Abstract
The control of floral organ identity by homeotic MADS box genes is well established in eudicots. However, grasses have highly specialized outer floral organs, and the identities of the genes that regulate the highly specialized outer floral organs of grasses remain unclear. In this study, we characterized a MIKC-type MADS box gene, CHIMERIC FLORAL ORGANS (CFO1), which plays a key role in the regulation of floral organ identity in rice (Oryza sativa). The cfo1 mutant displayed defective marginal regions of the palea, chimeric floral organs, and ectopic floral organs. Map-based cloning demonstrated that CFO1 encoded the OsMADS32 protein. Phylogenetic analysis revealed that CFO1/OsMADS32 belonged to a monocot-specific clade in the MIKC-type MADS box gene family. The expression domains of CFO1 were mainly restricted to the marginal region of the palea and inner floral organs. The floral organ identity gene DROOPING LEAF (DL) was expressed ectopically in all defective organs of cfo1 flowers. Double mutant analysis revealed that loss of DL function mitigated some of the defects of floral organs in cfo1 flowers. We propose that the CFO1 gene plays a pivotal role in maintaining floral organ identity through negative regulation of DL expression.
Most flowers consist of four distinct organ types arranged in concentric whorls: sepals (whorl 1), petals (whorl 2), stamens (whorl 3), and carpels and ovules (whorl 4). The well-established ABCDE model, which is mainly based on genetic and molecular studies involving eudicots, such as Arabidopsis (Arabidopsis thaliana), snapdragon (Antirrhinum majus), and petunia (Petunia hybrida), explains how floral organ identity is coordinately defined by A-, B-, C-, D-, and E-class MADS box genes in eudicots (Coen and Meyerowitz, 1991; Weigel and Meyerowitz, 1994; Theissen and Saedler, 2001; Ditta et al., 2004).
The Poaceae, one of the largest monocot families, includes many important crops, such as barley (Hordeum vulgare), maize (Zea mays), rice (Oryza sativa), and wheat (Triticum aestivum). The floral architecture of grass species is distinct from those of eudicots and other monocots. The spikelet, the basic unit of the grass inflorescence, consists of glumes and one to 40 florets that comprise the lemma and palea (possibly homologous to sepals), lodicules (homologous to petals), stamens, pistils, and ovules (Bommert et al., 2005; Itoh et al., 2005; Malcomber et al., 2006). The recent characterization of several MADS box genes that specify floral organ identity in rice suggests that the ABCDE model applies to the regulation of stamen, pistil, and ovule development in grass species, at least in part (Nagasawa et al., 2003; Yamaguchi et al., 2006; Dreni et al., 2007; Yao et al., 2008). However, the molecular mechanisms that control the specification of grass-specific floral organs (the lemma, palea, and lodicule) remain to be elucidated.
The B-class genes are well conserved between Arabidopsis and rice. The Arabidopsis B-class genes APETALA3 (AP3) and PISTILLATA (PI) are required to specify petal and stamen identities (Bowman et al., 1989; Goto and Meyerowitz, 1994; Jack et al., 1994). The OsMADS16/SUPERWOMAN1 (SPW1) and OsMADS4 genes, the orthologs of Arabidopsis AP3 and PI, determine the lodicule and stamen identities in rice (Nagasawa et al., 2003; Xiao et al., 2003; Yao et al., 2008). The OsMADS2 gene, a paralog of OsMADS4 in rice, mainly functions in lodicule specification (Prasad and Vijayraghavan, 2003; Yadav et al., 2007; Yao et al., 2008).
The C-class gene AGAMOUS (AG) is a key regulator of stamen and carpel identities and floral meristem determinacy in Arabidopsis (Bowman et al., 1991; Drews et al., 1991). Two C-class paralogs, OsMADS3 and OsMADS58, which are orthologous to AG, play distinct roles in developmental regulation of the lodicule, stamen, and pistil in rice (Yamaguchi et al., 2006). More intriguingly, DROOPING LEAF (DL), an ortholog of the Arabidopsis YABBY gene CRABS CLAW (CRC), plays pivotal roles in pistil specification and floral meristem determinacy and antagonizes the function of B-class genes in rice (Bowman and Smyth, 1999; Yamaguchi et al., 2004). In Arabidopsis, the D-class gene SEEDSTICK (STK) acts redundantly with the C-class genes AG and SHATTERPROOF1/2 to determine ovule identity (Pinyopich et al., 2003). In rice, two STK-like genes have been identified: whereas OsMADS13 is required for ovule specification, the function of OsMADS21 remains unclear (Dreni et al., 2007).
In Arabidopsis, the A-class genes AP1 and AP2 specify sepal and petal identities (Mandel et al., 1992; Jofuku et al., 1994). The E-class genes SEPALLATA1/2/3/4 (SEP1/2/3/4) redundantly specify all floral organ identities and floral meristem determination (Pelaz et al., 2000; Ditta et al., 2004). In rice, three AP1-like genes and five SEP-like genes are known, of which only three have been characterized (Fornara et al., 2003; Malcomber and Kellogg, 2004, 2005; Kater et al., 2006). The SEP-like gene OsMADS1/LEAFY HULL STERILE (LHS1) is required for the specification of lemma and palea identity (Jeon et al., 2000; Agrawal et al., 2005; Prasad et al., 2005; Chen et al., 2006). Recently, another SEP-like gene, OsMADS34/PANICLE PHYTOMER2, was found to control rudimentary glume and sterile lemma development (Kobayashi et al., 2010). An AP1/FRUITFULL-like gene, OsMADS15/DEGENERATIVE PALEA (DEP), specifies palea and sterile lemma identity (Wang et al., 2010).
All of the above ABCDE floral regulators, except Arabidopsis AP2, belong to several classes of MIKC-type MADS box genes. The role of a special class of MIKC-type MADS box genes, which includes rice OsMADS32 and its orthologs, still remains to be elucidated (Ma et al., 1991; Nam et al., 2004; Arora et al., 2007). In this study, we characterized a spontaneous mutant named chimeric floral organs1 (cfo1). Molecular cloning indicated that the underlying allele of CFO1 encoded the OsMADS32 protein. Phenotypic analysis revealed that the loss of function of CFO1 led to defective marginal regions of the palea, chimeric floral organs, and ectopic organs. CFO1 expression was detected throughout the meristematic region of the inflorescence and flower before initiation of the floral organs. Subsequently, the expression domains became mainly restricted to the marginal regions of the palea and inner floral organs. Expression and double mutant analysis suggested that CFO1 repressed DL transcription. We conclude that CFO1 plays a pivotal role in maintaining floral organ identity by negative regulation of DL expression.
RESULTS
Alteration of Floral Organ Identity in cfo1
The developmental stages of rice described by Ikeda et al. (2004) were applied in this study. The morphology of cfo1 and wild-type flowers was compared at the inflorescence 9 stage (heading stage). A typical wild-type rice flower is composed of a pistil in the central whorl (whorl 4), six stamens around the pistil in whorl 3, two lodicules adjacent to the lemma in whorl 2, and two interlocking organs (the palea and lemma, collectively termed the hull) that surround the inner floral organs in whorl 1 (Figs. 1, A, C, and I, and 2, A-1 and A-5).
Figure 1.
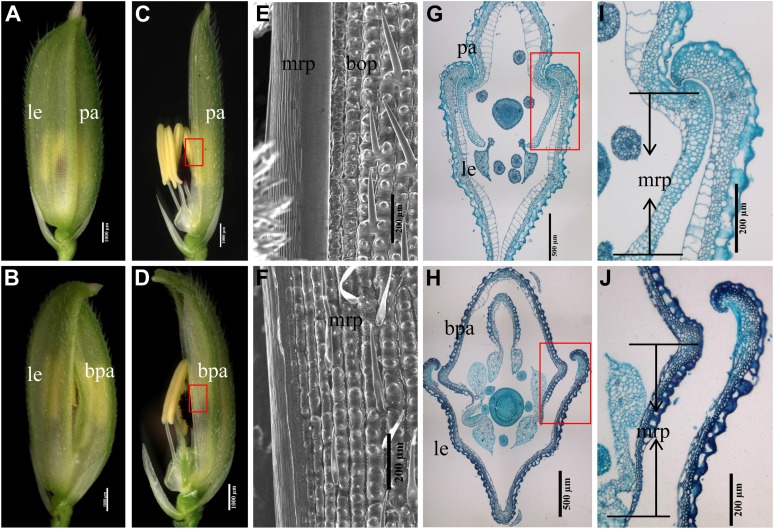
Palea identity in cfo1 and wild-type flowers. A and B, Spikelets of wild-type (A) and cfo1 (B) flowers. C and D, Spikelets of wild-type (C) and cfo1 (D) flowers. The lemmas have been removed. The mrp in cfo1 flowers is larger than that in wild-type flowers. E and F, Scanning electron micrographs of the mrp in wild-type (E) and cfo1 (F) flowers. G and H, Transverse sections of wild-type (G) and cfo1 (H) spikelets. I and J, Transverse sections through the margin between the lemma and palea in wild-type (I) and cfo1 (J) flowers. bpa, Bent palea; le, lemma; pa, palea. Bars = 1,000 μm in A to D, 500 μm in G and H, and 200 μm in E, F, I, and J. [See online article for color version of this figure.]
Figure 2.
