Abstract
The biosynthesis of gibberellic acid (GA3) by the fungus Fusarium fujikuroi is catalyzed by seven enzymes encoded in a gene cluster. While four of these enzymes are characterized as cytochrome P450 monooxygenases, the nature of a fifth oxidase, GA4 desaturase (DES), is unknown. DES converts GA4 to GA7 by the formation of a carbon-1,2 double bond in the penultimate step of the pathway. Here, we show by expression of the des complementary DNA in Escherichia coli that DES has the characteristics of a 2-oxoglutarate-dependent dioxygenase. Although it has low amino acid sequence homology with known 2-oxoglutarate-dependent dioxygenases, putative iron- and 2-oxoglutarate-binding residues, typical of such enzymes, are apparent in its primary sequence. A survey of sequence databases revealed that homologs of DES are widespread in the ascomycetes, although in most cases the homologs must participate in non-gibberellin (GA) pathways. Expression of des from the cauliflower mosaic virus 35S promoter in the plant species Solanum nigrum, Solanum dulcamara, and Nicotiana sylvestris resulted in substantial growth stimulation, with a 3-fold increase in height in S. dulcamara compared with controls. In S. nigrum, the height increase was accompanied by a 20-fold higher concentration of GA3 in the growing shoots than in controls, although GA1 content was reduced. Expression of des was also shown to partially restore growth in plants dwarfed by ectopic expression of a GA 2-oxidase (GA-deactivating) gene, consistent with GA3 being protected from 2-oxidation. Thus, des has the potential to enable substantial growth increases, with practical implications, for example, in biomass production.
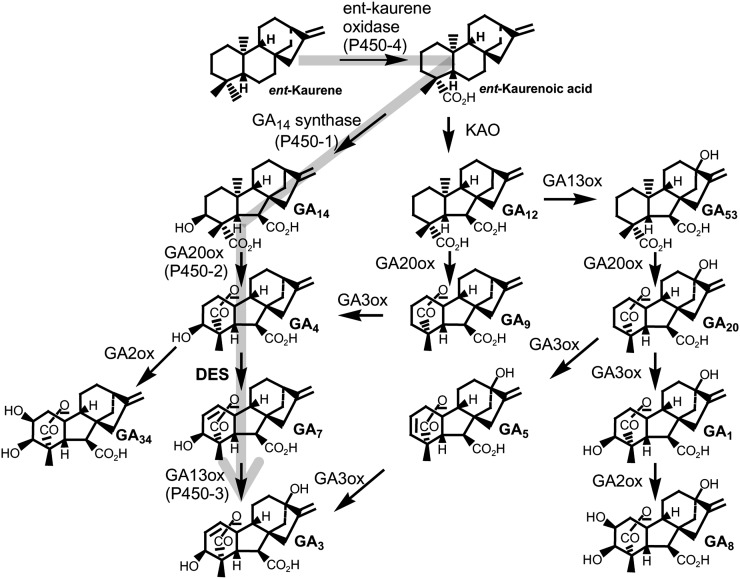
The GAs are a class of diterpenoid hormones that regulate many aspects of growth and development in plants, including stem extension (Thomas and Hedden, 2006). Despite being ubiquitous in higher plants, they were first discovered as secondary metabolites of the plant pathogenic fungus Gibberella fujikuroi, the causative agent of the bakanae disease of rice (Oryza sativa; Phinney, 1983). This fungus is now known to comprise a group of reproductively isolated species or mating populations, the rice pathogen belonging to mating group C and assigned the name Fusarium fujikuroi (Leslie and Summerell, 2006; Kvas et al., 2009). Details of the GA biosynthetic pathways in both plants and the fungus are known in considerable detail and have revealed that, although they give rise to common metabolites, the pathways utilize different types of enzymes for several steps and appear to have evolved independently (Hedden et al., 2001; Bömke and Tudzynski, 2009).
Higher plants differ from the GA-producing fungi by possessing the means for GA inactivation, which is necessary to allow precise regulation of their GA concentration. In contrast, the fungi are not dependent on GAs for their development but produce and secrete large quantities of the compounds to modify the behavior of their hosts. It has been shown that GAs interfere with plant defense by suppressing jasmonate signaling and may thus compromise the host’s ability to evade fungal infection (Navarro et al., 2008; Hou et al., 2010). An apparent ubiquitous inactivation mechanism involves 2β-hydroxylation (Thomas et al., 1999), the effect of which reduces binding of the GA within the active site of the GID1 receptor (Murase et al., 2008). However, GAs such as GA3 and GA5, which are unsaturated on C-2, are protected from 2β-hydroxylation and, as a consequence, would be expected to be turned over more slowly than their saturated analogs (King et al., 2008). In accordance with the requirement to regulate GA content, shoots of higher plants contain relatively little 1,2-unsaturated GAs, although developing seeds of some species contain substantial quantities. They are produced in a two-step reaction via a 2,3-dehydro intermediate, which is then hydroxylated on C-3β with rearrangement of the double bond from C-2,3 to C-1,2 (Albone et al., 1990). The reactions are catalyzed by GA 3-oxidase-type enzymes, with a single enzyme catalyzing both reactions in cereal shoots to produce GA3 from GA20 as a minor by-product of GA1 biosynthesis (Itoh et al., 2001; Appleford et al., 2006; Fig. 1). In developing seeds of Marah macrocarpus, which contain high concentrations of the 1,2-unsaturated GA, GA7, the formation of this GA from GA9 requires the activities of two functionally different GA 3-oxidases acting sequentially (Ward et al., 2010). However, direct formation of GA7 from GA4, such as occurs in F. fujikuroi, is not usual in higher plants.
Figure 1.
The GA biosynthetic pathway in plants and F. fujikuroi. The fungal pathway to GA3 is indicated by the thick gray arrow. DES catalyzes the conversion of GA4 to GA7.
