Abstract
In higher plants, cellulose is synthesized by so-called rosette protein complexes with cellulose synthases (CESAs) as catalytic subunits of the complex. The CESAs are divided into two distinct families, three of which are thought to be specialized for the primary cell wall and three for the secondary cell wall. In this article, the potential of primary and secondary CESAs forming a functional rosette complex has been investigated. The membrane-based yeast two-hybrid and biomolecular fluorescence systems were used to assess the interactions between three primary (CESA1, CESA3, CESA6), and three secondary (CESA4, CESA7, CESA8) Arabidopsis (Arabidopsis thaliana) CESAs. The results showed that all primary CESAs can physically interact both in vitro and in planta with all secondary CESAs. Although CESAs are broadly capable of interacting in pairwise combinations, they are not all able to form functional complexes in planta. Analysis of transgenic lines showed that CESA7 can partially rescue defects in the primary cell wall biosynthesis in a weak cesa3 mutant. Green fluorescent protein-CESA protein fusions revealed that when CESA3 was replaced by CESA7 in the primary rosette, the velocity of the mixed complexes was slightly faster than the native primary complexes. CESA1 in turn can partly rescue defects in secondary cell wall biosynthesis in a cesa8ko mutant, resulting in an increase of cellulose content relative to cesa8ko. These results demonstrate that sufficient parallels exist between the primary and secondary complexes for cross-functionality and open the possibility that mixed complexes of primary and secondary CESAs may occur at particular times.
Cellulose is the most abundant component of the biosphere, with more than 1011 tons estimated to be synthesized each year (Brown, 2004). This linear β-1,4-glucan polymer is synthesized by the membrane-embedded cellulose synthase (CESA), which is represented by 10 isoforms in Arabidopsis (Arabidopsis thaliana; Doblin et al., 2002; Somerville 2006). In higher plants, CESA proteins form a rosette complex 25 nm in diameter in the plasma membrane, proposed to consist of 36 CESA subunits (Giddings et al., 1980; Muller and Brown, 1980; Kimura et al., 1999; Scheible et al., 2001; Taylor et al., 2003). Genetic evidence shows that at least three isoforms are involved in the synthesis of primary walls in growing cells, CESA1, -3, and -6, and three other isoforms are involved in the deposition of secondary walls in xylem cells, CESA4, -7, and -8 (Fagard et al., 2000; Taylor et al., 2000; Scheible et al., 2001; Desprez et al., 2002, 2007; Ellis et al., 2002; Persson et al., 2007). Double and triple mutants and coimmunoprecipitation analysis in Arabidopsis demonstrate that the remaining CESA proteins, CESA2, -5, and -9, are partially redundant with CESA6 (Desprez et al., 2007; Persson et al., 2007), suggesting specialized functions for CESAs in certain developmental or environmental conditions (Mutwil et al., 2008).
Phylogenetic analysis revealed six distinct CESA clades found in seed plants, each corresponding to one of the six required components of the primary and secondary cellulose synthase complexes in Arabidopsis (Holland et al., 2000; Samuga and Joshi, 2002; Tanaka et al., 2003; Burton et al., 2004; Djerbi et al., 2005; Nairn and Haselkorn, 2005; Ranik and Myburg, 2006; Suzuki et al., 2006; Kumar et al., 2009; Carroll and Specht, 2011). The interaction between the different CESA proteins in the primary and secondary rosettes has been characterized previously by coimmunoprecipitation and yeast two-hybrid methods, showing interaction patterns with similarities between primary and secondary CESAs (Taylor et al., 2000; Desprez et al., 2007; Wang et al., 2008; Atanassov et al., 2009; Timmers et al., 2009). These results suggest that despite the ancient divergence of the families, the complexes may have retained the same positioning of the CESAs in the complex with respect to each other.
The primary and secondary cell walls are formed at different developmental stages. The primary cell wall is synthesized during cell division and expansion, while the secondary cell wall is deposited after the expansion phase. Primary CESAs do not appear to be coordinately expressed with secondary CESAs (Persson et al., 2005). The primary CESAs are thought to be expressed from the initial stages of cell formation until soon after the end of cell expansion, while the secondary CESA genes are assumed to be expressed from the last stages of cell expansion until cell death. Thus, there may be a limited period of time when both primary and secondary CESA genes are coexpressed.
GFP-labeled CESA complexes are seen by confocal microscopy as particles in the plasma membrane that move in linear tracks organized by cortical microtubules (Paredez et al., 2006). Fluorescently labeled CESAs are also seen in Golgi bodies and in small microtubule-associated compartments (SMaCCs), which are implicated in trafficking CESA from the Golgi to the plasma membrane (Crowell et al., 2009; Gutierrez et al., 2009). Although the association of CESA complexes with microtubules appears to be mediated by the cellulose synthase interactive protein 1 (Li et al., 2012), the timing and mechanism of CESA complex assembly remains an open question.
The localization of cellulose synthases is critical to their function. Cellulose is presumably only synthesized at the plasma membrane. Signal from GFP-labeled complexes at the membrane is rapidly lost following osmotic or mechanical shock and chemical inhibition through a number of inhibitors such as isoxaben (Crowell et al., 2009; Gutierrez et al., 2009). The timing of CESA complex assembly remains uncertain. Freeze-fracture images establish it at the membrane (Kimura et al., 1999). The only transmission electron microscopy images of immunolabeled CESA within the Golgi do not show apparent complexes at the stage of localization to the trans-Golgi network (Crowell et al., 2009).
In this report, we demonstrate limited interchangeability between primary and secondary CESAs, which suggests the retention of CESA positioning in the rosette complex and similarities in function across primary and secondary CESA complexes. The parallels between the primary and secondary CESA complexes were investigated by introducing primary CESA proteins in the secondary rosette and vice versa. The interactions between both primary and secondary CESA proteins in Arabidopsis were probed using the split-ubiquitin membrane-based yeast two-hybrid (MbYTH) and bimolecular fluorescence systems; these revealed that they are able to interact and form both homodimers and heterodimers. Through a series of promoter exchanges, we demonstrate that specific secondary CESA constructs are able to partially rescue mutants of certain primary CESAs and incorporate into the complex at the plasma membrane in these mutants. The functional incorporation of specific primary CESAs into the secondary walls is also shown. The incompleteness of the rescue suggests the development of some specialization in the function or regulation of CESA families. These results may also suggest that the synthesis of cellulose during the transition between the primary and secondary cell walls may involve the action of mixed primary-secondary complexes.
RESULTS
Primary CESAs Interact with Secondary CESAs in Vitro
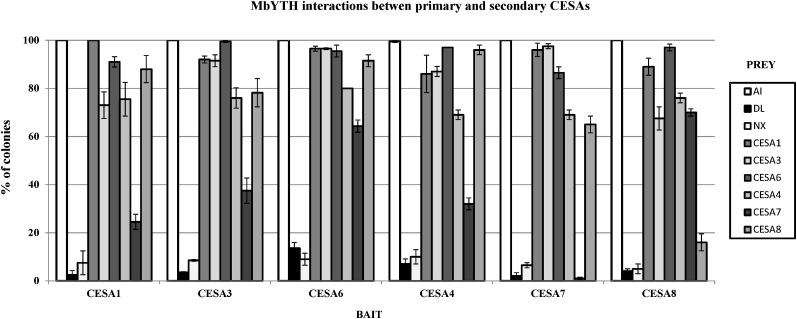
All possible combinations of one-to-one interactions between the primary CESAs (CESA1, CESA3, CESA6) and secondary CESAs (CESA4, CESA7, CESA8) were assessed using the split-ubiquitin MbYTH system (Dualsystems Biotech). Upon testing the interactions between the three primary CESA isoforms, the results show that all the primary CESAs were able to form both homodimers and heterodimers with all the other primary CESA isoforms (Fig. 1), confirming previous reports using bimolecular fluorescence (BiFC) analysis (Desprez et al., 2007). These protein interactions were carried out with each of the primary CESAs as bait and as prey, and both sets of experiments showed the same results (Fig. 1). The lack of growth in the negative controls indicated that the interactions were specific, as an unrelated protein expressed as prey and an empty prey vector (pADSL-Nx) were not able to activate the system.
Figure 1.
