Abstract
The potato cyst nematode Globodera rostochiensis invades roots of host plants where it transforms cells near the vascular cylinder into a permanent feeding site. The host cell modifications are most likely induced by a complex mixture of proteins in the stylet secretions of the nematodes. Resistance to nematodes conferred by nucleotide-binding-leucine-rich repeat (NB-LRR) proteins usually results in a programmed cell death in and around the feeding site, and is most likely triggered by the recognition of effectors in stylet secretions. However, the actual role of these secretions in the activation and suppression of effector-triggered immunity is largely unknown. Here we demonstrate that the effector SPRYSEC-19 of G. rostochiensis physically associates in planta with the LRR domain of a member of the SW5 resistance gene cluster in tomato (Lycopersicon esculentum). Unexpectedly, this interaction did not trigger defense-related programmed cell death and resistance to G. rostochiensis. By contrast, agroinfiltration assays showed that the coexpression of SPRYSEC-19 in leaves of Nicotiana benthamiana suppresses programmed cell death mediated by several coiled-coil (CC)-NB-LRR immune receptors. Furthermore, SPRYSEC-19 abrogated resistance to Potato virus X mediated by the CC-NB-LRR resistance protein Rx1, and resistance to Verticillium dahliae mediated by an unidentified resistance in potato (Solanum tuberosum). The suppression of cell death and disease resistance did not require a physical association of SPRYSEC-19 and the LRR domains of the CC-NB-LRR resistance proteins. Altogether, our data demonstrated that potato cyst nematodes secrete effectors that enable the suppression of programmed cell death and disease resistance mediated by several CC-NB-LRR proteins in plants.
The survival and reproduction of the potato cyst nematode Globodera rostochiensis relies on the successful establishment and maintenance of a feeding site inside the root of a host plant. Secretions produced by sedentary plant-parasitic nematodes such as G. rostochiensis are thought to be instrumental in the formation of the feeding site (Haegeman et al., 2012). The nematodes use an oral stylet to deliver these secretions into the apoplast and the cytoplasm of host cells (Hussey, 1989; Davis et al., 2008). In a susceptible host plant, a recipient host cell may respond by increasing its metabolic activity and by progressing through several cycles of endoreduplication. The concomitant local cell wall degradation and subsequent fusion with neighboring protoplasts transform the infected host cells into a multinucleate syncytium (Sobczak et al., 2009). Freshly hatched infective juveniles of G. rostochiensis are mobile, but as soon as feeding on the syncytium commences, they lose their body wall muscles and adopt a sedentary lifestyle (De Boer et al., 1992). The syncytium functions as a metabolic sink that transfers plant assimilates from the conductive tissues in the vascular cylinder to the sedentary nematode (Jones and Northcote, 1972). A failure in syncytium formation caused, for example, by host defense responses prevents development of the feeding nematode into its reproductive stage (Sobczak et al., 2009).
The majority of plant resistance proteins are members of the NB-LRR receptor family, which consist of a central nucleotide-binding (NB) domain and a Leu-rich repeat (LRR) domain at the carboxyl terminus (Eitas and Dangl, 2010). At their amino-termini, the NB-LRR plant immune receptors either carry a coiled-coil (CC) domain, or a Toll/IL-1 receptor like (TIR) domain. The NB domain, which is also referred to as the NB-ARC (nucleotide-binding adaptor shared by APAF-1, certain resistance proteins, and CED-4) domain, most likely changes from a closed ADP-bound state to an open ATP-bound state when the resistance protein detects a pathogen (Lukasik and Takken, 2009). The LRR domain is thought to act as the sensor in NB-LRR receptors, which in the absence of the cognate effector keeps the resistance protein in an autoinhibited “off” state. In this model, the recognition of a pathogen effector induces a conformational change in the LRR domain that lifts the inhibition of the NB domain in the core of the resistance protein. Artificially induced mutations in NB-LRR immune receptors suggest that the two functions of the LRR domain, pathogen recognition, and negative regulation of the NB domain, reside in different parts of the domain. Several sequence exchanges and deletions at the N terminus of the LRR domain switch NB-LRR immune receptors into a permanent effector-independent autoactive state (Rairdan and Moffett, 2006). By contrast, mutations in repeats at the C terminus of the LRR domain do not lift the autoinhibition, but instead change the recognition specificity of NB-LRR immune receptors (Farnham and Baulcombe, 2006).
