JS, Hahn TR. (2012) Manipulation of triose phosphate/phosphate translocator and cytosolic fructose-1,6-bisphosphatase, the key components in photosynthetic sucrose synthesis, enhances the source capacity of transgenic Arabidopsis plants. Photosynth Res 111: 261–268 [DOI] [PubMed] [Google Scholar]
- Deborde C, Maucourt M, Baldet P, Bernillon S, Biais B, Talon G, Ferrand C, Jacob D, Ferry Dumazet H, de Daruvar A, et al. (2009) Proton NMR quantitative profiling for quality assessment of greenhouse-grown tomato fruit. Metabolomics 5: 183–198 [Google Scholar]
- Delmar P, Robin S, Daudin JJ. (2005a) VarMixt: efficient variance modelling for the differential analysis of replicated gene expression data. Bioinformatics 21: 502–508 [DOI] [PubMed] [Google Scholar]
- Delmar P, Robin S, Tronik-Le Roux D, Daudin JJ. (2005b) Mixture model on the variance for differential analysis of gene abundance data. JR Stat Soc 54: 31–50 [Google Scholar]
- Demartini DG, Jain R, Agrawal G, Thelen JJ. (2011) A proteomic comparison of plastids from developing embryos and leaves of Brassica napus. J Prot Res 10: 2226–2237 [DOI] [PubMed] [Google Scholar]
- Dieterle F, Ross A, Schlotterbeck G, Senn H. (2006) Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures: application in 1H NMR metabonomics. Anal Chem 78: 4281–4290 [DOI] [PubMed] [Google Scholar]
- Egea I, Barsan C, Bian W, Purgatto E, Latché A, Chervin C, Bouzayen M, Pech JC. (2010) Chromoplast differentiation: current status and perspectives. Plant Cell Physiol 51: 1601–1611 [DOI] [PubMed] [Google Scholar]
- Egea I, Bian W, Barsan C, Jauneau A, Pech JC, Latché A, Li ZG, Chervin C. (2011) Chloroplast to chromoplast transition in tomato fruit: spectral confocal microscopy analyses of carotenoids and chlorophylls in isolated plastids and time-lapse recording on intact live tissue. Ann Bot (Lond) 108: 291–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Emes MJ, Neuhaus HE. (1997) Metabolism and transport in non-photosynthetic plastids. J Exp Bot 48: 1995–2005 [Google Scholar]
- Fan X, Mattheis JP, Fellman JK. (1998) A role for jasmonates in climacteric fruit ripening. Planta 204: 444–449 [Google Scholar]
- Faurobert M, Mihr C, Bertin N, Pawlowski T, Negroni L, Sommerer N, Causse M. (2007) Major proteome variations associated with cherry tomato pericarp development and ripening. Plant Physiol 143: 1327–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M, Brugière S, Salvi D, Seigneurin-Berny D, Court M, Moyet L, Ramus C, Miras S, Mellal M, Le Gall S, et al. (2010) AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol Cell Proteomics 9: 1063–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K, Kammerer B, Gutensohn M, Arbinger B, Weber A, Häusler RE, Flügge UI. (1997) A new class of plastidic phosphate translocators: a putative link between primary and secondary metabolism by the phosphoenolpyruvate/phosphate antiporter. Plant Cell 9: 453–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris WM, Spurr AR. (1969) Chromoplasts of tomato fruit. II. The red tomato. Am J Bot 56: 380–389 [Google Scholar]
- Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH. (2004) Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16: 241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. (2007) SUBA: the Arabidopsis subcellular database. Nucleic Acids Res (Database issue) 35: D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington SE, Smillie RM, Davies WJ. (1998) Photosynthetic activities of vegetative and fruiting tissues of tomato. J Exp Bot 49: 1173–1181 [Google Scholar]
- Huang J, Taylor JP, Chen JG, Uhrig JF, Schnell DJ, Nakagawa T, Korth KL, Jones AM. (2006) The plastid protein THYLAKOID FORMATION1 and the plasma membrane G-protein GPA1 interact in a novel sugar-signaling mechanism in Arabidopsis. Plant Cell 18: 1226–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issaq HJ, Van QN, Waybright TJ, Muschik GM, Veenstra TD. (2009) Analytical and statistical approaches to metabolomics research. J Sep Sci 32: 2183–2199 [DOI] [PubMed] [Google Scholar]
- Jebanathirajah JA, Coleman JR. (1998) Association of carbonic anhydrase with a Calvin cycle enzyme complex in Nicotiana tabacum. Planta 204: 177–182 [DOI] [PubMed] [Google Scholar]
