Abstract
The Medicago truncatula NIP/LATD (for Numerous Infections and Polyphenolics/Lateral root-organ Defective) gene encodes a protein found in a clade of nitrate transporters within the large NRT1(PTR) family that also encodes transporters of dipeptides and tripeptides, dicarboxylates, auxin, and abscisic acid. Of the NRT1(PTR) members known to transport nitrate, most are low-affinity transporters. Here, we show that M. truncatula nip/latd mutants are more defective in their lateral root responses to nitrate provided at low (250 μm) concentrations than at higher (5 mm) concentrations; however, nitrate uptake experiments showed no discernible differences in uptake in the mutants. Heterologous expression experiments showed that MtNIP/LATD encodes a nitrate transporter: expression in Xenopus laevis oocytes conferred upon the oocytes the ability to take up nitrate from the medium with high affinity, and expression of MtNIP/LATD in an Arabidopsis chl1(nrt1.1) mutant rescued the chlorate susceptibility phenotype. X. laevis oocytes expressing mutant Mtnip-1 and Mtlatd were unable to take up nitrate from the medium, but oocytes expressing the less severe Mtnip-3 allele were proficient in nitrate transport. M. truncatula nip/latd mutants have pleiotropic defects in nodulation and root architecture. Expression of the Arabidopsis NRT1.1 gene in mutant Mtnip-1 roots partially rescued Mtnip-1 for root architecture defects but not for nodulation defects. This suggests that the spectrum of activities inherent in AtNRT1.1 is different from that possessed by MtNIP/LATD, but it could also reflect stability differences of each protein in M. truncatula. Collectively, the data show that MtNIP/LATD is a high-affinity nitrate transporter and suggest that it could have another function.
All plants require nitrogen (N) as an essential nutrient and are able to acquire N from nitrate (NO3−) and ammonium (NH4+) in the soil. Nitrate acquisition begins with its transport into root cells, accomplished by NO3− transporters. Soil NO3− concentrations can vary by 5 orders of magnitude (Crawford, 1995), and to cope with the variability, plants have evolved both high-affinity (HATS) and low-affinity (LATS) transport systems. These are encoded by two gene families: the phylogenetically distinct NRT1(PTR) and NRT2 families. Members of these families also participate in the movement of NO3− throughout the plant and within plant cells (Miller et al., 2007; Segonzac et al., 2007; Tsay et al., 2007; Almagro et al., 2008; Lin et al., 2008; Fan et al., 2009; Li et al., 2010; Barbier-Brygoo et al., 2011; Wang and Tsay, 2011; Xu et al., 2012). Proteins in the Chloride Channel (CLC) transporter family also transport NO3−; these transporters are associated with cytosol-to-organelle NO3− movement (Zifarelli and Pusch, 2010).
NRT1(PTR) is a large family of transporters, comprising 53 members in Arabidopsis (Arabidopsis thaliana), 84 members in rice (Oryza sativa), with NRT1(PTR) members known in several other species (Tsay et al., 2007; Zhao et al., 2010). In addition to transporting NO3− coupled to H+ movement, members of the NRT1(PTR) family have been found to transport dipeptides or tripeptides, amino acids (Waterworth and Bray, 2006), dicarboxylic acids (Jeong et al., 2004), auxin (Krouk et al., 2010b), and/or abscisic acid (Kanno et al., 2012). Only a small number of NRT1(PTR) proteins have been functionally studied compared with the large number that exist in higher plants; thus, the number of biochemical functions ascribed to this family may expand.
Of the NRT1(PTR) members known to transport NO3−, most are LATS transporters. An important exception is Arabidopsis NRT1.1(CHL1), a dual-affinity transporter that is the most extensively studied NRT1(PTR) protein. AtNRT1.1(CHL1) was identified initially on the basis of its ability to confer chlorate toxicity resistance and was the first of this family to be cloned (Doddema et al., 1978; Tsay et al., 1993). AtNRT1.1(CHL1) is an essential component of NO3− transport and NO3− signaling pathways, with important roles regulating the expression of other NO3− transporters and root architecture (Remans et al., 2006; Walch-Liu et al., 2006; Walch-Liu and Forde, 2008). Its expression is inducible by NO3− (Huang et al., 1996). Reversible phosphorylation is essential to its ability to switch between LATS and HATS activity (Liu and Tsay, 2003). In addition, AtNRT1.1(CHL1) acts as a NO3− sensor (Muños et al., 2004; Ho et al., 2009; Wang et al., 2009) and has been shown to transport auxin in a NO3− concentration-dependent manner (Krouk et al., 2010b). It has been suggested that AtNRT1.1’s ability to transport auxin may be part of its NO3−-sensing mechanism (Krouk et al., 2010a, 2010b; Gojon et al., 2011).
Most legumes and actinorhizal plants have the additional ability to form symbiotic N-fixing root nodules with soil bacteria, enabling them to thrive in NO3−- and NH4+-depleted environments. Legume nodulation commences with signal exchange between the plant and rhizobia, followed by root cortical cell divisions, invasion of the root by rhizobia inside plant-derived infection threads, and subsequent endocytosis of rhizobia into newly divided plant host cells, forming symbiosomes. Within symbiosomes, the rhizobia differentiate into bacteroids that are capable of N fixation (Oldroyd and Downie, 2008; Kouchi et al., 2010). Before N fixation begins, nodules are a sink for the plant’s N; N is required to support nodule organogenesis and rhizobial proliferation and differentiation in nodules (Udvardi and Day, 1997). After N fixation begins, nodules are a source of bioavailable N and are a large carbon sink because of the energetic needs of the rhizobia that fix N (White et al., 2007).
NRT1(PTR) transporters have received less attention in legumes and actinorhizal plants than in other plants, but they are beginning to be investigated. Transcription patterns for several soybean (Glycine max) NRT1(PTR) transporter genes have been studied; they were predicted to encode transporters of NO3−, but were not characterized functionally (Yokoyama et al., 2001). Benedito et al. (2010) recognized 111 nonredundant Medicago truncatula sequences corresponding to genes in the 2.A.17 transporter class, containing NRT1(PTR) genes (Benedito et al., 2010). In Lotus japonicus, 37 members of the NRT1(PTR) family were identified by an in silico search (Criscuolo et al., 2012). The recent availability of three sequenced legume genomes will add to our knowledge of this important gene family (Sato et al., 2008; Schmutz et al., 2010; Young et al., 2011). In faba bean (Vicia faba), two NRT1(PTR) transporters have been studied; one was demonstrated to transport dipeptides in yeast, while the second was found to be phylogenetically close to a soybean NRT1(PTR) (Miranda et al., 2003). In alder (Alnus glutinosa) nodules, the NRT1(PTR) transporter AgDCAT localizes to the symbiotic interface and transports dicarboxylic acid from the cytosol toward its symbiotic partner, Frankia spp. (Jeong et al., 2004). The M. truncatula MtNRT1.3 transporter was shown to be a dual-affinity NO3− transporter; MtNRT1.3 is up-regulated by the absence of NO3− (Morère-Le Paven et al., 2011).
The M. truncatula NIP/LATD (for Numerous Infections and Polyphenolics/Lateral root-organ Defective) gene encodes a predicted NRT1(PTR) transporter (Harris and Dickstein, 2010; Yendrek et al., 2010), and the three known nip and latd mutants have pleiotropic defects in nodulation and root architecture. Mtnip-1, containing a missense (A497V) mutation in one of the NIP/LATD protein’s transmembrane domains, is well characterized with respect to nodulation phenotypes (Veereshlingam et al., 2004). Mtnip-1 develops nodules that initiate rhizobial invasion but fail to release rhizobia from infection threads. Its nodules lack meristems and accumulate polyphenolics, a sign of host defense. Mtnip-1 plants also have defective root architecture (Veereshlingam et al., 2004). The Mtlatd mutant has the most severe phenotype, caused by a stop codon (W341STOP) in the middle of the NIP/LATD putative protein (Yendrek et al., 2010). Mtlatd has serious defects in root architecture, with a nonpersistent primary root meristem, lateral roots (LRs) that fail to make the transition from LR primordia to LRs containing a meristem, and defects in root hairs. Mtlatd also has defective nodules (Bright et al., 2005). The missense Mtnip-3 mutant (E171K) is the least affected, forming invaded nodules with meristems and polyphenol accumulation. Mtnip-3 has nearly normal root architecture (Teillet et al., 2008; Yendrek et al., 2010).
Here, the function of the MtNIP/LATD protein is investigated. We find that MtNIP/LATD is a high-affinity NO3− transporter and provide evidence suggesting that it may have an additional biochemical function.
RESULTS
Mtnip Mutants’ Nitrate Phenotypes
Because of MtNIP/LATD’s similarity to low-affinity NO3− transporters in the NRT1(PTR) family (Yendrek et al., 2010), one hypothesis is that it may be a low-affinity NO3− transporter. Bioavailable N is known to suppress nodulation and to inhibit N fixation in mature N-fixing nodules (Streeter, 1988; Fei and Vessey, 2009). It is also possible that Mtnip/latd mutants would develop functional root nodules in conditions of NO3− sufficiency. To test whether Mtnip-1 was altered in the suppression of nodulation by bioavailable N, Mtnip-1 mutant plants were cultivated in 1 and 10 mm KNO3 and also in 5 mm NH4NO3 in the presence of Sinorhizobium meliloti. As shown in Table I, Mtnip-1 formed low numbers of nodules under these conditions compared with no-N conditions, similar to the wild type (A17), suggesting normal suppression by bioavailable N sources. The few nodules that formed in Mtnip-1 in 1 mm KNO3, 10 mm KNO3, or 5 mm NO3NH4 had an Mtnip-1 nodule phenotype.