Interactions between the primary and secondary CESAs visualized by yeast growth. The y axes represent the percentage of colonies that show visible growth after 5 d at 30°C on selective medium. Yeast expressing CESA1, CESA3, CESA6, CESA4, CESA7, and CESA8 as bait with N-terminal fusions of Nub and Cub to a CESA and with the ALG5 protein fused to NubI as positive control (AI) and NubG as negative control (DL) and an empty prey vector as another negative control (Nx) and different CESA proteins fused to NubG as prey are shown. sd is indicated by error bars.
In a second step, the interactions were determined between three members of the primary CESAs (CESA1, CESA3, CESA6) and the secondary CESAs (CESA4, CESA7, CESA8) using the same MbYTH system. Although with different interaction strength, the six primary and secondary CESAs all had the ability to form heterodimers in all possible combinations (Fig. 1).
Primary and Secondary CESAs Can Be Part of the Same Complex in Planta
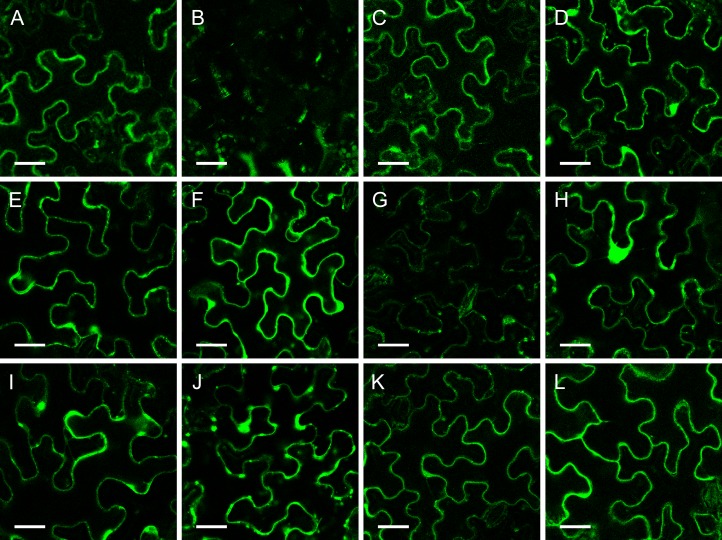
The BiFC technique offers the possibility of analyzing protein interactions in living plant cells (Walter et al., 2004). To analyze the interaction between the three primary CESAs and the secondary CESAs in planta, the BiFC assays were used, and the results are shown in Figure 2. It was observed that yellow fluorescent protein (YFP) fluorescence was reconstituted for all of the combinations, indicating that all isoforms from the primary CESAs (CESA1, CESA3, CESA6) can interact with those of the secondary CESAs (CESA4, CESA7, CESA8). The intensity of the YFP signals was not the same for all combinations. Upon interaction of CESA3 and CESA7, a weaker signal was observed, which may indicate that dimerization is less stable. All the pairwise CESA combinations were carried out with each of the CESAs fused with the N and C terminus of the YFP, and both sets of experiments showed the same results.
Figure 2.
BiFC analysis of the one-to-one interactions between the different primary and secondary CESA proteins. The proteins were transiently expressed in tobacco leaf epidermal cells. A, Positive control YN-PIP/YC-PIP. B, Negative control YN-PIP/YC-CESA7. C, YFP/N-CESA1/YFP/C-CESA4. D, YFP/N-CESA1/YFP/C-CESA7. E, YFP/N-CESA1/YFP/C-CESA8. F, YFP/N-CESA3/YFP/C-CESA4. G, YFP/N-CESA3/YFP/C-CESA7. H, YFP/N-CESA3/YFP/C-CESA8. I, YFP/N-CESA6/YFP/C-CESA4. J, YFP/N-CESA6/YFP/C-CESA7. K, YFP/N-CESA6/YFP/C-CESA8. L, YFP/N-CESA8/YFP/C-CESA6. Bars = 100 μm.
CESA7 Can Partially Rescue the Defects in the cesa3 Mutant je5
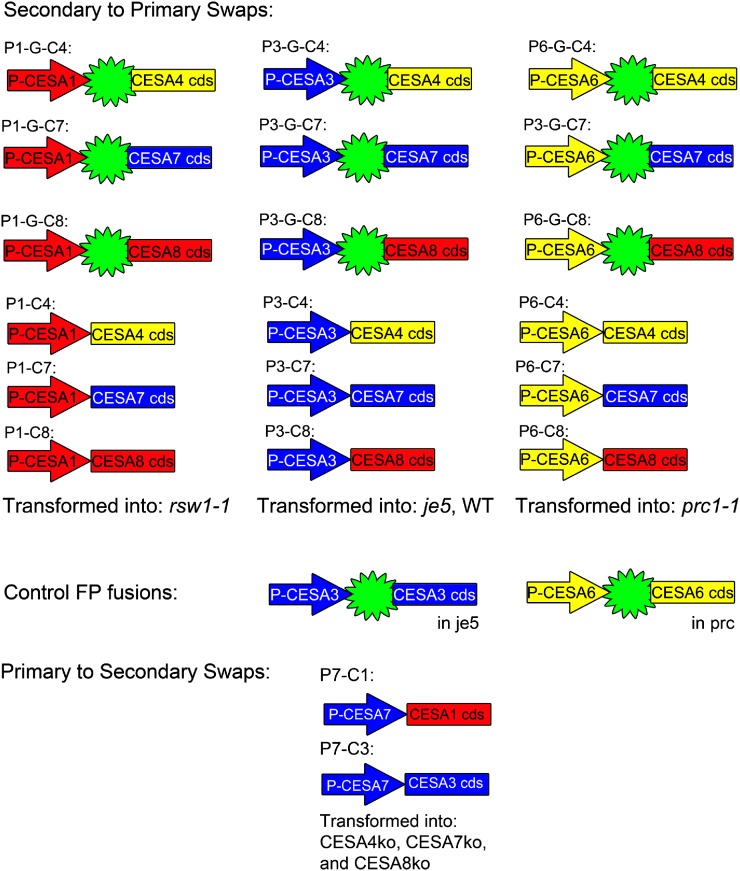
To determine whether CESAs from the secondary complex could enter and function in the primary complex, a series of promoter-swap constructs were generated. Combinations of each of the primary promoters were placed upstream of each of the secondary CESA coding sequences, both with and without an N-terminal GFP. We named these constructs Px-Cy based on the promoter and coding sequence used. A construct containing the CESA1 promoter is P1, while one containing the coding sequence of CESA4 is C4, giving the combination of the two the name P1C4. If GFP is N-terminally fused, we place the letter “G” before the coding sequence. The fusions with GFP (P1-G-C4, P1-G-C7, P1-G-C8, P3-G-C4, P3-G-C7, P3-G-C8, P6-G-C4, P6-G-C7, and P6-G-C8) and without GFP (P1C4, P1C7, P1C8, P3C4, P3C7, P3C8, P6C4, P6C7, and P6C8) were transformed into the mutant lines corresponding to the promoter used. The CESA1 promoter constructs were transformed into the temperature-sensitive (ts) cesa1 mutant rsw1-1 (line P1-G-CY; c1ts), CESA3 promoter constructs were transformed into the weak (w) cesa3 mutant je5 (P3-G-CY; c3w), and CESA6 promoter constructs were transformed into the cesa6 null line prc (P6-G-CY; c6ko). In addition, the weak cesa3 mutant je5 was transformed with the P3-G-C3 construct (Fig. 3).
Figure 3.
Promoter-swap constructs generated and transformed into plants. Arrows indicate promoter regions, and the presence of the star-like symbol indicates that the coding sequence of GFP is N-terminally fused in frame to the coding sequence of one of the secondary CESAs, indicated as a labeled rectangular box. Primary and secondary promoter and coding sequences are colored based on grouping of their sequence similarity at the C terminus. [See online article for color version of this figure.]
