Abstract
Most of the world’s natural fiber comes from cotton (Gossypium spp.), which is an important crop worldwide. Characterizing genes that regulate cotton yield and fiber quality is expected to benefit the sustainable production of natural fiber. Although a huge number of expressed sequence tag sequences are now available in the public database, large-scale gene function analysis has been hampered by the low-efficiency process of generating transgenic cotton plants. Tobacco rattle virus (TRV) has recently been reported to trigger virus-induced gene silencing (VIGS) in cotton leaves. Here, we extended the utility of this method by showing that TRV-VIGS can operate in reproductive organs as well. We used this method to investigate the function of KATANIN and WRINKLED1 in cotton plant development. Cotton plants with suppressed KATANIN expression produced shorter fibers and elevated weight ratio of seed oil to endosperm. By contrast, silencing of WRINKLED1 expression resulted in increased fiber length but reduced oil seed content, suggesting the possibility to increase fiber length by repartitioning carbon flow. Our results provide evidence that the TRV-VIGS system can be used for rapid functional analysis of genes involved in cotton fiber development.
Cotton (Gossypium spp.) is the most important fiber-producing plant in the world and also a significant oilseed crop. This crop is grown in more than 80 countries with a worldwide production of 123 million bales (480 pounds per bale) during the 2011/2012 growing season (United States Department of Agriculture, 2012).
As the most important agronomic traits of cotton are fiber quality and yield, it is important to improve our understanding of genes that directly or indirectly impact such traits. To this end, a public effort was initiated in 2007 to determine the complete cotton genomic sequence. While this effort is underway, there is an ever-expanding set of cotton EST sequences (about 400,000 now) being deposited in the public database. Notwithstanding the availability of such a huge amount of cotton gene sequences so far, only a relatively small number of genes have been shown to directly affect cotton fiber development. Most of these genes are involved either in cytoskeletal dynamics or in carbohydrate biosynthesis. In the case of cytoskeletal genes, the cotton actin gene has been shown to be important for fiber elongation but not fiber initiation (Li et al., 2005). Overexpression of a fiber-specific profilin (GhPFN2), an actin-binding protein, blocked cell elongation prematurely (Wang et al., 2010), whereas down-regulation of the actin-depolymerizing factor gene increased fiber length and fiber strength (Wang et al., 2009). Since cellulose is the major constituent of cotton fiber, it is not surprising that several carbohydrate biosynthetic genes have been shown to modulate fiber development. Transgenic overexpression of a SUCROSE SYNTHASE gene, a SUCROSE PHOSPHATE SYNTHASE gene, and cellulose synthesis genes improved cotton fiber length and strength (Li et al., 2004; Haigler et al., 2007; Jiang et al., 2012). Similarly, higher xyloglucan endotransglycosylase/hydrolase (XTH) activity can promote fiber cell elongation, and transgenic cotton overexpressing the xth gene has been shown to increase mature fiber length (Lee et al., 2010).
The major impediment to analyzing cotton gene function on a large scale is the laborious and time-consuming process of generating transgenic cotton. Moreover, many cotton cultivars and Gossypium species that contain important genes for cotton improvement are recalcitrant to genetic transformation. Therefore, there is an urgent need to develop a rapid method for functional analysis of cotton genes on a genomic scale. Virus-induced gene silencing (VIGS) offers such a possibility because it allows the investigation of gene functions without plant transformation (Ruiz et al., 1998; Burch-Smith et al., 2004). A partial fragment of a candidate gene is inserted into a virus vector to generate a recombinant virus. Inoculation with the recombinant virus leads to the production of virus-related small interfering RNAs (Baulcombe, 2004) in infected plants. The small interfering RNAs generated can mediate the degradation of related endogenous gene transcripts resulting in the silencing of candidate gene expression (Brigneti et al., 2004; Burch-Smith et al., 2004). The silencing effect on endogenous gene expression can usually be assayed 1 to 2 weeks after virus inoculation. VIGS has become one of the most widely used and indeed important reverse genetics tools, especially for nonmodel plants, such as tomato (Solanum lycopersicum; Liu et al., 2002), barley (Hordeum vulgare; Holzberg et al., 2002), soybean (Glycine max; Zhang and Ghabrial, 2006), Medicago truncatula (Grønlund et al., 2008), poppy (Papaver somniferum; Hileman et al., 2005), and the shrub tree jatropha (Jatropha curcas; Ye et al., 2009a). Using different infection and treatment protocols, VIGS has been shown to be effective in leaves (Liu et al., 2002), roots (Valentine et al., 2004), flowers (Liu et al., 2004), and even fruits (Fu et al., 2005).
Recently, two different VIGS systems have been shown to be effective in cotton. Using the geminivirus Cotton leaf crumple virus (CLCrV) as a vector, Tuttle et al. (2008) silenced two visible marker genes encoding magnesium chelatase subunit I and phytoene desaturase with an efficiency of 70%. Gao et al. (2011a, 2011b), on the other hand, used the Tobacco rattle virus (TRV) as a vector to silence another marker gene, chloroplastos alterados1, with an efficiency of 100%. The TRV-VIGS system appears to be better than the CLCrV-VIGS system because the former has a higher silencing efficiency and a more homogenous silencing phenotype with a milder virus symptom. Although both VIGS systems have been shown to work in cotton leaves, it was not known whether these methods can be used to assess gene function in reproductive tissues and particularly in developing cotton fibers. Nevertheless, using a GFP-expressing CLCrV, Tuttle et al. (2008) observed occasional GFP fluorescence in outer boll walls, the central column of the ovary, and ovule integuments.
Here, we used two anthocyanin biosynthetic genes, ANTHOCYANIDIN SYNTHASE (ANS) and ANTHOCYANIDIN REDUCTASE (ANR), as reporters to optimize the TRV-mediated VIGS system in cotton plants, and we showed that it can be used to silence gene expression in reproductive tissues and developing fibers. We used this method to characterize the functions of two genes, KATANIN (KTN) and WRINKLED1 (WRI1), in cotton fiber development.
RESULTS
Synthetic TRV-Induced Silencing of Proanthocyanidin Biosynthetic Genes ANS and ANR
We first chemically synthesized two TRV genomes, TRV1 and TRV2 (Ye et al., 2009b, 2010) to construct a synthetic TRV (sTRV) VIGS system. Our method for agroinfection of cotton plants was different from that of Gao et al. (2011a, 2011b), who used a syringe to infiltrate Agrobacterium tumefaciens culture into two fully expanded cotyledons. By contrast, we used cotton plants with two to four true leaves, vacuum infiltrated the leaves with A. tumefaciens culture, and subsequently drenched the root system using the same culture.
We used two proanthocyanidin (PA) biosynthetic genes as reporter genes to test whether the sTRV-induced silencing was effective throughout the whole cotton plant, especially in cotton fibers. PAs are a major class of flavonoids, one of the largest groups of plant secondary metabolites. Most genetic studies on the PA biosynthesis pathway were performed in Arabidopsis (Arabidopsis thaliana) and M. truncatula (Xie et al., 2003; Pang et al., 2007). Two enzymes, ANS and ANR, function at branches between anthocyanin and PA biosynthesis. ANS converts the substrate flavan-3,4-diol (leucoanthocyanidin) to anthocyanidin, which serves as a substrate for ANR to produce a major PA unit, 2,3-cis-flavan-3-ol (epicatechin), in Arabidopsis and M. truncatula (Xie et al., 2003; Pang et al., 2007). As far as we know, cotton genes involved in the PA biosynthesis pathway have not been well characterized. Here, we took advantage of the sTRV-VIGS system in cotton to confirm the function of two genes in the PA biosynthetic pathway.