While the late stages of GA biosynthesis in higher plants, including desaturation when it occurs and 2β-hydroxylation, are catalyzed by 2-oxoglutarate-dependent dioxygenases (ODDs), these enzymes have not been shown to be involved in GA biosynthesis in fungi. F. fujikuroi contains a cluster of seven genes for GA biosynthesis, including a geranylgeranyl diphosphate synthase that is specific to the GA pathway and a bifunctional terpene cyclase that converts geranylgeranyl diphosphate to ent-kaurene in two steps via ent-copalyl diphosphate (for review, see Hedden et al. [2001]; Bömke and Tudzynski [2009]). The formation of GA3 from ent-kaurene requires the activity of five oxidases (Fig. 1), four of which are cytochrome P450 monooxygenases: P450-4 (ent-kaurene oxidase) oxidizes ent-kaurene to ent-kaurenoic acid (Tudzynski et al., 2001), which is converted to GA14 by P450-1 (GA14 synthase; Rojas et al., 2001); P450-2 functions as a GA 20-oxidase, converting GA14 to GA4 (Tudzynski et al., 2002), while, in the final step of the pathway, P450-3 13-hydroxylates GA7 to form GA13 (Tudzynski et al., 2003). However, the nature of the desaturase (DES), which converts GA4 to GA7 (Fig. 1), is unknown. When first described, it was found to have closest, albeit weak, homology to a component of the 7α-cephem-methoxylase from Nocardia lactamdurans, giving little indication of its mechanism (Tudzynski et al., 2003). Besides F. fujikuroi, several other ascomycetes, including Sphaceloma manihoticola (Bömke et al., 2008), Phaeosphaeria spp. (Kawaide, 2006), and two other species of the G. fujikuroi species complex, Fusarium konzum (Malonek et al., 2005) and Fusarium sacchari (Troncoso et al., 2010), have been shown to synthesize GAs, although the first two species do not carry out the desaturation step and do not contain a desaturase gene.
The promotion of vegetative growth offers potential benefits, for example, in biomass production (Demura and Ye, 2010). In order to test the hypothesis that growth could be stimulated by increasing the shoot concentrations of GAs that are unsaturated on C-2 and therefore resistant to 2β-hydroxylation, we introduced the fungal desaturase gene into plants. The feasibility of this approach was reinforced by the demonstration that DES has the characteristics of an ODD and, therefore, would be expected to function in higher plants.
RESULTS
Characterization of GA4 Desaturase
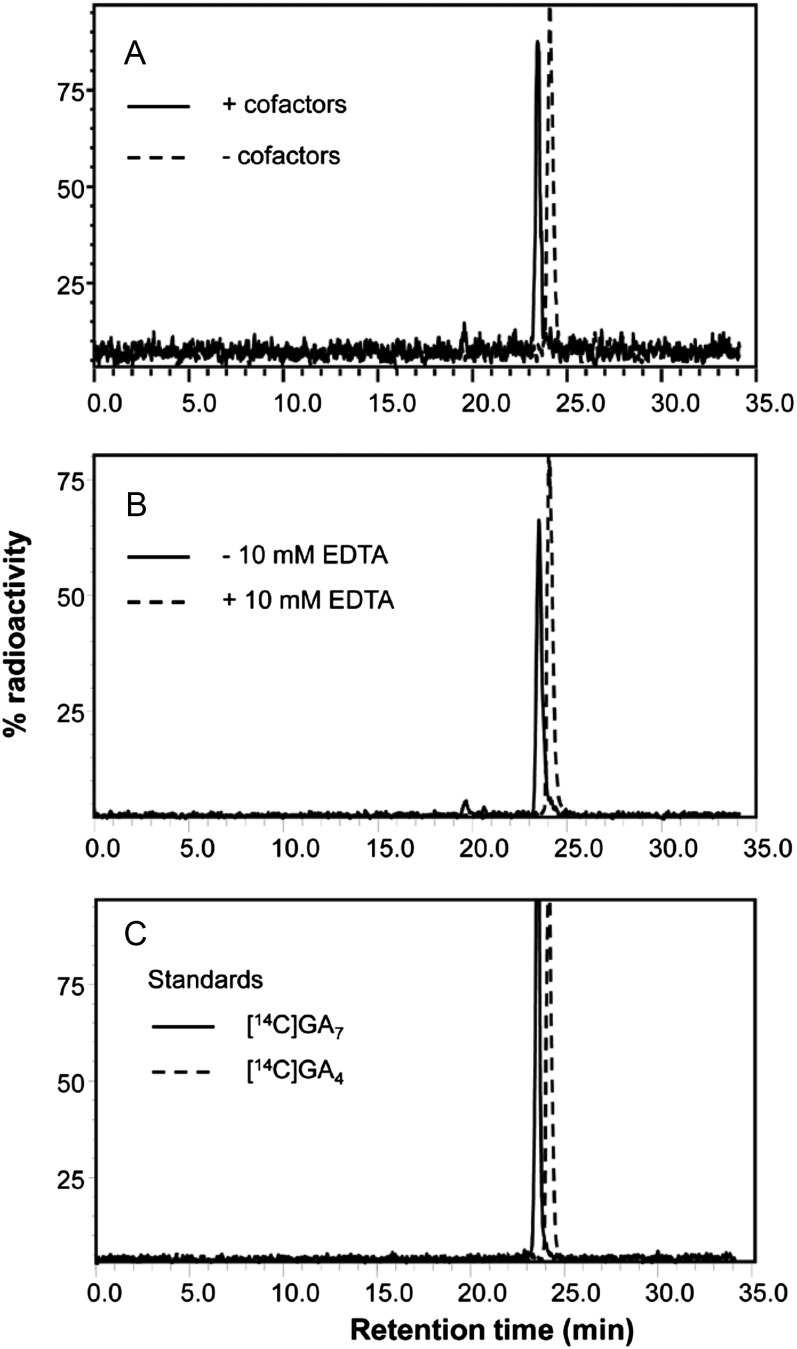
Although the derived amino acid sequence of DES has little overall homology with known ODDs (Tudzynski et al., 2003), in common with these enzymes it contains an HxD motif that may be involved in binding Fe2+ and an RxS motif that has been shown to bind 2-oxoglutarate in anthocyanidin synthase (Wilmouth et al., 2002; Fig. 2). Therefore, we decided to determine whether DES functions as an ODD; for this purpose, Ffdes complementary DNA (cDNA) was cloned into the pET32a vector for expression in Escherichia coli. After transformation of E. coli strain BL21 with the plasmid harboring Ffdes and induction of expression, bacterial lysates were incubated with [14C]GA4 and ODD cofactors (MacMillan et al., 1997). Separation of the products by HPLC radiochromatography followed by identification of the product by gas chromatography-mass spectrometry (GC-MS) demonstrated complete conversion to [14C]GA7 (Fig. 3, A and C; Supplemental Fig. S1). However, conversion of [14C]GA4 to [14C]GA7 was also obtained in the absence of added cofactors, providing no information on the nature of the enzyme. It is possible that the bacterial lysate contained sufficient concentrations of the cosubstrates and cofactors (2-oxoglutarate, Fe2+, and ascorbate) necessary to support ODD activity. Therefore, an aliquot of the lysate was filtered through a Sephadex G-50 NICK column to remove low-Mr components. The gel-filtered lysate was inactive in the absence of the cofactors but fully active when they were added to the incubation (Fig. 3A). The cofactor requirements for enzyme activity in the filtered lysate were then determined (Table I). Full activity (i.e. complete conversion of [14C]GA4 to [14C]GA7 when incubated for 5 h at 30°C) was obtained in the presence of 2-oxoglutarate and ascorbate, regardless of whether FeSO4 was present, whereas 2-oxoglutarate alone supported 19% of full activity. Addition of FeSO4 or ascorbate alone failed to support any enzyme activity. These cofactor requirements are indicative of ODDs (Prescott and John, 1996) and also suggest that Fe2+ is retained in the enzyme active site. The presence of 10 mm EDTA to remove bound Fe2+ in incubations with 2-oxoglutarate and ascorbate resulted in complete loss of enzyme activity (Fig. 3B).