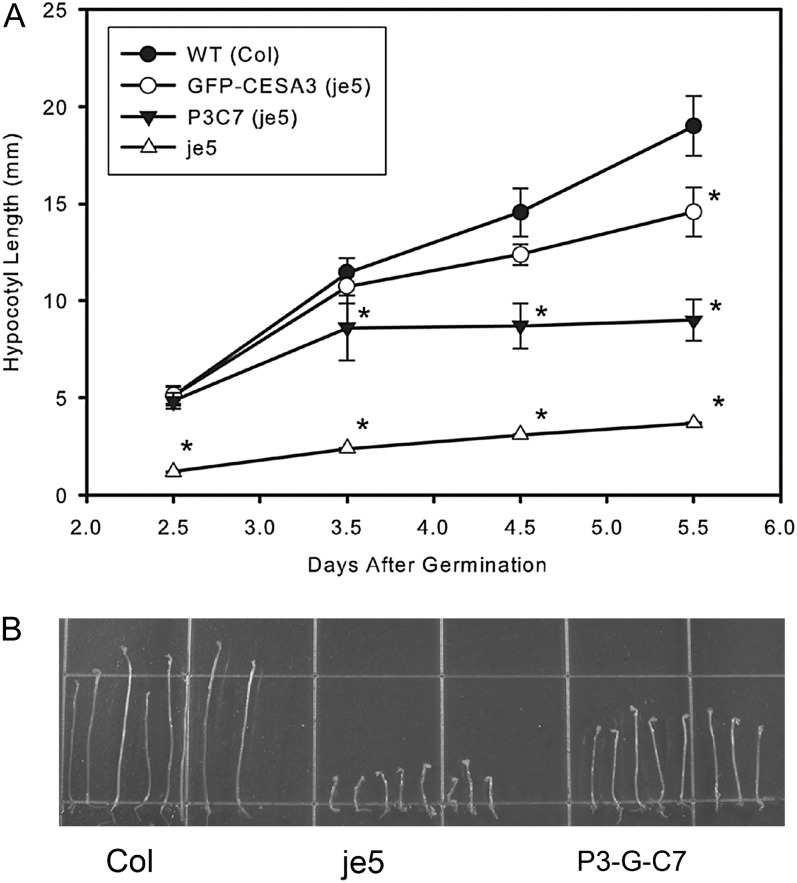
A partial rescue in the P3-G-C7 (c3w) line was observed (Fig. 4B). Etiolated seedlings of P3-G-C7 in je5 were not significantly different in hypocotyl length from Columbia plants or from P3-G-C3 (c3w) plants up to 2.5 d of growth. After 2.5 d, however, P3-G-C7 (c3w) does not elongate as rapidly as Columbia or P3-G-C3 (c3w; Fig. 4A). The CESA7 rescue of the cesa3 primary cell wall mutants without GFP was also incomplete (Supplemental Fig. S1). No rescue was apparent for either P6-G-C7 (c6ko) or P1-G-C7 (c1ts). The CESA4 and CESA8 constructs did not rescue any of the primary cell wall mutants, either with (data not shown) or without the N-terminal GFP (Supplemental Fig. S1).
Figure 4.
GFP-CESA7 is able to partially rescue the cesa3 mutant je5. A, A growth curve of hypocotyl elongation after various periods of etiolation revealed that early in growth the rescue of GFP-CESA7 is more complete, with elongation slowing after 3.5 d. Error bars represent sd, and asterisks indicate significant differences from the wild-type control (WT) at P < 0.001. B, At 5.5 d after germination in dark-grown conditions, the GFP-CESA7-containing line P3C7 in the je5 background is able to partially rescue the je5 phenotype of reduced hypocotyl elongation.
Reverse transcription (RT)-PCR analysis of GFP transcript revealed that expression of the CESA7 gene in the P3-G-C7 (c3w) mutant was similar to expression of the CESA3 gene in the rescue c3w mutant (P3-G-C3), as shown in Supplemental Figure S2.
Mixed Rosette Complexes Behave Differently from Primary Rosettes
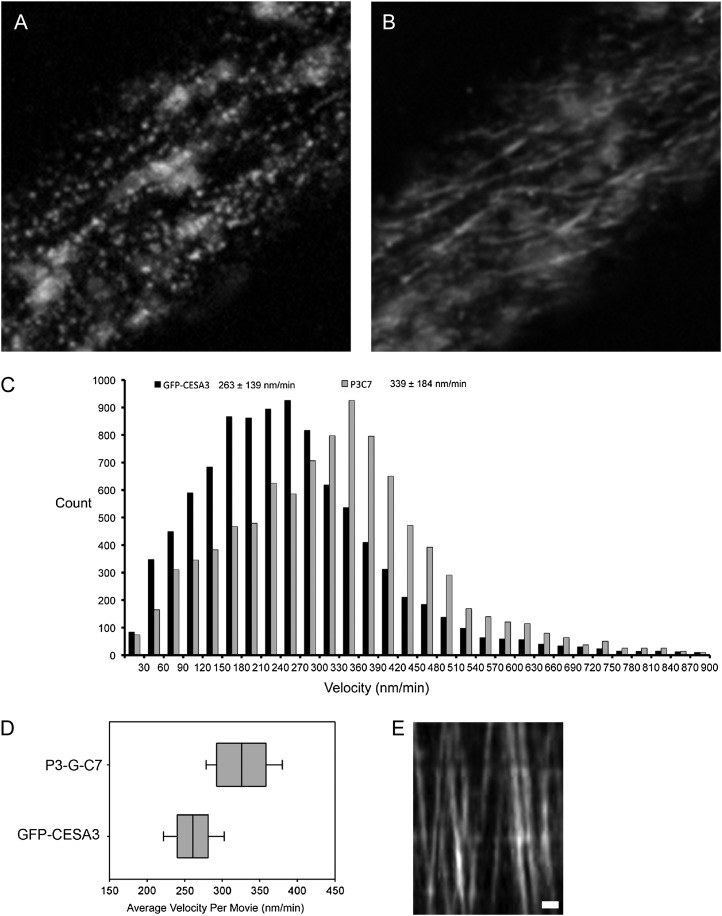
Spinning-disk confocal microscopy analysis in 2.5-d-old P3-G-C7 (c3w) and P3-G-C3 (c3w) etiolated seedlings revealed GFP signal in Golgi bodies and in membrane particles (Fig. 5, A and B; Supplemental Movies S1 and S2). The velocity distributions of both P3-G-C7 (c3w) and P3-G-C3 (c3w) were calculated by tracking individual particles in time-lapse movies. This revealed that membrane particles were approximately half as abundant in P3-G-C7 (c3w) compared with P3-G-C3 (c3w). Interestingly, complexes in the P3-G-C7 (c3w) line migrated about 30% faster than P3-G-C3 (c3w) complexes, a difference that is significant at P < 0.001 in a two-tailed t test (Fig. 5C). This phenomenon was consistently observed across biological replicates (26 GFP-CESA3 control and 15 P3-G-C7 [c3w] plants) acquired across 7 d, tracking around 40,000 GFP-CESA3- and 11,000 GFP-CESA7-labeled complexes (Fig. 5, D and E; Table I). The number of plasma membrane-localized particles decreased for both P3-G-C3 (c3w) and P3-G-C7 (c3w) lines after 3.5 d of etiolation, but the decrease in particle number was far more pronounced in P3-G-C7 (c3w), making it difficult to track enough particles for an adequate characterization of particle velocity in P3-G-C7 (c3w) after 2.5 d of growth. Using the total distance traveled by all CESA complexes observed in cells of the P3-G-C7 (c3w) and GFP-CESA3 lines, we estimated the relative cellulose produced in those lines over the course of the movies. Tracked complexes in GFP-CESA3 traveled an average of 572,252 pixels (77.3 mm) per cell, compared with 421,133 pixels (56.9 mm) in P3-G-C7 (c3w). This estimated the cellulose content of P3-G-C7 (c3w) at around 26% lower than the content in the GFP-CESA3 control. Chemical determination of the cellulose content showed similar results of lower cellulose content in the P3-G-C7 mutant relative to the GFP-CESA3 control (data not shown).
Figure 5.
GFP-CESA7 incorporates into CESA complexes in the je5 background. A, GFP-CESA7-containing puncta are visible at the plasma membrane focal plane and are arranged in linear tracks. Slightly out of focus Golgi bodies containing GFP-CESA7 near the membrane are also visible as large, circular areas of fluorescence. B, A time projection of a 3-min movie shows the motion of individual CESAs along tracks in the membrane. C, The distribution of particle velocity indicates that GFP-CESA7-containing complexes have a faster average velocity than those observed in GFP-CESA3-containing complexes. D, The average velocity in the 34 GFP-CESA3 and 15 P3-G-C7 are represented as box plots to show day-to-day variability. The whiskers show 1 sd from the mean, while the lines of the box indicate the first quartile, the median, and the third quartile. E, Kymograph of GFP-CESA7 particle movement in a track. Bar = 1 µm.
Table I. CESA complexes containing GFP-CESA7 are less abundant than complexes containing GFP-CESA3 in the je5 background.