The molecular mechanisms underlying effector recognition by plant immune receptors are not well understood. NB-LRR immune receptors may activate signaling pathways that lead to effector-triggered immunity when they physically associate with their cognate effectors (Krasileva et al., 2010). However, the fact that such direct interactions seem to be exceptional inspired the formulation of the “guard” model in which immune receptors activate host defenses by detecting effector-induced perturbations in other plant proteins (Van der Biezen and Jones, 1998). Plant immune receptors may thus efficiently expand the spectrum of disease resistances of a plant by guarding common virulence targets of multiple effectors (Chung et al., 2011). In the recently proposed intermediate “bait-and-switch” model, a pathogen effector may still directly interact with NB-LRR immune receptors, but only after binding to an accessory protein that functions as cofactor for the receptor (Collier and Moffett, 2009).
There are only a few examples of plant immune receptors that directly interact with their cognate pathogen effector (Jia et al., 2000; Deslandes et al., 2003; Ellis et al., 2007; Krasileva et al., 2010; Tasset et al., 2010; Chen et al., 2012). For only three of these resistance proteins, a physical association with the effector was demonstrated in planta. The TIR-NB-LRR resistance protein RPP1 of Arabidopsis (Arabidopsis thaliana) associates via its LRR domain with the effector ATR1 of Peronospora parasitica (Krasileva et al., 2010). This interaction results in a defense-related programmed cell death in leaves of Nicotiana tabacum. Also in Arabidopsis, the association of the TIR-NB-LRR resistance protein RRS1-R with the PopP2 effector of Ralstonia solanacearum results in immunity (Tasset et al., 2010). Similarly, the physical association of the CC domain of the resistance protein Rpi-blb1 (RB) from potato (Solanum tuberosum) with the IPI-O1 effector of Phytophthora infestans triggers a programmed cell death in Nicotiana benthamiana (Chen et al., 2012). Recently, we found that the effector SPRYSEC-19 of G. rostochiensis interacts in yeast (Saccharomyces cerevisiae) with the seven C-terminal repeats of the LRR domain of the CC-NB-LRR protein SW5F of tomato (Solanum lycopersicum; Rehman et al., 2009). The SW5 resistance gene cluster in tomato confers resistance to a broad range of tospoviruses (Boiteux and de Giordano, 1993). Five other SW5 resistance gene homologs have been identified in tomato. The homolog SW5B confers resistance to tomato spotted wilt virus, whereas the functions of SW5A and SW5C-F are currently unknown (Spassova et al., 2001).
SPRYSEC effectors are produced as secretory proteins in the dorsal esophageal gland of G. rostochiensis that is connected via the lumen of the esophagus to the oral stylet (Rehman et al., 2009). They only consist of a SPRY/B30.2 domain, which in many different eukaryotic proteins is involved in intermolecular interactions (Rhodes et al., 2005; Tae et al., 2009). The expression of the SPRYSEC effectors in G. rostochiensis is highly up-regulated in infective juveniles and during the first few days post invasion. The function of the SPRYSEC effectors in plant parasitism is not well understood. It has been shown that the coexpression of the SPRYSEC GpRBP1 from Globodera pallida and the CC-NB-LRR resistance protein Gpa2 from potato induces a programmed cell death in leaves of N. benthamiana (Sacco et al., 2009). This finding suggests that GpRBP1 triggers Gpa2-mediated nematode resistance. However, because both virulent and avirulent G. pallida populations harbor GpRBP1, its role in nematode resistance remains to be shown. Furthermore, it is also not clear if the Gpa2-mediated programmed cell death requires a physical association between Gpa2 and GpRBP1.