Figure 2.
Amino acid sequences of anthocyanidin synthase and DES, indicating in boldface the residues that are involved in iron and 2-oxoglutarate binding in anthocyanidin synthase and the putative binding residues in DES. The iron-binding residues are marked underneath with asterisks and those binding 2-oxoglutarate with carets.
Figure 3.
HPLC radiochromatograms of products from incubations of [14C]GA4 with DES. A, Gel-filtered bacterial lysate incubated without cofactors (dashed trace) or with full cofactors as indicated in Table I (continuous trace). B, Filtered lysate incubated with 4 mm 2-oxoglutarate and 4 mm ascorbate in the presence (dashed trace) or absence (continuous trace) of 10 mm EDTA. C, Standards [14C]GA4 (dashed trace) and [14C]GA7 (continuous trace).
Table I. The effect of ODD cofactors on the conversion of [14C]GA4 to [14C]GA7 by recombinant DES in vitro.
Final concentrations are as follows: 4 mm ascorbate, 4 mm 2-oxoglutarate, 5 mm FeSO4, 2 mg mL−1 bovine serum albumin, and 0.1 mg mL−1 catalase.
| Cofactor Composition | Conversion |
|---|---|
| % | |
| Full cofactor complement | 100 |
| No cofactors | 0 |
| 2-Oxoglutarate | 19 |
| Ascorbate | 0 |
| FeSO4 | 0 |
| Ascorbate, 2-oxoglutarate | 100 |
| 2-Oxoglutarate, FeSO4 | 82 |
| Ascorbate, 2-oxoglutarate, FeSO4 | 100 |
The substrate specificity of DES was investigated by incubating with 14C-labeled GA1, GA9, or GA12, separation by HPLC, and identification of 14C-labeled products by GC-MS (Supplemental Fig. S1). However, only the substrates were recovered in each case, indicating that DES is highly specific for GA4.
Heterologous Expression of des in Solanum nigrum, Solanum dulcamara, and Nicotiana sylvestris
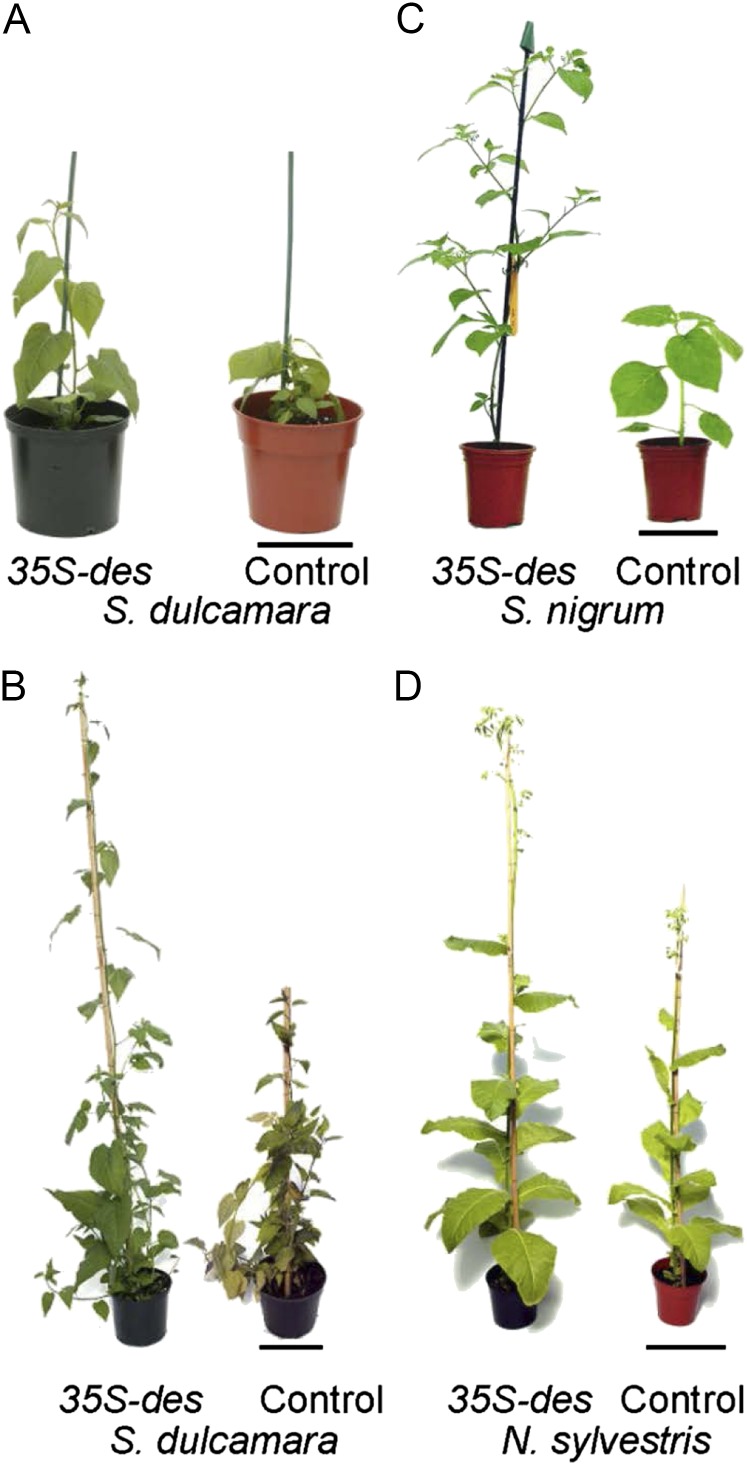
The identification of DES as an ODD indicated that it should be functional in higher plants. In order to test this and determine its effect on plant growth, S. nigrum, S. dulcamara, and the ornamental species N. sylvestris were transformed with des behind the cauliflower mosaic virus 35S promoter. Stem growth of these species was substantially promoted (Fig. 4; Table II), with the primary S. nigrum transformants having a mean height at 5 weeks after acclimation of 98.7 cm compared with 55.4 cm for the untransformed controls, an increase of 78% (Table II). This corresponded to a similar increase in internode length (70%). The increase in stem height of S. dulamara expressing des was even more pronounced, with an almost 3-fold (177%) increase in height measured 6 weeks after the plants were transplanted from tissue culture to the glasshouse (Fig. 4; Table II). In both species, there was a significant (P ≤ 0.003) reduction in stem width but no difference in leaf number. There was also no change in leaf size and shape in either species (Table II). The height of transformed N. sylvestris plants at 12 weeks after transfer from tissue culture was also significantly greater than for the nontransformed controls (P = 0.001), although the increase (28%) was not as great as for Solanum spp. (Fig. 3; Table II), and there was no difference in stem width.