Movies were selected in which the membrane of a single cell spans the field of view. Three-minute movies were taken with frames captured at 2-s time intervals. Particles tracked for longer than 30 s were counted. The difference between P3-G-C7 and GFP-CESA3 is significant at P < 0.003.
| Line | No. of Movies | Particles per Cell ± sd |
|---|---|---|
| GFP-CESA3 | 34 | 2,255 ± 1,145 |
| P3-G-C7 | 15 | 1,294 ± 629 |
In P1-G-C4 (c1ts) and P1-G-C8 (c1ts) plants, confocal microscopy revealed fluorescence in Golgi bodies, but no membrane complexes were detected (Supplemental Movie S3). Additionally, small fluorescent bodies were faintly visible in focal planes at or near the plasma membrane that did not behave like linearly moving complexes and whose behavior resembled previously reported subpopulations of SMaCCs (Supplemental Movie S4). In P1-G-C7 (c1ts), the GFP-CESA7 signal in SMaCCs was more apparent when plants were grown at the restrictive temperature of 30°C (Supplemental Movie S4). To determine whether the failure of GFP-CESA7 to reach membrane complexes was due to the compromised CESA6 and CESA1 proteins in these mutant lines, or to competition from the wild-type CESA3, the P3-G-C7 construct was transformed into the wild type, generating line P3-G-C7 (WT). These lines did not have any noticeable phenotype (Supplemental Fig. S1), indicating that the incompleteness of the rescue in P3-G-C7 (c3w) was most likely not due to a dominant-negative effect of CESA7 expression. P3-G-C7 (WT) plants had strong GFP-CESA7 fluorescence in Golgi bodies but no signal from membrane complexes (Supplemental Movie S5). The same fluorescence patterns were observed when GFP-CESA7 was transformed into either prc or rsw1-1 and fluorescence was strongly visible in Golgi bodies but not visible in membrane complexes (data not shown). These lines retained their phenotypes: prc was radially swollen and dwarfed, as was rsw1-1 when grown at the restrictive temperature. This is consistent with the hypothesis that GFP-CESA7 is excluded from membrane complexes in the presence of a wild-type copy of CESA3, as both prc and rsw1-1 retain wild-type copies of CESA3. One cannot exclude the possibility that the GFP-CESA7-containing rosettes are somehow blocked in transport to the plasma membrane.
Primary CESA1 Substitutes CESA8 in Secondary Walls
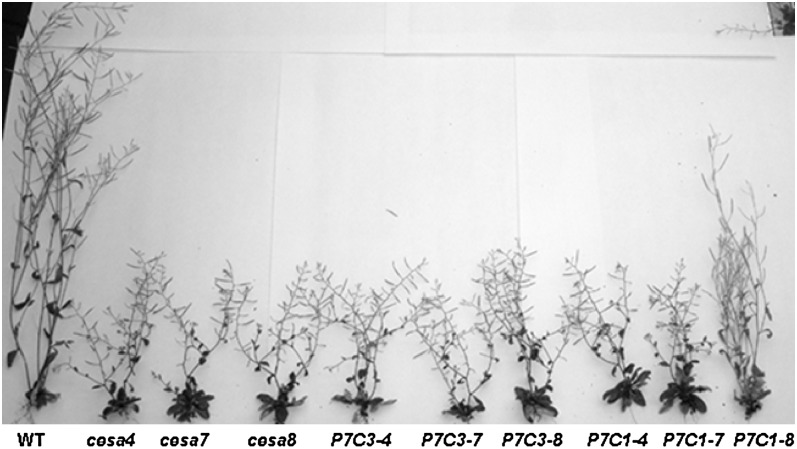
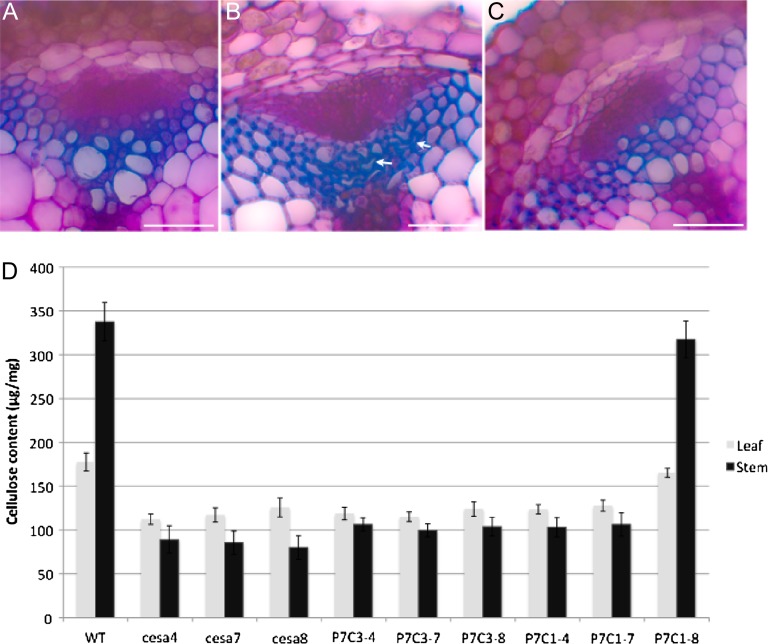
The expression profile comparison between primary CESAs and secondary CESAs indicates that secondary CESAs are more stringently controlled; therefore, the promoter of CESA7 was chosen to be used in the promoter-swap constructs. The null mutants of CESA4, CESA7, and CESA8 (cesa4ko, cesa7ko, cesa8ko) were identified by PCR identification of the T-DNA flanking regions (for primers, see Supplemental Table S1). All the secondary promoter-swap constructs (P7C1, P7C3) were transformed into cesa4ko, cesa7ko, and cesa8ko. Among all the possible combinations, only P7C1 partially complemented the cesa8ko phenotype. The leaf morphologies of cesa4ko, cesa7ko, and cesa8ko were indistinguishable from each other, all displaying dark green and reduced leaf size (Supplemental Fig. S3). The leaf of P7C1 (cesa8ko) was reverted to almost its wild-type size. However, the margin of the leaf was not as even as that of the wild type (data not shown). The adult homozygous plants of cesa4ko, cesa7ko, and cesa8ko were dwarfed, mainly due to shorter internodes (Fig. 6). In addition, cesa4ko, cesa7ko, and cesa8ko were almost completely sterile. P7C1 (cesa8ko) partially recovered the elongation defect in internodes, and these recoveries were more obvious in the main stem. In addition, P7C1 plants were fully fertile. A deficiency in secondary cell wall cellulose deposition leads to collapsed xylem cells, as shown in irx1, irx3, and irx5 plants (Taylor et al., 2000). Examination of the stem sections from cesa8ko showed its collapsed xylem phenotype. The xylem cells in P7C1 (cesa8ko) showed a similar phenotype to the wild type, indicating that P7C1 complemented the collapsed xylem phenotype in cesa8ko (Fig. 7, A–C). In both stems and leaves, the cellulose content in cesa4ko, cesa7ko, and cesa8ko was reduced, confirming the results of Taylor et al. (2000). Lesions in IRX1, IRX3, or IRX5 plants resulted in a decrease in cellulose of more than 70% in stems (Taylor et al., 2000). Correlating with the morphological recovery, the cellulose content of P7C1 (cesa8ko) was increased in both stems and leaves (Fig. 7B), indicating that P7C1 functionally incorporated into the secondary CESA complexes.
Figure 6.
Whole-plant morphology of secondary cesa mutants and various transgenic lines. Whole-plant morphology of various transgenic lines in secondary cesa mutants is shown. From left to right are the wild type (WT), cesa4ko, cesa7ko, cesa8ko, P7C3 in cesa4ko (P7C3-4), P7C3 in cesa7ko (P7C3-7), P7C3 in cesa8ko (P7C3-8), P7C1 in cesa4ko (P7C1-4), P7C1 in cesa7ko (P7C1-7), and P7C1 in cesa8ko (P7C1-8).
Figure 7.
P7C1 complements morphological and molecular defect in cesa8ko. A to C, Cross-sections of stem vascular bundles. Stem sections were stained with toluidine blue O. A, The wild type (WT). B, cesa8ko. C, P7C1 in cesa8ko (P7C1-8). Arrows indicate collapsed xylem vessels. Bars = 50 mm. D, Cellulose content in leaf or stem from the WT and various transgenic lines in secondary cesa mutants. Error bars represent se (n = 5).