In this paper, we report the functional characterization of the effector SPRYSEC-19 of G. rostochiensis, and its interaction with SW5F, in plants. We first tested the hypothesis that SPRYSEC-19 activates SW5F-dependent programmed cell death and nematode resistance. However, coexpression of SPRYSEC-19 and SW5F by agroinfiltration in leaves of N. benthamiana and in tomato did not trigger a defense-related programmed cell death. Moreover, nematode infection assays on tomato plants harboring SW5F showed no resistance to G. rostochiensis. Next, we tested the alternative hypothesis that SPRYSEC-19 modulates host defense responses in plants. Our data demonstrated that SPRYSEC-19 selectively suppresses CC-NB-LRR-mediated programmed cell death and disease resistance.
RESULTS
SPRYSEC-19 Does Not Trigger an SW5F-Mediated Programmed Cell Death
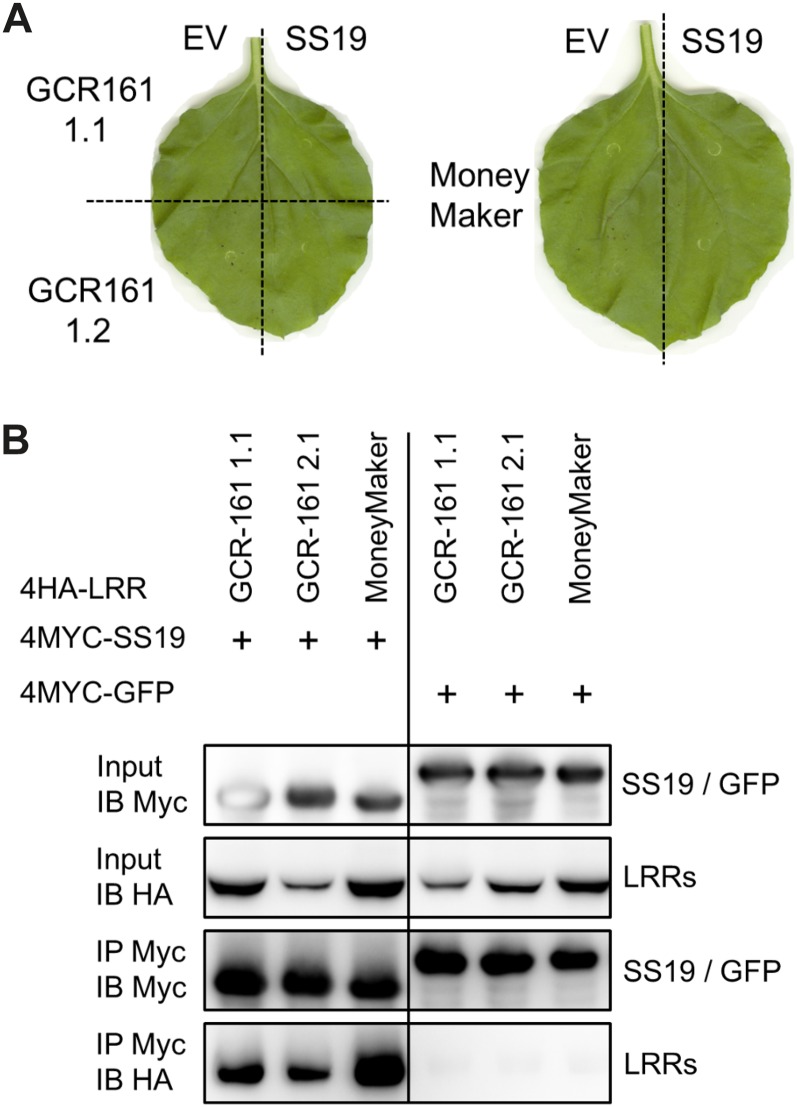
Previously, we showed that the effector SPRYSEC-19 of G. rostochiensis interacts with a C-terminal fragment of the LRR domain of SW5F (SW5F-LRR7-13) in a yeast two-hybrid screen on tomato root complementary DNA (cDNA) (Rehman et al., 2009). An in vitro pull-down assay confirmed that SPRYSEC-19 and SW5F-LRR can interact without cofactors (Rehman et al., 2009). This specific association of SPRYSEC-19 and SW5F was confirmed in planta by bimolecular fluorescence complementation (BiFC) and coimmunoprecipitations (Co-IPs; Supplemental Fig. S1). The only other known physical association of a pathogen effector and the LRR domain of a resistance protein in planta triggers a defense-related programmed cell death in N. tabacum leaves (Krasileva et al., 2010). We expected that coexpression of SPRYSEC-19 and SW5F would also trigger a cell death response in agroinfiltrated leaves of N. benthamiana. However, no local cell death was observed within 10 d after transient overexpression of SW5F with either 4MYC-tagged SPRYSEC-19 or untagged SPRYSEC-19 (Supplemental Fig. S2). The fragment of SW5F (SW5F-LRR7-13) that interacted with SPRYSEC-19 in the yeast-two-hybrid screen derived from the near-isogenic line CGR161 of tomato ‘MoneyMaker.’ We reasoned that other close homologs of SW5F either in CGR161 or in the parent cv MoneyMaker might be able to mediate a SPRYSEC-19-triggered cell death in N. benthamiana. A PCR using SW5F-specific primers resulted in the identification of three SW5F homologs (Supplemental Fig. S3). Transient coexpression of none of the SW5F homologs with either SPRYSEC-19 (Fig. 1A) or 4MYC-SPRYSEC-19 resulted in a local programmed cell death in agroinfiltrated areas of N. benthamiana leaves. The three SW5F variants are polymorphic at nine amino acid positions in the LRR region (Supplemental Fig. S3). Despite these differences, 4MYC-SPRYSEC-19 captured on anti-MYC beads pulled-down the transiently expressed LRR domain of all three SW5F variants (Fig. 1B). This demonstrated that the absence of programmed cell death is not caused by lack of a physical interaction between SPRYSEC-19 and the LRR domains of the SW5F homologs.
Figure 1.
The physical association of SPRYSEC-19 and three SW5F variants in planta does not trigger a programmed cell death. A, Transient expression of SW5F variants (GCR161-1.1 and -1.2, and cv MoneyMaker) by agroinfiltration in N. benthamiana leaves together with empty expression vector (EV; left side of leaves) or SPRYSEC-19 (SS19; right side of leaves). Photographs were taken at 10 d post infiltration. B, Co-IP of HA-tagged LRR domains of the SW5F variants by 4MYC-SPRYSEC-19 or 4-MYC-GFP. SPRYSEC-19 and GFP were captured on anti-MYC agarose beads (IP MYC/IB MYC) in total protein extracts of agroinfiltrated leaves of N. benthamiana transiently coexpressing the proteins. LRR domains pulled-down by either 4-MYC-SPRYSEC-19 or MYC-GFP (IP MYC/IB HA) were detected on western blots with anti-HA serum. [See online article for color version of this figure.]