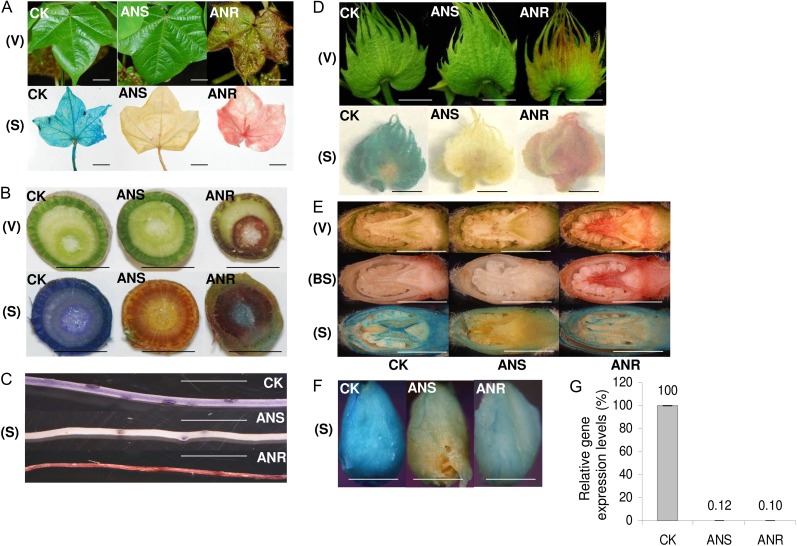
Two cotton gene sequences with homology to Arabidopsis genes encoding ANS and ANR were used for functional analysis. The partial fragments of putative cotton ANS and ANR were cloned into the psTRV vector to give psTRV:ANS and psTRV:ANR, respectively. Plants infiltrated with psTRV empty vector (psTRV1+psTRV2) were used as control plants in this study. All plants infiltrated with empty vector (henceforth referred to as vector control plants) grew normally and did not display any abnormal phenotype on vegetative and reproductive organs (data not shown). No phenotypic differences were observed between uninfected plants and vector control plants. This is one of the important advantages of sTRV-mediated gene silencing. At 7 to 10 d post inoculation, leaf veins and leaf margins around new systemic leaves began to appear brownish in all of the psTRV:ANR-treated plants. With time, the brownish phenotype became visible on leaves and leaf petioles (Fig. 1A) and stems (Fig. 1B). The brownish phenotype was also seen in more than 90% of floral buds in psTRV:ANR-treated plants (Fig. 1D). We surmised that this brownish phenotype was due to the anthocyanidin accumulation resulting from the blockage of ANR function. By contrast, all of the organs of psTRV:ANS plants (Fig. 1, A–E) showed no obvious visible differences from vector control plants. Nevertheless, the difference between psTRV:ANS and control plants can be uncovered by the use of the aromatic aldehyde agent p-dimethylaminocinnamaldehyde (DMACA), which stains PA (Fig. 1, A–E). After reaction with catechins, the major flavan-3-ols associated with cotton PAs, DMACA turned deep blue. Upon DMACA staining, all examined cotton organs of vector control plants turned blue, indicating a high level of PA accumulation. By contrast, organs from psTRV:ANS plants appeared yellow, indicating low anthocyanidin and PA levels. Organs from psTRV:ANR plants, on the other hand, appeared red because of reduced PA and anthocyanidin accumulation. With this staining method, the silencing effects of ANS and ANR expression could be detected in floral buds and reproductive organs (Fig. 1, D and E) and, most importantly, cotton fibers (Fig. 1F).
Figure 1.
sTRV-induced silencing of anthocyanidin and PA biosynthetic genes ANS and ANR. A to F, Plants infiltrated with empty vector control (CK), sTRV:ANS, and sTRV:ANR showed different phenotypes in systemic leaf (A), stem (B), root (C), flower bud (D), sliced flower bud (E), and fiber (F). V, Visual image (top); S, DMACA staining (bottom); BS, before staining but after decoloration (middle of E). Bars = 10 mm except in F, where bar = 2 mm. G, Relative transcript levels of ANS and ANR in systemic leaves of plants infiltrated with sTRV:ANS and sTRV:ANR. Control value was set as 1. Error bars represent sd (n = 3 biological replicates).
We collected three leaf samples from three independent cotton plants with prominent phenotypes (based on the visible phenotype and DMACA staining result) and further analyzed transcript levels of relevant genes in systemic leaves. We found a more than 99% reduction of ANS and ANR transcript levels in silenced plants compared with vector control plants (Fig. 1G). Results from the psTRV:ANR and psTRV:ANS plants confirmed the putative functions of these two cotton genes, and most importantly, they showed that the VIGS-mediated silencing effect could be detected not only in vegetative organs, including roots, but also in reproductive organs.
Effects of Suppression of KTN Expression on Reproductive Organs and Fiber Growth
We next used the sTRV-VIGS system to explore the function of the KTN gene in cotton development. KTN has been well studied in animals and the model plant Arabidopsis but not yet in cotton. KTN is a microtubule-severing protein that generates internal breaks along microtubules (McNally and Vale, 1993). This protein plays an important role in the release of microtubules from their centrosomal attachment in animal cells (Ahmad et al., 1999). In Arabidopsis, KTN mutation causes a delay in the disappearance of the perinuclear microtubule array, resulting in aberrant organization of microtubules in elongating cells (Burk et al., 2007).
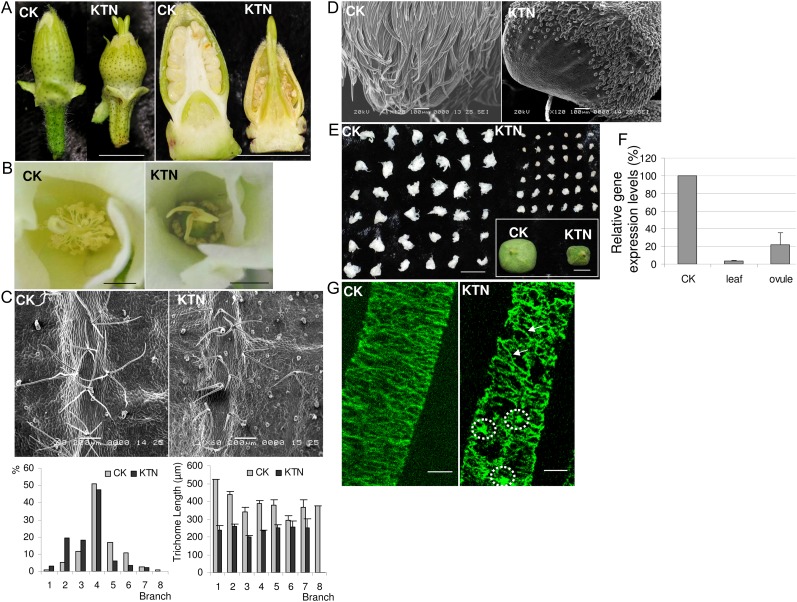
After infection with the psTRV:KTN recombinant virus, more than 95% of the treated plants appeared dwarf with shorter internodes, and the stems produced dark green and smaller leaf blades with shorter petioles (Supplemental Fig. S1). Similar to vegetative organs, the reproductive organs of KTN-silenced plants were also smaller in size. This phenotype of vegetative and reproductive organs was consistently seen in the same plant. Around 30% of psTRV:KTN plants showed a very severe phenotype in reproductive organs. The floral buds were first converted to pear shape from bullet shape. At a later stage, in addition to the reduced organ size, developing stigma divided and formed a tripod, which subsequently elongated and emerged from the clustered sepals in floral buds. Furthermore, there were obvious morphological and positional changes in ovaries, stamenal columns, and stamens (Fig. 2A). In KTN-silenced flowers, there was a wide separation between the anthers and the stigma, and the anthers were located at the bottom of the flower without any filaments (Fig. 2B). Trichomes on floral sepals of silenced plants were shorter and carried fewer branches compared with control trichomes (Fig. 2C). The severe defects in both male and female reproductive organs in psTRV:KTN flowers (Fig. 2, A and B) resulted in the abortion of cotton bolls at 2 to 3 d post anthesis (dpa), which corresponds to the end of the fiber initiation stage.