Table I. Nodulation in the absence and presence of N sources.
In each experiment, wild-type A17 and Mtnip-1 plants were grown in the same aeroponic chamber in the given N regime, as described in “Materials and Methods.” For each experiment, seven to 11 plants of each genotype were evaluated. Nodules were counted at 15 dpi. Data are presented as the mean number of nodules per plant ± se.
| Experiment | N Conditions | A17 Nodule No. | Mtnip-1 Nodule No. |
|---|---|---|---|
| 1 | No NO3 | 31. 9 ± 3.2 | 20.8 ± 1.6 |
| 2 | No NO3 | 19.9 ± 4.1 | 13.25 ± 0.8 |
| 3 | 1 mm KNO3 | 7.8 ± 0.7 | 2.2 ± 0.4 |
| 4 | 1 mm KNO3 | 5.3 ± 0.6 | 1.1 ± 0.3 |
| 5 | 5 mm NH4NO3 | 1.7 ± 0.6 | 0. 7 ± 0.2 |
| 6 | 5 mm NH4NO3 | 5.7 ± 0.6 | 2.8 ± 0.5 |
| 7 | 10 mm KNO3 | 3.4 ± 0.8 | 0.6 ± 0.3 |
| 8 | 10 mm KNO3 | 4.0 ± 1.0 | 0.7 ± 0.4 |
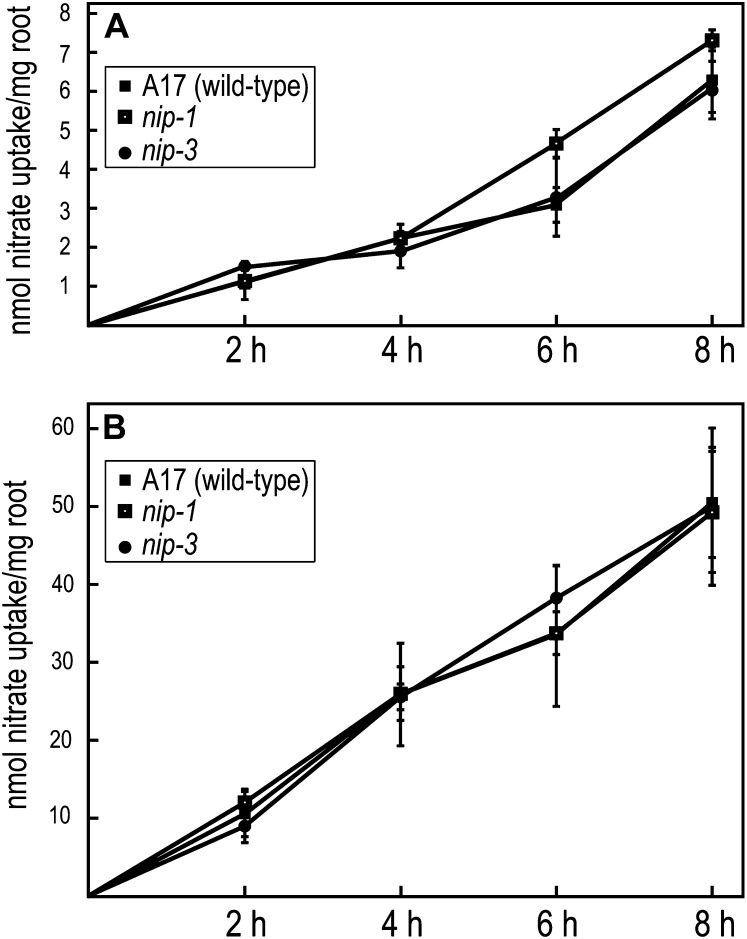
To examine whether the mutants had defects in NO3− uptake, we grew Mtnip-1 and Mtnip-3 mutants, with wild-type A17 as a control, in two different concentrations of KNO3: 250 μm and 5 mm. Uptake was measured by monitoring the depletion of NO3− from the medium. As can be seen in Figure 1, no differences in NO3− uptake were observed in the mutants, suggesting that NO3− LATS and HATS are functioning in these plants. However, because measurement of the depletion of NO3− from the medium is less sensitive than measuring NO3− influx, subtle changes in NO3− uptake may not have been detected in this experimental system.
Figure 1.
Nitrate uptake in M. truncatula. Wild-type A17, Mtnip-1, and Mtnip-3 plants were placed in solutions containing 250 μm (A) or 5 mm (B) nitrate. Nitrate uptake was monitored by its depletion from the medium at 2-h intervals. Data are plotted for one biological replicate ± se.
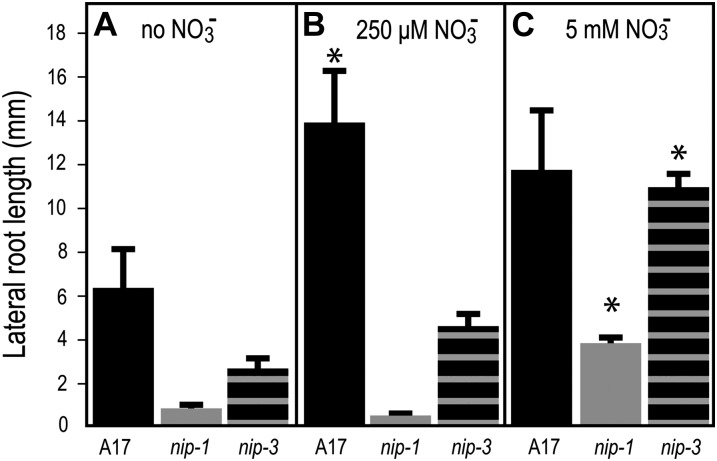
In Arabidopsis, growth of plants in different NO3− concentrations is known to affect root architecture (Zhang and Forde, 2000; Linkohr et al., 2002). To determine if M. truncatula nip mutants’ LR phenotype was affected by NO3−, we grew Mtnip-1, Mtnip-3, and A17 in the presence of 0 μm, 250 μm, and 5 mm KNO3 and examined root growth parameters after 2 weeks. Wild-type A17 produced longer LRs in both NO3− concentrations tested than it did at 0 NO3− and slightly shorter LRs in 5 mm as compared with 250 μm KNO3 (Fig. 2). LR lengths of both Mtnip-1 and Mtnip-3 were significantly shorter than those of the wild type in all conditions tested, with one exception; in 5 mm KNO3, the average LR lengths of Mtnip-3 were similar to those of A17. Both Mtnip-1 and Mtnip-3 mutants had longer LRs when grown in 5 mm KNO3 (Fig. 2C) than they did in 250 μm KNO3 (Fig. 2B) and when grown in the absence of NO3− (Fig. 2A). These data are consistent with an MtNIP/LATD function in high-affinity, low-concentration NO3− uptake or response.
Figure 2.
LR lengths of M. truncatula plants grown in different conditions. Wild-type A17 (black bars), Mtnip-1 (gray bars), and Mtnip-3 (horizontally striped bars) were grown in liquid buffered nodulation medium with no added NO3− (A), with 250 μm KNO3 (B), or with 5 mm KNO3 (C), with the medium changed every other day. LR lengths were measured after 2 weeks. Data are shown for one biological replicate ± se (n = 5). Replicates gave similar results. Asterisks mark LR lengths from plants grown at 250 μm KNO3 and 5 mm KNO3 that are significantly different from the same genotype grown at 0 mm KNO3, using Student’s t test at P < 0.05.