SW5F Does Not Confer Resistance to G. rostochiensis
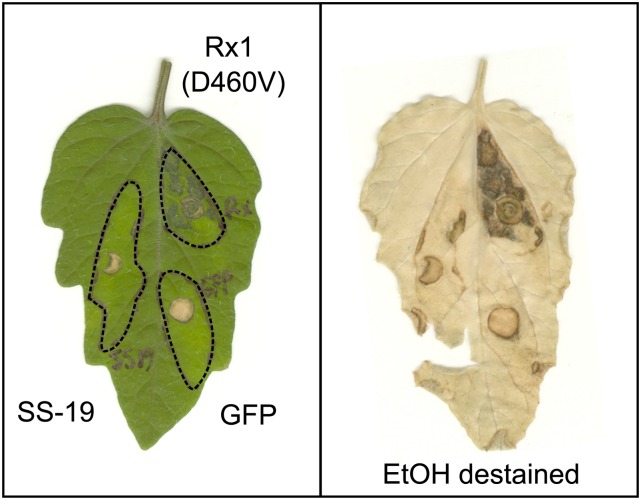
SW5F from tomato might not be able to mediate programmed-cell death in N. benthamiana because it requires accessory proteins that are absent in N. benthamiana. However, transient expression of SPRYSEC-19 by agroinfiltration in leaves of the tomato ‘MoneyMaker’ harboring the SW5F gene did not result in a local cell death either (Fig. 2). Not all functional disease resistance proteins trigger a local cell death at the infection site of avirulent pathogens (Bendahmane et al., 1999; Bulgarelli et al., 2010), and SW5F might therefore still confer resistance to G. rostochiensis in tomato. To test whether SW5F mediates resistance to the population of G. rostochiensis from which SPRYSEC-19 was isolated (pathotype Ro1-Mierenbos, line 19), we inoculated 7-d-old seedlings of the tomato cultivar from which SW5F was cloned (cv MoneyMaker) with infective second-stage juveniles. Three weeks post inoculation, on average, 29 (se ± 1.1) juveniles per tomato plant developed into the adult female stage, which is consistent with a normal susceptibility to G. rostochiensis in tomato (Sobczak et al., 2005).
Figure 2.
Transient expression of SPRYSEC-19 in tomato ‘MoneyMaker’ harboring the SW5F gene does not result in local cell death. SPRYSEC-19, GFP, and an autoactive mutant of the resistance gene Rx1 (i.e. Rx1[D460V]) were transiently expressed by agroinfiltration in leaves of tomato plants. Photograph was taken 7 d post infiltration.
SPRYSEC-19 Suppresses Programmed Cell Death Mediated by an SW5 Homolog in N. benthamiana Leaves
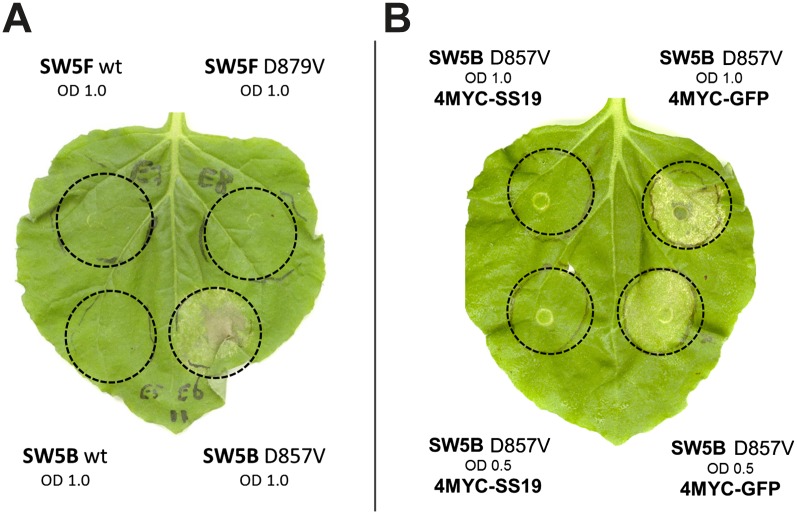
Next, we reasoned that SPRYSEC-19 interacts with SW5F to suppress effector-triggered activation of SW5F-mediated immune signaling. The SW5F gene has not been linked to a particular disease resistance trait, and by consequence, the elicitor of the pathogen that might activate SW5F-mediated signaling is also not known. The tomato spotted wilt virus resistance mediated by SW5B is currently the only phenotype linked to the SW5 cluster in tomato. However, the elicitor of the virus that activates SW5B has not been identified either. To be able to test if SPRYSEC-19 suppresses SW5-mediated programmed cell death, we introduced a D-to-V mutation at position 879 in SW5F and at position 857 in SW5B to make the proteins autoactive (Bendahmane et al., 2002; de la Fuente van Bentem et al., 2005; Tameling et al., 2006; van Ooijen et al., 2008). Only the expression of SW5B-D857V resulted in an effector-independent cell death response following agroinfiltration of N. benthamiana leaves (Fig. 3A). Coexpression of 4MYC-SPRYSEC-19 suppressed the effector-independent cell death response mediated by the SW5B-D857V mutant protein in agroinfiltrated leaves of N. benthamiana (Fig. 3B). This outcome suggested that SPRYSEC-19 suppresses SW5B-mediated activation of effector-triggered immunity.