Figure 2.
psTRV-induced silencing of KTN. A and B, Plants infiltrated with empty vector control (CK) and psTRV:KTN showed different phenotypes in flower bud (A) and flower (B). Bars = 10 mm. C, Scanning electron micrograph of CK and psTRV:KTN sepals (top) and analysis of trichome branch numbers and trichome length in CK and psTRV:KTN sepals (bottom). Bars = 200 µm. D, Scanning electron micrograph of CK and psTRV:KTN ovule at 4 dpa. Bars = 100 µm. E, CK and psTRV:KTN ovules from cotton bolls (inset at right bottom) at 8 dpa. Bars = 10 mm. F, Relative KTN transcript levels in systemic leaves and ovules of plants infiltrated with CK and psTRV:KTN. CK value was set as 1. Error bars represent sd (n = 3 biological replicates). G, Immunolabeling of microtubule cytoskeleton of CK and psTRV:KTN fiber at 12 dpa. Arrows and circles delineate the disorganization of cortical microtubules. Bars = 10 µm.
Around 60% of psTRV:KTN plants showed a less severe phenotype in reproductive organs, and in these plants the cotton bolls could develop into the fiber elongation stage. Using scanning electron microscopy, we found much shorter fiber length and fewer fiber numbers in psTRV:KTN ovules at 4 dpa, indicating a reduced fiber initiation and growth rate (Fig. 2D). At 8 dpa, psTRV:KTN bolls were smaller in size and produced shorter fibers compared with vector control bolls (Fig. 2E). Compared with the vector control, KTN transcript levels in psTRV:KTN plants were reduced to 3.3% in systemic leaves and 22.1% in ovules at 8 dpa (Fig. 2F). In contrast to the parallel array of cortical microtubules (CMTs) seen in control fibers, immunolabeling of microtubules showed random CMT orientation (arrows) and microtubule aggregation points (circles) in psTRV:KTN fibers at 12 dpa (Fig. 2G). These defects in the organization of CMTs in the KTN-silenced cotton fibers indicated that KTN plays important roles in fiber elongation by affecting the microtubule structure. CMTs are essential for normal plant morphogenesis because they affect the axes of cell elongation. The abnormal arrangement of CMTs, especially the multiple sites of microtubule convergence in fibers, led to defects in fiber elongation and impacted the normal growth and development of cotton fibers.
To further understand the molecular basis of the developmental defects induced by KTN deficiency, we designed a custom array to investigate possible changes of gene expression in fiber-bearing ovules of KTN-silenced plants at 8 dpa. We found that in fiber-bearing ovules of KTN-silenced plants (one-way ANOVA, P < 0.05), the expression of 37 probe sets was down-regulated by 1.5-fold or more and that of 31 probe sets was up-regulated by 1.5-fold or more. The expression of some of these probes was confirmed with real-time PCR (Table I).
Table I. Microarray analysis and real-time PCR validation for major pathways and genes affected in psTRV:KTN fiber-bearing ovules (KTN) compared with those in psTRV control ovules (CK).
N.D., Not done.
| Pathway | The Arabidopsis Information Resource 10 | Annotation | Gene Chip |
Quantitative PCR | EST | |
|---|---|---|---|---|---|---|
| KTN/CK | P Value | KTN/CK | ||||
| Cytoskeleton | AT1G80350 | Katanin (KTN) | 0.28 | 0.014 | 0.22 ± 0.13 | TC193948 |
| 0.30 | 0.021 | TC179681 | ||||
| Auxin transporting and response | AT1G04240 | Auxin-responsive protein IAA3 | 0.26 | 0.042 | 0.19 ± 0.18 | TC214591 |
| AT1G70940 | PIN3, a regulator of auxin efflux involved in differential growth | 0.56 | 0.037 | 0.10 ± 0.02 | TC199285 | |
| AT4G34760 | Auxin-responsive protein | 0.06 | 0.016 | 0.03 ± 0.03 | TC192062 | |
| AT1G80490 | TOPLESS-related1 | 0.53 | 0.030 | N.D. | TC187522 | |
| Carbohydrate metabolism | AT2G36460 | Fru-bisP aldolase | 0.62 | 0.011 | N.D. | TC188344 |
| AT5G23530 | Plastidic hexokinase | 0.36 | 0.029 | 0.25 ± 0.14 | TC225147 | |
| AT1G47960 | Pectin esterase inhibitor | 3.60 | 0.018 | N.D. | TC192343 | |
| AT3G29360 | UDP-d-Glc dehydrogenase | 0.41 | 0.009 | 0.03 ± 0.003 | TC182299 | |
| Fatty acid biosynthesis and metabolism | AT5G13640 | Phospholipid:diacylglycerol acyltransferase1 | 1.50 | 0.045 | N.D. | TC224822 |
| AT3G05970 | Peroxisomal long-chain acyl-CoA synthetase isozymes | 1.50 | 0.021 | N.D. | TC188948 | |
| AT4G28570 | Long-chain fatty alcohol oxidase | 2.00 | 0.036 | N.D. | TC222705 | |
| AT1G19440 | 3-Ketoacyl-CoA synthase4 | 2.30 | 0.021 | N.D. | TC227779 | |
| RNA binding and mRNA splicing | AT2G21660.1 | Gly-rich RNA-binding protein | 0.46 | 0.020 | 0.39 ± 0.25 | TC191640 |
| AT2G21660.1 | Gly-rich RNA-binding protein | 0.52 | 0.004 | TC190791 | ||
| AT2G21660.1 | Gly-rich RNA-binding protein | 0.49 | 0.012 | TC205820 | ||
| AT2G21660.1 | Gly-rich RNA-binding protein | 0.50 | 0.025 | TC191837 | ||
| AT2G21660.1 | Gly-rich RNA-binding protein | 0.48 | 0.017 | TC191004 | ||
| AT2G21660.1 | Gly-rich RNA-binding protein | 0.49 | 0.019 | TC228437 | ||
| AT2G21660.2 | Gly-rich RNA-binding protein | 0.45 | 0.037 | N.D. | TC223833 | |
| AT2G21660.2 | Gly-rich RNA-binding protein | 0.43 | 0.015 | N.D. | TC224420 | |
| AT2G21660.2 | Gly-rich RNA-binding protein | 0.50 | 0.007 | N.D. | TC228154 | |
| AT3G54770 | RNA recognition motif (RRM) domain-containing RNA binding | 0.66 | 0.010 | 0.24 ± 0.16 | TC203219 | |
| AT3G54770 | RRM domain-containing RNA binding | 0.65 | 0.008 | TC186377 | ||
| AT3G16780 | Ribosomal protein L19 | 0.66 | 0.025 | N.D. | TC182163 | |
| AT5G20160 | U4/U6 small nuclear ribonucleoprotein | 0.66 | 0.023 | N.D. | TC182279 | |
| Transcription factor | AT3G54320 | WRI1 | N.D | – | 4.8 ± 2.1 | TC200263 |
Among the 37 probe sets that were down-regulated in fiber-bearing ovules of psTRV:KTN plants, two different probe sets (TC193948 and TC179681) were annotated to be the KTN gene. This result provided independent confirmation of the silencing of KTN expression in the examined ovules. The remaining 35 probe sets included genes of three major categories. The first category encoded proteins related to RNA binding and mRNA splicing (13 of 35). Most of the RNA-binding proteins were Gly-rich RNA-binding protein. One of the main Gly-rich RNA-binding protein genes, GhGRP1, is the homolog of AtGRP7, which plays a key role in alternative mRNA exporting/splicing in Arabidopsis (Schöning et al., 2008). This result suggested possible expression changes associated with alternative splicing. The second category of down-regulated genes was related to auxin transport and response (four of 35), such as genes for the auxin efflux carrier PIN3 (TC199285) and the auxin-responsive protein IAA3 (TC214591). The third category of down-regulated genes was related to carbohydrate metabolism (three of 35), including those encoding a plastidic hexokinase (TC225147), a Fru-bisP aldolase (TC188344), and a positive regulator of pectic precursor synthesis (TC182299; Table I).