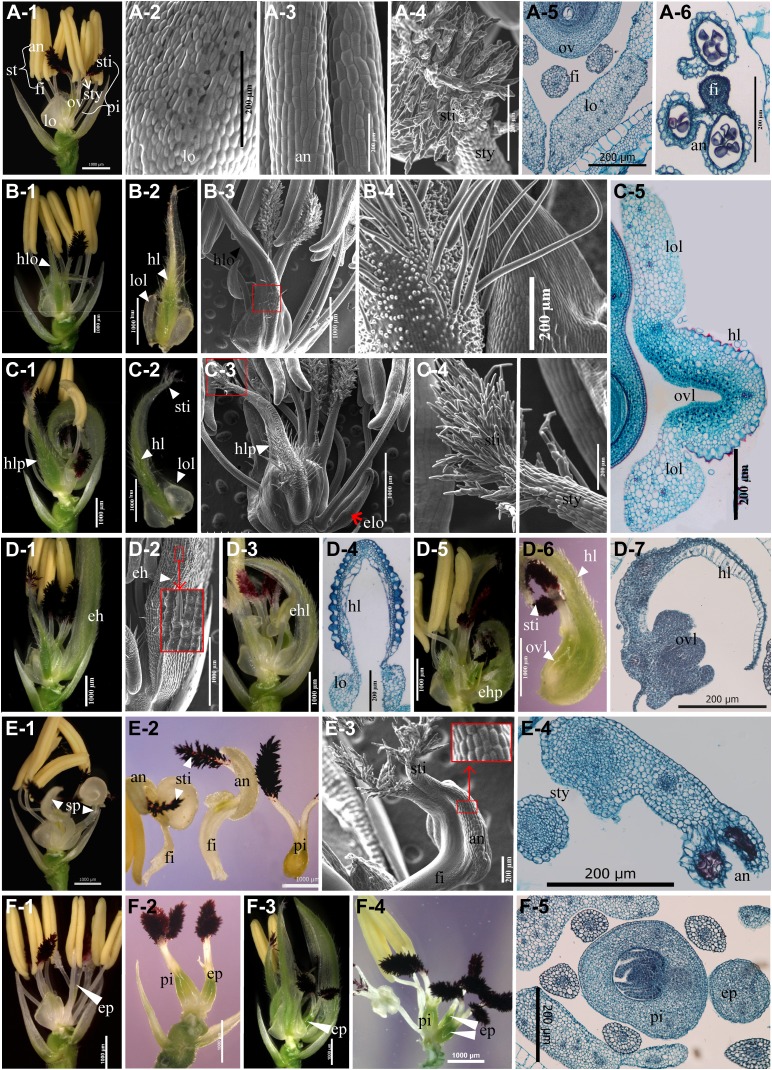
Inner floral organs of cfo1 flowers. A, Inner floral organs in the wild type. A-1, A wild-type spikelet; the lemma and palea were removed. A-2 to A-4, Scanning electron micrographs of the lodicule, stamen, and stigma, respectively. A-5 and A-6, Transverse sections of the inner floral organs. B, Hull-lodicule chimeras (hlo) in cfo1 flowers. B-1, A cfo1 spikelet with a hlo (indicated by the white triangle); the lemma and palea were removed. B-2, The hlo shown in B-1. The white triangles indicate lodicule- and hull-like tissues. B-3 and B-4, Scanning electron micrographs of a hlo. B-4, Higher magnification of the portion of B-3 enclosed in the red frame. Trichomes and protrusions were observed on the epidermal cells of the hlo. B-5, Transverse section of a hlo. Hull- and lodicule-like cell structures were observed. C, Hull-lodicule-pistil chimeras (hlp) in cfo1 flowers. C-1, A cfo1 spikelet with hlp (indicated by the white triangle); the lemma and palea were removed. C-2, The hlp shown in C-1. The white triangles indicate lodicule-, hull-, and stigma-like tissues. C-3 and C-4, Scanning electron micrographs of a hlp. C-4 shows a higher power magnification of the portion of C-3 enclosed in the red frame. Stigma-like cells were observed at the top of the hlp. D, The efo in cfo1 flowers. D-1, A cfo1 spikelet with an ectopic hull (eh; indicated by white triangles); the lemma and palea were removed. D-2, Scanning electron micrographs of an eh. The large red frame shows a higher power magnification of the portion enclosed in the small red frame. D-3, A cfo1 spikelet with an ectopic hull-lodicule chimera (ehl). D-4, Transverse section of an ehl. D-5, A cfo1 spikelet with an ectopic hull-pistil chimera (ehp). D-6, The ehp shown in D-5. D-7, Transverse section of an eh. Hull- and ovary-like cell structures were observed. E, Stamen-pistil chimeras (sp) in cfo1 flowers. E-1, A cfo1 spikelet with two sp; the lemma and palea were removed. E-2, The two sp and the pistil in whorls 3 and 4 shown in E-1. E-3, Scanning electron micrograph of a sp. E-4, Transverse section of a sp. Style- and anther-like cell structures were observed. F, Extra pistils in whorl 4 of cfo1 flowers. F-1, A cfo1 flower with six stamens and two pistils; the lemma and palea were removed. F-2, A cfo1 flower with normal pistil and the extra pistil in whorl 4; the other floral organs were removed. F-3, A cfo1 flower with extra pistils; the lemma and palea were removed. F-4, One normal pistil and two extra pistils shown in F-3; the other floral organs were removed. F-5, Transverse section of a cfo1 flower with one extra pistil in whorl 4. an, Anther; fi, filament; elo, ectopic lodicule-like organ; ep, extra pistil; hl, hull-like tissue; lo, lodicule; lol, lodicule-like tissue; ov, ovary; ovl; ovary-like tissue; pi, pistil; st, stamen; sti, stigma; sty, style. Bars = 1,000 μm in A-1, B-1 to B-3, C-1 to C-3, D-1 to D-3, D-5, D-6, E-1, E-2, and F-1 to F5; bars = 200 μm in all other images.
Figure 5.
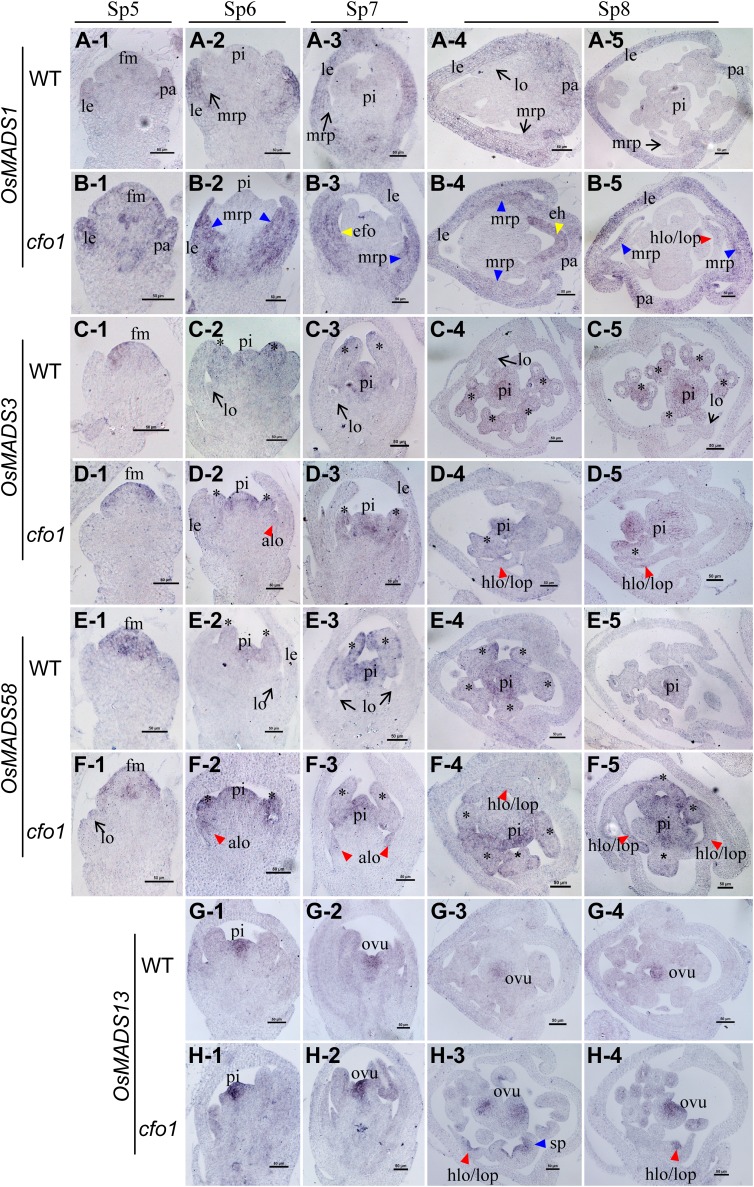
Expression of OsMADS1 and C- and D-class genes in cfo1 and wild-type flowers at early stages of flower development. Expression of OsMADS1 (A), OsMADS3 (C), OsMADS58 (E), and OsMADS13 (G) is shown in wild-type flowers, and expression of OsMADS1 (B), OsMADS3 (D), OsMADS58 (F), and OsMADS13 (H) is shown in cfo1 flowers. Rows 1 to 3 show longitudinal sections of flowers at stages Sp5 to Sp7, respectively, and rows 4 and 5 show transverse sections of flowers at the Sp8 stage. Black arrows indicate normal gene expression in the lodicules or mrp. Asterisks indicate normal gene expression in stamens. Blue, red, and yellow triangles indicate abnormal gene expression in the mrp, abnormal lodicule/hull-lodicule chimera/lodicule-pistil chimera, and efo, respectively. alo, Abnormal lodicule; eh, ectopic hull-like organ; fm, floral meristem; hlo, hull-lodicule chimera; le, lemma; lo, lodicule; lop, lodicule-pistil chimera; ovu, ovule; pa, palea; pi, pistil; sp, stamen-pistil chimera. Bars = 50 μm.
In cfo1 flowers, the most obvious alterations were split florets and bent paleas with broad marginal regions. Loss of the hook from the palea disrupted the mechanism by which this structure normally interlocks with the counterpart hook of the lemma (Fig. 1, B and H; Supplemental Fig. S1A). Further examination showed that the identity of the marginal region of the palea (mrp) of cfo1 flowers was not well established. The wild-type palea consists of two parts: the body of the palea (bop) and two mrp (Ohmori et al., 2009; Fig. 1, C, E, and G). The bop, which has a texture similar to that of the lemma, was composed of a silicified upper epidermis that bore trichomes and protrusions, fibrous sclerenchyma, spongy parenchyma, and a vacuolated inner epidermis (Fig. 1I; Prasad et al., 2005). The mrp had a distinct cellular structure that contained a nonsilicified upper epidermis without trichomes and protrusions (Fig. 1E), a large number of spongy parenchyma cells, a few fibrous sclerenchyma cells, and a nonvacuolated inner epidermis (Fig. 1I; Prasad et al., 2005). However, in cfo1 flowers, the mrp was converted to a lemma-like structure with silicified upper epidermal cells (Fig. 1F) and vacuolated inner epidermal cells (Fig. 1J). In addition, the cfo1 palea was significantly wider than that of the wild type because of its expanded mrp, which was similar in width to that of the lemma (Supplemental Fig. S1B).
Expression of the ABCDE-class genes and the CRC-like gene DL was evaluated in the lemmas and paleas of wild-type and cfo1 flowers with quantitative reverse transcription (qRT)-PCR. In wild-type flowers, DL was expressed in the lemma but not in the palea (Yamaguchi et al., 2004; Supplemental Fig. S2A). However, in cfo1 flowers, significant expression of DL was detected in the palea (Supplemental Fig. S2A), which suggested that the cfo1 palea acquired some degree of lemma identity. The expression of the other genes was similar in the paleas of wild-type and cfo1 flowers.
The lemma and palea were removed for analysis of the phenotypes of the inner floral organs. Severely defective morphogenesis of the inner floral organs was observed in cfo1 flowers.
In whorl 2, the wild-type lodicule was a small, fleshy, cup-shaped, nonphotosynthetic organ (Fig. 2A-1) with smooth epidermal cells (Fig. 2A-2). The lodicule contained parenchymatous cells interspersed with tracheal elements (Fig. 2A-5; Yadav et al., 2007). However, elongated lodicules that carried hull- and/or pistil-like tissues were observed in the majority of cfo1 flowers (Fig. 2, B and C; Supplemental Fig. S1A). Figure 2B shows the hull-lodicule chimeras in cfo1 flowers. Lodicule-like tissues were observed in the marginal regions of the base, whereas hull-like tissues developed along the medial axis (Fig. 2, B-1 and B-2). Trichomes and protrusions were borne on the upper epidermal cells of hull-like tissues (Fig. 2, B-3 and B-4). Figure 2C shows the hull-lodicule-pistil chimeras in cfo1 flowers. In addition to the differentiated lodicule- and hull-like tissues, stigma-like tissues (Fig. 2, C-3 and C-4) were observed at the top of these hull-lodicule-pistil chimeras. Transverse sections of the hull-lodicule-pistil chimeras revealed the presence of inner pistil-like cell layers in the middle segment, together with silicified upper epidermal cells that resembled those of the lemma or bop, whereas lodicule-like cell layers were observed in the marginal region (Fig. 2C-5).
Figure 3.