- Jeong SY, Rose A, Meier I. (2003) MFP1 is a thylakoid-associated, nucleoid-binding protein with a coiled-coil structure. Nucleic Acids Res 31: 5175–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez A, Creissen G, Kular B, Firmin J, Robinson S, Verhoeyen M, Mullineaux P. (2002) Changes in oxidative processes and components of the antioxidant system during tomato fruit ripening. Planta 214: 751–758 [DOI] [PubMed] [Google Scholar]
- Kahlau S, Bock R. (2008) Plastid transcriptomics and translatomics of tomato fruit development and chloroplast-to-chromoplast differentiation: chromoplast gene expression largely serves the production of a single protein. Plant Cell 20: 856–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato BS, Nicholson G, Neiman M, Rantalainen M, Holmes CC, Barrett A, Uhlén M, Nilsson P, Spector TD, Schwenk JM. (2011) Variance decomposition of protein profiles from antibody arrays using a longitudinal twin model. Proteome Sci 9: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Kouso T, Sakamoto W. (2012) Variegated tobacco leaves generated by chloroplast FtsH suppression: implication of FtsH function in the maintenance of thylakoid membranes. Plant Cell Physiol 53: 391–404 [DOI] [PubMed] [Google Scholar]
- Kleffmann T, Hirsch-Hoffmann M, Gruissem W, Baginsky S. (2006) plprot: a comprehensive proteome database for different plastid types. Plant Cell Physiol 47: 432–436 [DOI] [PubMed] [Google Scholar]
- Krech K, Ruf S, Masduki FF, Thiele W, Bednarczyk D, Albus CA, Tiller N, Hasse C, Schöttler MA, Bock R. (2012) The plastid genome-encoded Ycf4 protein functions as a nonessential assembly factor for photosystem I in higher plants. Plant Physiol 159: 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee U, Rioflorido I, Hong SW, Larkindale J, Waters ER, Vierling E. (2007) The Arabidopsis ClpB/Hsp100 family of proteins: chaperones for stress and chloroplast development. Plant J 49: 115–127 [DOI] [PubMed] [Google Scholar]
- Li L, Van Eck J. (2007) Metabolic engineering of carotenoid accumulation by creating a metabolic sink. Transgenic Res 16: 581–585 [DOI] [PubMed] [Google Scholar]
- Li X, Henry R, Yuan J, Cline K, Hoffman NE. (1995) A chloroplast homologue of the signal recognition particle subunit SRP54 is involved in the posttranslational integration of a protein into thylakoid membranes. Proc Natl Acad Sci USA 92: 3789–3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Gong Q, Ma Y, Li P, Li J, Yang S, Yuan L, Yu Y, Pan D, Xu F, et al. (2010a) cpSecA, a thylakoid protein translocase subunit, is essential for photosynthetic development in Arabidopsis. J Exp Bot 61: 1655–1669 [DOI] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR., III (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem 76: 4193–4201 [DOI] [PubMed] [Google Scholar]
- Liu X, Yu F, Rodermel S. (2010b) Arabidopsis chloroplast FtsH, var2 and suppressors of var2 leaf variegation: a review. J Integr Plant Biol 52: 750–761 [DOI] [PubMed] [Google Scholar]
- Ljubesić N, Wrischer M, Devidé Z. (1991) Chromoplasts—the last stages in plastid development. Int J Dev Biol 35: 251–258 [PubMed] [Google Scholar]
- Lopez-Juez E. (2007) Plastid biogenesis, between shadow and light. J Exp Biol 58: 11–26 [DOI] [PubMed] [Google Scholar]
- Lundquist PK, Poliakov A, Bhuiyan NH, Zybailov B, Sun Q, van Wijk KJ. (2012) The functional network of the Arabidopsis plastoglobule proteome based on quantitative proteomics and genome-wide coexpression analysis. Plant Physiol 158: 1172–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytovchenko A, Eickmeier I, Pons C, Osorio S, Szecowka M, Lehmberg K, Arrivault S, Tohge T, Pineda B, Anton MT, et al. (2011) Tomato fruit photosynthesis is seemingly unimportant in primary metabolism and ripening but plays a considerable role in seed development. Plant Physiol 157: 1650–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maple J, Chua NH, Møller SG. (2002) The topological specificity factor AtMinE1 is essential for correct plastid division site placement in Arabidopsis. Plant J 31: 269–277 [DOI] [PubMed] [Google Scholar]
- Maple J, Møller SG. (2007) Interdependency of formation and localisation of the Min complex controls symmetric plastid division. J Cell Sci 120: 3446–3456 [DOI] [PubMed] [Google Scholar]
- Marano MR, Serra EC, Orellano G, Carrillo N. (1993) The path of homoplast development in fruits and flowers. Plant Sci 94: 1–17 [Google Scholar]