Among the major up-regulated probe sets were genes related to fatty acid biosynthesis and metabolism (four of 31), such as a positive regulator of triacylglycerol and unusual fatty acid accumulation level (TC224822). Another up-regulated gene was an inhibitor of pectic precursor synthesis (TC192343), which was related to carbohydrate metabolism (Table I). Pang et al. (2010) have shown that the biosynthesis of pectic precursors is important for cotton fiber elongation.
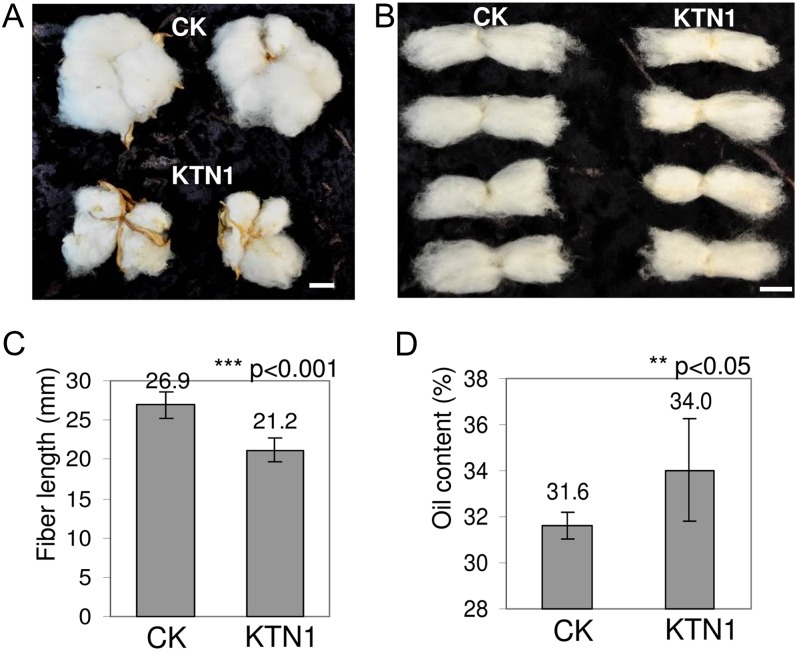
The up-regulated fatty acid biosynthetic genes, down-regulated carbohydrate biosynthetic genes, and up-regulated carbohydrate biosynthesis inhibitor gene suggested that in KTN fiber-bearing ovule inhibition of carbohydrate biosynthesis in fibers may be accompanied by increased fatty acid accumulation. We next measured the seed oil content in KTN-silenced plants. Because severe silencing of KTN resulted in the abortion of cotton bolls at an early stage, only those with mild phenotype (less than 10%) could develop into mature cotton bolls. These KTN-silenced mature bolls were smaller in size (Fig. 3A) and contained shorter cotton fiber (Fig. 3, B and C). The weight ratio of seed oil to endosperm in seed of KTN-silenced plants increased by 7.6% compared with the empty vector control (Fig. 3D). The shorter fiber length in KTN-silenced plants was inversely correlated with fatty acid accumulation in seed. This result suggests a competition of fixed carbon between lipid accumulation and fiber growth. It is possible that, upon inhibition of fiber growth, there was a diversion of fixed carbon into fatty acid biosynthesis.
Figure 3.
Silencing of KTN caused shorter cotton fibers and increased seed oil content. A, Phenotypes of empty vector control (CK) and KTN-silenced cotton bolls. Bar = 10 mm. B, Phenotypes of CK and KTN-silenced cotton fibers. Bar = 10 mm. C, Analysis of fiber length of CK and KTN-silenced cotton bolls (P < 0.001). D, Analysis of seed oil content (the ratio of raw oil weight to dry endosperm weight) in CK and KTN-silenced cotton seed (P < 0.05). [See online article for color version of this figure.]
Silencing of WRI1 Is a Positive Regulator of Lipid Biosynthesis
The results obtained with the silencing of KTN led us to hypothesize that we may be able to increase fiber growth by reducing fatty acid accumulation in cotton seed. We first suppressed the expression of several key cotton genes related to fatty acid biosynthesis. However, the infected plants invariably displayed a very severe phenotype and could not develop too much further post inoculation (data not shown). Next, we attempted to identify transcriptional factors that can regulate carbon flow between lipids and carbohydrates in reproductive organs. Work with Arabidopsis has shown that overexpression of an Arabidopsis WRI1 complementary DNA under the control of the cauliflower mosaic virus 35S promoter led to increased seed oil content (Cernac and Benning, 2004). On the other hand, seed oil accumulation in an Arabidopsis splicing mutant allele, wri1-1, was reduced. Glycolysis was compromised in this mutant, rendering developing embryos unable to efficiently convert Suc into precursors of triacylglycerol biosynthesis (Cernac and Benning, 2004). Interestingly, we found that WRI1 transcript levels were increased 4.8-fold in fiber-bearing ovules of KTN-silenced plants compared with that of empty vector control plants (Table I). This observation prompted us to silence the WRI1 expression in developing cotton fibers and seed to examine its effect on fiber growth.