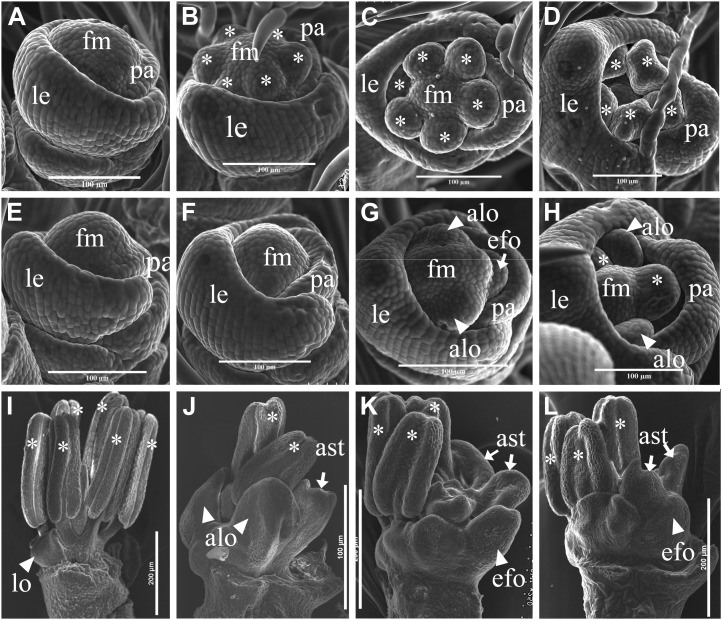
Scanning electron micrographs of early cfo1 flowers. A to D, Scanning electron microscopy of wild-type flowers at stages Sp5 (A), early Sp6 (B), later Sp6 (C), and Sp7 (D). E to H, Scanning electron microscopy of cfo1 flowers at stages Sp5 (E), early Sp6 (F), later Sp6 (G), and Sp7 (H). F to H show delayed initiation of stamen primordia. Abnormal lodicule primordia were observed in G and H, and efo primordia were observed in G. I, Inner floral organs during stage Sp8 in a wild-type flower. J to L, Inner floral organs during stage Sp8 in cfo1 flowers. Abnormal lodicules were observed in J, and efo and abnormal stamens were observed in K and L. alo, Abnormal lodicule; ast, abnormal stamen; fm, floral meristem; le, lemma; lo, lodicule; pa, palea. Bars = 100 μm in A to H and 200 μm in I to L.
Figure 4.
OsMADS6 and B-class gene expression in cfo1 and wild-type flowers at early stages of development of cfo1 flowers. Expression of OsMADS6 (A), OsMADS2 (C), OsMADS4 (E), and OsMADS16 (G) is shown in wild-type flowers, and expression of OsMADS6 (B), OsMADS2 (D), OsMADS4 (F), and OsMADS16 (H) is shown in cfo1 flowers. Rows 1 to 3 show longitudinal sections of flowers at stages Sp5 to Sp7, respectively, and rows 4 and 5 show transverse sections of flowers at the Sp8 stage. Black arrows indicate normal gene expression in the lodicules or mrp. Asterisks indicate normal gene expression in the stamens. Blue, red, yellow, and green triangles indicate abnormal gene expression in the mrp, abnormal lodicule/hull-lodicule chimera/lodicule-pistil chimera, efo, and stamen-pistil chimera, respectively. alo, Abnormal lodicule; eh, ectopic hull-like organ; elo, elongated lodicule; fm, floral meristem; hlo, hull-lodicule chimera; lo, lodicule; lop, lodicule-pistil chimera; pi, pistil; sp, stamen-pistil chimera. Bars = 50 μm.
Most cfo1 flowers displayed ectopic floral organs (efo) with different types of identities on the palea side of whorl 2 (Supplemental Fig. S1A), where the wild-type flowers developed no organs. Most of the efo resembled hulls with lemma- or bop-like epidermal cells (Fig. 2, D-1 and D-2). Some of the efo showed lodicule-like identity (Fig. 2C-3, red arrow). Others developed into types of chimeras, such as an ectopic hull-lodicule chimera (Fig. 2, D-3 and D-4) and ectopic hull-pistil chimera (Fig. 2, D-5–D-7).
Figure 7.
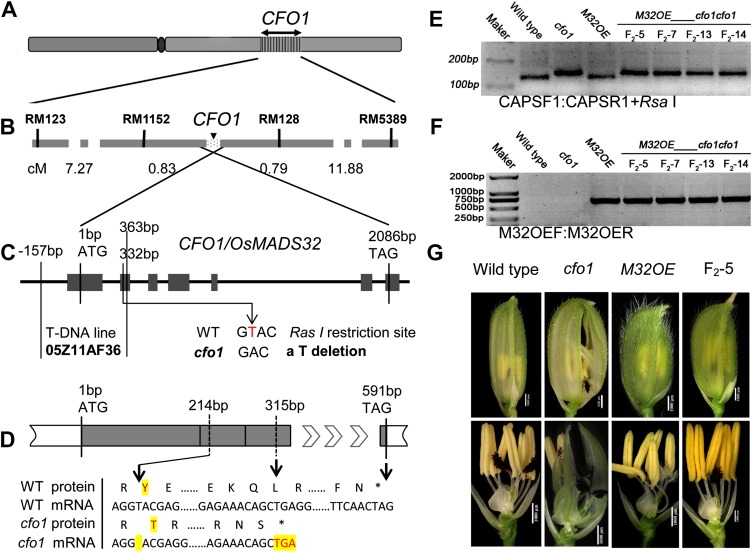
Map-based cloning of CFO1. A, Location of CFO1 on chromosome 1. B, Fine-mapping of the CFO1 locus. C, Genomic structure of CFO1/OsMADS32. The site of the mutation in cfo1 and the T-DNA insertion site of AF36 are shown. The cfo1 mutation lies within an RsaI restriction site. D, The cfo1 mutation caused a premature translation stop. E, Verification of the cfo1 mutation site in the F2 progeny of cfo1 and M32OE plants using a CAPS marker. F, Verification of the M32OE site in the F2 progeny of cfo1 and M32OE plants using specific markers. G, Flower phenotypes of wild-type, cfo1, M32OE, and F2 progeny with genotype M32OE_cfo1cfo1. WT, Wild type. Bars = 1,000 μm in G. [See online article for color version of this figure.]
In whorl 3, stamen number was reduced in 39% (n = 100) of the cfo1 flowers examined (Supplemental Fig. S1A). Stamen-pistil chimeras were visualized in 16% (n = 100) of the cfo1 flowers examined (Fig. 2E). These stamen-pistil chimeras generally consisted of a basal thick filament, middle vestigial anthers, and apical styles plus stigmas (Fig. 2, E-2–E-4), whereas the wild-type stamen was composed of a thin filament and an anther with four sacs (Fig. 2, A-1, A-3, and A-6).
Figure 6.
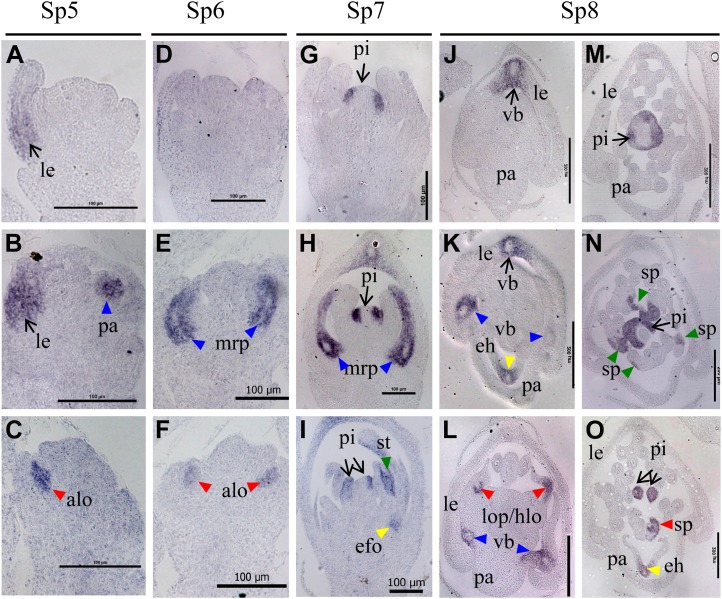
Expression of DL during early stages of development of cfo1 and wild-type flowers. Expression in flowers at the Sp5 (A–C), Sp6 (D–F), Sp7 (G–I), and Sp8 (J–O) stages in wild-type flowers (A, D, G, J, and M) and cfo1 flowers (B, C, E, F, H, I, K, L, N, and O) is shown. The domain of DL expression in the lemma and pistil is indicated by arrows, whereas its ectopic expression in other organs is indicated by triangles. Blue, red, yellow, and green triangles indicate the expression domain in abnormal palea/mrp, lodicule, efo, and stamens, respectively. alo, Abnormal lodicule; eh, ectopic hull-like organ; hlo, hull-lodicule chimera; le, lemma; lo, lodicule; lop, lodicule-pistil chimera; pa, palea; pi, pistil; sp, stamen-pistil chimera; vb, vascular bundle. Bars = 100 μm in A to I and 200 μm in J to O.
In whorl 4, the wild-type pistil contained one ovary and bore a double-plumed stigma on a short style (Fig. 2, A-1, A-4, and A-5). In cfo1 flowers, the pistil usually appeared morphologically normal and fertile. However, one or occasionally more additional pistils were produced at the side of the normal pistil in half of the cfo1 flowers (Fig. 2, F-1–F-5; Supplemental Fig. S1A). The extra pistils appeared to have a normal ovary and normal stigmas, but they might fail to develop normal ovules and thus be sterile (Fig. 2F-5), given that no multiseeded spikelets were observed among the mutants.
To determine further the identities of these mutated organs, the expression levels of known floral organ development genes were determined in different organs using qRT-PCR (Supplemental Fig. S2). In cfo1 lodicules, the A-class genes OsMADS14 and OsMADS15, C-class genes OsMADS3 and OsMADS58, D-class gene OsMADS13, E-class gene OsMADS1, and DL all showed ectopic expression. The B-class gene OsMADS2 and the AGL6-like gene OsMADS6 were expressed at lower levels than in wild-type lodicules, whereas the expression levels of two other B-class genes, OsMADS4 and OsMADS16, were not obviously affected. In cfo1 stamens, the OsMADS58, OsMADS13, DL, and OsMADS6 genes showed ectopic expression, and the expression of OsMADS2, OsMADS4, and OsMADS3 was up-regulated. The expression levels of other genes were not visibly altered. In the efo of cfo1 flowers, all detected genes except OsMADS58 showed a high level of expression. In cfo1 pistils, the expression of most detected genes other than OsMADS2 and DL was unaffected. These results suggest that in cfo1 flowers, the lodicule acquired hull- and pistil-like identities, the efo acquired other types of identities, and the stamens acquired pistil-like identities. These results are consistent with the results of the phenotype analysis.
Abnormal Early Flower Development in cfo1
Scanning electron microscopy analysis of floral organ morphogenesis during early developmental stages was undertaken. No significant morphological differences were observed during the spikelet 5 stage (Sp5; formation of lodicule primordia; Fig. 3, A and E). During the Sp6 stage, the wild-type flower formed six hemispherical stamen primordia (Fig. 3, B and C). The stamen primordia were not observed in cfo1 flowers (Fig. 3, F and G). Early in Sp6, no inner floral organ primordia appeared in cfo1 flowers (Fig. 3F). Later in Sp6, two zygomorphic crescent-shaped primordia began to develop on the lemma side, where wild-type lodicule primordia develop (Fig. 3G). The efo primordia were formed on the palea side of cfo1 flowers during this stage. At the Sp7 stage, with formation of the pistil, wild-type stamen primordia differentiated into quadrangular anthers and filaments (Fig. 3D), whereas cfo1 flowers displayed expanding lodicule-like organ primordia in whorl 2 and hemispherical stamen primordia in whorl 3 (Fig. 3H). During Sp8, with formation of the ovules and pollen, inner floral organs underwent normal development in wild-type flowers (Fig. 3I), whereas cfo1 flowers displayed elongated lodicules (Fig. 3J) and efo in whorl 2 (Fig. 3, K and L) and irregular-shaped organs in whorl 3 (Fig. 3, J–K). All of these results indicated that floral organ identities were altered during early flower development in cfo1 mutants.