MtNIP/LATD Protein Transports Nitrate, But Not His, in Xenopus laevis Oocytes
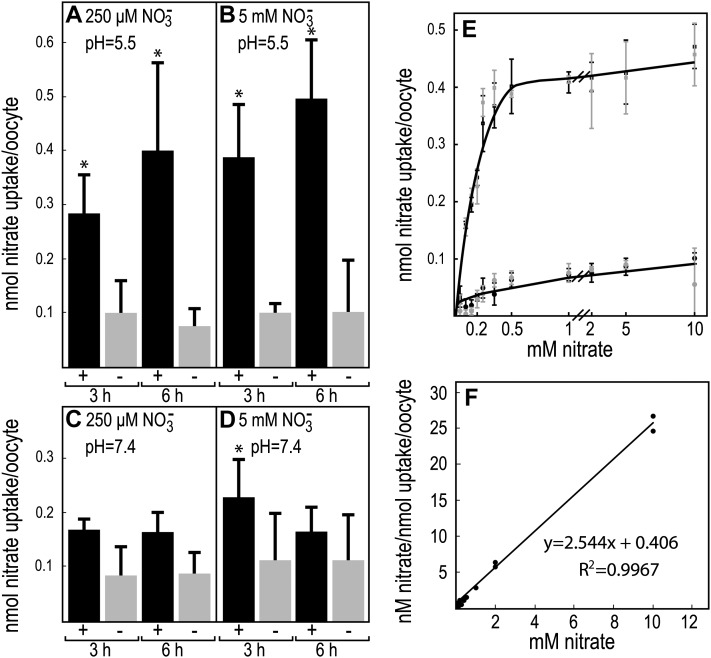
To test whether the MtNIP/LATD protein transports NO3−, we expressed MtNIP/LATD in X. laevis oocytes and assayed them for the acquisition of NO3− transport activity. Transport activity was initially assessed at two different NO3− concentrations to categorize transporter affinity and at pH 5.5 and 7.4 to test pH dependence. As shown in Figure 3, A and B, oocytes expressing MtNIP/LATD were capable of significant NO3− uptake above the water-injected control oocytes at both low (250 μm) and high (5 mm) NO3− concentrations at pH 5.5. However, at pH 7.4, there was no significant NO3− uptake (Fig. 3, C and D), indicating that transport is H+ coupled. We compared MtNIP/LATD NO3− transport with that of the dual-affinity AtNRT1.1 transporter and found that MtNIP/LATD had slightly lower nitrate transport than AtNRT1.1 at 250 μm NO3−. At 5 mm NO3−, MtNIP/LATD transported approximately 20% as much nitrate as did AtNRT1.1 (Supplemental Fig. S1), suggesting that MtNIP/LATD is not dual affinity. To determine MtNIP/LATD’s Km, we measured NO3− uptake in MtNIP/LATD-injected oocytes compared with water-injected control oocytes over NO3− concentrations ranging from 50 μm to 10 mm. MtNIP/LATD displays saturable kinetics for NO3− uptake, with a Km of 160 μm (Fig. 3, E and F). The data show only one saturation point (Fig. 3E) and thus indicate that MtNIP/LATD has single, high-affinity NO3− transport.
Figure 3.
Nitrate uptake in X. laevis oocytes expressing MtNIP/LATD. Oocytes were microinjected with MtNIP/LATD mRNA (black bars, +) or water as a negative control (gray bars, −), incubated for 3 d, and then placed for the indicated times in medium containing 250 μm or 5 mm NO3− at pH 5.5 or 7.4. The oocytes were rinsed, lysed, and assayed for NO3− content. A and C, Treatment with 250 μm NO3−. B and D, Treatment with 5 mm NO3−. A and B, pH = 5.5. C and D, pH = 7.4. Data are shown for one biological replicate ± sd (n = 3–5 batches of 4–6 oocytes per batch). Asterisks mark NO3− uptake significantly different from the negative control, using Student’s t test at P < 0.05. Similar results were obtained in more than five repetitions of the experiment. E, Michaelis-Menten plot of oocyte NO3− uptake. MtNIP/LATD-injected oocytes (squares) or water-injected oocytes (circles) as control were incubated for 3 h in 50 μm to 10 mm NO3− in batches of five and assayed for NO3− uptake. Results for two biological replicates are indicated by the black and gray symbols, with error bars showing sd. All NO3− uptake was significantly different from the negative control, using Student’s t test at P < 0.05, except for that at 50 μm. F, Hanes-Woolf plot of averaged NO3− uptake data, in MtNIP/LATD-injected oocytes minus water-injected oocytes, presented in E. These data were used to calculate the Km of 160 μm.
Because the NRT1(PTR) family BnNRT1.2 NO3− transporter also transports His (Zhou et al., 1998), a rat NRT1(PTR) family member transports both peptides and His (Yamashita et al., 1997), and Arabidopsis AtPTR1 and AtPTR2 peptide transporters also transport His (Tsay et al., 2007), we assessed His uptake in MtNIP/LATD-injected oocytes compared with water-injected control oocytes. At pH 5.5 and 1 mm His, we observed no His transport (Supplemental Table S1).
Proteins Encoded by Two MtNIP/LATD Mutant Alleles But Not a Third Allele Are Defective in Nitrate Transport in the Oocyte System
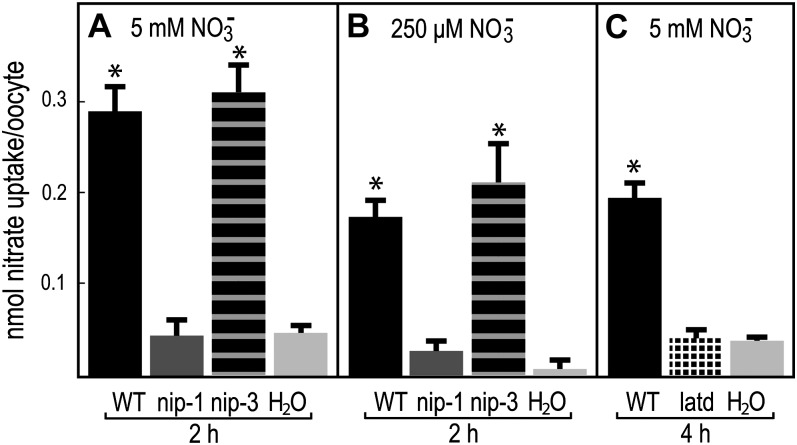
The finding that MtNIP/LATD transports NO3− opens the possibility that the defects observed in the Mtnip/latd mutants result from defective NO3− transport. To determine if the proteins encoded by the available defective MtNIP/LATD genes are capable of NO3− transport, we tested them in the X. laevis oocyte system. The results demonstrate that the missense Mtnip-1 and truncated Mtlatd proteins are defective in NO3− transport at 5 mm NO3−, while the Mtnip-3 protein is capable of transport at this higher concentration (Fig. 4). Oocytes expressing Mtnip-3 protein as well as Mtnip-1 protein were assessed for high-affinity transport at 250 μm NO3−; since the Mtlatd protein, encoded by a gene with a nonsense codon in the middle of MtNIP/LATD, failed to transport NO3− at 5 mm NO3−, it was not tested for transport at 250 μm NO3−. Mtnip-3 was capable of transport at 250 μm, while Mtnip-1 was not. Because the Mtnip-3 mutant has defective root architecture, aberrant nodulation, and fixes far less N than the wild type (Teillet et al., 2008), it may be defective in another function besides NO3− transport.
Figure 4.
Nitrate uptake in X. laevis oocytes expressing MtNIP/LATD or mutant Mtnip/latd mRNAs. Oocytes were microinjected with MtNIP/LATD mRNA, mutant mRNA, or water as a negative control, incubated for 3 d, and then placed for the indicated times in medium containing 5 mm or 250 μm nitrate, at pH 5.5, and assayed for nitrate uptake. A, Treatment with 5 mm nitrate. B, Treatment with 250 μm nitrate. C, Treatment with 5 mm nitrate. Oocytes expressing wild-type (WT) MtNIP/LATD (black bars), Mtnip-1 (dark gray bars), Mtnip-3 (horizontally striped bars), Mtlatd (hatched bars), or water as a negative control (gray bars) are shown. Data are shown for one biological replicate ± sd (n = 3–5 batches of 4–6 oocytes per batch). Asterisks mark nitrate uptake that is significantly different from the negative control, using Student’s t test at P < 0.05. Similar results were obtained in more than three repetitions of the experiment.
MtNIP/LATD Expression in the Arabidopsis chl1-5 Mutant Restores Chlorate Sensitivity
Although MtNIP/LATD transports NO3− in oocytes, it could be argued that it may not function as a NO3− transporter in planta. The NO3− transporter activity of MtNIP/LATD was further tested by studying its ability to complement the well-characterized Arabidopsis chl1-5 mutant, containing a large deletion in the AtNRT1.1 gene (Muños et al., 2004). This mutant was originally isolated on the basis of its resistance to the herbicide chlorate, which is taken up through the AtNRT1.1(AtCHL1) NO3− transporter and reduced by NO3− reductase to toxic chlorite (Doddema et al., 1978; Tsay et al., 1993). A construct containing MtNIP/LATD cDNA under the control of the constitutive Arabidopsis EF1α promoter (pAtEF1α-MtNIP/LATD) was introduced into Atchl1-5 plants, with plants transformed by AtNRT1.1 cDNA regulated by the same promoter (pAtEF1α-AtNRT1.1) serving as a positive control. Atchl1-5/pAtEF1α-MtNIP/LATD was found to be sensitive to chlorate, similar to the positive control Atchl1-5 plants transformed with pAtEF1α-AtNRT1.1 and wild-type ecotype Columbia (Col-0). Negative control mutant Atchl1-5 plants were resistant to chlorate, as expected (Fig. 5; Supplemental Fig. S2). Wild-type Col-0 and the Atchl1-5 plants constitutively expressing either AtNRT1.1 or MtNIP/LATD showed reductions in fresh weight and chlorophyll content after chlorate treatment compared with the resistant Atchl1-5 plants (Table II). A second independent transformed line of Atchl1-5 transformed with pAtEF1α-MtNIP/LATD was also constructed and found to be chlorate sensitive as well (Supplemental Fig. S2). Therefore, we conclude that since MtNIP/LATD transports the NO3− analog chlorate in planta, it is extremely likely to transport NO3− in planta as well.
Figure 5.