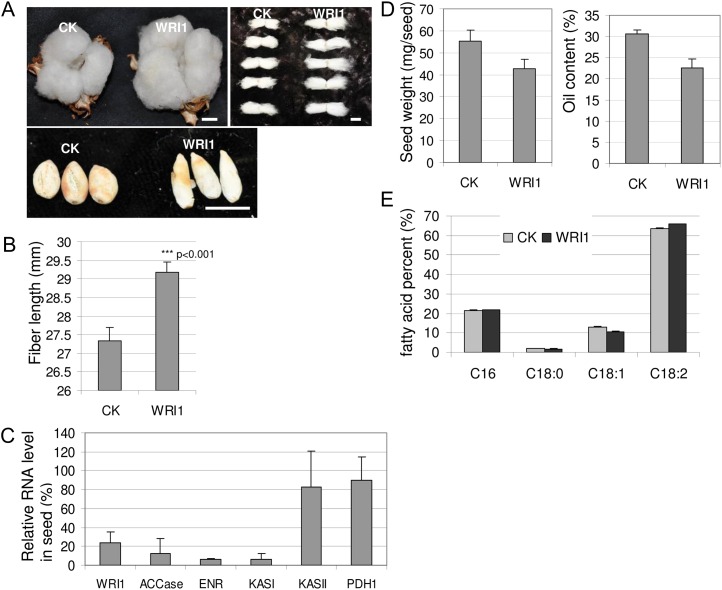
A homolog of WRI1 in cotton was identified using the Arabidopsis WRI1 gene (At3g54320.3) sequence to BLAST against the cotton EST database. No obvious phenotypes were seen in vegetative organs of psTRV:WRI1 plants (data not shown). However, the mature fiber length of psTRV:WRI1 plants was on average 2 mm longer than that of vector control plants (Fig. 4, A and B). Furthermore, there were obvious changes in seed shape (Fig. 4A). Real-time PCR analysis showed an average 76.6% reduction in WRI1 transcript levels in psTRV:WRI1 seed compared with vector control seed (Fig. 4C). WRI1 suppression down-regulated a set of genes involved in fatty acid synthesis (FAS), including genes encoding homomeric acetyl-CoA carboxylase (ACCase), enoyl-acyl carrier protein reductase (ENR), and ketoacyl-acyl carrier protein synthase (KASI; Fig. 4C). ACCase catalyzes the carboxylation of acetyl-CoA to malonyl-CoA, which is the first committed step in fatty acid biosynthesis. KASI encodes the main enzyme for the fatty acid condensation reaction, and ENR is the last enzyme in the fatty acid elongation cycle. By contrast, there were no obvious changes of transcript levels for other FAS genes, such as genes encoding KASII and pyruvate dehydrogenase (PDH1; Fig. 4C).
Figure 4.
Silencing of WRI1 is a positive regulator of oil biosynthesis. A, Phenotypes of empty vector control (CK) and WRI1-silenced cotton bolls and seeds. Bars = 10 mm. B, Analysis of fiber length of CK and WRI1-silenced cotton bolls (P < 0.001). C, Relative gene transcript levels in CK and WRI1-silenced cotton seeds. Error bars represent sd (n = 3 biological replicates). D, Seed weight (left) and oil content (the ratio of raw oil weight to dry endosperm weight; right) of CK and WRI1-silenced cotton seeds. E, Fatty acid composition of CK and WRI1-silenced cotton seeds. [See online article for color version of this figure.]
Further analysis showed that both seed weight and oil content were reduced in psTRV:WRI1 seed as compared with control seed (Fig. 4D), indicating the impact of changes in gene expression on seed oil accumulation. Gas chromatographic analysis showed that the relative amount of the end product of fatty acid desaturation, linoleic acid (18:2), was strongly increased, whereas the relative amount of the precursor oleic acid (18:1) was most obviously decreased in psTRV:WRI1 seed (Fig. 4E). In summary, the down-regulation of WRI1 reduced carbon flow into fatty acid biosynthesis while channeling the fixed carbon into fiber growth, resulting in longer fibers.
DISCUSSION
Cotton is an important and sustainable source of fiber and oil. Because cotton transformation is laborious and technically challenging, the development of a rapid method for gene functional analysis would greatly extend the power of molecular analysis. Gao et al. (2011a, 2011b) have shown that TRV-VIGS functions in cotton leaves with a 100% silencing efficiency. Extending the utility of this method, we showed here that sTRV-VIGS can also be effective in suppressing gene expression in reproductive organs.
We first used PA biosynthetic genes as markers to obtain evidence that VIGS can indeed be detected in reproductive organs, such as flower buds and fibers. Our main objective was to test whether we could use sTRV-VIGS to investigate gene functions related to fiber yield and quality. We silenced the KTN gene and studied the consequences of its suppression on cotton development. The Arabidopsis katanin homolog has been shown to possess ATP-dependent microtubule-severing activity in vitro, and Arabidopsis mutants deficient in KTN produce shorter organs (Burk et al., 2001). This organ length reduction was also observed in KTN-silenced cotton plants. In addition to the phenotypes of smaller vegetative and reproductive organ size, flowers of KTN-silenced cotton plants showed dramatic morphological and positional changes in ovaries, stamenal columns, and stamens, suggesting an essential role of KTN in flower development. KTN may play an important role in the competence of a cell to adapt its growth parameters to that of its neighbors (Uyttewaal et al., 2012). The silencing of KTN increased the growth variability of male and female floral organs. In floral buds of silenced plants, the stigma continued to extend and emerged from the clustered sepals. Defects of KTN led to reproductive failure, and cotton bolls developed from these flowers usually aborted at 2 to 3 dpa. Further analysis of KTN bolls with a less severe phenotype showed that knockdown of KTN expression caused an abnormal arrangement of cortical microtubules resulting in shorter cotton fibers. Cytoskeleton assembly has been known to play a key role in determining cotton fiber cell length (Li et al., 2005).
In contrast to the dramatic phenotypes in both vegetative and reproductive organs of KTN-silenced cotton plants, relatively few up-regulated or down-regulated genes were uncovered by microarray analysis. One explanation is that microRNA (miRNA)-mediated cleavage was not affected by the KTN deficiency in ovules of KTN-silenced plants. Since miRNAs are known to be expressed at high levels during cotton ovule and fiber development (Kwak et al., 2009; Pang et al., 2009), we designed around 270 miRNA targets as probes on our microarrays. Our microarray results showed that no putative miRNA target transcript was significantly up- or down-regulated in fiber-bearing ovules of KTN-silenced plants (data not shown). This result indicated that the cytoskeleton defects caused by KTN suppression did not affect miRNA-mediated cleavage of target mRNAs. This is consistent with a previous report that cytoskeleton dynamics in Arabidopsis is not important for miRNA-guided cleavage but may play a role in miRNA-mediated translational regulation (Brodersen et al., 2008). Their data also help to explain the apparent contradiction of a dramatic phenotype but expression changes in a relatively small number of genes. Since cytoskeleton dynamics is important for miRNA-guided translational regulation, the dramatic phenotype of KTN ovules might be caused by interference with mRNA translation, which cannot be detected by the microarray analysis. Furthermore, cotton fiber is the longest single cell in the plant kingdom, and nothing is known about how RNA is transported from the cell nucleus to the polar growth point. The cytoskeleton may be important for such polar RNA transport within fibers, which would be impaired by defects in cytoskeletal structure. This may be another explanation for the contradiction of a dramatic phenotype with few gene expression changes.