Figure 3.
SPRYSEC-19 suppresses SW5B activated programmed cell death. Leaves of N. benthamiana were agroinfiltrated with wild-type and autoactive mutant SW5 genes under 35S CaMV promoter and monitored for the initiation of cell death over 10 d. The suspensions of the bacteria carrying the constructs were infiltrated in a 1:1 ratio with a combined OD600 as indicated. Photographs were taken 5 d post infiltration. A, Wild-type (wt) SW5B and -F genes overexpressed next to SW5 mutants with a D-to-V mutation. B, Autoactive mutant SW5B(D857V) coinfiltrated with equal amounts of 4MYC-SPRYSEC-19 (left side of leaf) or 4MYC-GFP (right side of leaf).
SPRYSEC-19 Selectively Suppresses CC-NB-LRR-Mediated Programmed Cell Death in N. benthamiana Leaves
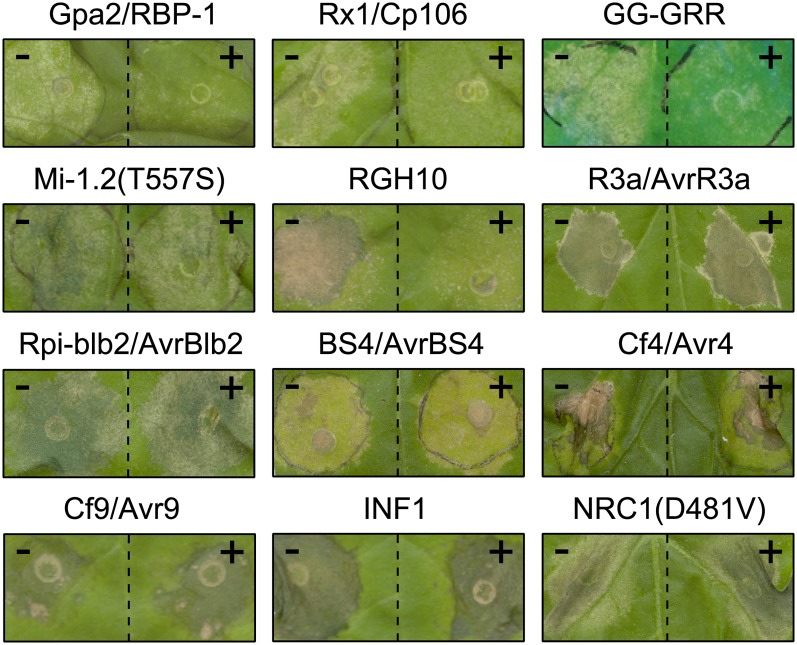
Next, we investigated whether SPRYSEC-19 also suppresses the programmed cell death mediated by other CC-NB-LRR resistance proteins. The SPRYSEC effector GpRBP-1 of the white potato cyst nematode G. pallida triggers a Gpa2-mediated cell death in N. benthamiana (Sacco et al., 2009). To investigate a possible SPRYSEC-19 controlled suppression of Gpa2-mediated programmed cell death, we coexpressed 4MYC-SPRYSEC-19 together with GpRBP-1 and Gpa2 by agroinfiltration in leaves of N. benthamiana. GpRBP-1 transiently expressed with Gpa2 and 4MYC-GFP triggered a strong cell death response in the infiltrated leaf areas within 4 to 7 d post infiltration. By contrast, no local cell death was observed following the coexpression of GpRBP-1, Gpa2, and 4MYC-SPRYSEC-19 in N. benthamiana. We therefore concluded that SPRYSEC-19 suppressed elicitor-dependent programmed cell death mediated by Gpa2. Gpa2 is highly similar to the virus resistance protein Rx1 that recognizes the coat protein of the avirulent Potato virus X (PVX) strain UK106 (Cp106; Bendahmane et al., 1995). Cp106 shares no sequence similarity with GpRBP-1 or with other SPRYSEC effectors. We used the Rx1-mediated cell death response in N. benthamiana to investigate whether SPRYSEC-19 suppresses the action of a homologous CC-NB-LRR protein that is not triggered by a SPRYSEC. As expected, coexpression of Rx1, Cp106, and 4MYC-GFP resulted in a local cell death response in agroinfiltrated leaf areas of N. benthamiana (Fig. 4). By contrast, replacing 4MYC-GFP with 4MYC-SPRYSEC-19 completely abrogated the Rx1/Cp106-triggered cell death response in N. benthamiana leaves. SPRYSEC-19 of G. rostochiensis thus also suppresses programmed cell death mediated by the CC-NB-LRR resistance proteins Gpa2 and Rx1.