MtNIP/LATD complements the chlorate-insensitivity phenotype of the Arabidopsis chl1-5 mutant. Arabidopsis chl1-5 plants were transformed with a construct containing MtNIP/LATD cDNA under the control of the Arabidopsis EF1α promoter, pAtEF1α-MtNIP/LATD, or a positive control construct containing the Arabidopsis AtNRT1.1 gene under the control of the same promoter, pAtEF1α-AtNRT1.1. The plants were treated with chlorate, a NO3− analog that can be converted to toxic chlorite after uptake, as described by Tsay et al. (1993). A, Arabidopsis Col-0 plant. B, Arabidopsis chl1-5 plant. C, Arabidopsis chl1-5/pAtEF1α-AtNRT1.1 plant. D, Arabidopsis chl1-5/pAtEF1α-MtNIP/LATD plant. Bars = one-quarter inch. The MtNIP/LATD gene was able to confer chlorate sensitivity on Arabidopsis chl1-5 plants, similar to the AtNRT1.1 gene.
Table II. Fresh weight and chlorophyll content of chlorate-treated Arabidopsis plants.
Plants were grown in vermiculite:perlite (1:1) and irrigated with medium containing 5 mm NO3− for 5 to 7 d. Plants were then irrigated with ClO3−-containing medium without NO3− for 3 d and subsequently with medium lacking both ClO3− and NO3−. Fresh weight and chlorophyll content were obtained from plants 7 to 10 d after ClO3− treatment. Each data point represents seven to nine plants. Two independent replicates gave similar results.
| Genotype | Fresh Weight per Plant | Chlorophyll Content |
|---|---|---|
| mg | mg g−1 fresh wt | |
| Col-0 | 18.4 ± 1.6 | 1.05 ± 0.07 |
| chl1-5 | 150.4 ± 6.7 | 2.08 ± 0.08 |
| chl1-5/pAtEF1α-AtNRT1.1 | 20.9 ± 1.5 | 0.92 ± 0.06 |
| chl1-5/pAtEF1α-MtNIP/LATD | 21.7 ± 0.9 | 1.15 ± 0.15 |
AtNRT1.1, But Not AgDCAT1, Partially Rescues the M. truncatula nip-1 Phenotype
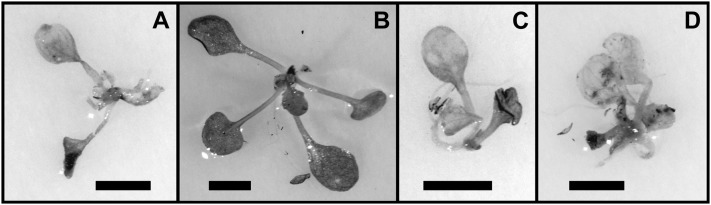
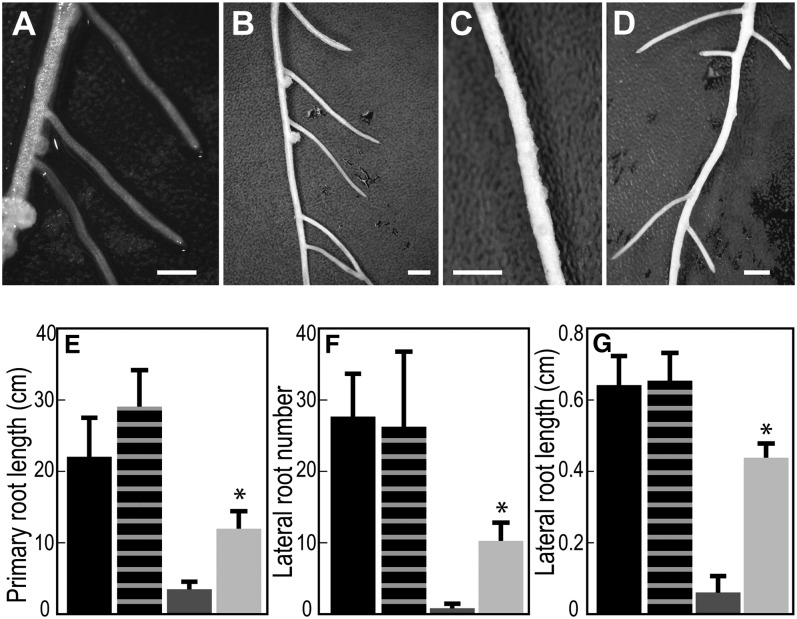
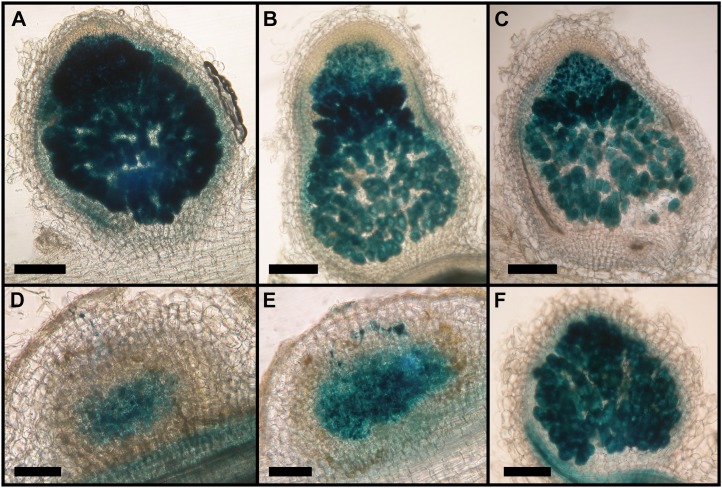
Since MtNIP/LATD restored chlorate sensitivity to the Arabidopsis chl1-5 mutant, we tested whether AtNRT1.1 would restore the M. truncatula nip-1 mutant to its wild-type phenotype. At the time that this experiment was performed, AtNRT1.1 was the only NRT1(PTR) member known to be a high-affinity (dual-affinity) NO3− transporter (Tsay et al., 2007). We used composite M. truncatula plant hairy roots (Boisson-Dernier et al., 2001) transformed with pAtEF1α-AtNRT1.1 as our test system. Composite plants were grown in aeroponic chambers in the absence of NO3−, inoculated with S. meliloti/phemA::lacZ (Boivin et al., 1990), and root architecture and nodulation were evaluated at 15 d post inoculation (dpi). The results showed that AtNRT1.1 partially restored Mtnip-1 root architecture, increasing primary root length, LR number, and LR length to 53%, 37%, and 67% of the wild-type values, respectively (Fig. 6). However, the nodules in Mtnip-1 plants transformed with the pAtEF1α-AtNRT1.1 construct had an Mtnip-1 phenotype, with the presence of brown pigments, no obvious meristem, and did not differentiate into the zones that are the hallmark of N-fixing nodules (Fig. 7). Control plants transformed with pAtEF1α-MtNIP/LATD had wild-type phenotype nodules (Fig. 7F) and showed comparable rescue of nodulation and root architecture phenotypes as those transformed with pMtNIP/LATD-MtNIP/LATD (Supplemental Fig. S3), showing that use of the Arabidopsis EF1α promoter is not a limiting factor for complementation of the Mtnip-1 phenotype. Overall, the experiment shows that although AtNRT1.1 partially restores the root architecture phenotype, it is not able to restore normal nodulation to the Mtnip-1 plants.
Figure 6.
Arabidopsis NRT1.1 partially complements the M. truncatula nip-1 mutant for its root architecture phenotype. M. truncatula nip-1 and control wild-type composite plants transformed with pAtEF1α-AtNRT1.1 or empty pCAMBIA2301 vector, as a control, were grown in aeroponic chambers, inoculated with S. meliloti containing a constitutive lacZ gene, and grown in a 16/8-h light/dark cycle at 22°C. At 15 dpi, root architecture characteristics were evaluated. A to D, Appearance of the roots. Bars = 10 mm. A, A17, empty vector. B, A17, pAtEF1α-AtNRT1.1. C, Mtnip-1, empty vector. D, Mtnip-1, pAtEF1α-AtNRT1.1. E to G, Quantitation of primary root length (E), LR number (F), and LR length (G). Averaged values with sd are plotted (n = 5). Asterisks mark root attributes that are significantly different from the negative control, using Student’s t test at P < 0.01. Black bars, A17, empty vector; vertically striped bars, A17, pAtEF1α-AtNRT1.1; dark gray bars, Mtnip-1, empty vector; light gray bars, Mtnip-1, pAtEF1α-AtNRT1.1.
Figure 7.
Arabidopsis NRT1.1 does not complement the M. truncatula nip-1 nodule phenotype. M. truncatula nip-1 and control wild-type composite plants transformed with pAtEF1α-AtNRT1.1, empty vector pCAMBIA2301 as a negative control, or pAtEF1α-MtNIP/LATD were grown as in Figure 5 with S. meliloti containing a constitutive lacZ gene. At 15 dpi, nodule characteristics were evaluated after staining with X-Gal for localization of rhizobia, which stain blue. A, A17 transformed with empty vector. B, A17 transformed with pAtEF1α-AtNRT1.1. C, A17 transformed with pAtEF1α-MtNIP/LATD. D, Mtnip-1 transformed with empty vector. E, Mtnip-1 transformed with pAtEF1α-AtNRT1.1. F, Mtnip-1 transformed with pAtEF1α-MtNIP/LATD. Bars = 200 μm.
We also tested whether the gene encoding the alder symbiotic AgDCAT1 dicarboxylate transporter could restore normal root development or nodulation to Mtnip-1 plants, using a similar approach as for AtNRT1.1, in composite transformed plants. Mtnip-1 plants expressing AgDCAT1 had a phenotype indistinguishable from Mtnip-1 plants transformed with an empty vector; in contrast, Mtnip-1 plants expressing MtNIP/LATD were restored to the wild type. Wild-type A17 plant phenotypes were unaffected by AgDCAT1 expression (Supplemental Fig. S3).