Transcriptome analysis of KTN fiber-bearing ovules showed that fatty acid biosynthetic genes are up-regulated whereas carbohydrate biosynthetic genes are down-regulated. We further explored the notion to increase fiber length by repartitioning carbon flow. In nature, the partitioning of fixed carbon between lipids and carbohydrates can vary widely. For example, oil palm (Elaeis guineensis) can accumulate up to 90% lipids in its mesocarp, the highest level observed in the plant kingdom, whereas the closely related date palm (Phoenix dactylifera) accumulates almost exclusively sugars (90%; Bourgis et al., 2011). The concept of channeling more carbon flow into triacylglycerol by reducing the amount of carbohydrates and protein has been proven in Arabidopsis. The transcript level of an ortholog of the transcription factor gene WRI1, which controls the conversion of other components into lipids, is positively correlated with lipid content but negatively correlated with sugar content (Bourgis et al., 2011). Therefore, we hypothesized that WRI1 expression level is positively correlated with lipid content but negatively correlated with fiber length. With the rapid gene function analysis set up in this study, we have validated this hypothesis by down-regulation of WRI1 expression in seed. Our results are consistent with the data of Zhang et al. (2011) that the increased fiber yield is associated with the cost of a smaller seed size, resulting from the repartition of seed oil into carbohydrate fibers. This result is consistent with previous reports that transgenic overexpression of carbohydrate biosynthesis genes improved cotton fiber length and strength (Li et al., 2004; Haigler et al., 2007; Jiang et al., 2012). All these studies suggest the possibility to increase fiber length by diverting the flow of fixed carbon from seed oil into carbohydrate fibers.
The microarray results provide clues for several candidate genes that may be involved in regulating fiber growth. For example, an auxin efflux carrier, the PIN family gene PIN3 (TC199285), was shown to be down-regulated in fiber-bearing ovules of KTN-silenced plants. In developing cotton ovules, auxin accumulates before anthesis, and the accumulation of this hormone peaks at +2 dpa and gradually decreases to basal levels by +10 dpa (Zhang et al., 2011). A steady supply of indole-3-acetic acid to ovule epidermis from −2 to +10 dpa increased the number of fiber cell initials (14%–19%) and reduced the number of fuzzy fibers. The overexpression of the PIN3 gene might increase the transport of endogenous auxin to ovule epidermis, thus enhancing fiber quality and quantity. In fact, genes related to phytohormone biosynthesis have been used to increase cotton fiber yield and quality. For example, targeted expression of the indole-3-acetic acid biosynthetic gene iaaM increased the number of lint fibers and fiber fineness (Zhang et al., 2011). Transgenic cotton, overexpressing gibberellin 20-oxidase, produced more fiber initials per ovule and longer fibers (Xiao et al., 2010). Another up-regulated gene in fiber-bearing ovules of KTN-silenced plants was a MPK4 ortholog (TC182568). Ectopic and branched root hairs and increased root width were seen in the Arabidopsis mpk4 mutant. Defects in MAPK4 signaling result in the misregulation of members of the microtubule-associated protein (MAP65) family, and cortical microtubules appeared heavily bundled and were randomly oriented (Beck et al., 2010). It is possible that the MAPK4 signaling pathway plays an important role in cotton fiber development and that KTN acts as a negative regulator of MAKP4 signaling pathways.
CONCLUSION
In summary, we have used the TRV-VIGS system to elucidate the functions of KTN and WRI1 in cotton fiber development. We believe that this system can be used to rapidly identify functions of genes that play a role in cotton fiber growth and elongation.
MATERIALS AND METHODS
Plant Materials
Seeds of upland cotton (Gossypium hirsutum ‘Coker 312’) were germinated in a greenhouse, and seedlings with two to four true leaves were used for VIGS assays. Infiltrated plants were grown in a growth chamber at 25°C with a 16-h-light/8-h-dark photoperiod cycle. Cotton bolls on treated plants were tagged on the day of anthesis (0 dpa).
Gene Cloning and Generation of Recombinant VIGS Vectors
psTRV1 and psTRV2 (Ye et al., 2009b, 2010) were used for all VIGS assays. Cotton gene sequences were obtained from the EST database of GenBank. Candidate gene fragments were cloned by PCR and inserted into psTRV2 to generate psTRV2 derivatives. The gene fragment sequences are listed in Supplemental File S1.
Agrobacterium Infiltration
psTRV1 and psTRV2 or psTRV2 derivatives were introduced into Agrobacterium tumefaciens strain AGL1 by electroporation. Agrobacterial cells were grown, collected, and resuspended in 10 mm MES, 10 mm MgCl2, and 200 µm acetylsyringone solution to a final optical density at 600 nm of 1.5 and then left at room temperature for 3 to 4 h without shaking. Before infiltration, A. tumefaciens cultures containing psTRV1 and psTRV2 or its derivatives were mixed in a 1:1 ratio. Cotton plants were agroinoculated by vacuum infiltration. Whole plants were submerged in agrobacterial inoculum and subjected to 80 to 100 kPa vacuum for 1 min, and the vacuum was quickly released to allow the inoculum to rapidly enter plant tissues. After infiltration, the excess agroinoculum was used to drench the root system of infiltrated plants. Note that TRV is transmitted by a nematode vector, mechanical inoculation, and grafting but not by contact between plants (http://www.agls.uidaho.edu/ebi/vdie/descr808.htm). We used separate pots of plants for different TRV treatments to ensure that the treated plants were not contaminated by a different TRV vector. After completion of the experiment, the plants and soil were autoclaved before disposal to prevent release of the virus into the environment.
RNA Extraction and Quantitative Real-Time PCR Analysis
Leaf, ovule, fiber, or seeds (fresh weight, less than 100 mg) were ground in liquid N2 and extracted with the plant RNA purification kit following the manufacturer’s protocol (Invitrogen). RNAs for microarray experiments were prepared by a similar method. Harvested bolls (7–8 dpa) of empty vector control, KTN-silenced, and WRI1-silenced cotton plants were stored on ice, and carefully dissected ovules were further frozen in liquid nitrogen and stored at −80°C until RNA extraction. RNA concentration was measured by Nanodrop (Thermo). Moloney murine leukemia virus reverse transcriptase (Promega) was used for reverse transcription reactions. Real-time PCR was performed with Power SYBR Green PCR Master (Applied Biosystems) and run in ABI7900HT. For each construct, we collected three samples from three plants with significant phenotypes (one sample from each plant). All samples were run in triplicate, and data were analyzed with RQ manager at a preset cycle threshold value (Applied Biosystems). The internal control was rbcL mRNA for leaf samples and UBQ mRNA for ovule samples. Cycle threshold values included in the analyses were based on three independent biological samples with three technical replicates for each biological sample. sd was calculated based on three biological replicates. For real-time PCR primer sequences, see Supplemental File S2.
Histochemical Assay
The DMACA-HCl protocol for PA staining was adapted from Li et al. (1999). Fresh organs were immediately soaked in ethanol:glacial acetic acid solution (3:1, v/v) for decoloration. The decolorized organs were stained for about 20 min with 0.3% (w/v) DMACA in a cold mixture of methanol and 6 m HCl (1:1, v/v), rinsed with several changes of 70% (v/v) ethanol, and the organs were observed with a dissecting microscope. Proanthocyanin-containing cells stained blue.
Scanning Electron Microscopy
Organs harvested from treated cotton plants were dissected and fixed with tape inside a sample chamber. Samples were frozen in liquid N2 and visualized with a scanning electron microscope (JSM-6360LV; JEOL).