Figure 4.
Comparison of T0 transgenic plants expressing des and controls. A and B, S. dulcamara at 3 weeks (A) and 6 weeks (B) post acclimation. Bars = 14 cm (A) and 13 cm (B). C, S. nigrum at 3 weeks. Bar = 12 cm. D, N. sylvestris at 8 weeks. Bar = 21.7 cm.
Table II. Phenotypic parameters for control and T0 transgenic S. nigrum (measured 5 weeks after transfer to the glasshouse), S. dulcamara (6 weeks), and N. sylvestris (12 weeks) expressing 35S:des.
Mean values are shown with replication given as n. Significance of differences between the means for control and transgenic lines for each species is given as a P value following a two-sample t test, along with the se of the difference (SED) between means and the degrees of freedom (df). Means of natural log-transformed stem height for statistical comparison using the SED are given in parentheses. Significant (P < 0.05) results are given in boldface. Dashes indicate not measured.
| Species | Stem Height | Stem Girth | Internode Length | Leaf No. | Leaf Length | Leaf Width |
|---|---|---|---|---|---|---|
| cm | mm | cm | cm | cm | ||
| S. nigrum | ||||||
| Genotype | ||||||
| 35S:des (n = 24) | 98.70 | 7.25 | 10.36 | 59.25 | 6.55 | 5.73 |
| (4.59) | ||||||
| Control (n = 10) | 55.35 | 8.70 | 6.13 | 58.20 | 6.46 | 6.21 |
| (4.01) | ||||||
| SED (32 df) | 0.019 | 0.236 | 0.491 | 1.085 | 0.313 | 0.424 |
| P value | <0.001 | <0.001 | <0.001 | 0.341 | 0.765 | 0.269 |
| S. dulcamara | ||||||
| Genotype | ||||||
| 35S:des (n = 19) | 180.30 | 5.89 | 10.99 | 55.11 | 6.72 | 5.08 |
| (5.19) | ||||||
| Control (n = 3) | 65.10 | 8.00 | 6.13 | 52.67 | 6.43 | 4.83 |
| (4.17) | ||||||
| SED (20 df) | 0.021 | 0.618 | 0.562 | 2.525 | 0.217 | 0.144 |
| P value | <0.001 | 0.003 | <0.001 | 0.346 | 0.207 | 0.096 |
| N. sylvestris | ||||||
| Genotype | ||||||
| 35S:des (n = 8) | 171.6 | 11.88 | 15.59 | – | – | – |
| (5.14) | ||||||
| Control (n = 3) | 133.8 | 12.00 | 11.93 | – | – | – |
| (4.90) | ||||||
| SED (9 df) | 0.052 | 0.211 | 1.025 | – | – | – |
| P value | 0.001 | 0.568 | 0.006 | – | – | – |
In view of the large height increases found for Solanum spp. transformed with 35S:des, the effect of the transgene on GA content was determined in S. nigrum. The uppermost internodes and leaves of plants from transformed and control lines taken 5 weeks after acclimation were analyzed for the bioactive GAs GA1, GA3, GA4, and GA7 as well as some biosynthetic precursors and 2β-hydroxylated metabolites (Table III). The concentrations of GA3 and GA7 increased in the des transformants by about 20- and 3-fold, respectively, from very low levels in the untransformed plants. However, the concentrations of GA1 and GA4 were substantially less in the transformed plants, such that the combined concentration of bioactive GAs was similar to that of the controls. The concentrations of precursors for the 13-hydroxy GA pathway (GA53 to GA20) were all less in the transgenic lines than in the controls, although the change in GA20 content was not significant, while the 2β-hydroxy GAs GA8 and GA34 were below the level of detection.
Table III. Mean stem height (cm) and GA concentrations (ng g−1 dry weight) in shoots of control (n = 4) and T0 transgenic S. nigrum plants containing 35S:des (n = 10).
Plants were measured and analyzed 5 weeks after transfer to the glasshouse. Significance of differences between the means of control and transgenic lines for each species is given as a P value following a two-sample t test, along with the se of the difference (SED) between means and the degrees of freedom (df). Means of natural log-transformed stem height for statistical comparison using the SED are given in parentheses. Significant (P < 0.05) results are given in boldface. GA8 and GA34 were also analyzed but were below the levels of detection.
| Genotype |
||||
|---|---|---|---|---|
| Parameter | 35S:des | Control | SED (12 df) | P Value |
| Stem height | 98.48 (4.59) | 59.88 (4.09) | 0.020 | <0.001 |
| GA1 | 3.09 | 8.96 | 1.821 | 0.007 |
| GA3 | 3.43 | 0.17 | 0.920 | 0.004 |
| GA4 | 0.03 | 1.87 | 0.721 | 0.025 |
| GA7 | 0.47 | 0.16 | 0.066 | <0.001 |
| GA19 | 6.10 | 17.36 | 2.065 | <0.001 |
| GA20 | 12.50 | 20.80 | 5.640 | 0.165 |
| GA29 | 0.12 | 0.19 | 0.055 | 0.272 |
| GA44 | 0.02 | 0.30 | 0.070 | 0.002 |
| GA53 | 0.99 | 1.76 | 0.167 | <0.001 |
The presence of the C-1,2 double bond in GA3 and GA7 prevents 2β-hydroxylation. Thus, the ectopically expressed desaturase should protect against high levels of 2β-hydroxylase activity, which has been shown to cause severe dwarfism in plants in which GA2ox genes are overexpressed (for review, see Phillips, 2004). This was tested by comparing N. sylvestris and S. nigrum plants transformed with the 35S:PcGA2ox1 gene (Dijkstra et al., 2008) with plants transformed also with 35S:des. Whereas in S. nigrum, only one of 10 plants expressing both transgenes showed recovery from the severely dwarfed phenotype, in N. sylvestris, all seven doubly transformed plants showed substantial growth restoration, although not to the height of nontransformed controls (Fig. 5; Table IV).