DISCUSSION
The major finding of the work reported here is that MtNIP/LATD protein is a NO3− transporter, and its NO3− transport activity partially correlates with root architecture development. Our results also suggest that MtNIP/LATD has another unknown activity that is important to MtNIP/LATD’s biological roles in nodule development and in the modulation of root architecture.
Data for MtNIP/LATD NO3− transport come from two complementary experimental approaches. When MtNIP/LATD is expressed in the heterologous X. laevis oocyte system, it enables the oocytes to transport NO3− in a pH-dependent manner, demonstrating that NO3− transport is H+ driven (Fig. 3), similar to other NO3− transporters in the NRT1(PTR) family (Tsay et al., 1993). NO3− uptake was characterized as having a Km of 160 μm, making it a HATS. A second line of evidence that MtNIP/LATD is a NO3− transporter is that its gene’s constitutive expression can complement the Arabidopsis chl1-5 mutant, containing a deletion spanning the AtNRT1.1 gene (Muños et al., 2004). Atchl1-5 mutants expressing MtNIP/LATD are susceptible to chlorate, indicating that MtNIP/LATD confers on them the ability to take up the herbicide chlorate, a NO3− analog, from the medium (Fig. 5). Both approaches showing that MtNIP/LATD transports NO3− indicate that the direction of NO3− transport is from outside to inside cells.
One might expect Mtnip/latd mutants to exhibit defects in NO3− uptake. We measured NO3− uptake from medium by Mtnip-1 and Mtnip-3 mutants and control wild-type M. truncatula A17 and observed no differences between the plants in NO3− uptake at either 250 μm or 5 mm NO3−, representative of HATS and LATS, respectively (Fig. 1). This suggests that MtNIP/LATD is not a rate-limiting transporter for NO3− uptake into plant tissue. MtNIP/LATD is expressed in primary and LR tips (Yendrek et al., 2010); if MtNIP/LATD’s primary biological role is to transport NO3−, it may constitute only a small portion of NO3− transport in M. truncatula roots. It is also possible that in Mtnip/latd mutants, the plant compensates by up-regulating the activity of another transporter. Another possibility is that MtNIP/LATD’s transport function may be critical for redistribution of NO3− within the plant. We found that Mtnip-1 is apparently normal for NO3− suppression of nodulation (Table I), implying that MtNIP/LATD may not be involved in this pathway (Streeter, 1988; Fei and Vessey, 2009), and/or that there are other NO3− transporters that can compensate for this function.
Previously, the effects of 10 and 50 mm KNO3 on primary root length and LR density in Mtlatd mutants were monitored; Mtlatd plants, like the wild type, do not show altered LR density in response to global increases in NO3− (Yendrek et al., 2010). Here, we examined LR lengths of Mtnip-1 and Mtnip-3 plants grown in 0 μm, 250 μm, or 5 mm KNO3 and found that the LR length phenotype was rescued for Mtnip-3 and partially rescued for Mtnip-1 at the 5 mm KNO3 level but not at 250 μm KNO3 (Fig. 2), suggesting that MtNIP/LATD might have a more biologically important role at lower NO3− concentrations than at higher ones. However, these experiments did not control for the effects of salt concentration on root architecture, which could have affected abscisic acid levels, leading to changes in LR lengths (Liang and Harris, 2005; Liang et al., 2007), and thus are only suggestive.
Even though NO3− uptake was not limited in Mtnip/latd mutants compared with wild-type plants, NO3− uptake differed when these alleles were expressed in oocytes and assayed for nitrate transport. Nitrate uptake experiments in the oocyte system showed that the protein encoded by the weakest allele, Mtnip-3, transported NO3− indistinguishably from the wild type, while the proteins encoded by the two more severe alleles, Mtnip-1 and Mtlatd, did not (Fig. 4). The Mtnip-3 mutant has a phenotype: it forms Fix+/− nodules that accumulate polyphenolics and has minor defects in root architecture, in primary root length (Teillet et al., 2008; Yendrek et al., 2010) and LR length (Fig. 2). Thus, there is a correlation between MtNIP/LATD’s ability to transport NO3− and Mtnip/latd mutants’ abilities to form and maintain nodule and root meristems and to form nodules invaded intracellularly by rhizobia. Despite that NO3− transport by Mtnip-3 protein is indistinguishable from the transport by wild-type MtNIP/LATD, Mtnip-3 has a root and nodule phenotype. This indicates that MtNIP/LATD may have another function besides NO3− transport.
To further address the possible link between MtNIP/LATD and NO3− transport, we expressed AtNRT1.1 in Mtnip-1 roots. We observed that the AtNRT1.1-transformed Mtnip-1 plants are partially restored for their root architecture phenotype but not for the nodulation phenotype. The use of pAtEF1α is not a limiting factor for phenotype rescue, since MtNIP/LATD, expressed under the control of the same promoter, was able to restore both the root architecture phenotype (data not shown) and the nodulation phenotype (Fig. 7). No effect was observed when AtNRT1.1 was expressed in wild-type A17 plants (Figs. 6 and 7). This suggests that AtNRT1.1 protein’s dual-affinity NO3− transport activity affects Mtnip-1 root architecture and supports the idea that MtNIP/LATD’s NO3− transport activity has a role in modulating root developmental responses. Additionally, the lack of full complementation of the root architecture defects by AtNRT1.1 and the noncomplementation of the nodulation phenotype by AtNRT1.1 suggest that there is a function of MtNIP/LATD that is different from that of AtNRT1.1. Alternatively, it is possible that posttranslational regulation of AtNRT1.1 in M. truncatula differs from that of MtNIP/LATD, especially in nodules, and that this is the cause of AtNRT1.1’s inability to complement Mtnip-1’s nodulation phenotype. Because AtNRT1.1 has been demonstrated to transport auxin in the absence of NO3− (Krouk et al., 2010b) and NO3− was only provided during plant transformation in these experiments, another possible explanation for the observed effects on Mtnip’s roots is that the partial restoration of normal root architecture was brought about by AtNRT1.1-induced changes in auxin concentration. When normally expressed in Arabidopsis, AtNRT1.1 is thought to prevent auxin accumulation at LR tips by mediating basipetal auxin transport in LRs, thus halting LR growth (Krouk et al., 2010b); this is the opposite of what we observed in the Mtnip-1 mutant expressing AtNRT1.1 (Fig. 6). Because our experiments used a constitutive promoter to express AtNRT1.1, it is possible that the perturbation of auxin gradients within the roots caused the observed changes in root architecture. It is also curious that the nodule phenotype of Mtnip-1 plants transformed with AtNRT1.1 is different from that of Mtnip-3. If the ability of Mtnip-3 nodules to form a meristem and allow rhizobia to invade intracellularly is related to Mtnip-3 protein’s ability to transport NO3−, one would expect to find the same phenotype in Mtnip-1 plants transformed with AtNRT1.1 as in Mtnip-3, which is not the case. This finding further supports the idea that MtNIP/LATD protein’s activity is more than simply NO3− transport and is also not explained by the spectrum of activities inherent in AtNRT1.1.
What role could MtNIP/LATD-mediated NO3− transport play in nodulation, root architecture development, or their regulation? MtNIP/LATD NO3− transport could supply N at low NO3−concentrations to dividing plant and bacterial cells early in nodulation, and also to differentiated nodules, at the dividing and endoreduplicating cells present in zones I and II, where MtNIP/LATD’s promoter is active (Yendrek et al., 2010). The MtNIP/LATD promoter is also active in primary root meristems, LR meristems and surrounding tissue, and MtNIP/LATD NO3− transport could have a similar role there. In this case, the supply of NO3− to these tissues would provide the N required for basic cellular functions required by dividing cells, possibly facilitating the transition from primordium to self-sustaining meristem by these nascent LR organs.
Alternately, MtNIP/LATD could transport NO3− as a precursor to the potent signaling molecule nitric oxide (NO) early in nodulation and LR development. NO has been detected in M. truncatula nodule primordia not containing intracellular rhizobia, suggesting an active NO pathway in these cells, as well as in infection threads, where NO could come from either symbiotic partner (del Giudice et al., 2011). NO has also been detected in LR primordia in tomato (Solanum lycopersicum; Correa-Aragunde et al., 2004). We note, however, that nodulation occurs in environmental conditions where N is limiting; indeed, our laboratory conditions for nodulation occur in N starvation. We supply 0 μm NO3− during nodulation, and only the trace NO3−, expected to be in the micromolar range, present as contaminants in nutrient media and glassware are available. If NO3− is supplied to dividing nodule cells, it must come from seed NO3− stores, which should be close to depleted by the time nodules are forming or be remobilized from other N-rich components within the plant. Another possibility is that MtNIP/LATD NO3− transport could participate in a proposed NO3−-NO respiration pathway in nodules (Horchani et al., 2011; Meilhoc et al., 2011). There are several observations that argue against this: Mtnip-3, with functional NO3− transport (Fig. 4), has abnormal nodules (Teillet et al., 2008), and MtNIP/LATD’s promoter is active in nodule meristems and in the invasion zone but not in the N-fixing zone (Yendrek et al., 2010), suggesting that MtNIP/LATD may be absent in the N-fixing zone.