Immunolabeling of Microtubules
Immunolabeling of microtubules was performed according to Zhao et al. (2010). Briefly, ovules were fixed in PEM buffer (50 mm PIPES, 5 mm EGTA, and 1 mm MgSO4, pH 6.9) containing 4% (w/v) paraformaldehyde, 0.1% Triton X-100, and 0.3 m mannitol for 1 h at room temperature. Fixed ovules were washed three times with PEM buffer. Fibers were cut off from the ovules and attached to slides coated with poly-l-Lys (Sigma-Aldrich). The fibers were incubated with 1% (w/v) Cellulase R-10 and 0.1% (w/v) Macerozyme in PEM buffer containing 0.3 m mannitol and 1 mm phenylmethylsulfonyl fluoride for 10 to 20 min at room temperature and then washed in phosphate-buffered saline (PBS) three times. After a preincubation in PBS buffer containing 1% (w/v) bovine serum albumin, the slides were incubated with mouse monoclonal anti-tubulin antibody (1:1,000) at 4°C overnight. The fibers were then washed three times in PBS buffer and incubated for 1 h with fluorescein isothiocyanate-labeled goat anti-mouse secondary antibody (1:200). The fibers were washed with PBS buffer and observed using a Zeiss LSM 5 EXCITER upright confocal microscope.
Custom Microarray and Data Analysis
We used Agilent GE 8x60K arrays provided by the full Custom Microarray Service of Genomax. Forty-four thousand Tentative Consensus sequences from Cotton Gene Index 10.1 cotton (http://compbio.dfci.harvard.edu/cgi-bin/tgi/gimain.pl?gudb=cotton; March 4, 2010) were used for Custom eARRAY probe design. RNA labeling, hybridization to custom arrays, washing, and scanning were all performed by the Custom Microarray. Feature extraction and application support were according to Agilent protocols. Briefly, to perform the comparative analysis of various samples, we applied total RNA samples from the cotton fiber-bearing ovules onto the custom microarrays. Total RNA from the fibers was labeled and applied separately. Briefly, 0.1 μg of total RNA from each sample was used to synthesize fluorescence-labeled complementary RNA using Cyanine 3-CTP according to the manufacturer’s protocol (Agilent Quick Amp Labeling Kit, one-color). Labeled RNA was hybridized for 16 h at 65°C on the custom array. After hybridization, the microarray slides were washed according to the standard protocol (Agilent Technologies) and scanned on an Agilent microarray scanner. Raw data were filtered, normalized, and further analyzed using the Agilent Feature Extraction Software (version 10.7). Statistics analysis was performed with one-way ANOVA, and probes with P < 0.05 between empty vector control and treated samples were used for ontology and pathway analysis. All Tentative Consensus numbers of the probes generated after significance analysis were retrieved from Tentative Consensus annotator (http://compbio.dfci.harvard.edu/cgi-bin/tgi/gimain.pl?gudb=cotton). BLAST similarity search using BLASTX (E-value < 10−10) against The Arabidopsis Information Resource 10 and RefSeq (National Center for Biotechnology Information) databases was performed to assign an annotation to the transcripts.
Cotton Fiber Length Measurement
Dry and mature fiber length was manually measured against a ruler. The mean fiber length of three measurements from one seed, 10 seeds from one boll, and five typical size bolls from each treated cotton group were recorded and used for Student’s t test analysis. The effect of silencing of KTN1 and WRI1 on cotton fiber length was repeated in two independent experiments.
Fatty Acid Analysis
Oil content and fatty acid profile analysis were performed according to Qu et al. (2012). Briefly, the outer seed coat was removed from dried cotton seeds. The dry endosperm part was ground to a fine powder, and the lipids were extracted with hexane three times. The combined supernatant was transferred to a glass vial, and the hexane was evaporated with a flow of dry nitrogen gas at 50°C. The weight of the raw oil was determined, and the oil content was recorded as the ratio of raw oil weight to dry endosperm weight.
Total lipid, extracted from cotton seed, was transmethylated with 3 n methanolic-HCl (Sigma) plus 400 μL of 2,2-dimethoxypropane (Sigma). The fatty acid methyl ester was analyzed using GC Agilent 6890 employing helium as the carrier gas and DB-23 columns for component separation. The fatty acid composition value included in the analyses was calculated based on peak areas in three biological replicates and presented as mean ± sd.
Sequence information for the gene fragment used for VIGS is listed in Supplemental File S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. psTRV:KTN plants with severe vegetative phenotypes.
Supplemental File S1. Sequence information of the gene fragment used for VIGS.
Supplemental File S2. Real-time PCR primer sequences.
Acknowledgments
We thank Mr. Khar Meng Ng for his help in taking care of cotton plants. We thank Mr. Xue-Zhi Ouyang, Dr. Meredith Calvert, and Ms. Fiona Chia for their help in scanning electron microscopy and confocal observation.
Glossary
- VIGS
virus-induced gene silencing
- CLCrV
Cotton leaf crumple virus
- TRV
Tobacco rattle virus
- sTRV
synthetic Tobacco rattle virus
- PA
proanthocyanidin
- DMACA
p-dimethylaminocinnamaldehyde
- dpa
days post anthesis
- CMT
cortical microtubules
- miRNA
microRNA
- PBS
phosphate-buffered saline
- PEM
50 mm PIPES, 5 mm EGTA, and 1 mm MgSO4, pH 6.9
References
- Ahmad FJ, Yu W, McNally FJ, Baas PW. (1999) An essential role for katanin in severing microtubules in the neuron. J Cell Biol 145: 305–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D. (2004) RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Beck M, Komis G, Müller J, Menzel D, Samaj J. (2010) Arabidopsis homologs of nucleus- and phragmoplast-localized kinase 2 and 3 and mitogen-activated protein kinase 4 are essential for microtubule organization. Plant Cell 22: 755–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgis F, Kilaru A, Cao X, Ngando-Ebongue GF, Drira N, Ohlrogge JB, Arondel V. (2011) Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proc Natl Acad Sci USA 108: 12527–12532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigneti G, Martín-Hernández AM, Jin H, Chen J, Baulcombe DC, Baker B, Jones JD. (2004) Virus-induced gene silencing in Solanum species. Plant J 39: 264–272 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP. (2004) Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J 39: 734–746 [DOI] [PubMed] [Google Scholar]
- Burk DH, Liu B, Zhong R, Morrison WH, Ye ZH. (2001) A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13: 807–827 [PMC free article] [PubMed] [Google Scholar]
- Burk DH, Zhong R, Ye Z-H. (2007) The katanin microtubule severing protein in plants. J Integr Plant Biol 49: 1174–1182 [Google Scholar]
- Cernac A, Benning C. (2004) WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J 40: 575–585 [DOI] [PubMed] [Google Scholar]
- Fu DQ, Zhu BZ, Zhu HL, Jiang WB, Luo YB. (2005) Virus-induced gene silencing in tomato fruit. Plant J 43: 299–308 [DOI] [PubMed] [Google Scholar]