Effects of Mutation of CFO1 on Expression Patterns of Floral Organ Identity Genes during Early Stages of Flower Development
To determine further the identity of chimeric organs in cfo1, the expression patterns of known genes for floral organ identity were investigated during early stages of flower development (stages Sp5–Sp8) with in situ hybridization.
During stages Sp5 to Sp7, OsMADS6 expression was detected in precursor cells and primordia of the mrp, lodicule, and pistil of both wild-type (Fig. 4, A-1–A-3) and cfo1 (Fig. 4, B-1–B-3) flowers. However, abnormal expression was detected in transverse sections of Sp8 flowers. In the cfo1 mrp, the OsMADS6 expression signal decreased gradually from the lateral vascular bundle to the margin (blue triangles in Fig. 4, B-4 and B-5), whereas its expression was uniformly distributed in the wild-type mrp (arrows in Figure 4, A-4 and A-5). In potential cfo1 hull-pistil-lodicule chimeras, OsMADS6 was only expressed in both marginal tissues (potential lodicule tissues) and not in the central portion containing potential hull tissues (red triangles in Fig. 4B-4). In some efo, an obvious OsMADS6 expression signal was detected (yellow triangles in Fig. 4, B-4 and B-5).
Expression signals of the B-class genes OsMADS2, OsMADS4, and OsMADS16 were detected in the lodicules and stamens of both wild-type and cfo1 flowers during stages Sp5 to Sp8 (Fig. 4, B-1–H-5). However, differences between wild-type and cfo1 flowers were obvious. In longitudinal sections of Sp6- and Sp7-stage cfo1 flowers, OsMADS2 and OsMADS16 signals were absent in parts of the lodicule primordium (red triangles in Fig. 4, D-2, D-3, H-2, and H-3). In transverse sections of Sp6- and Sp7-stage cfo1 flowers, OsMADS2, OsMADS4, and OsMADS16 signals were absent in the central region of hull-pistil-lodicule chimeras of cfo1 flowers, whereas signals were detected in both sides of these organs (red triangles in Figure 4, D-4, F-4, and H-4). In addition, OsMADS16 was only expressed in potential stamen tissues of cfo1 stamen-pistil chimeras (green triangles in Fig. 4, H-3 and H-5). The OsMADS2 and OsMADS4 genes were expressed in efo that carried potential lodicule tissues (yellow triangles in Fig. 4, D-4 and F-4) but not in ectopic hull organs (yellow triangles in Fig. 4, D-5 and F-5).
After stage Sp5, strong OsMADS1 expression was detected in the lemma and bop, whereas a weak OsMADS1 expression signal was detected in the mrp and pistil in wild-type flowers (Fig. 5, A-1–A-5). However, in cfo1 flowers, OsMADS1 was expressed in the mrp (blue triangles in Fig. 5, B-2 –B-5) and ectopic hull-like organ (yellow triangle in Fig. 5B-4), with a similar level of signal detected in the lemma and bop (Fig. 5, B-1–B-5). In addition, a distinct expression signal was detected in the central regions (potential hull tissues) of hull-pistil-lodicule chimeras in cfo1 flowers during stage Sp8 (red triangle in Fig. 5B-5), whereas expression was difficult to detect in wild-type lodicules and pistils at the same stage (Fig. 5, A-4 and A-5).
Two C-class genes, OsMADS3 and OsMADS58, were expressed in incipient cells and primordia of the stamens and pistil in both wild-type and cfo1 flowers during stages Sp5 to Sp8 (Fig. 5, B-1–F-5). However, ectopic expression signals in abnormal lodicules were detected in longitudinal sections of Sp6- and Sp7-stage cfo1 flowers (red triangles in Fig. 5, D-2, F-2, and F-3). The ectopic expression signal was observed in partial regions (potential pistil tissues) of hull-pistil-lodicule chimeras of Sp8-stage cfo1 flowers (red triangles in Fig. 5, D-4, D-5, F-4, and F-5). The D-class gene OsMADS13 was expressed in ovule primordia in both wild-type and cfo1 flowers during stages Sp6 to Sp8 (Fig. 5, D-4, D-5, F-4, and F-5). However, an ectopic OsMADS13 expression signal was detected in lower layer cells (potential ovary tissues) of the central portion of hull-pistil-lodicule chimeras of cfo1 flowers at the Sp8 stage (Fig. 5, H-3 and H-4).
During stages Sp5 to Sp6, DL expression was only detected in lemma primordia (arrow in Fig. 6A) and not in pistil incipient cells or primordia in wild-type flowers (Fig. 6, A and D). Ectopic expression signals of DL were detected in abnormal mrp (blue triangles in Fig. 6, B and E) and abnormal lodicule primordia (red triangles in Fig. 6, C and F) in cfo1 flowers. During the Sp7 stage, DL was expressed in pistil primordia in wild-type (arrow in Fig. 6G) and cfo1 (arrows in Fig. 6, H and I) flowers. In addition, DL expression was detected in abnormal primordia of mrp, efo, and stamens (triangles in Fig. 6, H and I) in cfo1 flowers. During the Sp8 stage, DL expression was observed in the peripheral domain of the medial vascular bundle of the lemma (arrows in Figure 6, J and K) and the pistil (arrows in Fig. 6, M–O) in wild-type and cfo1 flowers. In addition, strong DL transcription signals in cfo1 flowers were detected in the peripheral domains of lateral vascular bundles of the palea (blue triangles in Fig. 6, K and L), the peripheral domains of the vascular bundle of ectopic hull-like organs (yellow triangles in Fig. 6, K and O), central regions of hull-pistil-lodicule chimeras (red triangles in Fig. 6L), and stamen-pistil chimeras (green triangles in Fig. 6, N and O).
In the cfo1 mrp, the ectopic expression of DL, increased expression of OsMADS1, and decreased expression of OsMADS6 implied that the cfo1 mrp primordia developed a lemma identity at an early stage of flower development. In the cfo1 lodicule, the ectopic expression of DL, OsMADS1, and C/D-class genes, and the lack of expression of OsMADS6 and B-class genes, suggested that cfo1 lodicules acquired hull and pistil identities in the primordial development stage. The ectopic expression of DL and OsMADS13 in cfo1 stamen primordia implied that pistil-like tissues were produced in whorl 3 at an early stage of flower development. In particular, we noted that DL is expressed ectopically throughout all of the abnormal floral organs in early-stage cfo1 flowers during the initiation and formation of organ primordia. Thus, we speculated that the expression of DL is probably regulated negatively by CFO1.
CFO1 Encodes OsMADS32, a MADS-Box Protein
A map-based cloning strategy was used to isolate the CFO1 gene. Genetic analysis demonstrated that the cfo1 trait was controlled by a single recessive gene (Supplemental Table S1). The cfo1 locus was mapped on the short arm of chromosome 1 within an approximately 645-kb region between the simple sequence repeat markers RM1152 and RM128 (Fig. 7, A and B). Fortunately, a gene that encodes a 196-residue MADS box transcription factor, OsMADS32, occurs in this region. Sequence comparison revealed that OsMADS32 in the cfo1 mutant was missing a single nucleotide (T) at position 214 of the open reading frame, which caused a premature translation stop (Fig. 7, C and D). An RsaI restriction site in the wild-type allele was abolished in the cfo1 mutant allele because of this single-nucleotide deletion (Fig. 7C). A cleavage amplification polymorphism site (CAPS) marker was designed to distinguish the wild-type and mutant alleles in the segregating population. Results of RsaI digestion of PCR products suggested significant cosegregation of the CAPS marker with the cfo1 allele (Supplemental Fig. S3).
Complement testing was conducted as follows. First, OsMADS32 overexpression plants (M32OE) that expressed OsMADS32 under the control of the cauliflower mosaic virus 35S promoter in the japonica rice ‘Zhonghua11’ were generated. These plants displayed no defects except reduced palea size (Supplemental Fig. S4). Next, we crossed the cfo1 mutant with M32OE homozygotes. Six F1 individuals with the M32OE phenotype were planted to generate F2 populations, and a 3:12:1 (normal phenotype:M32OE phenotype:cfo1 mutant phenotype) ratio was observed in these F2 plants (Supplemental Table S2). The genotypes of 25 random F2 lines were identified using two specific molecular markers. Four M32OE_cfo1cfo1 lines showed normal floral organ development, except for reduced palea size (Fig. 7, E–G). This indicated that OsMADS32 expression in the cfo1 mutant background can recreate the cfo1 phenotype. In addition, we identified one T-DNA insertion line at the OsMADS32 locus (no. 05Z11AF36 [AF36]) from the Rice Mutant Database. In AF36, the T-DNA insertion resulted in severe DNA recombination of OsMADS32 genome sequences from −157 bp (upstream promoter) to 363 bp (the second exon; Fig. 7C; Supplemental Fig. S5, A–C; Zhang et al., 2006). This suggested that AF36 is a complete loss-of-function mutant of OsMADS32. Next, allelic verification between AF36 and cfo1 was performed. F1 individuals derived from the cross between cfo1 and AF36 were identified (Supplemental Fig. S5, D and E). The cfo1, AF36, and F1 progeny displayed very similar flower phenotypes (Supplemental Fig. S5F), which indicated that cfo1 and AF36 are allelic. Taken together, these results demonstrated that CFO1 and OsMADS32 are the same gene.
CFO1 Belongs to a Monocot-Specific Class
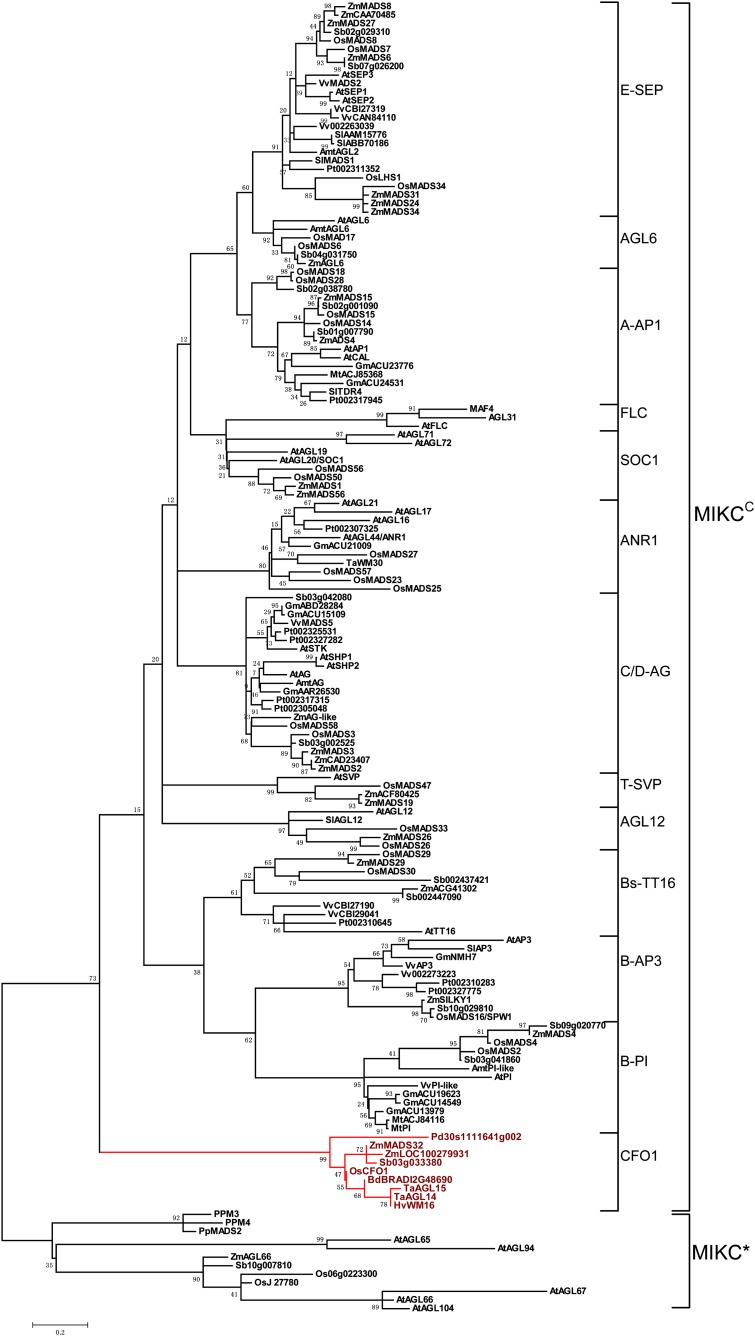
The MIKC-type genes are classified as a plant-specific subfamily of the MADS box gene family. This subfamily contains a well-conserved MADS (M) domain and three additional plant-specific domains: an intervening (I) domain, a keratin-like coiled-coil (K) domain, and a C-terminal (C) domain (Theissen et al., 1996; Alvarez-Buylla et al., 2000). The MIKC-type genes can be further divided into two subgroups, MIKCC and MIKC*, based on the intron-exon structure. Although OsMADS32/CFO1 is indicated to be a typical MIKCC-type protein, its phylogenetic relationships remain unclear (Ma et al., 1991; Nam et al., 2004; Arora et al., 2007).