- McAndrew RS, Olson BJ, Kadirjan-Kalbach DK, Chi-Ham CL, Vitha S, Froehlich JE, Osteryoung KW. (2008) In vivo quantitative relationship between plastid division proteins FtsZ1 and FtsZ2 and identification of ARC6 and ARC3 in a native FtsZ complex. Biochem J 412: 367–378 [DOI] [PubMed] [Google Scholar]
- Millar AH, Whelan J, Small I. (2006) Recent surprises in protein targeting to mitochondria and plastids. Curr Opin Plant Biol 9: 610–615 [DOI] [PubMed] [Google Scholar]
- Moing A, Aharoni A, Biais B, Rogachev I, Meir S, Brodsky L, Allwood JW, Erban A, Dunn WB, Kay L, et al. (2011) Extensive metabolic cross-talk in melon fruit revealed by spatial and developmental combinatorial metabolomics. New Phytol 190: 683–696 [DOI] [PubMed] [Google Scholar]
- Neta-Sharir I, Isaacson T, Lurie S, Weiss D. (2005) Dual role for tomato heat shock protein 21: protecting photosystem II from oxidative stress and promoting color changes during fruit maturation. Plant Cell 17: 1829–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomski P, Knappe S, Geimer S, Fischer K, Schulz B, Unte US, Rosso MG, Ache P, Flügge UI, Schneider A. (2005) The Arabidopsis plastidic glucose 6-phosphate/phosphate translocator GPT1 is essential for pollen maturation and embryo sac development. Plant Cell 17: 760–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi T, Takahashi Y. (2009) Effects of site-directed mutations in the chloroplast-encoded Ycf4 gene on PSI complex assembly in the green alga Chlamydomonas reinhardtii. Plant Cell Physiol 50: 1750–1760 [DOI] [PubMed] [Google Scholar]
- Osorio S, Alba R, Damasceno CMB, Lopez-Casado G, Lohse M, Zanor MI, Tohge T, Usadel B, Rose JKC, Fei Z, et al. (2011) Systems biology of tomato fruit development: combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol 157: 405–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma JM, Corpas FJ, del Río LA. (2011) Proteomics as an approach to the understanding of the molecular physiology of fruit development and ripening. J Proteomics 74: 1230–1243 [DOI] [PubMed] [Google Scholar]
- Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, Conaway JW, Florens L, Washburn MP. (2006) Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc Natl Acad Sci USA 103: 18928–18933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pech JC, Purgatto E, Bouzayen M, Latché A. (2012) Ethylene and fruit ripening. In MT McManus, ed, Annual Plant Reviews, Vol 44. The Plant Hormone Ethylene. Wiley-Blackwell, Oxford, UK, pp 275–304
- Peng L, Ma J, Chi W, Guo J, Zhu S, Lu Q, Lu C, Zhang L. (2006) LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18: 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierleoni A, Martelli PL, Fariselli P, Casadio R. (2007) eSLDB: eukaryotic subcellular localization database. Nucleic Acids Res (Database issue) 35: D208–D212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrello J, Regad F, Latché A, Pech JC, Bouzayen M. (2009) Regulation of tomato fruit ripening. CAB Reviews 4: 1–14 [Google Scholar]
- Plücken H, Müller B, Grohmann D, Westhoff P, Eichacker LA. (2002) The HCF136 protein is essential for assembly of the photosystem II reaction center in Arabidopsis thaliana. FEBS Lett 532: 85–90 [DOI] [PubMed] [Google Scholar]
- Ren G, An K, Liao Y, Zhou X, Cao Y, Zhao H, Ge X, Kuai B. (2007) Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol 144: 1429–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly E, Leister D. (2004) An improved prediction of chloroplast proteins reveals diversities and commonalities in the chloroplast proteomes of Arabidopsis and rice. Gene 329: 11–16 [DOI] [PubMed] [Google Scholar]
- Rocco M, D’Ambrosio C, Arena S, Faurobert M, Scaloni A, Marra M. (2006) Proteomic analysis of tomato fruits from two ecotypes during ripening. Proteomics 6: 3781–3791 [DOI] [PubMed] [Google Scholar]
- Rutschow H, Ytterberg AJ, Friso G, Nilsson R, van Wijk KJ. (2008) Quantitative proteomics of a chloroplast SRP54 sorting mutant and its genetic interactions with CLPC1 in Arabidopsis. Plant Physiol 148: 156–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Roessner U, Gur A, Balbo I, Carrari F, Pleban T, Perez-Melis A, Bruedigam C, Kopka J, et al. (2006) Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat Biotechnol 24: 447–454 [DOI] [PubMed] [Google Scholar]