- Gao X, Britt RC, Jr, Shan L, He P. (2011a) Agrobacterium-mediated virus-induced gene silencing assay in cotton. J Vis Exp 54: e2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Wheeler T, Li Z, Kenerley CM, He P, Shan L. (2011b) Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J 66: 293–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grønlund M, Constantin G, Piednoir E, Kovacev J, Johansen IE, Lund OS. (2008) Virus-induced gene silencing in Medicago truncatula and Lathyrus odorata. Virus Res 135: 345–349 [DOI] [PubMed] [Google Scholar]
- Haigler CH, Singh B, Zhang D, Hwang S, Wu C, Cai WX, Hozain M, Kang W, Kiedaisch B, Strauss RE, et al. (2007) Transgenic cotton over-producing spinach sucrose phosphate synthase showed enhanced leaf sucrose synthesis and improved fiber quality under controlled environmental conditions. Plant Mol Biol 63: 815–832 [DOI] [PubMed] [Google Scholar]
- Hileman LC, Drea S, Martino G, Litt A, Irish VF. (2005) Virus-induced gene silencing is an effective tool for assaying gene function in the basal eudicot species Papaver somniferum (opium poppy). Plant J 44: 334–341 [DOI] [PubMed] [Google Scholar]
- Holzberg S, Brosio P, Gross C, Pogue GP. (2002) Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J 30: 315–327 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Guo W, Zhu H, Ruan YL, Zhang T. (2012) Overexpression of GhSusA1 increases plant biomass and improves cotton fiber yield and quality. Plant Biotechnol J 10: 301–312 [DOI] [PubMed] [Google Scholar]
- Kwak PB, Wang QQ, Chen XS, Qiu CX, Yang ZM. (2009) Enrichment of a set of microRNAs during the cotton fiber development. BMC Genomics 10: 457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Burns TH, Light G, Sun Y, Fokar M, Kasukabe Y, Fujisawa K, Maekawa Y, Allen RD. (2010) Xyloglucan endotransglycosylase/hydrolase genes in cotton and their role in fiber elongation. Planta 232: 1191–1205 [DOI] [PubMed] [Google Scholar]
- Li X, Wang XD, Zhao X, Dutt Y. (2004) Improvement of cotton fiber quality by transforming the acsA and acsB genes into Gossypium hirsutum L. by means of vacuum infiltration. Plant Cell Rep 22: 691–697 [DOI] [PubMed] [Google Scholar]
- Li XB, Fan XP, Wang XL, Cai L, Yang WC. (2005) The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 17: 859–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YG, Tanner G, Larkin P. (1999) The DMACA-HCl protocol and the threshold proanthocyanidin content for bloat safety in forage legumes. J Sci Food Agric 70: 89–101 [Google Scholar]
- Liu Y, Nakayama N, Schiff M, Litt A, Irish VF, Dinesh-Kumar SP. (2004) Virus induced gene silencing of a DEFICIENS ortholog in Nicotiana benthamiana. Plant Mol Biol 54: 701–711 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP. (2002) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- McNally FJ, Vale RD. (1993) Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell 75: 419–429 [DOI] [PubMed] [Google Scholar]
- Pang CY, Wang H, Pang Y, Xu C, Jiao Y, Qin YM, Western TL, Yu SX, Zhu YX. (2010) Comparative proteomics indicates that biosynthesis of pectic precursors is important for cotton fiber and Arabidopsis root hair elongation. Mol Cell Proteomics 9: 2019–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang M, Woodward AW, Agarwal V, Guan X, Ha M, Ramachandran V, Chen X, Triplett BA, Stelly DM, Chen ZJ. (2009) Genome-wide analysis reveals rapid and dynamic changes in miRNA and siRNA sequence and expression during ovule and fiber development in allotetraploid cotton (Gossypium hirsutum L.). Genome Biol 10: R122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y, Peel GJ, Wright E, Wang Z, Dixon RA. (2007) Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula. Plant Physiol 145: 601–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J, Mao HZ, Chen W, Gao SQ, Bai YN, Sun YW, Geng YF, Ye J. (2012) Development of marker-free transgenic Jatropha plants with increased levels of seed oleic acid. Biotechnol Biofuels 5: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz MT, Voinnet O, Baulcombe DC. (1998) Initiation and maintenance of virus-induced gene silencing. Plant Cell 10: 937–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöning JC, Streitner C, Meyer IM, Gao Y, Staiger D. (2008) Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res 36: 6977–6987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle JR, Idris AM, Brown JK, Haigler CH, Robertson D. (2008) Geminivirus-mediated gene silencing from Cotton leaf crumple virus is enhanced by low temperature in cotton. Plant Physiol 148: 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States Department of Agriculture. (2012) Cotton: World Markets and Trade. http://www.fas.usda.gov/cotton/Current/ (June 8, 2012)
- Uyttewaal M, Burian A, Alim K, Landrein B, Borowska-Wykręt D, Dedieu A, Peaucelle A, Ludynia M, Traas J, Boudaoud A, et al. (2012) Mechanical stress acts via katanin to amplify differences in growth rate between adjacent cells in Arabidopsis. Cell 149: 439–451 [DOI] [PubMed] [Google Scholar]
- Valentine T, Shaw J, Blok VC, Phillips MS, Oparka KJ, Lacomme C. (2004) Efficient virus-induced gene silencing in roots using a modified tobacco rattle virus vector. Plant Physiol 136: 3999–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Wang J, Gao P, Jiao GL, Zhao PM, Li Y, Wang GL, Xia GX. (2009) Down-regulation of GhADF1 gene expression affects cotton fibre properties. Plant Biotechnol J 7: 13–23 [DOI] [PubMed] [Google Scholar]
- Wang J, Wang HY, Zhao PM, Han LB, Jiao GL, Zheng YY, Huang SJ, Xia GX. (2010) Overexpression of a profilin (GhPFN2) promotes the progression of developmental phases in cotton fibers. Plant Cell Physiol 51: 1276–1290 [DOI] [PubMed] [Google Scholar]
- Xiao YH, Li DM, Yin MH, Li XB, Zhang M, Wang YJ, Dong J, Zhao J, Luo M, Luo XY, et al. (2010) Gibberellin 20-oxidase promotes initiation and elongation of cotton fibers by regulating gibberellin synthesis. J Plant Physiol 167: 829–837 [DOI] [PubMed] [Google Scholar]
- Xie DY, Sharma SB, Paiva NL, Ferreira D, Dixon RA. (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299: 396–399 [DOI] [PubMed] [Google Scholar]
- Ye J, Chua N-H, Qu J, Geng Y-F, Bu Y-P, inventors June 10, 2010. Virus induced gene silencing (VIGS) for functional analysis of genes in cotton. United States Patent Application No. 13/376,902
- Ye J, Qu J, Bui HT, Chua N-H. (2009a) Rapid analysis of Jatropha curcas gene functions by virus-induced gene silencing. Plant Biotechnol J 7: 964–976 [DOI] [PubMed] [Google Scholar]
- Ye J, Qu J, Chua N-H, inventors December 16, 2009b. Functional analysis of Jatropha curcas genes. United States Patent Application no. 13/141,752
- Zhang C, Ghabrial SA. (2006) Development of Bean pod mottle virus-based vectors for stable protein expression and sequence-specific virus-induced gene silencing in soybean. Virology 344: 401–411 [DOI] [PubMed] [Google Scholar]
- Zhang M, Zheng X, Song S, Zeng Q, Hou L, Li D, Zhao J, Wei Y, Li X, Luo M, et al. (2011) Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat Biotechnol 29: 453–458 [DOI] [PubMed] [Google Scholar]
- Zhao PM, Wang LL, Han LB, Wang J, Yao Y, Wang HY, Du XM, Luo YM, Xia GX. (2010) Proteomic identification of differentially expressed proteins in the Ligon lintless mutant of upland cotton (Gossypium hirsutum L.). J Proteome Res 9: 1076–1087 [DOI] [PubMed] [Google Scholar]