DISCUSSION
Several studies have shown an absolute requirement of six unique CESA proteins, AtCESA1, AtCESA3, and AtCESA6-like, which form primary complexes (Desprez et al., 2002, 2007), and AtCESA4, AtCESA7, and AtCESA8, which form secondary complexes (Taylor, 2008; Timmers et al., 2009), for normal deposition of cellulose in the primary and secondary cell walls, respectively. Phylogenetic analysis reveals that these unique components, in the primary and secondary cell walls, represent distinct gene families that diverged early in the evolution of land plants (Holland et al., 2000; Samuga and Joshi, 2002; Tanaka et al., 2003; Burton et al., 2004; Djerbi et al., 2005; Nairn and Haselkorn, 2005; Ranik and Myburg, 2006; Suzuki et al., 2006; Kumar et al., 2009; Carroll and Specht, 2011).
Primary and Secondary CESAs Can Be Part of the Same Protein Complex
The yeast two-hybrid and bimolecular fluorescence results indicated that the CESAs can broadly interact with each other and that this interaction can be observed both in vitro and in planta. In contrast to the secondary cell wall, all primary wall CESAs are able to homodimerize, supporting previous BiFC data (Desprez et al., 2007). This result suggests that there is more flexibility in the positioning of the individual CESAs in the primary rosette complex than in the secondary complex, where only CESA4 is able to form homodimers (Timmers et al., 2009).
Previous reports have shown that primary and secondary CESAs are mainly expressed at different developmental stages in plants (Persson et al., 2005). Detailed gene expression analysis of single cells in Arabidopsis roots confirmed these results; however, they also revealed that primary and secondary CESAs can be coexpressed in specific cell types at certain time points (Birnbaum et al., 2003). The ability of primary and secondary CESAs to interact in all combinations indicates that these CESAs have the potential to be part of the same rosette complex, provided that they are colocalizing.
Although the existence of CESA mixed complexes has not been possible to resolve in vivo with the methods currently available, there are several reports supporting the idea that primary and secondary wall formation are interrelated. Overexpression of a mutant allele of the Arabidopsis CESA7 gene, named fra5, resulted in changes in cellulose synthesis during primary wall formation (reduced thickness of the cell wall and cell elongation) as well as causing a dominant-negative effect on cellulose synthesis during secondary wall formation (Zhong et al., 2003), as was also suggested in the case of the widely recognized secondary wall-specific AtCESA7 (MUR10), being required for normal primary cell wall carbohydrate composition in mature leaves, normal plant growth, hypocotyl strength, and fertility (Bosca et al., 2006). Another study shows that despite CESA9 having already been classified as a primary cell wall CESA (Desprez et al., 2007; Persson et al., 2007), a nonredundant role was shown in secondary cell wall thickening in the seed coat (Stork et al., 2010). The rice (Oryza sativa) brittle culm11 mutant has shown both altered primary (increased callose, pectic arabinan, and xylan) and secondary (brittleness of the culm, abnormal secondary structure, decreased wall thickness, and reduced cellulose content) wall composition (Zhang et al., 2009), further supporting the possibility of cross-talk and overlapping functions between the primary and secondary CESAs. In addition, the putative ability of primary and secondary CESAs to change roles through evolution appears more dynamic than was once believed. Recent results have shown that the secondary complexes produce secondary thickenings of cotton (Gossypium hirsutum) fibers, while the primary complexes have acquired this role in the analogous Arabidopsis structure of trichomes.
CESA7 Partially Rescues the Defects in the Primary cesa3 Mutant (je5)
Although the MbYTH and BiFC systems revealed that all primary and secondary CESAs can be part of the same protein complex, mutant complementation analysis revealed that the mixed complex where CESA3 has been replaced by CESA7 is functional in the primary cell wall and can partially rescue the cesa3 knockout mutant. At the same time, CESA7 could not rescue cesa1 and cesa6 mutants, indicating that the rescue occurs due to CESA7’s ability to substitute for CESA3. The exclusion of GFP-CESA7 from the plasma membrane of WT-CESA3 plants suggests that WT-CESA3 outcompetes CESA7 for inclusion in the cellulose synthase complex, indicating that there has been a small degree of shift in the interactions required to place a protein into the complex at the CESA3 position. Shifts in the affinity of CESA-CESA interactions over time could also explain the inability of CESA4 and CESA8 to rescue any of the primary cesa mutants. One interpretation of these results is that individual isoforms within the CESA complex can be thought of as having assigned “positions.” These positions could be consistent and distinct spatial locations in the structure of the complex, or they could instead arise more loosely from stronger interaction affinities between CESA classes during assembly of the complex. From these results, it appears the CESA3 and CESA7 can gain access to the same position in the complex.
Another possible explanation is that CESA7 incorporates into the complex as efficiently as CESA3, but the transfer of these complexes to the membrane is deficient. This could occur because the process responsible for transport or fusion involves a check on the integrity of the complex. CESA7-containing complexes are slightly deficient in this check, not so much that they cannot be transferred but enough that CESA3-containing complexes outcompete for access to the transfer process and saturate transfer to the membrane.
Since the je5 line is a weak allele of CESA3, we also cannot exclude the possibility that some mutant copies of CESA3 are able to help CESA7 enter the complex or otherwise facilitate complex formation.
The faster movement of GFP-CESA7 raises a number of questions. The general explanation would be that activity at the CESA3/CESA7 position is the rate-limiting process for complex mobility. A biochemical perspective may provide a better general explanation. The process of cellulose synthesis may be rate limited by steps in addition to catalysis; for example, the nascent cellulose chain may have to crystallize before synthesis can continue. If the substitution of CESA7 for CESA3 in the complex changes some property of the cellulose produced, this could produce an effect that could propagate through to the complex as a whole. Mutations in CESA1 and CESA3 were recently described that caused the complex to move faster and also altered cellulose crystallinity (Harris et al., 2012). It is also possible that the faster rate may reflect a compensatory mechanism to the lower abundance of complexes visible in the GFP-CESA7 line. Substantially higher rates of CESA compartment movement have been reported previously (Wightman et al., 2009).
CESA1 Partially Rescues the Defects of cesa8 Knockout
Lesions in the secondary CESAs, CESA4, CESA7, and CESA8, result in deficiency in the deposition of cellulose in secondary cell walls and in collapsed xylem cells. These mutants are also known as irregular xylem mutants, irx5 (CESA4), irx3 (CESA7), and irx1 (CESA8). Reverse genetic approaches have identified additional alleles of the irx mutants that were used in this study, including irx1-5, irx3-4, and irx5-4 (Brown et al., 2005). In addition to defects in xylem cells, these T-DNA mutants also display defects in overall morphology, such as dwarf stature, slow growth, dark green and reduced leaf size, short siliques, and reduced fertility (Brown et al., 2005). CESA1 was able to completely rescue the collapsed xylem cells in the irx1-5 mutant, which is consistent with the recovered cellulose content. In terms of overall morphology, CESA1 was also able to rescue defects in leaf color and fertility in the irx1-5 mutant. CESA1 was partially able to recover the overall stature of the plant in irx1-5. The nonredundant phenotype of secondary cesa mutants supports the hypothesis that CESA4, CESA7, and CESA8 constitute the secondary cellulose synthase complex. CESA1 apparently is able to take over the role of secondary CESA when it is expressed in the secondary cell walls.
A C-Terminal Sequence Separates Primary and Secondary CESAs into Three Groups
In order to further analyze the similarities and differences between the primary and secondary CESAs making up the complex, the sites of the C-terminal rsw5 mutation implicated in disrupting the incorporation of CESA3 into the primary cellulose synthase complex were compared (Wang et al., 2006; Carroll and Specht, 2011). The C terminus is a putatively cytosolic region of approximately 20 amino acids that follows the eighth transmembrane domain. The C-terminal region contains two strongly conserved Cys residues, and we speculate that the formation of disulfide bonds between the C terminus of one CESA and one of the other Cys-rich regions in another CESA might help mediate complex assembly. Chimeric CESA and CESA/CSLD proteins exchanging the N-terminal region (Wang et al., 2006) and catalytic domain (Park et al., 2011) have both retained the identity of the genetic position or localization of the C-terminal domain. This site was absolutely conserved in CESA families 3, 4, 6, and 7 but not in CESA families 1 and 8, with CESA families 3 and 7 showing more similarity to each other than with the other CESAs (Carroll and Specht, 2011). These observations are in agreement with the rescues of the primary and secondary knockout mutants, where CESA7 can partially rescue the defects in the cesa3 mutant and CESA1 can partially rescue the cesa8 mutant. These results, and the fact that most primary and secondary CESA proteins are not able to rescue CESAs, demonstrate that additional selectivity exists within the plant cell, either through directed assembly or competition for interacting partners. This also supports the possibility that CESAs have distinct functions in the rosette, either structurally and/or enzymatically related.