- Seigneurin-Berny D, Rolland N, Garin J, Joyard J. (1999) Technical advance: differential extraction of hydrophobic proteins from chloroplast envelope membranes: a subcellular-specific proteomic approach to identify rare intrinsic membrane proteins. Plant J 19: 217–228 [DOI] [PubMed] [Google Scholar]
- Shipman-Roston RL, Ruppel NJ, Damoc C, Phinney BS, Inoue K. (2010) The significance of protein maturation by plastidic type I signal peptidase 1 for thylakoid development in Arabidopsis chloroplasts. Plant Physiol 152: 1297–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique MA, Grossmann J, Gruissem W, Baginsky S. (2006) Proteome analysis of bell pepper (Capsicum annuum L.) chromoplasts. Plant Cell Physiol 47: 1663–1673 [DOI] [PubMed] [Google Scholar]
- Simkin AJ, Gaffé J, Alcaraz JP, Carde JP, Bramley PM, Fraser PD, Kuntz M. (2007) Fibrillin influence on plastid ultrastructure and pigment content in tomato fruit. Phytochemistry 68: 1545–1556 [DOI] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C. (2004) Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4: 1581–1590 [DOI] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. (1985) Measurement of protein using bicinchoninic acid. Anal Biochem 150: 76–85 [DOI] [PubMed] [Google Scholar]
- Spurr AR, Harris WM. (1968) Ultrastructure of chloroplasts and chromoplasts in Capsicum annuum. I. Thylakoid membrane changes during fruit ripening. Am J Bot 55: 1210–1224 [Google Scholar]
- Su PH, Li HM. (2008) Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermotolerance of germinating seeds. Plant Physiol 146: 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zybailov B, Majeran W, Friso G, Olinares PD, van Wijk KJ. (2009) PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res (Database issue) 37: D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Koussevitzky S, Mittler R, Miller G. (2012) ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ 35: 259–270 [DOI] [PubMed] [Google Scholar]
- The Uniprot Consortium (2010) The universal protein resource (UniProt) in 2010. Nucleic Acids Res (Web Server issue) 38: D142–D148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen JJ, Peck SC. (2007) Quantitative proteomics in plants: choices in abundance. Plant Cell 19: 3339–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Thomson WW, Whatley JM. (1980) Development of non-green plastids. Annu Rev Plant Physiol 31: 375–394 [Google Scholar]
- Tiwari K, Paliyath G. (2011) Microarray analysis of ripening-regulated gene expression and its modulation by 1-MCP and hexanal. Plant Physiol Biochem 49: 329–340 [DOI] [PubMed] [Google Scholar]
- Torgrip RJO, Aberg KM, Alm E, Schuppe-Koistinen I, Lindberg J. (2008) A note on normalization of biofluid 1D 1H-NMR data. Metabolomics 4: 114–121 [Google Scholar]
- van Wijk KJ, Baginsky S. (2011) Plastid proteomics in higher plants: current state and future goals. Plant Physiol 155: 1578–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S, Froehlich JE, Koksharova O, Pyke KA, van Erp H, Osteryoung KW. (2003) ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 15: 1918–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zychlinski A, Kleffmann T, Krishnamurthy N, Sjölander K, Baginsky S, Gruissem W. (2005) Proteome analysis of the rice etioplast: metabolic and regulatory networks and novel protein functions. Mol Cell Proteomics 4: 1072–1084 [DOI] [PubMed] [Google Scholar]
- Waters MT, Pyke KA. (2004) Plastid development and differentiation. In SG Møller, ed, Plastids. Blackwell, Oxford, pp 30–59
- Yakir-Tamang L, Gerst JE. (2009) A phosphatidylinositol-transfer protein and phosphatidylinositol-4-phosphate 5-kinase control Cdc42 to regulate the actin cytoskeleton and secretory pathway in yeast. Mol Biol Cell 20: 3583–3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Pan Z, Ding Y, Zhu A, Cao H, Xu Q, Deng X. (2011) A proteomic analysis of the chromoplasts isolated from sweet orange fruits [Citrus sinensis (L.) Osbeck]. J Exp Bot 62: 5297–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Yuan B, Leng P. (2009) The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit. J Exp Bot 60: 1579–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Smith JW, Huang CM. (November 10, 2009) Mass-spectrometry-based label-free quantitative proteomics. J Biomed Biotech http://dx.doi.org/10.1155/2010/840518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ. (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One 3: e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]