We constructed a phylogenetic tree with 153 selected MIKC protein sequences from clubmosses, mosses, basal angiosperms, core eudicots, and monocots, including several genome-sequenced species, such as Arabidopsis, soybean (Glycine max), Medicago truncatula, poplar (Populus trichocarpa), Phoenix dactylifera, grape (Vitis vinifera), rice, Sorghum bicolor, and maize (Fig. 8). The 153 MIKCC-type genes were divided into 13 distinct classes: A or AP1-like class (A-AP1), B-AP3, B-PI, Bs-TT16, C/D-AG, E-SEP, F-SOC1, T-SVP, AGL6-like, AGL12-like, ANR1-like, FLC-like, and CFO1-like (Fig. 8). Interestingly, CFO1 and its orthologs from grasses and P. dactylifera constituted the earliest diverging CFO1-like class in the MIKCC family (Fig. 8). This class included Pd from P. dactylifera, TaAGL14 and TaAGL15 from wheat, ZmMADS32 and ZmLOC100279931 from maize, HvWM16 from barley, Sb03g03380 from S. bicolor, Bd12g48690 from Brachypodium distachyon, and CFO1 from rice (Fig. 8). These results suggest that the CFO1-like gene class is monocot specific.
Figure 8.
Evolutionary relationships among 153 MIKC genes. The phylogenetic tree was constructed using the maximum likelihood method based on the Jones-Taylor-Thornton matrix-based model. The tree was rooted using 12 MIKC*-type genes as the outgroup. Simplified class names were used (Nam et al., 2004) except for the CFO1 class. Species names are abbreviated as follows. Mosses: Physcomitrella patens (Pp); clubmosses: Lycopodium annotinum (La); basal angiosperm: Amborella trichopoda (Amt); eudicots: Arabidopsis (At), soybean (Gm), M. truncatula (Mt), poplar (Pt), grape (Vv); monocots: P. dactylifera (Pd), B. distachyon (Bd), wheat (Ta), rice (Os), barley (Hv), S. bicolor (Sb), maize (Zm). [See online article for color version of this figure.]
Sequence analysis indicated that CFO1-like proteins are well conserved, except that Sb03g03380 contains a distinct C domain (Supplemental Fig. S6). Although the M and K domains were conserved in other MIKCC-type proteins, CFO1-like proteins possessed some unique sites (Ser-6, Ser-19, Asp-26, and Phe-29) and motifs (DLLLLL42–47 and MTVDDG109–114) in these two domains (Supplemental Fig. S6). In addition, distinct differences were observed in the I and C domains across all classes (Supplemental Fig. S6).
Temporal and Spatial Expression Patterns of CFO1
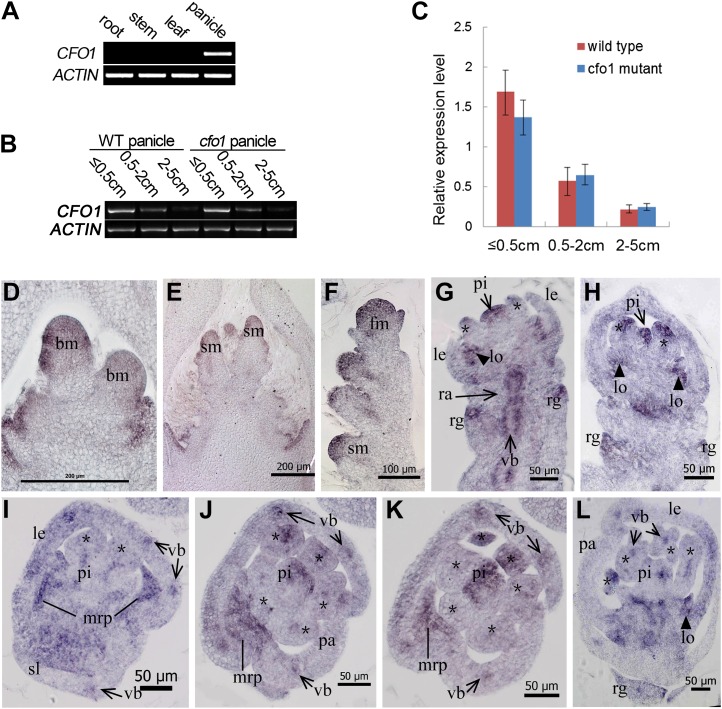
As already described, the expression of defective CFO1 disturbs floral organ identities in rice. To determine the pattern of CFO1 expression, semiquantitative reverse transcription (RT)-PCR and qRT-PCR analysis were conducted. No CFO1 transcripts were detectable in vegetative organs, namely roots, stems, or leaves (Fig. 8A). However, CFO1 transcription was observed in inflorescences at stages 1 to 7 (Fig. 9, B and C). After inflorescence stage 6, when the floral organs start to differentiate, CFO1 transcription levels decreased gradually (Fig. 9, B and C).
Figure 9.
Expression pattern of CFO1. A, CFO1 expression in different tissues as shown by RT-PCR. B, RT-PCR analysis of CFO1 in developing wild-type (WT) and cfo1 panicles at different stages. C, qRT-PCR analysis of CFO1 in developing wild-type and cfo1 panicles at different stages. D to L, In situ hybridization in wild-type panicles and flowers using a CFO1 antisense probe. Transcription of CFO1 was detected in the inflorescence meristem (D and E), spikelet (F), and florets (G–L) at an early development stage. Serial longitudinal sections of one floret are shown in G and H, and serial transverse sections of a different floret are shown in I to K. Asterisks in G to L indicate stamens. bm, Branch meristem; fm, flower meristem; le, lemma; lo, lodicule; pa, palea; pi, pistil; rg, rudimentary glume; sm, spikelet meristem; st, stamen; vb, vascular bundle. Bars = 200 μm in D and E, 100 μm in F, and 50 μm in G to L.
We investigated CFO1 expression in wild-type plants using in situ hybridization. The CFO1 gene was expressed initially in the primary and secondary branch meristems (Fig. 9D). Abundant CFO1 transcripts were detected subsequently in the meristems of spikelets and florets (Fig. 9, E and F). During stages Sp6 and Sp7, when the stamen primordia were initiated, analysis of serial longitudinal and transverse sections indicated that the expression signals of CFO1 were focused on several specific domains and organs (Fig. 9, G–K). First, CFO1 was expressed strongly in organ primordia, such as rudimentary glumes (Fig. 9, G and H), the mrp (lines in Fig. 9, I–K), lodicules (triangles in Fig. 9, G, H, and L), upper and central regions of stamens (asterisks in Fig. 9, I–K), and the upper region of the pistil (Fig. 9, G–K). Second, CFO1 was expressed in vascular bundles of floral organ primordia, such as the rachilla (arrows in Fig. 9G), sterile lemma (arrows in Fig. 6I), and lemma and palea (arrows in Fig. 9, I–K), which are normal in cfo1 flowers. During stage Sp8, CFO1 was expressed at high levels in the lodicules and rudimentary glumes and at low levels in the stamens and pistils (Fig. 9L). Although CFO1 function was not obvious in rudimentary glumes and vascular bundles, the data strongly suggested that CFO1 expression was associated with its function in floral organ specification.
Genetic Interactions between CFO1 and DL
The expression pattern analysis indicated that CFO1 negatively regulates DL expression. In order to examine the genetic interactions between CFO1 and DL, we generated cfo1+dl-sup1 double mutants and analyzed the phenotypes of their floral organs.
The dl-sup1 mutant is a loss-of-function mutant in which DL expression is not observed at any stage of flower development (Yamaguchi et al., 2004). The lemma and pistil in cfo1+dl-sup1 flowers displayed similar phenotypes to those of dl-sup1 flowers but not those of cfo1 flowers. The lemma is smaller than the palea (Fig. 10, A and B), and the pistil is transformed into stamen-like organs or repeat lodicule- and stamen-like structures in both cfo1+dl-sup1 and dl-sup1 flowers (Fig. 10, G and H), whereas the lemma and pistil identities in cfo1 flowers showed no obvious changes compared with the wild type (Figs. 1B and 2E-2). Therefore, the phenotypes of the lemmas and pistils in cfo1+dl-sup1 flowers were as expected. However, interesting changes were found in the mrp, lodicules, efo, and stamens of each cfo1+dl-sup1 flower. First, the palea showed a normal phenotype in cfo1+dl-sup1 flowers because the mrp possessed normal identity (Fig. 10, D and F), whereas a lemma-like identity was exhibited in the cfo1 mrp (Fig. 1, B and D). Second, the cfo1+dl-sup1 flowers had elongated lodicules but no hull- or pistil-like tissues (Fig. 10, H, J, and L), whereas cfo1 flowers showed hull- and pistil-like lodicules in whorl 2 (Fig. 2, B and C). Third, although efo were still produced in whorl 2 on the palea side of cfo1+dl-sup1 flowers, all of these appeared to be identical to lodicules (Fig. 10, J and M). In the cfo1 flowers, hull- and pistil-like tissues were also observed in efo, together with lodicule-like tissues (Fig. 2D). Finally, no stamen-pistil chimeras were found in whorl 3 of cfo1+dl-sup1 flowers, although they often occurred in cfo1 flowers (Fig. 2E). Therefore, these results indicated that DL inactivation reverses the defects seen in cfo1 floral organs. Given the ectopic expression of DL in cfo1 flowers, these results suggested that DL activation was an important cause of the defective mrp and ectopic formation of hull- and pistil-like tissues in whorls 2 and 3.
Figure 10.