Figure 5.
Comparison of T0 transgenic plants expressing PcGA2ox1 or PcGA2ox1 and des. A, S. nigrum at 3 weeks post acclimation. Bar = 5.2 cm. B, N. sylvestris at 6 weeks post acclimation. Bar = 11.7 cm.
Table IV. Mean stem height (cm) of transgenic N. sylvestris plants expressing both 35S:PcGA2ox1 and 35S:des (n = 7) compared with plants expressing only 35S:PcGA2ox1 (n = 3) and untransformed control plants (n = 5), numbered as (1), (2), and (3), respectively.
Overall significance of differences between means is given as a P value (F test) following one-way ANOVA, along with the se of the difference (SED) between means and the degrees of freedom (df). Means of natural log-transformed data for statistical comparison using the SED are given in parentheses. Significant (P < 0.05) results are given in boldface. The lsd values at the 5% and 1% levels of significance are also given. The height of 35S:GA2ox + 35S:des plants was significantly different from that of 35S:GA2ox (P < 0.01; lsd) and of the control (P < 0.05; lsd).
| 35S:GA2ox + 35S:des (1) | 35S:GA2ox (2) | Control(3) | SED (12 df) | lsd (5%, 1%) | P Value |
|---|---|---|---|---|---|
| 82.97 (4.36) | 12.47 (2.52) | 121.40 (4.80) | (1) versus (2), 0.176 | (1) versus (2), 0.384, 0.539 | <0.001 |
| (1) versus (3), 0.150 | (1) versus (3), 0.326, 0.458 | ||||
| (2) versus (3), 0.187 | (2) versus (3), 0.407, 0.571 |
DISCUSSION
The recombinant F. fujikuroi DES expressed in E. coli demonstrated an absolute requirement for 2-oxoglutarate and a partial requirement for ascorbate, which are properties characteristic of ODDs. The gel-filtered protein maintained full activity in the absence of added Fe2+, although this activity was lost completely in the presence of the iron chelator, EDTA. This suggests that the iron remains bound at the enzyme active site during gel filtration. Whereas in higher plants, two ODDs, GA 20-oxidase and GA 3-oxidase, participate in the biosynthesis of bioactive GAs, DES is the only ODD encoded from the GA biosynthesis gene cluster in F. fujikuroi, all other oxidative steps in the pathway being catalyzed by cytochrome P450s (Bömke and Tudzynski, 2009). Although DES has low overall homology with other previously characterized monomeric ODDs, putative iron-binding (His-175, Asp-177, and His-317) and 2-oxoglutarate-binding (Arg-331 and Ser-333) residues (Roach et al., 1997; Wilmouth et al., 2002) are present in the protein (Fig. 2). Since the initial characterization of des and the function of its encoded protein (Tudzynski et al., 2003), genome sequences have become available for a number a fungal species. A BLAST search with the DES protein sequence detected sequences in many such species with more than 25% amino acid identity with DES, although these species do not contain GA biosynthetic gene clusters, such as Verticillium albo-atrum VaMs.102 and Fusarium graminearum (Broad Institute Genome Sequencing Platform), the latter species not belonging to the G. fujikuroi species complex. The DES homologs presumably participate in other biosynthetic pathways. Despite the obvious functional differences, the putative iron- and 2-oxoglutarate-dependent-binding residues are completely conserved in these proteins (Supplemental Fig. S2), providing support for this assignment and indicating that these enzymes also function as ODDs. This finding should aid in the future characterization of these enzymes.
Due to the necessity of regulating GA concentrations precisely, plant shoots generally contain low levels of 1,2-unsaturated GAs, such as GA3, which are protected from deactivation by 2β-hydroxylation. By producing GA3, F. fujikuroi has developed an effective strategy to bypass one of the host’s protective mechanisms against hyper GA signaling, whereas some other GA-producing fungi, such as S. manihoticola, which produces GA4 (Rademacher and Graebe, 1979), and Phaeosphaeria spp., which produce GA1 (Kawaide and Sassa, 1993), have not evolved this strategy. ODDs are involved in numerous primary and secondary biosynthetic pathways in plants; therefore, it could be anticipated that DES would be functional if expressed in plant tissues. Introduction of des into higher plants to promote the accumulation of 1,2-unsaturated GAs would thus appear to be a potentially effective method for promoting plant growth.
Constitutive expression of des in three species resulted in height increases ranging from 28% in N. sylvestris to 177% in S. dulcamara relative to nontransformed controls (Table II). Consistent with 1,2-desaturation serving to protect from 2β-hydroxylation, expression of des gave greater growth promotion in a high-GA 2-oxidase background, with N. sylvestris plants expressing both PcGA2ox1 and des being on average 6-fold taller than plants expressing PcGA2ox1 alone (Table IV). In S. nigrum, it was shown that enhanced growth was associated with higher concentrations of GA7 and particularly GA3 but lower concentrations of their saturated analogs GA4 and GA1, respectively, consistent with these last GAs undergoing desaturation. However, DES was found to have a high substrate specificity and did not metabolize GA1 in vitro. Furthermore, GA1 is not converted to GA3 by fungal cultures (Bearder et al., 1975), so it is possible that the higher concentrations of GA3 are formed from GA7 by 13-hydroxylation. Although 13-hydroxylation in plants is thought to occur early in the GA biosynthetic pathway (Yamaguchi, 2008), evidence for late 13-hydroxylation has been reported (Rood and Hedden, 1994). Indeed, some GA 3-oxidases are capable of 13-hydroxylating C19-GAs (Appleford et al., 2006; Ward et al., 2010). However, it is also possible that in the environment of the plant cell, DES is capable of desaturating GA1, if at a low rate. The concentrations of all 13-hydroxylated GA precursors (concentrations of non-13-hydroxylated intermediates were too low to be measured accurately) were lower in the S. nigrum des lines than in controls, which could be due, in part, to homeostasis mechanisms (Yamaguchi, 2008).
Despite containing higher amounts of 1,2-unsaturated GAs, overall concentrations of bioactive GAs in actively growing shoots of the 35S:des plants were similar to those in controls (Table III). On the basis of their binding affinities for the GA receptor (Ueguchi-Tanaka et al., 2005), there is no evidence that 1,2-unsaturated GAs are intrinsically more active than their saturated analogs. However, in bioassays involving application to intact seedlings, GA3 is consistently more active than GA1 (Crozier et al., 1970), presumably due to it being inactivated more slowly. Although there is no detailed information on the relative stability in planta of GA3 and GA1, GA5, which is also unsaturated on C-2 and therefore resistant to 2β-hydroxylation, was shown to be metabolized more slowly than the C-2-saturated GA4 and GA20 in vegetative shoots of Lolium temulentum (King et al., 2008). The enhanced growth of the 35S:des plants may be due to the greater persistence of GA3/GA7 at or en route to their sites of action and would require these to be separate from the major sites of biosynthesis.