MATERIALS AND METHODS
Constructs for the Split-Ubiquitin MbYTH System
The full-length complementary DNAs (cDNAs) from Arabidopsis (Arabidopsis thaliana) were obtained from the RIKEN Bioresource Center (Seki et al., 1998, 2002): AtCESA1 (RAFL09-89-G08), AtCESA3 (RAFL05-19-M03), and AtCESA6 (RAFL05-02-P19) as well as AtCESA4 (RAFL15-30-K05), AtCESA7 (RAFL09-35-F05), and AtCESA8 (RAFL09-65-M12; Timmers et al., 2009). The cDNAs of the CESA genes were amplified by PCR using the Phusion DNA Polymerase (Finnzymes) with the primers listed in Supplemental Table S1. The resulting PCR products were digested and ligated into the pTFB1 vector (bait) and the pADSL-Nx vector (prey; Dualsystems Biotech). Bait and prey expression was regulated by the TEF1 and ADH1 promoters, respectively. The sequences of the inserts were confirmed by Sanger sequence analysis. The bait and prey proteins were fused N terminally to the Cub transcription factor reporter cassette of the vector pTFB1 and the NubG cassette of the vector pADSL-Nx, respectively.
The Split-Ubiquitin MbYTH Screen
The interactions between the CESA proteins were assayed using the split-ubiquitin MbYTH system (Johnsson and Varshavsky, 1994; Reinders et al., 2002) with the yeast strain NYM51 in the Split Ubiquitin System kit (Dualsystems Biotech). The assays were performed according to supplier instructions (DUAL Membrane Kit 1). This system (Stagljar et al., 1998; Stagljar and te Heesen, 2000) was used to detect interaction between the CESAs, in which each CESA was fused to the Cub-coding sequence of vector pTFB1 (bait), the Cub transcription factor, and the NubG-coding sequence of vector pADSL-Nx (prey; Fetchko and Stagljar, 2004). The yeast resident endoplasmic reticulum protein ALG5 fused to NubG was used as a negative control. Coexpression of the bait proteins with prey protein ALG5-NubG should not result in an interaction, and therefore not in activation of the system, as it is not involved in the pathways of interest. As a positive control, the ALG5 protein was fused to the wild-type ubiquitin domain. In contrast to the I13G mutant (NubG), the wild-type N-terminal ubiquitin domain (NubI) can readily interact with the C-terminal ubiquitin domain. Thus, the coexpression of the bait, containing CUB, with a prey fused to NubI will lead to an interaction and therefore may be used to test for bait expression and accessibility without the need for the fused proteins to interact. Interactions were quantified by 100 colonies spotted on synthetic dextrose medium (lacking Leu, Trp, His, and adenine) containing the appropriate concentration of 3-ammonium-triazole, as reported by Timmers et al. (2009), and grown at 30°C for 5 d, after which the number of spots grown was scored. The bait was also screened using the inhibitor (3-ammonium-triazole) in the selection medium to rule out autoactivation. Detection of β-galactosidase activity was performed with the filter-lift assay. All experiments have been performed in quadruplicate for independent biological replicates. Having two different auxotrophic markers for selection increased the reliability of the system in that the prey had to circumvent two different pathways to autoactivate the system as well as a colorimetric marker.
Constructs for Split-YFP
The full-length cDNAs of the CESA genes were generated through Phusion DNA Polymerase (Finnzymes) with suitable primers (Desprez et al., 2007; Timmers et al., 2009; Supplemental Table S1). Coding sequences of the CESAs were cloned into the Gateway-compatible destination vectors pBIFc-2 and pBIFc-3 plasmids regulated by the constitutive 35S promoter (Hu et al., 2002). The N-terminal and C-Terminal fragments of YFP were both fused to the N terminus of the coding sequences of the CESAs. As a positive control, the aquaporin PIP2-1 (Boursiac et al., 2005; Desprez et al., 2007) was used, as aquaporins are known to form homotetramers in the plasma membrane (Murata et al., 2000). As a negative control, the PIP2-1 chimera was coexpressed with the corresponding CESA constructs.
Split-YFP Screen
The BiFC screen was used to analyze in planta the interaction between the different CESA proteins. All possible combinations between the three primary and three secondary CESAs were analyzed with this method: YFP/N-CESA1/YFP/C-CESA4, YFP/N-CESA1/YFP/C-CESA7, YFP/N-CESA1/YFP/C-CESA8, YFP/N-CESA3/YFP/C-CESA4, YFP/N-CESA3/YFP/C-CESA7, YFP/N-CESA3/YFP/C-CESA8, YFP/N-CESA6/YFP/C-CESA4, YFP/N-CESA6/YFP/C-CESA7, and YFP/N-CESA6/YFP/C-CESA8. These interactions were also tested in the reverse combination, with both the C and N termini of the YFP. Leaves of 3-week-old tobacco (Nicotiana benthamiana) plants were infiltrated following transformation with Agrobacterium tumefaciens strain GV3101 pMP90 (Koncz and Schell, 1986) by transient coexpression of the desired protein pairs (Desprez et al., 2007). YFP fluorescence was detected 3 d after infiltration using the 514-nm laser line of a SP2 AOBS confocal laser scanning microscope (Leica) equipped with an argon laser. To check the YFP reconstitution, spectral analysis was performed with the 496-nm laser line. All experiments were carried out in triplicate.
Promoter-Swap Constructs
Using the same full-length cDNA genes previously indicated, the coding sequence for each CESA (CESA4, -7, and -8) was amplified using Phusion DNA Polymerase (Finnzymes) with primers suitable for the Gateway BP cloning reaction. These were inserted into pDONR207 through a BP reaction. CESA7 was amplified with Phusion DNA Polymerase, an adenine overhang added through 30 min of incubation with Taq polymerase at 72°C, and inserted into the PCR8 TOPO vector from Invitrogen. All three pDONR vectors were inserted into destination vectors carrying the 2-kb upstream promoter region of each of the primary CESA and the coding sequence from GFP immediately prior to the attR recombination sites (Desprez et al., 2007). The final vectors were sequenced over the entire length of their CESA coding region to confirm that no point mutations were present and to confirm that the GFP-CESA fusion was in frame. These constructs were made with N-terminal GFP fusions as well as untagged versions of the constructs. This resulted in two sets of nine constructs that were termed PX-G-CY for the fusion of the promoter for CESA X to the GFP-fused coding sequence of CESA Y (P1-G-C4, P1-G-C7, P1-G-C8, P3-G-C3, P3-G-C4, P3-G-C7, P3-G-C8, P6-G-C4, P6-G-C7, and P6-G-C8) and PX-CY for the untagged constructs (P1C4, P1C7, P1C8, P3C4, P3C7, P3C8, P6C4, P6C7, and P6C8) to designate the promoter (P) driving the coding sequence (C) in each construct. CESA1 promoter constructs were transformed into the temperature-sensitive cesa1 mutant rsw1-1 [line P1-G-CY (c1ts)], CESA3 promoter constructs were transformed into the weak cesa3 mutant je5 [P3-G-CY (c3w)], and CESA6 promoter constructs were transformed into the CESA6 null line prc [P6-G-CY (c6ko)] through the floral dip method (Clough and Bent, 1998). These constructs and lines are illustrated in Figure 1.
Fourteen transgenic P3-G-C7 and 27 P6-G-C7 lines were identified by genotyping. Ten lines from each type of transformant were investigated for the presence of fluorescence, with seven P3-G-C7 lines and 10 P6-G-C7 lines having visible fluorescence. Two lines of each were selected for further in-depth analysis. We identified 30 transgenic lines for each construct by genotyping; all were investigated for fluorescence, with only a few lines found to display weak fluorescence for each construct.