Phenotypes of dl-sup1 and dl-sup1+cfo1 flowers at the heading stage. A and B, Spikelet of dl-sup1 (A) and dl-sup1+cfo1 (B). C and D, Spikelet of dl-sup1 (C) and dl-sup1+cfo1 (D); the lemma has been removed. E and F, Scanning electron micrographs of the mrp in dl-sup1 (E) and dl-sup1+cfo1 (F). G and H, Spikelet of dl-sup1 (G) and dl-sup1+cfo1 (H); the lemma and palea have been removed. In H, elongated lodicules and efo are indicated by triangles and arrows, respectively. I and J, Scanning electron microscopy of lodicule in a dl-sup1 flower (I) and elongated lodicule and efo in a dl-sup1+cfo1 flower (J). K to M, Epidermal cells of lodicule in a dl-sup1 flower (K), elongated lodicule (L), and efo in a dl-sup1+cfo1 flower (M). The cells in K are part of the lodicule illustrated in I highlighted in the red box. The cells in L and M are parts of the lodicule in J highlighted in the red and yellow boxes, respectively. N, Relative expression levels of floral organ identity genes in cfo1 and cfo1+dl-sup1 floral organs. Error bars indicate sd. elo, Elongated lodicule; le, lemma; lo, lodicule; pa, palea; wt, wild type. Bars = 1,000 μm in A to D, G, H, and J, 500 μm in E, F, and I, 200 μm in K, and 100 μm in L and M. [See online article for color version of this figure.]
The expression levels of known genes for floral organ development were analyzed in cfo1 and cfo1+dl-sup1 floral organs during the heading stage (Fig. 10, N–P). The OsMADS14 and OsMADS3 genes maintained expression levels comparable to those of the ectopically expressed genes in cfo1 lodicules. However, compared with cfo1 lodicules, the expression of OsMADS58 and OsMADS13 was drastically decreased in cfo1+dl-sup1 lodicules, whereas the expression of OsMADS2 and OsMADS16 was drastically increased. Expression of OsMADS1 was not detected in cfo1+dl-sup1 lodicules but was clearly evident in cfo1 lodicules. Compared with the ectopically expressed genes in the cfo1 efo, OsMADS14, OsMADS15, OsMADS13, OsMADS1, and OsMADS6 showed clearly decreased expression in the cfo1+dl-sup1 efo. Absent or diminished expression of these genes, which control hull and pistil development, may result in cfo1+dl-sup1 lodicules and efo that lack hull- and pistil-like identities. This conclusion is consistent with the results of the phenotype analysis.
We also investigated floral organ morphogenesis in cfo1+dl-sup1 and dl-sup1 flowers during early developmental stages. During stages Sp6 and Sp7, the dl-sup1 and wild-type flowers were characterized by six spherical stamen primordia (Fig. 3, C and D; Supplemental Fig. S7, B and C), whereas the initiation and development of stamen primordia in cfo1+dl-sup1 (Supplemental Fig. S7, D–F) occurred later than in dl-sup1 and the wild type. The stamen primordia in cfo1+dl-sup1 resembled those of cfo1 flowers (Fig. 3, F–H). In addition, similar to cfo1, the cfo1+dl-sup1 mutant exhibited expanding lodicule-like primordia and efo primordia in whorl 2 (Fig. 3G; Supplemental Fig. S7, D–F). Therefore, these results suggested that the occurrence of ectopic organs, lodicule elongation, and delay in the development of the stamen primordia were not dependent on ectopic DL activation.
DISCUSSION
CFO1 Is an Important Regulator of Floral Organ Identity in Rice
The rice floret consists of one lemma, one palea, two lodicules, six stamens, and one central pistil that contains one ovule. The lemma, palea, and lodicule are monocot-specific organs, whereas the stamen, pistil, and ovule are highly conserved across all angiosperms (Kellogg, 2001). In this study, map-based cloning and functional characterization demonstrated that CFO1, a monocot-specific MIKCC-type gene, is a key regulator in the specification of palea and lodicule identities in rice. Several important genes for floral organ identity have been characterized in rice. The SEP-like gene OsMADS1 is required for determination of the identities of the lemma and palea (Jeon et al., 2000; Agrawal et al., 2005; Prasad et al., 2005; Chen et al., 2006). The B-class gene OsMADS16, C-class genes OsMADS3 and OsMADS58, and D-class gene OsMADS13 play critical roles in the specification of stamen and pistil/ovule identities (Nagasawa et al., 2003; Yamaguchi et al., 2006; Dreni et al., 2007). Recently, the AGL6-like gene OsMADS6/MOSAIC FLORAL ORGANS1 was shown to specify palea, lodicule, and stamen identities (Ohmori et al., 2009; Li et al., 2010). All of these genes are MIKCC-type members of the MADS box gene family. The B-, C-, and D-class genes mainly specify conserved organs, whereas the OsMADS1, OsMADS6, and CFO1 genes determine grass-specific organs.
CFO1 Is Required for mrp Identity
In grass flowers, the palea and lemma are thought to have different origins (Kellogg, 2001). The palea is considered to be homologous to the prophyll (the first leaf produced by the axillary meristem), which is formed on a floret axis, whereas the lemma corresponds to the bract (the leaf that subtends the axillary meristem), which is formed on a spikelet axis (Kellogg, 2001; Ohmori et al., 2009). However, some evidence indicates that the rice palea might be derived from fusion of the mrp and bop. First, the cellular structure of the palea is very similar to that of the lemma in the body region but is distinct from that of the lemma in the marginal region (Fig. 1I; Prasad et al., 2005). Second, in plants with nonfunctional B-class genes, lodicules are transformed into organs that resemble the mrp but not the bop. Moreover, Arabidopsis plants with a mutated B class undergo homeotic transformation of petals (equivalent to lodicules) into sepals (Nagasawa et al., 2003; Yadav et al., 2007; Yao et al., 2008). This finding suggests that only the mrp, and not the whole palea, is equivalent to the sepal. In this study, the mrp of cfo1 developed a lemma- or bop-like identity, and CFO1 was expressed abundantly in the wild-type mrp. Recent studies have shown that OsMADS6 is also expressed predominantly in the mrp, and mutations in OsMADS6 lead to conversion of the mrp into lemma- or bop-like structures (Ohmori et al., 2009; Li et al., 2010). These results suggest that CFO1 and OsMADS6 confer important functions in the regulation of mrp identity but not bop identity. In accordance with these studies, we speculate that the mrp, bop, and lemma are equivalent to the sepal, prophyll, and bract, respectively.
In addition, in rice with the mutations depressed palea1 (dp1) and osmads15/dep, the bop is lost or depressed, whereas the mrp remains (Luo et al., 2005; Wang et al., 2010; Jin et al., 2011). Therefore, it is likely that CFO1 and OsMADS6 are required for the determination of mrp identity, and DP1 and OsMADS15/DEP might be involved in the regulation of bop identity. However, differences in the mrp phenotype occur between cfo1 and osmads6 flowers. In osmads6 flowers, the vascular bundle forms in the broadened mrp (Ohmori et al., 2009; Li et al., 2010). Analysis of osmads6 mutant flowers using RT-PCR indicated that DL is expressed ectopically in the lemma-like palea at the heading stage (Ohmori et al., 2009). Further study using in situ hybridization showed that DL is expressed in the midrib of palea in osmads6 mutant flowers (Li et al., 2011a). These results suggested that homeotic conversion of the palea into the lemma occurs in osmads6 mutant flowers. In contrast, the cfo1 mrp contained no vascular bundle, despite its lemma-like cellular structure. In addition, in cfo1 flowers, ectopic DL expression was detected in the peripheral domains of lateral vascular bundles of the palea, whereas normal DL expression was detected in the peripheral domains of medial vascular bundles in the wild-type lemma. These findings indicate that the palea is not equivalent to the lemma in cfo1 flowers. In addition, they imply that the cfo1 mrp defects arise because cells in the peripheral domains of lateral vascular bundles follow a similar cellular developmental program to cells in the peripheral domains of medial vascular bundles in the lemma.
Role of CFO1 in Lodicule Development
Lodicules are grass-specific organs that are considered to be homologous to dicot petals (Bommert et al., 2005; Whipple et al., 2007). The rice B-class genes OsMADS2, OsMADS4, and OsMADS16 determine lodicule identity (Nagasawa et al., 2003; Prasad and Vijayraghavan, 2003; Xiao et al., 2003; Yadav et al., 2007; Yao et al., 2008). In plants that have nonfunctional copies of these genes, the lodicules are elongated or transformed into mrp-like structures. The OsMADS6 gene also determines lodicule identity. Some osmads6 flowers displayed hull-like lodicules, in which OsMADS1, OsMADS14, and OsMADS15 were expressed ectopically at the heading stage (Ohmori et al., 2009; Li et al., 2010).
Our results here show that the lodicules are transformed into hull-lodicule or pistil-lodicule chimeras in the majority of cfo1 flowers. In addition, the expression of B-class genes and OsMADS6 was absent in partial domains of cfo1 lodicules, whereas OsMADS1, OsMADS13, and DL were expressed ectopically in similar domains at an early stage of flower development. Given the phenotype of hull-lodicule or pistil-lodicule chimeras, the domains are possibly hull-like or pistil-like tissues. Thus, CFO1 was shown to maintain proper lodicule identity by prevention of the establishment of hull- and pistil-like identities in lodicules, whereas OsMADS6 prevented hull-like tissue formation, and B-class genes prevented mrp-like tissue formation, in whorl 2.
Hull-like lodicules developed in both the osmads6 and cfo1 mutants, which suggested that OSMADS6 and CFO1 contribute to prevent the establishment of hull-like identities in lodicules. However, the osmads6 lodicule displayed complete hull-like histological identity in all of the cell layers (Ohmori et al., 2009). In contrast, the cfo1 lodicule showed hull-like identity only in the silicified upper epidermis and not in the other tissues, of which the inner cell layers often displayed pistil-like identity. Therefore, CFO1 and OsMADS6 may function in different regulatory pathways to control lodicule identity.
CFO1 Regulates Asymmetrical Development in Whorl 2
In this investigation, efo were often observed on the palea side in whorl 2 of most cfo1 flowers. In the same position, ectopic hull-like organs are observed in plants with loss of OsMADS6 gene function (Yamaguchi et al., 2006). Ectopic lodicules develop in plants with loss of OsMADS3 and OsMADS58 gene function (Yamaguchi et al., 2006; Ohmori et al., 2009; Li et al., 2010). In wild-type rice flowers, two lodicules are located interior to the lemma instead of the palea, which is indicative of asymmetrical lodicule development in rice. Although the identities of the ectopic organs differ among these mutants, the formation of these organs strongly implies that the four MADS-box genes play an important role in the repression of ectopic organ initiation in whorl 2 and maintenance of the asymmetrical development of lodicules.
Rice flowers might have evolved from an ancestral species with flowers that possessed three lodicules (Clifford, 1987; Grass Phylogeny Working Group, 2001; Yamaguchi et al., 2006). Grasses comprise the basal grasses and BEP and PACMAD clades (Grass Phylogeny Working Group II, 2012). Flowers with three lodicules occur among the basal grasses and many species of Bambusoideae, a subfamily in the BEP clade, whereas other species in the BEP and PACMAD clades develop flowers with two lodicules. Therefore, it is hypothesized that flowers with two lodicules evolved near the base of the BEP+PACMAD clade. It is possible that CFO1-like, OsMADS6-like, OsMADS3-like, and OsMADS58-like genes evolved new functions involved in the regulation of the asymmetrical development of lodicules near the base of the BEP+PACMAD clade, with a subsequent reversal in Bambusoideae species. However, it remains unclear whether the four MADS box genes function in the same regulatory pathway. In addition, it would be interesting to determine the correlation between the asymmetrical arrangement of lodicules and the functions of related genes such as CFO1-like, AGL6-like, and C-class genes in additional grass species.