The potential practical benefits of increasing plant biomass through manipulating GA metabolism have been recognized (Phillips, 2004; Salas Fernandez et al., 2009; Bhattacharya et al., 2010). For example, ectopic expression of GA 20-oxidase has been shown to increase biomass in hybrid aspen (Populus tremula × Populus tremuloides; Eriksson et al., 2000) and tobacco (Nicotiana tabacum; Biemelt et al., 2004). The height increases obtained by the expression of des are exceptional, particularly in S. dulcamara, in which the transgenic lines were almost three times the height of controls. However, there was no effect on leaf number or size in transgenic Solanum spp. and there was a decrease in stem girth in these species. Nevertheless, stem volumes increased in the S. nigrum and S. dulcamara des lines by 19% and 56%, respectively, with a corresponding 28% increase for N. sylvestris, for which there was a negligible reduction in stem girth. The species specificity of the height increases obtained by expressing des are likely to depend on the efficiency of mechanisms for GA homeostasis and the degree to which GA signaling is normally saturated. Therefore, it is important to determine the effectiveness of this technology in a range of species, particularly trees, in which higher biomass would have practical benefits.
MATERIALS AND METHODS
Preparation of DES Plant Transformation Vector
The des open reading frame was amplified from the cDNA clone in pUC19 (Tudzynski et al., 2003) using a sense primer incorporating an EcoRI site (5′-ACGAATTCATGCCTCATAAAGAT-3′) and an antisense primer incorporating a HindIII site (5′-GTAAGCTTCTACCAGAATGCAAT-3′). The reactions contained 50 ng of cDNA, 25 pmol of each primer, 0.5 mm deoxyribonucleotide triphosphates, 0.5 units of Taq polymerase (Promega), 2.5 mm MgCl2, and PCR buffer (Promega) in a total volume of 10 μL. The reaction was heated to 94°C for 2 min and then subjected to 30 cycles of 94°C for 20 s, 55°C for 20 s, 65°C for 1 min 15 s, and finally 65°C for 4 min. The PCR products were purified by 1% (w/v) agarose gel electrophoresis, and the band of the anticipated size (1,045 bp) was excised and cloned into pGEM-T (Promega) according to the manufacturer’s instructions. After confirming that the insert had the correct nucleotide sequence, it was excised with EcoRI/HindIII and ligated into the shuttle vector pART7, allowing transcriptional fusion to the cauliflower mosaic virus 35S promoter (Gleave, 1992). The 35S::des construct was excised with NotI and ligated into the binary vector pBJ40 (provided by Bart Janssen, Horticultural and Food Research Institute), which contained the neomycin phosphotransferase (nptII) gene.
Bacterial Expression and Enzyme Assays
The des open reading frame, obtained by digestion of des-containing pBJ40 with EcoRI/HindIII, was ligated into the pET32a expression vector (Novagen, Merck Chemical). Expression of the cDNAs in Escherichia coli strain BL21 and recovery of the recombinant protein were as described previously (Williams et al., 1998), except that the cultures were grown at 20°C after induction with isopropylthio-β-galactoside and the cells were harvested after 4 h. Bacterial lysates (90 µL) were incubated at 30°C for 3 to 5 h with 14C-labeled GA substrates (obtained from Prof. L. Mander, Australian National University) and dioxygenase cofactors in a total volume of 100 µL, after which the products were separated by HPLC (Shimadzu) and detected with an online radioactivity monitor (Berthold Technologies). The identity of products was confirmed by GC-MS as described by MacMillan et al. (1997). Gel filtration of the lysate was achieved by application in 100 µL to NICK columns (GE Healthcare UK) and elution with 400 µL of 100 mm Tris-HCl, pH 7.5. The gel-filtered lysate (90 µL) was incubated with [14C]GA4 (0.3 kBq) with different combinations of cofactors as given in Table I.
Plant Material
Nicotiana sylvestris, Solanum nigrum, and Solanum dulcamara were grown from seed in the glasshouse in a 6:6:1:1 (by volume) mixture of Levington M3 compost (Scotts UK), John Innes No. 3 compost (J. Bentley), Perlite (Silvaperl), and vermiculite (Silvaperl). Natural light was supplemented with 16 h of fluorescent illumination (195 µmol m−2 s−1; TLD/58W 35V “Daylight” fluorescent tubes; Phillips) with day and night temperatures of 25°C ± 1°C.
Plant Transformation
Agrobacterium tumefaciens strain AGL1 was transformed by electroporation with pBJ40 containing 35S:des. Transformation of leaf explants and plant regeneration were as described by Dijkstra et al. (2008). Control nontransformed plants were obtained from tissue culture, but without incubation of leaf explants with A. tumefaciens and antibiotic selection. In order to obtain plants transformed with both 35S::des and 35S::PcGA2ox1, S. nigrum and N. sylvestris plants were transformed with A. tumefaciens strain LBA4404 harboring 35S::PcGA2ox1 in pLARS120 using the protocol described by Dijkstra et al. (2008). Homozygous T2 35S::PcGA2ox1 plants obtained from T1 lines that demonstrated 3:1 segregation for the transgene were transformed with 35S::des, as described above. All T0 des lines were analyzed by PCR for the presence of des, nptII, and, where appropriate, PcGA2ox1 genes. Positive lines were analyzed by reverse transcription-PCR for expression of the transgenes.
Molecular Characterization of Transgenic Plants
The presence of the transgene in regenerated plants was confirmed by PCR using the following primers: Gfdes sense primer (5′-GCCTCATAAAGATAATCTTC-3′) and antisense primer (5′-GTACATCCGACTCAAATGTC-3′); PcGA2ox1 sense primer (5′-AAAGGTACCCAACCATGGTTGTTCTGTCTCA-3′) and antisense primer (5′-AAATCTAGATGGTTAATCAGCAGCAGATT-3′); nptII sense primer (5′-AGACAATCGGCTGCTCTGA-3′) and antisense primer (5′-ATACTTTCTCGGCAGGAGCA-3′).