Because MtNIP/LATD may have another function besides NO3− transport, it is plausible that the other function is responsible for some of the Mtnip/latd mutants’ phenotypes. Here, we have presented data suggesting that neither His nor dicarboxylates are substrates for MtNIP/LATD transport (Supplemental Table S1; Supplemental Fig. S3). We and others have speculated that MtNIP/LATD may be a NO3− transceptor or sensor (Harris and Dickstein, 2010; Yendrek et al., 2010; Gojon et al., 2011). If it is a NO3− transceptor or sensor, we predict that it may be responsible for high-affinity NO3− sensing. This is because it is a high-affinity transporter and because the root architecture phenotypes are partially rescued by high, but not low, NO3− concentrations (Fig. 2). It is also possible that MtNIP/LATD is able to transport hormones like AtNRT1.1 (Krouk et al., 2010b) and AtNRT1.2 (Kanno et al., 2012) and that this is responsible for the other function(s) of MtNIP/LATD.
MATERIALS AND METHODS
Plant Growth Conditions
Medicago truncatula A17 (wild type) and nodulation mutants were grown in aeroponic chambers with Lullien medium (Lullien et al., 1987), starved for N for 5 d, and inoculated with Sinorhizobium medicae strain ABS7 (Bekki et al., 1987) or Sinorhizobium meliloti strain Rm2011 (Rosenberg et al., 1981) harboring pXLGD4 (Boivin et al., 1990; Penmetsa and Cook, 1997), as described previously (Veereshlingam et al., 2004; Pislariu and Dickstein, 2007). For experiments where M. truncatula plants were grown in the presence of N sources, the relevant N sources were added to Lullien medium from the beginning of the experiment and inoculations were done with ABS7/pXLGD4. At 15 dpi, plants were stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside acid (X-Gal) to identify nodules. Arabidopsis (Arabidopsis thaliana) plants were grown as described (Srivastava et al., 2008) at 22°C with a 16/8-h light/dark regime.
Nitrate Uptake Studies in M. truncatula
A17, Mtnip-1, and Mtnip-3 seedlings were surface sterilized, germinated, placed on buffered nodulation medium (Ehrhardt et al., 1992) solidified with agar, pH 5.8, supplemented with 5 mm NH4NO3, and grown for 7 d at 22°C with a 16/8-h light/dark regime, with roots shielded from the light. Plants were transferred to N-free buffered nodulation medium agar and grown for 3 d to starve them for N. Plants were placed in liquid buffered nodulation medium supplemented with either 250 μm or 5 mm KNO3. Samples from the medium were collected at the indicated times and assayed for NO3− using the Cayman (no. 780001) NO3−/NO2− assay kit following the manufacturer’s instructions.
Construction of Oocyte Expression Vectors
RNA was extracted from M. truncatula A17 plants and mutant Mtnip-1, Mtlatd, and Mtnip-3 using the RNeasy kit (Qiagen). First-strand cDNA was transcribed using oligo(dT) and SuperScript III reverse transcriptase (Invitrogen), and the wild-type MtNIP/LATD and mutant Mtnip/latd cDNAs were made using MtNIP/LATD-specific primers NIPcEXP1R, NIPcExp1F, and NIPcDNANheI_F (Supplemental Table S2) via PCR with Phusion high-fidelity DNA polymerase (New England Biolabs). The resulting 1,776-bp cDNAs were cloned into vectors pSP64T (Krieg and Melton, 1984) and pcDNA3.1(−) (Invitrogen). AtNRT1.1 cDNA was amplified from pMS008 (see below) using primers Chl_1F and Chl_1R, cloned into PCR8/GW/TOPO, and then moved into pOO2/GW (a gift from Dr. John Ward) downstream of an SP6 promoter. All clones were verified by DNA sequencing.
Binary Vectors
MtNIP/LATD cDNA was amplified with NIPC2F and NIPCODBst1R, digested with NcoI and BstEII, and ligated into a vector containing the AtEF1α promoter engineered to contain a 5′ BamHI site and a 3′ NcoI site, creating pMS004. Subsequently, the AtEF1α -MtNIP/LATD cDNA fragment was cloned into the BamHI and BstEII sites of binary vector pCAMBIA2301. AtNRT1.1 cDNA was amplified from total cDNA made from wild-type Col-0 Arabidopsis mRNA using primers CHL11F and CHL11R, digested with BspHI and BstEII, and cloned into NcoI- and BstEII-digested pMS004 to obtain pMS008. The pAtEF1α-AtNRT1.1 fragment was then subcloned into the BamHI and BstEII sites of pCAMBIA2301. For the pMtNIP/LATD-MtNIP/LATD construct, the pAtEF1α in pMS004 was replaced by a 3-kb upstream region of MtNIP/LATD amplified using NIP 10F and NIP P1R primers, followed by EcoRI and NcoI/BspHI digestion to create the pMS006 clone. Then, the pMS006 EcoRI/BstEII fragment was moved into the EcoRI/BstEII site of pCAMBIA2301. AgDCAT1 cDNA was PCR amplified from pJET1.2-AgDCAT1 (a gift of Dr. K. Pawlowski) using DC-BglII and DC-NheI primers and cloned into the BamHI and NheI sites of pMS014 vector, to create pMS015. Then, the pMS015 EcoRI/BstEII fragment was moved into the EcoRI/BstEII site of pCAMBIA2301 to create the pMtNIP/LATD-AgDCAT1 construct. Supplemental Table S2 contains the primer sequences.
Nitrate and His Uptake in Xenopus laevis Oocytes
Capped mRNA was transcribed in vitro from linearized plasmid using SP6 or T7 RNA polymerase (mMESSAGE mMACHINE; Ambion). Collagenase-treated oocytes were isolated and microinjected with approximately 50 ng of RNA in 50 nL of sterile water. Oocytes microinjected with 50 nL of sterile water were used as negative controls. Oocytes were incubated in ND96 solution (Liu and Tsay, 2003) for 24 to 40 h at 18°C. For NO3− uptake, oocytes were incubated in a solution containing 230 mm mannitol, 15 mm CaCl2, and 10 mm HEPES (pH 7.4), containing various KNO3 concentrations at pH 5.5 or 7.4, at 16°C for the indicated times. After incubation, batches of four to six oocytes were lysed in 40 μL of water, centrifuged at 13,500g for 20 min, and the supernatant was analyzed for NO3− content as above. For His uptake, oocytes were incubated in ND96 medium, pH 5.5, at 25°C containing 1 mm uniformly 13C-labeled His (Cambridge Isotope). After incubation, oocytes were washed five times with the same medium containing 10 mm unlabeled His (Sigma-Aldrich), and batches of eight oocytes were lysed in 100 μL of water and centrifuged at 13,500g for 20 min. The supernatant was lyophilized to 15 μL and analyzed using ultraperformance liquid chromatography-electrospray ionization-tandem mass spectrometry (UPLC-ESI-MS/MS).
UPLC-ESI-MS/MS Analysis
[13C6]His was quantified using a precolumn derivatization method with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate (AQC) combined with UPLC-ESI-MS/MS. AQC derivatization was performed using the AccQ•Tag derivatization kit (Waters) according to the manufacturer’s protocol. UPLC-ESI-MS/MS analysis was carried out on a Waters Acquity UPLC system interfaced to a Waters Xevo TQ mass spectrometer as described (Salazar et al., 2012). Briefly, the AQC-derivatized [13C6]His was separated on a Waters AccQ•Tag Ultra column (2.1 mm i.d. × 100 mm, 1.7-μm particles) using AccQ•Tag Ultra eluents (Waters) and a gradient described earlier (Salazar et al., 2012). The sample injection volume was 1 μL, the UPLC column flow rate was 0.7 mL min−1, and the column temperature was 55°C. Mass spectra were acquired using positive ESI and the multiple reaction monitoring mode, with the following ionization source settings: capillary voltage, 1.99 kV (ESI+); desolvation temperature, 650°C; desolvation gas flow rate, 1,000 L h−1; source temperature, 150°C. Argon was used as the collision gas at a flow rate of 0.15 mL min−1. The collision energy and cone voltage were optimized for the derivatized [13C6]His using the IntelliStart software (collision energy = 26 eV; cone voltage = 20 V). The most sensitive parent-daughter ion transition of derivatized His (mass-to-charge ratio 332.1 > 171.0) was selected for quantitation. The mass spectrometer response was calibrated by injecting AQC-derivatized [13C6]His standard solutions of known concentration. The UPLC-ESI-MS/MS system control and data acquisition were performed with Waters MassLynx software. Data analysis was conducted with TargetLynx software (Waters).
Transformation of Atchl1-5 Plants with the MtNIP/LATD Expression Construct
The MtNIP/LATD expression construct (pAtEF1α-MtNIP/LATD) was transformed into the Agrobacterium tumefaciens GV3101(pMP90) strain by electroporation. Positive colonies were selected by colony PCR, verified by restriction digestion, and transformed into the Atchl1-5 mutant (Tsay et al., 1993) by the floral dip method (Clough and Bent, 1998). Seeds were collected, and transformed plants were selected on kanamycin medium. Two independent homozygous transformed lines of Atchl1-5 transformed with pAtEF1α-MtNIP/LATD were selected.