The CESA7 promoter was amplified using the primers indicated in Supplemental Table S1. The amplified CESA7 promoter was inserted into PCR8 TOPO (Invitrogen). Sequence-confirmed PCR8-pCESA7 was digested using SmaI/XbaI and inserted into pGW2 vector (Nakagawa et al., 2007) to replace the 35S promoter. The full-length cDNAs of CESA1 and CESA3 were PCR amplified and cloned into pDONR-zeo using the primers listed in Supplemental Table S1. CESA1 and CESA3 were then inserted to destination vectors containing the 2-kb CESA7 promoter using LR clonase II (Invitrogen).
Isolation of a T-DNA Insertion Line
The identification of secondary cesa knockout lines from the SIGNAL collection (http://signal.salk.edu/cgi-bin/tdnaexpress) was based on a combination of database searches and PCR amplification of T-DNA flanking regions. For T-DNA lines identified from the SIGNAL collection, seeds were obtained from the Arabidopsis Biological Resource Center. PCR was carried out to identify single plants for the T-DNA insertion. Primers used for T-DNA genotyping of CESA alleles are listed in Supplemental Table S1.
The secondary cesa Arabidopsis homozygous mutants used in this study included irx1-5, irx3-4, and irx5-4 (Brown et al., 2005).
Plant Growth Conditions
Arabidopsis ecotype Columbia seeds and various mutant lines were sterilized and germinated on Murashige and Skoog (MS) plates (one-half-strength MS salts, 0.8% agar, and 0.05% MES, pH 5.7). Seedlings were then grown vertically on the agar at 22°C under continuous light for 5 d before being transferred to pots in a greenhouse at 22°C under a 16-h-light/8-h-dark cycle.
RT-PCR Analysis
Total RNA was isolated from Arabidopsis seedlings using the RNAeasy Mini Kit (Qiagen). RT and PCR amplification were performed. For GFP amplification, 30 cycles of PCR amplification (94°C for 30 s, 54°C for 30 s, and 72°C for 1 min) were performed using the primers shown below. As PCR amplification and loading controls, the same template cDNA was amplified using primers for the constitutive ACTIN2 (ACT2) gene. The primers used for RT-PCR analysis were as follows: 5′GFP, 5′-ATGGTGAGCAAGGGCGAGGA-3′; 3′GFP, 5′-TACAGCTCGTCCATGCCGTGA-3′; 5′ACT2, 5′-ATGGCTGAGGCTGATGATAT-3′; 3′ACT2, 5′-TTAGAAACATTTTCTGTGAAC-3′.
Cellulose Measurement
Rosette leaves or stems were harvested and ground in liquid nitrogen. After overnight extraction in 80% ethanol at 65°C in a water bath, tissues were exchanged with acetone. Dry cell wall materials were ball milled to a fine powder. Cellulose was measured as described by Updegraff (1969). Data were collected from five technical replicates for each tissue sample. Experiments were repeated twice.
Xylem Staining
Stems from Arabidopsis were hand cut by a razor blade and stained in 0.02% toluidine blue O as described previously (Persson et al., 2005). Stem sections were rinsed, mounted in water, and viewed with a compound microscope (Leitz DMRB; Leica). Around five individual plants were examined for each line.
Confocal Microscopy
For analyses of GFP-CESA proteins expressed in the promoter-swap lines, seeds were germinated on MS agar plates and grown vertically in darkness for 3 d at 22°C. Seedlings were mounted between two coverslips in water. Imaging was performed on a Yokogawa CSUX1 spinning-disk system featuring the DMI6000 Leica motorized microscope and a Leica 100×/1.4 numerical aperture oil objective. GFP was excited at 488 nm, and a band-pass filter (520/50 nm) was used for emission filtering. Image analysis was performed using Metamorph (Molecular Devices) and Imaris (Bitplane) software.
Movies were collected on 7 d, without a consistent pattern regarding which lines were imaged first. Movies were taken at ambient temperatures. On none of the 7 d did the average recorded GFP-CESA3 control velocity exceed the velocity of the P3C7 lines recorded on that day. The lower number of P3-G-C7 movies compared with GFP-CESA3 occurs because the weaker signal makes it more difficult to maintain the focal plane appropriately. An approximately equal number of acquisitions were attempted for each, with poor-focal-quality movies discarded during postprocessing.
Image analysis was performed with ImageJ (Magelhaes et al., 2004) and Imaris software. Movies were first contrast enhanced in ImageJ, and a walking average of four frames was taken using the kymograph plugin for ImageJ. These steps were performed to improve the accuracy of automated particle recognition performed in subsequent steps. These images were then opened in Imaris 6.2.1 and switched from Z-series to time series. The voxel size was set to 135 nm per voxel based on measurements from the scope, and the time interval was set to 5 s. The particle-recognition algorithm in Imaris was performed with a spot size of 250 nm. High-intensity signal was filtered to eliminate Golgi signal. Following this, the connected components program was run, which determines particle identity over several frames and converts a particle’s movement into tracks. All tracks present for less than 60 s (12 frames) were discarded. The displacement and duration of the remaining tracks were exported to a spreadsheet, and their average velocity, distribution of velocities, and any directional bias were calculated.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Rescues from secondary swap constructs without N-terminal GFP fusion.
Supplemental Figure S2. RT-PCR analysis of GFP expression in P3-G-C7 plants.
Supplemental Figure S3. Leaf morphology of secondary cesa mutants and various transgenic lines.
Supplemental Table S1. DNA primers used in the study.
Supplemental Movie S1. GFP-CESA3 particles observed at the plasma membrane of a P3-G-C3 (c3w) plant.
Supplemental Movie S2. GFP-CESA7 particles observed at the plasma membrane of a P3-G-C7 (c3w) plant.
Supplemental Movie S3. Incorporation of CESA4 and CESA8 protein into complexes is minimal.
Supplemental Movie S4. GFP-CESA7 is observed in Golgi bodies and SMaCCs of a P1-G-C7 (c1ts) plant imaged at the restrictive temperature of 30°C.
Supplemental Movie S5. GFP-CESA7 is observed in Golgi bodies of a P3-G-C7 (WT) plant.
Acknowledgments
We gratefully acknowledge the assistance of Thierry Desprez (Laboratoire de Biologie Cellulaire, Institute Jean-Pierre Bourgin, Institut National de la Recherche Agronomique) with the BiFC experiments and also the assistance of Kian Hématy (Laboratoire de Biologie Cellulaire, Institute Jean-Pierre Bourgin, Institut National de la Recherche Agronomique) with the conceptual design of the experiments.