Genomic DNA was extracted from plants using a GenElute Plant Miniprep kit (Sigma-Aldrich). PCR was performed using RED Taq Ready Mix (Sigma-Aldrich) according to the manufacturer’s instructions. Amplification was performed in a DNA Thermal Cycler 480 (Perkin-Elmer) with the following PCR conditions: 50 to 200 ng of genomic DNA, 100 pmol of each primer, Sigma Ready Mix RED Taq (Sigma-Aldrich; 8 μL) in a 25-μL total volume. The reaction was heated to 94°C for 2 min and then subjected to 35 cycles of 94°C for 20 s, 47°C/53°C/53°C (des/PcGA2ox1/nptII) for 30 s, 72°C for 1 min, with a final extension phase at 72°C for 9 min.
For reverse transcription-PCR, RNA was extracted from leaves of 4-week-old plants using the RNeasy Plant Mini Kit (Qiagen) and treated with DNase (RQ1 RNase-Free DNase kit; Promega). cDNA was synthesized using the Reverse-iT Synthesis Kit (ABgene) following the manufacturer’s protocol. PCR was carried out with the same primers and reaction conditions as given above. PCR products were separated on 1% (w/v) agarose gels containing ethidium bromide (0.5 μL mL−1) with a 100-bp or a 1-kb ladder. Gels were visualized using a UV transilluminator (Appligene).
Plant Phenotypic Analysis
The phenotypic characteristics of primary transformed and control plants (height, internode length, leaf number, length, and width) were measured at 5 weeks after acclimation for S. nigrum, 6 weeks for S. dulcamara, and 12 weeks for N. sylvestris. The phenotypic and GA analytical data (see below) were analyzed by two-sample t test or ANOVA, as appropriate, using the GenStat statistical software package (VSN International) and given a completely randomized design for all experiments. A natural log (to base e) transformation was used for stem height to account for some heterogeneity of variance in these data, but all other data did not require transformation. Following ANOVA of stem height data for the three genotypes of N. sylvestris (expressing both 35S:PcGA2ox1 and 35S:des, only 35S:PcGA2ox1, or untransformed control), genotype means were compared using the lsd at the 5% and 1% levels of significance.
Quantitative GA Analysis
Quantitative analysis of GAs in S. nigrum was performed on young shoot tips (including the youngest three to four leaves of each branch) of plants sampled 5 weeks after acclimation using combined GC-MS as described previously (Rieu et al., 2008). Biological replicates consisted of samples taken from individual plants.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers F. fujikuroi GA4 Desaturase (AJ417493); Ajellomyces dermatitidis (XP_002626943); Aspergillus terreus (XP_001215403); Botryotinia fuckeliana (CCD45190); Chaetomium globosum (XP_001227273); F. graminearum (FGSG_11397.3); Glomerella graminicola (EFQ25195); Metarhizium acridum (EFY93227); V. albo-atrum, (XP_003006826); Arabidopsis (Arabidopsis thaliana) anthocyanidin synthase (AEI99590.1); and Phaseolus coccineus GA2ox1 (AJ132438).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. HPLC radiochromatograms and GC-MS identification of products from incubation of recombinant DES with different 14C-labeled GAs.
Supplemental Figure S2. Alignment of the predicted F. fujikuroi DES amino acid sequence with those of related sequences from other fungi.
Acknowledgments
We thank Simon Vaughan for assistance with the statistical analysis.
Glossary
- ODD
2-oxoglutarate-dependent dioxygenase
- GC
gas chromatography
- MS
mass spectrometry
- cDNA
complementary DNA
References
- Albone KS, Gaskin P, Macmillan J, Phinney BO, Willis CL. (1990) Biosynthetic origin of gibberellin A3 and gibberellin A7 in cell-free preparations from seeds of Marah macrocarpus and Malus domestica. Plant Physiol 94: 132–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleford NEJ, Evans DJ, Lenton JR, Gaskin P, Croker SJ, Devos KM, Phillips AL, Hedden P. (2006) Function and transcript analysis of gibberellin-biosynthetic enzymes in wheat. Planta 223: 568–582 [DOI] [PubMed] [Google Scholar]
- Bearder JR, Macmillan J, Phinney BO. (1975) Fungal products. Part XIV. Metabolic pathways from ent-kaurenoic acid to fungal gibberellins in mutant B1-41a of Gibberella fujikuroi. J Chem Soc Perkin Trans 1 721–726 [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Kourmpetli S, Davey MR. (2010) Practical applications of manipulating plant architecture by regulating gibberellin metabolism. J Plant Growth Regul 29: 249–256 [Google Scholar]
- Biemelt S, Tschiersch H, Sonnewald U. (2004) Impact of altered gibberellin metabolism on biomass accumulation, lignin biosynthesis, and photosynthesis in transgenic tobacco plants. Plant Physiol 135: 254–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bömke C, Rojas MC, Gong F, Hedden P, Tudzynski B. (2008) Isolation and characterization of the gibberellin biosynthetic gene cluster in Sphaceloma manihoticola. Appl Environ Microbiol 74: 5325–5339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bömke C, Tudzynski B. (2009) Diversity, regulation, and evolution of the gibberellin biosynthetic pathway in fungi compared to plants and bacteria. Phytochemistry 70: 1876–1893 [DOI] [PubMed] [Google Scholar]
- Crozier A, Kuo CC, Durley RC, Pharis RP. (1970) The biological activities of 26 gibberellins in nine plant bioassays. Can J Bot 48: 867–877 [Google Scholar]
- Demura T, Ye Z-H. (2010) Regulation of plant biomass production. Curr Opin Plant Biol 13: 299–304 [DOI] [PubMed] [Google Scholar]
- Dijkstra C, Adams E, Bhattacharya A, Page AF, Anthony P, Kourmpetli S, Power JB, Lowe KC, Thomas SG, Hedden P, et al (2008) Over-expression of a gibberellin 2-oxidase gene from Phaseolus coccineus L. enhances gibberellin inactivation and induces dwarfism in Solanum species. Plant Cell Rep 27: 463–470 [DOI] [PubMed] [Google Scholar]
- Eriksson ME, Israelsson M, Olsson O, Moritz T. (2000) Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat Biotechnol 18: 784–788 [DOI] [PubMed] [Google Scholar]
- Gleave AP. (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL, Rojas MC, Carrera E, Tudzynski B. (2001) Gibberellin biosynthesis in plants and fungi: a case of convergent evolution? J Plant Growth Regul 20: 319–331 [DOI] [PubMed] [Google Scholar]