Chlorate Sensitivity Test
Plants were grown in vermiculite:perlite (1:1 mixture) under continuous illumination at 25°C to 27°C. Plants were irrigated every 2 to 3 d with medium containing 10 mm KH2PO4 (pH 5.3), 5 mm KNO3, 2 mm MgSO4, 1 mm CaCl2, 0.1 mm FeEDTA, 50 pm H3BO3, 12 pm MnSO4, 1 pm ZnCl, 1 pm CuSO4, and 0.2 pm Na2MoO4. At 5 to 7 d post germination, plants were irrigated twice with medium containing 2 mm NaClO3 without NO3−. Three days after ClO3− treatment, plants were switched to irrigation medium lacking ClO3− and NO3−. Plants were examined 7 to 10 d after ClO3− treatment for necrosis and bleaching symptoms characteristic of chlorate toxicity (Wilkinson and Crawford, 1991), and their fresh weights and chlorophyll contents were obtained. Digital color photographs of the plants were obtained, corrected for color balance (Supplemental Fig. S2), converted to grayscale, and corrected for contrast (Fig. 5) using Photoshop (Adobe Software).
Chlorophyll content was determined from approximately 10 mg of fresh leaves that were weighed, frozen in liquid N2, ground to a fine powder, added to tubes with 100 µL of water and 8 mL of 96% ethanol, and mixed. The tubes were kept at 25°C overnight, mixed again, and the particulates were allowed to settle. The absorbance was recorded at 648.6 and 664.2 nm. Total chlorophyll was calculated as described (Lichtenthaler, 1987).
M. truncatula Root Transformation
Vectors containing the expression constructs were transformed into Agrobacterium rhizogenes ARqua1 strain (Quandt et al., 1993) by the freeze-thaw method (Höfgen and Willmitzer, 1988). Positive ARqua1 colonies were transformed into Mtnip-1 and A17 plants by the needle-poking method (Mortier et al., 2010).
Analysis of LRs and Nodules in Transformed Plants
Root nodules were analyzed at 12 dpi. They were stained with X-Gal for lacZ, present in pXLGD4 plasmid in S. medicae ABS7 and mounted in 2.5% low melting point agarose; 50-µm sections were obtained using a 1000 Plus Vibratome (Vibratome) and observed by light microscopy. LRs were inspected visually and with a dissecting microscope.
The MtNIP/LATD sequence may be found under GenBank accession no. GQ401665.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparison of nitrate uptake in oocytes expressing MtNIP/LATD versus AtNRT1.1.
Supplemental Figure S2. MtNIP/LATD complements the chlorate-insensitivity phenotype of the Arabidopsis chl1-5 mutant.
Supplemental Figure S3. AgDCAT1 does not complement the Mtnip-1 mutant.
Supplemental Table S1. MtNIP/LATD-expressing oocytes do not take up His.
Supplemental Table S2. Sequences of oligonucleotide primers used in this study.
Acknowledgments
We thank Pudur Jagadeeswaran, Tina Machu, and their laboratories for help with the X. laevis oocyte experiments, John Ward for vector pOO2/GW and suggestions about oocytes, Jeanne Harris for helpful discussions, Brian Ayre and his laboratory for help with Arabidopsis transformation, Nigel Crawford for Arabidopsis chl1-5 seeds, Katharina Pawlowski for the AgDCAT1 cDNA, and Jyoti Shah and Jeremy Murray for critical reading of the manuscript.
Glossary
- HATS
high-affinity transport system
- LATS
low-affinity transport system
- N
nitrogen
- LR
lateral root
- Col-0
ecotype Columbia
- dpi
d post inoculation
- NO
nitric oxide
- UPLC
ultraperformance liquid chromatography
- ESI
electrospray ionization
- MS/MS
tandem mass spectrometry
- AQC
6-aminoquinolyl-N-hydroxysuccinimidyl carbamate
- cDNA
complementary DNA
- X-gal
5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside acid
References
- Almagro A, Lin SH, Tsay Y-F. (2008) Characterization of the Arabidopsis nitrate transporter NRT1.6 reveals a role of nitrate in early embryo development. Plant Cell 20: 3289–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier-Brygoo H, De Angeli A, Filleur S, Frachisse J-M, Gambale F, Thomine S, Wege S. (2011) Anion channels/transporters in plants: from molecular bases to regulatory networks. Annu Rev Plant Biol 62: 25–51 [DOI] [PubMed] [Google Scholar]
- Bekki A, Trinchant J-C, Rigaud J. (1987) Nitrogen fixation (C2H2 reduction) by Medicago nodules and bacteroids under sodium chloride stress. Physiol Plant 71: 61–67 [Google Scholar]
- Benedito VA, Li H, Dai X, Wandrey M, He J, Kaundal R, Torres-Jerez I, Gomez SK, Harrison MJ, Tang Y, et al. (2010) Genomic inventory and transcriptional analysis of Medicago truncatula transporters. Plant Physiol 152: 1716–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A, Chabaud M, Garcia F, Bécard G, Rosenberg C, Barker DG. (2001) Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact 14: 695–700 [DOI] [PubMed] [Google Scholar]
- Boivin C, Camut S, Malpica CA, Truchet G, Rosenberg C. (1990) Rhizobium meliloti genes encoding catabolism of trigonelline are induced under symbiotic conditions. Plant Cell 2: 1157–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright L, Liang Y, Mitchell DM, Harris JM. (2005) The LATD gene of Medicago truncatula is required for both nodule and root development. Mol Plant Microbe Interact 18: 521–532 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Correa-Aragunde N, Graziano M, Lamattina L. (2004) Nitric oxide plays a central role in determining lateral root development in tomato. Planta 218: 900–905 [DOI] [PubMed] [Google Scholar]
- Crawford NM. (1995) Nitrate: nutrient and signal for plant growth. Plant Cell 7: 859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo G, Valkov VT, Parlati A, Alves LM, Chiurazzi M. (2012) Molecular characterization of the Lotus japonicus NRT1(PTR) and NRT2 families. Plant Cell Environ 35: 1567–1581 [DOI] [PubMed] [Google Scholar]
- del Giudice J, Cam Y, Damiani I, Fung-Chat F, Meilhoc E, Bruand C, Brouquisse R, Puppo A, Boscari A. (2011) Nitric oxide is required for an optimal establishment of the Medicago truncatula-Sinorhizobium meliloti symbiosis. New Phytol 191: 405–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doddema H, Hofstra JJ, Feenstra WJ. (1978) Uptake of nitrate by mutants of Arabidopsis thaliana, disturbed in uptake or reduction of nitrate. Physiol Plant 43: 343–350 [Google Scholar]
- Ehrhardt DW, Atkinson EM, Long SR. (1992) Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science 256: 998–1000 [DOI] [PubMed] [Google Scholar]
- Fan S-C, Lin C-S, Hsu PK, Lin S-H, Tsay Y-F. (2009) The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate. Plant Cell 21: 2750–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei H, Vessey JK. (2009) Stimulation of nodulation in Medicago truncatula by low concentrations of ammonium: quantitative reverse transcription PCR analysis of selected genes. Physiol Plant 135: 317–330 [DOI] [PubMed] [Google Scholar]
- Gojon A, Krouk G, Perrine-Walker F, Laugier E. (2011) Nitrate transceptor(s) in plants. J Exp Bot 62: 2299–2308 [DOI] [PubMed] [Google Scholar]
- Harris JM, Dickstein R. (2010) Control of root architecture and nodulation by the LATD/NIP transporter. Plant Signal Behav 5: 1365–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C-H, Lin S-H, Hu H-C, Tsay Y-F. (2009) CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Höfgen R, Willmitzer L. (1988) Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res 16: 9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horchani F, Prévot M, Boscari A, Evangelisti E, Meilhoc E, Bruand C, Raymond P, Boncompagni E, Aschi-Smiti S, Puppo A, et al. (2011) Both plant and bacterial nitrate reductases contribute to nitric oxide production in Medicago truncatula nitrogen-fixing nodules. Plant Physiol 155: 1023–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N-C, Chiang C-S, Crawford NM, Tsay Y-F. (1996) CHL1 encodes a component of the low-affinity nitrate uptake system in Arabidopsis and shows cell type-specific expression in roots. Plant Cell 8: 2183–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Suh S, Guan C, Tsay Y-F, Moran N, Oh CJ, An C-S, Demchenko KN, Pawlowski K, Lee Y. (2004) A nodule-specific dicarboxylate transporter from alder is a member of the peptide transporter family. Plant Physiol 134: 969–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Hanada A, Chiba Y, Ichikawa T, Nakazawa M, Matsui M, Koshiba T, Kamiya Y, Seo M. (2012) Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc Natl Acad Sci USA 109: 9653–9658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi H, Imaizumi-Anraku H, Hayashi M, Hakoyama T, Nakagawa T, Umehara Y, Suganuma N, Kawaguchi M. (2010) How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol 51: 1381–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg PA, Melton DA. (1984) Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res 12: 7057–7070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Crawford NM, Coruzzi GM, Tsay YF. (2010a) Nitrate signaling: adaptation to fluctuating environments. Curr Opin Plant Biol 13: 266–273 [DOI] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, et al. (2010b) Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell 18: 927–937 [DOI] [PubMed] [Google Scholar]