Glossary
- CESA
cellulose synthase
- SMaCCs
small microtubule-associated compartments
- MbYTH
membrane-based yeast two-hybrid
- BiFC
bimolecular fluorescence
- YFP
yellow fluorescent protein
- RT
reverse transcription
- MS
Murashige and Skoog
- cDNA
complementary DNA
References
- Atanassov II, Pittman JK, Turner SR. (2009) Elucidating the mechanisms of assembly and subunit interaction of the cellulose synthase complex of Arabidopsis secondary cell walls. J Biol Chem 284: 3833–3841 [DOI] [PubMed] [Google Scholar]
- Betancur L, Singh B, Rapp RA, Wendel JF, Marks MD, Roberts AW, Haigler CH. (2010) Phylogenetically distinct cellulose synthase genes support secondary wall thickening in Arabidopsis shoot trichomes and cotton fiber. J. Integr. Plant Biol.52, 205–220 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Bosca S, Barton CJ, Taylor NG, Ryden P, Neumetzler L, Pauly M, Roberts K, Seifert GJ. (2006) Interactions between MUR10/CesA7-dependent secondary cellulose biosynthesis and primary cell wall structure. Plant Physiol 142: 1353–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursiac Y, Chen S, Luu DT, Sorieul M, van den Dries N, Maurel C. (2005) Early effects of salinity on water transport in Arabidopsis roots: molecular and cellular features of aquaporin expression. Plant Physiol 139: 790–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, Zeef LAH, Ellis J, Goodacre R, Turner SR. (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RM. (2004) Cellulose structure and biosynthesis: what is in store for the 21st century? J Polym Sci A Polym Chem 42: 487–495 [Google Scholar]
- Burton RA, Shirley NJ, King BJ, Harvey AJ, Fincher GB. (2004) The CesA gene family of barley. Quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiol 134: 224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll A, Specht C. (2011) Understanding plant cellulose synthases through a comprehensive investigation of the cellulose synthase family sequences. Front Plant Sci 2: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [Google Scholar]
- Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof YD, Schumacher K, Gonneau M, Höfte H, Vernhettes S. (2009) Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21: 1141–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S. (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T, Vernhettes S, Fagard M, Refrégier G, Desnos T, Aletti E, Py N, Pelletier S, Höfte H. (2002) Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol 128: 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djerbi S, Lindskog M, Arvestad L, Sterky F, Teeri TT. (2005) The genome sequence of black cottonwood (Populus trichocarpa) reveals 18 conserved cellulose synthase (CesA) genes. Planta 221: 739–746 [DOI] [PubMed] [Google Scholar]
- Doblin MS, Kurek I, Jacob-Wilk D, Delmer DP. (2002) Cellulose biosynthesis in plants: from genes to rosettes. Plant Cell Physiol 43: 1407–1420 [DOI] [PubMed] [Google Scholar]
- Ellis C, Karafyllidis I, Wasternack C, Turner JG. (2002) The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Höfte H. (2000) PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12: 2409–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetchko M, Stagljar I. (2004) Application of the split-ubiquitin membrane yeast two-hybrid system to investigate membrane protein interactions. Methods 32: 349–362 [DOI] [PubMed] [Google Scholar]
- Giddings TH, Jr, Brower DL, Staehelin LA. (1980) Visualization of particle complexes in the plasma membrane of Micrasterias denticulata associated with the formation of cellulose fibrils in primary and secondary cell walls. J Cell Biol 84: 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R, Lindeboom JJ, Paredez AR, Emons AM, Ehrhardt DW. (2009) Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol 11: 797–806 [DOI] [PubMed] [Google Scholar]
- Harris DM, Corbin K, Wang T, Gutierrez R, Bertolo AL, Petti C, Smilgies DM, Estevez JM, Bonetta D, Urbanowicz BR, et al. (2012) Cellulose microfibril crystallinity is reduced by mutating C-terminal transmembrane region residues CESA1A903V and CESA3T942I of cellulose synthase. Proc Natl Acad Sci USA 109: 4098–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland N, Holland D, Helentjaris T, Dhugga KS, Xoconostle-Cazares B, Delmer DP. (2000) A comparative analysis of the plant cellulose synthase (CesA) gene family. Plant Physiol 123: 1313–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK. (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Johnsson N, Varshavsky A. (1994) Ubiquitin-assisted dissection of protein transport across membranes. EMBO J 13: 2686–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Laosinchai W, Itoh T, Cui X, Linder CR, Brown RM., Jr (1999) Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. Plant Cell 11: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J. (1986) The promoter of gene 5 controls the tissue specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Kumar M, Thammannagowda S, Bulone V, Chiang V, Han K-H, Joshi CP, Mansfield SD, Mellerowicz E, Sundberg B, Teeri T, et al. (2009) An update on the nomenclature for the cellulose synthase genes in Populus. Trends Plant Sci 14: 248–254 [DOI] [PubMed] [Google Scholar]
- Li S, Lei L, Somerville CR, Gu Y. (2012) Cellulose synthase interactive protein 1 (CSI1) links microtubules and cellulose synthase complexes. Proc Natl Acad Sci USA 109: 185–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magelhaes PJ, Ram SJ, Abramoff MD. (2004) Image processing with ImageJ. Biophotonics International 11: 36–42 [Google Scholar]
- Mueller SC, Brown RM., Jr (1980) Evidence for an intramembrane component associated with a cellulose microfibril-synthesizing complex in higher plants. J Cell Biol 84: 315–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, Heymann JB, Engel A, Fujiyoshi Y. (2000) Structural determinants of water permeation through aquaporin-1. Nature 407: 599–605 [DOI] [PubMed] [Google Scholar]
- Mutwil M, Debolt S, Persson S. (2008) Cellulose synthesis: a complex complex. Curr Opin Plant Biol 11: 252–257 [DOI] [PubMed] [Google Scholar]
- Nairn CJ, Haselkorn T. (2005) Three loblolly pine CesA genes expressed in developing xylem are orthologous to secondary cell wall CesA genes of angiosperms. New Phytol 166: 907–915 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW. (2006) Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Park S, Szumlanski AL, Gu F, Guo F, Nielsen E. (2011) A role for CSLD3 during cell-wall synthesis in apical plasma membranes of tip-growing root-hair cells. Nat Cell Biol 13: 973–980 [DOI] [PubMed] [Google Scholar]
- Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR. (2007) Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci USA 104: 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Wei H, Milne J, Page GP, Somerville CR. (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA 102: 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranik M, Myburg AA. (2006) Six new cellulose synthase genes from Eucalyptus are associated with primary and secondary cell wall biosynthesis. Tree Physiol 26: 545–556 [DOI] [PubMed] [Google Scholar]
- Reinders A, Schulze W, Thaminy S, Stagljar I, Frommer WB, Ward JM. (2002) Intra- and intermolecular interactions in sucrose transporters at the plasma membrane detected by the split-ubiquitin system and functional assays. Structure 10: 763–772 [DOI] [PubMed] [Google Scholar]
- Samuga A, Joshi CP. (2002) A new cellulose synthase gene (PtrCesA2) from aspen xylem is orthologous to Arabidopsis AtCesA7 (irx3) gene associated with secondary cell wall synthesis. Gene 296: 37–44 [DOI] [PubMed] [Google Scholar]
- Scheible WR, Eshed R, Richmond T, Delmer D, Somerville C. (2001) Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proc Natl Acad Sci USA 98: 10079–10084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Carninci P, Nishiyama Y, Hayashizaki Y, Shinozaki K. (1998) High-efficiency cloning of Arabidopsis full-length cDNA by biotinylated CAP trapper. Plant J 15: 707–720 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y, et al. (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145 [DOI] [PubMed] [Google Scholar]
- Somerville C. (2006) Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol 22: 53–78 [DOI] [PubMed] [Google Scholar]
- Stagljar I, Korostensky C, Johnsson N, te Heesen S. (1998) A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc Natl Acad Sci USA 95: 5187–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagljar I, te Heesen S. (2000) Detecting interactions between membrane proteins in vivo using chimeras. Methods Enzymol 327: 190–198 [DOI] [PubMed] [Google Scholar]
- Stork J, Harris D, Griffiths J, Williams B, Beisson F, Li-Beisson Y, Mendu V, Haughn G, Debolt S. (2010) CELLULOSE SYNTHASE9 serves a nonredundant role in secondary cell wall synthesis in Arabidopsis epidermal testa cells. Plant Physiol 153: 580–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Li L, Sun Y-H, Chiang VL. (2006) The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiol 142: 1233–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Murata K, Yamazaki M, Onosato K, Miyao A, Hirochika H. (2003) Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiol 133: 73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG. (2008) Cellulose biosynthesis and deposition in higher plants. New Phytol 178: 239–252 [DOI] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR. (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100: 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Laurie S, Turner SR. (2000) Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12: 2529–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers J, Vernhettes S, Desprez T, Vincken JP, Visser RG, Trindade LM. (2009) Interactions between membrane-bound cellulose synthases involved in the synthesis of the secondary cell wall. FEBS Lett 583: 978–982 [DOI] [PubMed] [Google Scholar]
- Updegraff DM. (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32: 420–424 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang J, Howles PA, Cork AH, Birch RJ, Williamson RE. (2006) Chimeric proteins suggest that the catalytic and/or C-terminal domains give CesA1 and CesA3 access to their specific sites in the cellulose synthase of primary walls. Plant Physiol 142: 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Elliott JE, Williamson RE. 2008.. Features of the primary wall CESA complex in wild type and cellulose-deficient mutants of Arabidopsis thaliana.J. Exp. Bot. 59 (10): 2627–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman R, Marshall R, Turner SR. (2009) A cellulose synthase-containing compartment moves rapidly beneath sites of secondary wall synthesis. Plant Cell Physiol 50: 584–594 [DOI] [PubMed] [Google Scholar]
- Zhang B, Deng L, Qian Q, Xiong G, Zeng D, Li R, Guo L, Li J, Zhou Y. (2009) A missense mutation in the transmembrane domain of CESA4 affects protein abundance in the plasma membrane and results in abnormal cell wall biosynthesis in rice. Plant Mol Biol 71: 509–524 [DOI] [PubMed] [Google Scholar]
- Zhong R, Morrison WH, III, Freshour GD, Hahn MG, Ye ZH. (2003) Expression of a mutant form of cellulose synthase AtCesA7 causes dominant negative effect on cellulose biosynthesis. Plant Physiol 132: 786–795 [DOI] [PMC free article] [PubMed] [Google Scholar]