CFO1 Negatively Regulates DL
The DL gene, which is expressed in whorl 4 and specifies pistil identity, antagonizes OsMADS16 gene function between whorls 3 and 4 (Nagasawa et al., 2003; Yamaguchi et al., 2004). In spw1 mutants, ectopic expression of DL causes complete transformation of stamens into pistils in whorl 3 (Nagasawa et al., 2003; Yamaguchi et al., 2004). Whorls 2 and 3 of cfo1 flowers contained various chimeras with fused pistil-like tissue and ectopic DL expression. These results imply that ectopic DL expression results in ectopic formation of pistil-like tissue.
The DL gene is also expressed in the peripheral domain of the medial vascular bundle of the lemma, but it is not clear whether DL controls lemma development (Fig. 6J; Yamaguchi et al., 2004). Recently, a defective lemma was reported in dl-sup6, an allelic mutant of DL (Li et al., 2011b). The dl-sup6 lemmas displayed alternate numbers of vascular tissues (either three or four vascular bundles), whereas the wild-type lemma contains a characteristic five vascular bundles (Li et al., 2011b). This result suggested that DL has a function in the specification of lemma vascular bundles and their peripheral tissues. In our study, abundant DL transcripts were detected in the peripheral domain of lateral vascular bundles of the cfo1 palea. On the basis of these results, it is reasonable to speculate that the ectopic DL activation in peripheral domains of lateral vascular bundles of the cfo1 palea resulted in the domains undergoing a similar development pattern with the domains in the lemma, thereby causing transformation of the mrp into a lemma-like tissue. Similarly, the formation of hull-like tissues in the lodicule and efo of the cfo1 flowers is probably caused by ectopic DL activation.
The mrp in the flower of the cfo1+dl-sup1 double mutant has a normal phenotype, with no ectopic hull- or pistil-like tissues observed in whorls 2 and 3. This further confirms that ectopic DL activation causes the defects observed in the mrp, lodicule, and stamen of the cfo1 flower. Thus, it is possible that CFO1 plays a pivotal role in the maintenance of floral organ identity through repression of DL transcription in the mrp, lodicule, and stamen (Fig. 11A). When CFO1 is dysfunctional, DL expression is extended, which results in a lemma-like mrp and chimeric organs in whorls 2 and 3 (Fig. 11B). We also observed that overexpression of CFO1 did not result in dl phenotypes in the leaf, lemma, and pistil. Moreover, CFO1 is expressed not only in the mrp in whorls 2 and 3 but also in the lemma and pistil. These results suggest that CFO1 restriction of DL expression should depend on interaction with other factors in the mrp in whorls 2 and 3. Therefore, characterization of these factors will improve our understanding of the regulation of floral organ identity in rice.
Figure 11.
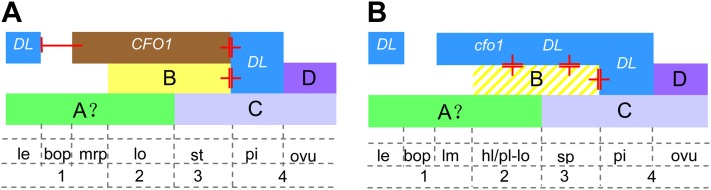
Roles of CFO1 in the specification of rice floral organ identity. A, In wild-type rice flowers, B-, C-, and D-class genes, CFO1 MADS box genes, and the DL gene determine floral organ identities. Although DL is mainly responsible for the specification of pistil identity, it also regulates lemma development. The CFO1 and B-class genes repress DL expression in whorls 2 and 3, whereas CFO1 also acts as a repressor of DL expression in the mrp. B, In cfo1 flowers, DL is expressed ectopically in the mrp, which results in mrp identity conversion resembling the lemma, and the ectopic expression of DL leads to chimeric organs in whorls 2 and 3. hl/pl-lo, Hull- or pistil-like lodicule; le, lemma; lm, lemma-like mrp; lo, lodicule; ovu, ovule; sp, stamen-pistil chimera; st, stamen.
CFO1 Is Also Required for Floral Meristem Development
In flowering plants, it is generally reported that many C- and E-class MADS box genes regulate the determination of the floral meristem in addition to floral organ identity. In general, mutation of these genes results in repeated organ occurrence in whorl 4 in severe cases or occasionally an increase in organ number (Mizukami and Ma, 1997; Jeon et al., 2000; Ditta et al., 2004; Ferrario et al., 2006; Yamaguchi et al., 2006; Ohmori et al., 2009; Li et al., 2010). In rice, loss of function of the C-class gene OsMADS58 causes a complete loss of floral meristem determinacy, resulting in the indeterminate development of flowers that consist of lodicules, stamens, and a carpel-like organ in whorl 4. These results suggest that OsMADS58 is crucial for the regulation of floral meristem determinacy in rice (Yamaguchi et al., 2006). Another C-class gene, OsMADS3, contributes weakly to this function, because multiple carpels developed in whorl 4 of osmads3 flowers. The AGL6-like gene OsMADS6 might be another major regulator of floral meristem determinacy. The osmads6 mutant displayed repetitive formation of abnormal carpels and occasional immature spikelets in whorl 4, which suggests that floral meristem determinacy was affected seriously (Ohmori et al., 2009; Li et al., 2010). The SEP-like gene LHS1/OsMADS1 also contributes weakly to the regulation of floral meristem determinacy. The lhs1 mutant showed additional carpels and florets occasionally (Jeon et al., 2000).
In cfo1 flowers, the initiation and development of floral organ primordia in whorls 3 and 4 were delayed, and the numbers of stamens and pistils were altered, which indicated that normal floral meristem development was disturbed. However, compared with the above-mentioned mutants, floral meristem determinacy was affected less by mutation of CFO1, because no repeated organs developed and the floral meristem was consumed by the pistils in cfo1 flowers. These results suggest that CFO1 is required for normal floral meristem development but contributes very weakly to the regulation of floral meristem determinacy.
CONCLUSION
In this study, we characterized the function of an ancient MIKC gene, CFO1/OsMADS32, in the regulation of floral organ identity in rice. The cfo1 mutant displayed a lemma-like mrp, chimeric floral organs, and efo. Almost all of the known floral organ identity genes showed defective expression in cfo1 flowers during the early and heading stages of flower development. In particular, we showed that the floral organ identity gene DL was expressed ectopically in all defective organs of cfo1 flowers. Double mutant analysis revealed that loss of DL function mitigated most of the defects in floral organs of cfo1 flowers. These results suggested that CFO1 plays a pivotal role in the maintenance of floral organ identity through the repression of DL transcription in the mrp, lodicule, and stamens. Our findings extend the current understanding of the origin and diversification of the MADS box genes that contribute to the development of grass-specific floral organs.
MATERIALS AND METHODS
Plant Materials
Rice (Oryza sativa) cfo1 is a spontaneous mutant obtained from the indica line 2B. Four F1 populations were produced by crossing the cfo1 mutant with four indica rice cultivars: Luhui17, R2727, Ce64, and Luhui602. The crosses were performed in Chongqing, China, during the summer of 2004. The F1 populations were raised in Hainan, China, during the autumn of 2004. F2 populations were planted in Chongqing, China, during the summer of 2005.
Map-Based Cloning of CFO1
Approximately 5,000 F2 plants were obtained from crosses between cfo1 and Luhui602, and 1,206 homozygotic mutants were selected for high-resolution mapping. Gene mapping was conducted using genetic markers obtained from publicly available rice databases, including Gramene (http://www.gramene.org) and RGP (http://rgp.dna.affrc.go.jp/publicdata/caps/index.html). Analysis of candidate genes was performed using a CAPS marker, sequencing, and complement testing. The sequences of primers used in the mapping and candidate gene analysis are listed in Supplemental Table S3.
Microscopy Analysis
Panicles were collected at different developmental stages and fixed in 50% ethanol, 0.9 m glacial acetic acid, and 3.7% formaldehyde overnight at 4°C, dehydrated with a graded ethanol series, infiltrated with xylene, and embedded in paraffin (Sigma). The 8-µm-thick sections were transferred onto poly-l-Lys-coated glass slides, deparaffinized in xylene, and dehydrated through an ethanol series. The sections were stained sequentially with 1% safranine (Amresco) and 1% Fast Green (Amresco), then dehydrated through an ethanol series, infiltrated with xylene, and finally mounted beneath a coverslip. Light microscopy was performed using a Nikon E600 microscope. For scanning electron microscopy, fresh samples were examined using a Hitachi S-3400 scanning electron microscope with a cool stage.
RT-PCR and qRT-PCR Analysis
Total RNA was isolated from various tissue samples using the RNeasy Plant Mini Kit (Waston). The first strand of cDNA was synthesized from 2 μg of total RNA using oligo(dT)18 primers in a 25-μL reaction volume using the SuperScript III Reverse Transcriptase Kit (Invitrogen). One-half microliter of the reverse-transcribed RNA was used as a PCR template with gene-specific primers (Supplemental Table S3). ACTIN was used as an endogenous control. For RT-PCR, the PCR products were separated in 2% agarose containing 1× Tris-acetate-EDTA buffer. The qRT-PCR analysis was performed with an ABI Prism 7000 Sequence Detection System and the SYBR Supermix Kit (Bio-Rad). At least three replicates were performed to produce the mean values of each expression level.
Vector Construction
To construct the OsMADS32 overexpression plasmids, the OsMADS32 cDNA that contained the whole open reading frame and partial untranslated region was amplified using the primers M32OE-F and M32OE-R. The cDNA were inserted into the expression cassette pCA-35S-NOS. The constructs were transferred into calli of japonica rice ‘Zhonghua11’ by Agrobacterium tumefaciens-mediated T-DNA transformation as described previously (Xiao et al., 2009). The primer sequences are listed in Supplemental Table S3.
In Situ Hybridization
The 419-bp gene-specific CFO1 probe was amplified with the primers M32H-F and M32H-R and labeled using the DIG RNA Labeling Kit (SP6/T7; Roche) in accordance with the vendor’s recommendations. Probes for the known floral organ genes were prepared using the same method. Pretreatment of sections, hybridization, and immunological detection were performed as described previously (Xiao et al., 2009). The primer sequences are listed in Supplemental Table S3.
Phylogenetic Analysis
A phylogenetic tree derived from 153 MIKC protein sequences was constructed using MEGA version 5 (Tamura et al., 2011). One class that consisted of 12 MIKC*-type genes was regarded as the outgroup. The tree was constructed using the maximum likelihood method based on the Jones-Taylor-Thornton matrix-based model with the lowest Bayesian Information Criterion scores (Jones et al., 1992; Tamura et al., 2011). Bootstrap support values for each node from 500 replicates are shown next to the branches (Felsenstein, 1985). Initial trees for the heuristic search were obtained automatically as follows. When the number of common sites was less than 100 or less than one-fourth of the total number of sites, the maximum parsimony method was used; otherwise, the Bio-neighbor-joining method with Markov Cluster distance matrix was used. A discrete γ-distribution was used to model evolutionary rate differences among sites (five categories [+G]; parameter = 1.9631). The rate variation model allowed for some sites to be evolutionarily invariable ([+I]; 5.8060% sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 153 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 109 positions in the final data set.
The GenBank accession numbers for the cDNA sequences of CFO1, DL, OsMADS1, OsMADS2, OsMADS3, OsMADS4, OsMADS6, OsMADS13, OsMADS14, OsMADS15, OsMADS16, and OsMADS58 are HQ711850, AB106553, NM_001055911, AK070894, L37528, NM_001062125, FJ666318, NM_001072917, NM_001057835, NM_001065255, NM_001065095, and NM_001061424, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Statistical analysis of cfo1 flowers.