- Li J-Y, Fu Y-L, Pike SM, Bao J, Tian W, Zhang Y, Chen C-Z, Zhang Y, Li H-M, Huang J, et al. (2010) The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22: 1633–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Harris JM. (2005) Response of root branching to abscisic acid is correlated with nodule formation both in legumes and nonlegumes. Am J Bot 92: 1675–1683 [DOI] [PubMed] [Google Scholar]
- Liang Y, Mitchell DM, Harris JM. (2007) Abscisic acid rescues the root meristem defects of the Medicago truncatula latd mutant. Dev Biol 304: 297–307 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Lin S-H, Kuo H-F, Canivenc G, Lin C-S, Lepetit M, Hsu P-K, Tillard P, Lin H-L, Wang Y-Y, Tsai CB, et al (2008) Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20: 2514–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkohr BI, Williamson LC, Fitter AH, Leyser HMO. (2002) Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J 29: 751–760 [DOI] [PubMed] [Google Scholar]
- Liu K-H, Tsay Y-F. (2003) Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J 22: 1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lullien V, Barker DG, de Lajudie P, Huguet T. (1987) Plant gene expression in effective and ineffective root nodules of alfalfa (Medicago sativa). Plant Mol Biol 9: 469–478 [DOI] [PubMed] [Google Scholar]
- Meilhoc E, Boscari A, Bruand C, Puppo A, Brouquisse R. (2011) Nitric oxide in legume-rhizobium symbiosis. Plant Sci 181: 573–581 [DOI] [PubMed] [Google Scholar]
- Miller AJ, Fan X, Orsel M, Smith SJ, Wells DM. (2007) Nitrate transport and signalling. J Exp Bot 58: 2297–2306 [DOI] [PubMed] [Google Scholar]
- Miranda M, Borisjuk L, Tewes A, Dietrich D, Rentsch D, Weber H, Wobus U. (2003) Peptide and amino acid transporters are differentially regulated during seed development and germination in faba bean. Plant Physiol 132: 1950–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morère-Le Paven M-C, Viau L, Hamon A, Vandecasteele C, Pellizzaro A, Bourdin C, Laffont C, Lapied B, Lepetit M, Frugier F, et al. (2011) Characterization of a dual-affinity nitrate transporter MtNRT1.3 in the model legume Medicago truncatula. J Exp Bot 62: 5595–5605 [DOI] [PubMed] [Google Scholar]
- Mortier V, Den Herder G, Whitford R, Van de Velde W, Rombauts S, D’Haeseleer K, Holsters M, Goormachtig S. (2010) CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol 153: 222–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muños S, Cazettes C, Fizames C, Gaymard F, Tillard P, Lepetit M, Lejay L, Gojon A. (2004) Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. Plant Cell 16: 2433–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JA. (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59: 519–546 [DOI] [PubMed] [Google Scholar]
- Penmetsa RV, Cook DR. (1997) A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275: 527–530 [DOI] [PubMed] [Google Scholar]
- Pislariu CI, Dickstein R. (2007) An IRE-like AGC kinase gene, MtIRE, has unique expression in the invasion zone of developing root nodules in Medicago truncatula. Plant Physiol 144: 682–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt HJ, Puhler A, Broer I. (1993) Transgenic root nodules of Vicia hirsuta: a fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol Plant Microbe Interact 6: 699–706 [Google Scholar]
- Remans T, Nacry P, Pervent M, Filleur S, Diatloff E, Mounier E, Tillard P, Forde BG, Gojon A. (2006) The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc Natl Acad Sci USA 103: 19206–19211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg C, Boistard P, Dénarié J, Casse-Delbart F. (1981) Genes controlling early and late functions in symbiosis are located on a megaplasmid in Rhizobium meliloti. Mol Gen Genet 184: 326–333 [DOI] [PubMed] [Google Scholar]
- Salazar C, Armenta JM, Cortés DF, Shulaev V. (2012) Combination of an AccQ·Tag-ultra performance liquid chromatographic method with tandem mass spectrometry for the analysis of amino acids. Methods Mol Biol 828: 13–28 [DOI] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, Sasamoto S, Watanabe A, Ono A, Kawashima K, et al. (2008) Genome structure of the legume, Lotus japonicus. DNA Res 15: 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Segonzac C, Boyer J-C, Ipotesi E, Szponarski W, Tillard P, Touraine B, Sommerer N, Rossignol M, Gibrat R. (2007) Nitrate efflux at the root plasma membrane: identification of an Arabidopsis excretion transporter. Plant Cell 19: 3760–3777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AC, Ganesan S, Ismail IO, Ayre BG. (2008) Functional characterization of the Arabidopsis AtSUC2 sucrose/H+ symporter by tissue-specific complementation reveals an essential role in phloem loading but not in long-distance transport. Plant Physiol 148: 200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeter J. (1988) Inhibition of legume nodule formation and N2 fixation by nitrate. CRC Crit Rev Plant Sci 7: 1–23 [Google Scholar]
- Teillet A, Garcia J, de Billy F, Gherardi M, Huguet T, Barker DG, de Carvalho-Niebel F, Journet E-P. (2008) api, a novel Medicago truncatula symbiotic mutant impaired in nodule primordium invasion. Mol Plant Microbe Interact 21: 535–546 [DOI] [PubMed] [Google Scholar]
- Tsay Y-F, Chiu C-C, Tsai C-B, Ho C-H, Hsu P-K. (2007) Nitrate transporters and peptide transporters. FEBS Lett 581: 2290–2300 [DOI] [PubMed] [Google Scholar]
- Tsay Y-F, Schroeder JI, Feldmann KA, Crawford NM. (1993) The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72: 705–713 [DOI] [PubMed] [Google Scholar]
- Udvardi MK, Day DA. (1997) Metabolite transport across symbiotic membranes of legume nodules. Annu Rev Plant Physiol Plant Mol Biol 48: 493–523 [DOI] [PubMed] [Google Scholar]
- Veereshlingam H, Haynes JG, Penmetsa RV, Cook DR, Sherrier DJ, Dickstein R. (2004) nip, a symbiotic Medicago truncatula mutant that forms root nodules with aberrant infection threads and plant defense-like response. Plant Physiol 136: 3692–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Liu P, Forde BG. (2008) Nitrate signalling mediated by the NRT1.1 nitrate transporter antagonises L-glutamate-induced changes in root architecture. Plant J 54: 820–828 [DOI] [PubMed] [Google Scholar]
- Walch-Liu P, Liu L-H, Remans T, Tester M, Forde BG. (2006) Evidence that L-glutamate can act as an exogenous signal to modulate root growth and branching in Arabidopsis thaliana. Plant Cell Physiol 47: 1045–1057 [DOI] [PubMed] [Google Scholar]
- Wang R, Xing X, Wang Y, Tran A, Crawford NM. (2009) A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiol 151: 472–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-Y, Tsay Y-F. (2011) Arabidopsis nitrate transporter NRT1.9 is important in phloem nitrate transport. Plant Cell 23: 1945–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterworth WM, Bray CM. (2006) Enigma variations for peptides and their transporters in higher plants. Ann Bot (Lond) 98: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Prell J, James EK, Poole P. (2007) Nutrient sharing between symbionts. Plant Physiol 144: 604–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson JQ, Crawford NM. (1991) Identification of the Arabidopsis CHL3 gene as the nitrate reductase structural gene NIA2. Plant Cell 3: 461–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Fan X, Miller AJ. (2012) Plant nitrogen assimilation and use efficiency. Annu Rev Plant Biol 63: 5.1–5.30 [DOI] [PubMed] [Google Scholar]
- Yamashita T, Shimada S, Guo W, Sato K, Kohmura E, Hayakawa T, Takagi T, Tohyama M. (1997) Cloning and functional expression of a brain peptide/histidine transporter. J Biol Chem 272: 10205–10211 [DOI] [PubMed] [Google Scholar]
- Yendrek CR, Lee Y-C, Morris V, Liang Y, Pislariu CI, Burkart G, Meckfessel MH, Salehin M, Kessler H, Wessler H, et al. (2010) A putative transporter is essential for integrating nutrient and hormone signaling with lateral root growth and nodule development in Medicago truncatula. Plant J 62: 100–112 [DOI] [PubMed] [Google Scholar]
- Yokoyama T, Kodama N, Aoshima H, Izu H, Matsushita K, Yamada M. (2001) Cloning of a cDNA for a constitutive NRT1 transporter from soybean and comparison of gene expression of soybean NRT1 transporters. Biochim Biophys Acta 1518: 79–86 [DOI] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GED, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KFX, Gouzy J, Schoof H, et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Forde BG. (2000) Regulation of Arabidopsis root development by nitrate availability. J Exp Bot 51: 51–59 [PubMed] [Google Scholar]
- Zhao X, Huang J, Yu H, Wang L, Xie W. (2010) Genomic survey, characterization and expression profile analysis of the peptide transporter family in rice (Oryza sativa L.). BMC Plant Biol 10: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J-J, Theodoulou FL, Muldin I, Ingemarsson B, Miller AJ. (1998) Cloning and functional characterization of a Brassica napus transporter that is able to transport nitrate and histidine. J Biol Chem 273: 12017–12023 [DOI] [PubMed] [Google Scholar]
- Zifarelli G, Pusch M. (2010) CLC transport proteins in plants. FEBS Lett 584: 2122–2127 [DOI] [PubMed] [Google Scholar]