- Hou X, Lee LYC, Xia K, Yan Y, Yu H. (2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sentoku N, Kitano H, Matsuoka M, Kobayashi M. (2001) Cloning and functional analysis of two gibberellin 3 beta-hydroxylase genes that are differently expressed during the growth of rice. Proc Natl Acad Sci USA 98: 8909–8914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaide H. (2006) Biochemical and molecular analyses of gibberellin biosynthesis in fungi. Biosci Biotechnol Biochem 70: 583–590 [DOI] [PubMed] [Google Scholar]
- Kawaide H, Sassa T. (1993) Accumulation of gibberellin A1 and the metabolism of gibberellin A9 to gibberellin A1 in a Phaeosphaeria sp l487 culture. Biosci Biotechnol Biochem 57: 1403–1405 [Google Scholar]
- King RW, Mander LN, Asp T, MacMillan CP, Blundell CA, Evans LT. (2008) Selective deactivation of gibberellins below the shoot apex is critical to flowering but not to stem elongation of Lolium. Mol Plant 1: 295–307 [DOI] [PubMed] [Google Scholar]
- Kvas M, Marasas WFO, Wingfield BD, Wingfield MJ, Steenkamp ET. (2009) Diversity and evolution of Fusarium species in the Gibberella fujikuroi complex. Fungal Divers 34: 1–21 [Google Scholar]
- Leslie JF, Summerell BA. (2006) The Fusarium Laboratory Manual. Blackwell, Oxford
- MacMillan J, Ward DA, Phillips AL, Sánchez-Beltrán MJ, Gaskin P, Lange T, Hedden P. (1997) Gibberellin biosynthesis from gibberellin A12-aldehyde in endosperm and embryos of Marah macrocarpus. Plant Physiol 113: 1369–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malonek S, Bömke C, Bornberg-Bauer E, Rojas MC, Hedden P, Hopkins P, Tudzynski B. (2005) Distribution of gibberellin biosynthetic genes and gibberellin production in the Gibberella fujikuroi species complex. Phytochemistry 66: 1296–1311 [DOI] [PubMed] [Google Scholar]
- Murase K, Hirano Y, Sun T-P, Hakoshima T. (2008) Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456: 459–463 [DOI] [PubMed] [Google Scholar]
- Navarro L, Bari R, Achard P, Lisón P, Nemri A, Harberd NP, Jones JDG. (2008) DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol 18: 650–655 [DOI] [PubMed] [Google Scholar]
- Phillips AL (2004) Genetic and transgenic approaches to improving crop performance. In PJ Davies, ed, Plant Hormones: Biosynthesis, Signal Transduction, Action! Kluwer, Dordrecht, The Netherlands, pp 582–609 [Google Scholar]
- Phinney BO. (1983) The history of gibberellins. In A Crozier, ed, The Biochemistry and Physiology of Gibberellins. Praeger, New York, pp 19–52
- Prescott AG, John P. (1996) Dioxygenases: molecular structure and role in plant metabolism. Annu Rev Plant Physiol Plant Mol Biol 47: 245–271 [DOI] [PubMed] [Google Scholar]
- Rademacher W, Graebe JE. (1979) Gibberellin A4 produced by Sphaceloma manihoticola, the cause of the superelongation disease of cassava (Manihot esculenta). Biochem Biophys Res Commun 91: 35–40 [DOI] [PubMed] [Google Scholar]
- Rieu I, Ruiz-Rivero O, Fernandez-Garcia N, Griffiths J, Powers SJ, Gong F, Linhartova T, Eriksson S, Nilsson O, Thomas SG, et al. (2008) The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J 53: 488–504 [DOI] [PubMed] [Google Scholar]
- Roach PL, Clifton IJ, Hensgens CMH, Shibata N, Schofield CJ, Hajdu J, Baldwin JE. (1997) Structure of isopenicillin N synthase complexed with substrate and the mechanism of penicillin formation. Nature 387: 827–830 [DOI] [PubMed] [Google Scholar]
- Rojas MC, Hedden P, Gaskin P, Tudzynski B. (2001) The P450-1 gene of Gibberella fujikuroi encodes a multifunctional enzyme in gibberellin biosynthesis. Proc Natl Acad Sci USA 98: 5838–5843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rood S, Hedden P. (1994) Convergent pathways of gibberellin A1 biosynthesis in Brassica. Plant Growth Regul 15: 241–246 [Google Scholar]
- Salas Fernandez MG, Becraft PW, Yin Y, Lübberstedt T. (2009) From dwarves to giants? Plant height manipulation for biomass yield. Trends Plant Sci 14: 454–461 [DOI] [PubMed] [Google Scholar]
- Thomas SG, Hedden P. (2006) Gibberellin metabolism and signal transduction. In P Hedden, SG Thomas, eds, Plant Hormone Signaling. Blackwell, Oxford, pp 147–184
- Thomas SG, Phillips AL, Hedden P. (1999) Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA 96: 4698–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troncoso C, González X, Bömke C, Tudzynski B, Gong F, Hedden P, Rojas MC. (2010) Gibberellin biosynthesis and gibberellin oxidase activities in Fusarium sacchari, Fusarium konzum and Fusarium subglutinans strains. Phytochemistry 71: 1322–1331 [DOI] [PubMed] [Google Scholar]
- Tudzynski B, Hedden P, Carrera E, Gaskin P. (2001) The P450-4 gene of Gibberella fujikuroi encodes ent-kaurene oxidase in the gibberellin biosynthesis pathway. Appl Environ Microbiol 67: 3514–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudzynski B, Mihlan M, Rojas MC, Linnemannstöns P, Gaskin P, Hedden P. (2003) Characterization of the final two genes of the gibberellin biosynthesis gene cluster of Gibberella fujikuroi: des and P450-3 encode GA4 desaturase and the 13-hydroxylase, respectively. J Biol Chem 278: 28635–28643 [DOI] [PubMed] [Google Scholar]
- Tudzynski B, Rojas MC, Gaskin P, Hedden P. (2002) The gibberellin 20-oxidase of Gibberella fujikuroi is a multifunctional monooxygenase. J Biol Chem 277: 21246–21253 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YIC, Kitano H, Yamaguchi I, et al (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Ward DA, MacMillan J, Gong F, Phillips AL, Hedden P. (2010) Gibberellin 3-oxidases in developing embryos of the southern wild cucumber, Marah macrocarpus. Phytochemistry 71: 2010–2018 [DOI] [PubMed] [Google Scholar]
- Williams J, Phillips AL, Gaskin P, Hedden P. (1998) Function and substrate specificity of the gibberellin 3β-hydroxylase encoded by the Arabidopsis GA4 gene. Plant Physiol 117: 559–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmouth RC, Turnbull JJ, Welford RWD, Clifton IJ, Prescott AG, Schofield CJ. (2002) Structure and mechanism of anthocyanidin synthase from Arabidopsis thaliana. Structure 10: 93–103 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251 [DOI] [PubMed] [Google Scholar]