Supplemental Figure S2. qRT-PCR analysis of floral organ identity gene expression in the inner floral organs of wild-type and cfo1 flowers.
Supplemental Figure S3. Results of a CAPS assay of 32 recombinants from 1,206 F2 individuals.
Supplemental Figure S4. Analysis of OsMADS32 overexpression plants.
Supplemental Figure S5. Molecular characterization of T-DNA insertion line AF36 and verification of allelism with cfo1.
Supplemental Figure S6. Protein sequence alignment of MIKCC genes in grasses.
Supplemental Figure S7. Scanning electron microscopy analysis of dl-sup1 and dl-sup1+cfo1 flowers at early stages.
Supplemental Table S1. Segregation of the cfo1 allele in four F2 populations.
Supplemental Table S2. Segregation of F1 progeny with the M32OEm32oeCFO1cfo1 genotype in F2 populations.
Supplemental Table S3. Primers used in the study.
Acknowledgments
We gratefully acknowledge Dr. Changyin Wu from the National Center of Plant Gene Research (Wuhan, China) for providing the OsMADS32 T-DNA insertion lines as well as Dr. Yasuo Nagato of the University of Tokyo and Prof. Lihuang Zhu from the Institute of Genetics and Developmental Biology (Beijing, China) for providing the dl-sup1 mutants. We also thank Prof. Yan Pei from Southwest University (Chongqing, China) for providing the rice transformation vectors.
Glossary
- mrp
marginal region of the palea
- bp
body of the palea
- qRT
quantitative reverse transcription
- RT
reverse transcription
- efo
ectopic floral organs
- CAPS
cleavage amplification polymorphism site
- bop
body of the palea
References
- Agrawal GK, Abe K, Yamazaki M, Miyao A, Hirochika H. (2005) Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene. Plant Mol Biol 59: 125–135 [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla ER, Pelaz S, Liljegren SJ, Gold SE, Burgeff C, Ditta GS, Ribas de Pouplana L, Martínez-Castilla L, Yanofsky MF. (2000) An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc Natl Acad Sci USA 97: 5328–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S. (2007) MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommert P, Satoh-Nagasawa N, Jackson D, Hirano HY. (2005) Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol 46: 69–78 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Drews GN, Meyerowitz EM. (1991) Expression of the Arabidopsis floral homeotic gene AGAMOUS is restricted to specific cell types late in flower development. Plant Cell 3: 749–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR. (1999) CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126: 2387–2396 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. (1989) Genes directing flower development in Arabidopsis. Plant Cell 1: 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZX, Wu JG, Ding WN, Chen HM, Wu P, Shi CH. (2006) Morphogenesis and molecular basis on naked seed rice, a novel homeotic mutation of OsMADS1 regulating transcript level of AP3 homologue in rice. Planta 223: 882–890 [DOI] [PubMed] [Google Scholar]
- Clifford HT. (1987) Spikelet and floral morphology. In TR Soderstrom, KW Hilu, CS Campbell, ME Barkworth, eds, Grass Systematics and Evolution. Smithsonian Institution Press, Washington, DC, pp 21–30.
- Coen ES, Meyerowitz EM. (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF. (2004) The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 14: 1935–1940 [DOI] [PubMed] [Google Scholar]
- Dreni L, Jacchia S, Fornara F, Fornari M, Ouwerkerk PB, An G, Colombo L, Kater MM. (2007) The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J 52: 690–699 [DOI] [PubMed] [Google Scholar]
- Drews GN, Bowman JL, Meyerowitz EM. (1991) Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65: 991–1002 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Ferrario S, Shchennikova AV, Franken J, Immink RGH, Angenent GC. (2006) Control of floral meristem determinacy in petunia by MADS-box transcription factors. Plant Physiol 140: 890–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, Marziani G, Mizzi L, Kater M, Colombo L. (2003) MADS-box genes controlling flower development in rice. Plant Biol 5: 16–22 [Google Scholar]
- Goto K, Meyerowitz EM. (1994) Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev 8: 1548–1560 [DOI] [PubMed] [Google Scholar]
- Grass Phylogeny Working Group (2001) Phylogeny and subfamilial classification of the grasses (Poaceae). Ann Mo Bot Gard 88: 373–457 [Google Scholar]
- Grass Phylogeny Working Group II (2012) New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytol 193: 304–312 [DOI] [PubMed] [Google Scholar]
- Ikeda K, Sunohara H, Nagato Y. (2004) Developmental course of inflorescence and spikelet in rice. Breed Sci 54: 147–156 [Google Scholar]
- Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y. (2005) Rice plant development: from zygote to spikelet. Plant Cell Physiol 46: 23–47 [DOI] [PubMed] [Google Scholar]
- Jack T, Fox GL, Meyerowitz EM. (1994) Arabidopsis homeotic gene APETALA3 ectopic expression: transcriptional and posttranscriptional regulation determine floral organ identity. Cell 76: 703–716 [DOI] [PubMed] [Google Scholar]
- Jeon JS, Jang S, Lee S, Nam J, Kim C, Lee SH, Chung YY, Kim SR, Lee YH, Cho YG, et al. (2000) leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Luo Q, Tong HN, Wang AJ, Cheng ZJ, Tang JF, Li DY, Zhao XF, Li XB, Wan JM, et al. (2011) An AT-hook gene is required for palea formation and floral organ number control in rice. Dev Biol 359: 277–288 [DOI] [PubMed] [Google Scholar]
- Jofuku KD, den Boer BGW, Van Montagu M, Okamuro JK. (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6: 1211–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275–282 [DOI] [PubMed] [Google Scholar]
- Kater MM, Dreni L, Colombo L. (2006) Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J Exp Bot 57: 3433–3444 [DOI] [PubMed] [Google Scholar]
- Kellogg EA. (2001) Evolutionary history of the grasses. Plant Physiol 125: 1198–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Maekawa M, Miyao A, Hirochika H, Kyozuka J. (2010) PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol 51: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HF, Liang WQ, Hu Y, Zhu L, Yin CS, Xu J, Dreni L, Kater MM, Zhang DB. (2011a) Rice MADS6 interacts with the floral homeotic genes SUPERWOMAN1, MADS3, MADS58, MADS13, and DROOPING LEAF in specifying floral organ identities and meristem fate. Plant Cell 23: 2536–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HF, Liang WQ, Jia RD, Yin CS, Zong J, Kong HZ, Zhang DB. (2010) The AGL6-like gene OsMADS6 regulates floral organ and meristem identities in rice. Cell Res 20: 299–313 [DOI] [PubMed] [Google Scholar]
- Li HF, Liang WQ, Yin CS, Zhu L, Zhang DB. (2011b) Genetic interaction of OsMADS3, DROOPING LEAF, and OsMADS13 in specifying rice floral organ identities and meristem determinacy. Plant Physiol 156: 263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q, Zhou KD, Zhao XF, Zeng QC, Xia HG, Zhai WX, Xu JC, Wu XJ, Yang HS, Zhu LH. (2005) Identification and fine mapping of a mutant gene for palealess spikelet in rice. Planta 221: 222–230 [DOI] [PubMed] [Google Scholar]
- Ma H, Yanofsky MF, Meyerowitz EM. (1991) AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev 5: 484–495 [DOI] [PubMed] [Google Scholar]
- Malcomber ST, Kellogg EA. (2004) Heterogeneous expression patterns and separate roles of the SEPALLATA gene LEAFY HULL STERILE1 in grasses. Plant Cell 16: 1692–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcomber ST, Kellogg EA. (2005) SEPALLATA gene diversification: brave new whorls. Trends Plant Sci 10: 427–435 [DOI] [PubMed] [Google Scholar]
- Malcomber ST, Preston JC, Reinheimer R, Kossuth J, Kellogg EA. (2006) Developmental gene evolution and the origin of grass inflorescence diversity. Adv Bot Res 44: 425–481 [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360: 273–277 [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Ma H. (1997) Determination of Arabidopsis floral meristem identity by AGAMOUS. Plant Cell 9: 393–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa N, Miyoshi M, Sano Y, Satoh H, Hirano H, Sakai H, Nagato Y. (2003) SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130: 705–718 [DOI] [PubMed] [Google Scholar]
- Nam J, Kim J, Lee S, An GH, Ma H, Nei MS. (2004) Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc Natl Acad Sci USA 101: 1910–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori S, Kimizu M, Sugita M, Miyao A, Hirochika H, Uchida E, Nagato Y, Yoshida H. (2009) MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 21: 3008–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203 [DOI] [PubMed] [Google Scholar]
- Pinyopich A, Ditta GS, Savidge B, Liljegren SJ, Baumann E, Wisman E, Yanofsky MF. (2003) Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424: 85–88 [DOI] [PubMed] [Google Scholar]
- Prasad K, Parameswaran S, Vijayraghavan U. (2005) OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J 43: 915–928 [DOI] [PubMed] [Google Scholar]
- Prasad K, Vijayraghavan U. (2003) Double-stranded RNA interference of a rice PI/GLO paralog, OsMADS2, uncovers its second-whorl-specific function in floral organ patterning. Genetics 165: 2301–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G, Kim JT, Saedler H. (1996) Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J Mol Evol 43: 484–516 [DOI] [PubMed] [Google Scholar]
- Theissen G, Saedler H. (2001) Plant biology: floral quartets. Nature 409: 469–471 [DOI] [PubMed] [Google Scholar]
- Wang K, Tang D, Hong L, Xu W, Huang J, Li M, Gu M, Xue Y, Cheng Z. (2010) DEP and AFO regulate reproductive habit in rice. PLoS Genet 6: e1000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Meyerowitz EM. (1994) The ABCs of floral homeotic genes. Cell 78: 203–209 [DOI] [PubMed] [Google Scholar]
- Whipple CJ, Zanis MJ, Kellogg EA, Schmidt RJ. (2007) Conservation of B class gene expression in the second whorl of a basal grass and outgroups links the origin of lodicules and petals. Proc Natl Acad Sci USA 104: 1081–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Tang JF, Li YF, Wang WM, Li XB, Jin L, Xie R, Luo HF, Zhao XF, Meng Z, et al. (2009) STAMENLESS 1, encoding a single C2H2 zinc finger protein, regulates floral organ identity in rice. Plant J 59: 789–801 [DOI] [PubMed] [Google Scholar]
- Xiao H, Wang Y, Liu DF, Wang WM, Li XB, Zhao XF, Xu JC, Zhai WX, Zhu LH. (2003) Functional analysis of the rice AP3 homologue OsMADS16 by RNA interference. Plant Mol Biol 52: 957–966 [DOI] [PubMed] [Google Scholar]
- Yadav SR, Prasad K, Vijayraghavan U. (2007) Divergent regulatory OsMADS2 functions control size, shape and differentiation of the highly derived rice floret second-whorl organ. Genetics 176: 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Lee DY, Miyao A, Hirochika H, An GH, Hirano HY. (2006) Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell 18: 15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Nagasawa N, Kawasaki S, Matsuoka M, Nagato Y, Hirano HY. (2004) The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16: 500–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao SG, Ohmori S, Kimizu M, Yoshida H. (2008) Unequal genetic redundancy of rice PISTILLATA orthologs, OsMADS2 and OsMADS4, in lodicule and stamen development. Plant Cell Physiol 49: 853–857 [DOI] [PubMed] [Google Scholar]
- Zhang J, Li C, Wu C, Xiong L, Chen G, Zhang Q, Wang S. (2006) RMD: a rice mutant database for functional analysis of the rice genome. Nucleic Acids Res 34: D745–D748 [DOI] [PMC free article] [PubMed] [Google Scholar]