Figure 4.
SPRYSEC-19 suppresses programmed cell death mediated by a subset of CC-NB-LRR proteins. Coexpression of 4MYC-SPRYSEC-19 (+) or 4MYC-GFP (–) with several programmed cell death inducing pairs of resistance proteins and their cognate effectors or autoactive resistance proteins (see “Results” section for details) after agroinfiltration in N. benthamiana leaves. Photographs were taken 3 to 5 d post infiltration.
To investigate whether the SPRYSEC-19-induced suppression of CC-NB-LRR mediated programmed cell death involves a disturbed effector recognition, we coexpressed SPRYSEC-19 in N. benthamiana leaves with an autoactive Gpa2-Rx1 chimera (GG-GRR; Rairdan and Moffett, 2006), Mi-1.2 mutant [Mi-1.2(T557S); Gabriëls et al., 2007], and natural resistance gene homolog 10 from the H1 locus in potato (RGH10; Finkers-Tomczak et al., 2011). The transient coexpression of these proteins with 4MYC-GFP led to a local cell death response in agroinfiltrated leaf areas. However, replacing 4MYC-GFP with 4MYC-SPRYSEC-19 abrogated the effector-independent cell death response mediated by GG-GRR and RGH10, but not by Mi-1.2(T557S) (Fig. 4). We therefore concluded that SPRYSEC-19 selectively suppresses cell death signaling of a subset of CC-NB-LRR resistance proteins.
We also coexpressed SPRYSEC-19 with R3a (Huang et al., 2005) and Rpi-blb2 (van der Vossen et al., 2005; Oh et al., 2009) from potato and their cognate elicitors from P. infestans in N. benthamiana leaves to test whether SPRYSEC-19 also modulates the cell death responses mediated by more distantly related CC-NB-LRR resistance proteins (see Supplemental Fig. S4 for an identity matrix). The coexpression of R3a and Rpi-blb-2 and their cognate elicitors resulted in a local cell death response in N. benthamiana, which was not suppressed in the presence of SPRYSEC-19 (Fig. 4). Similarly, the effector-triggered cell death response mediated by a resistance protein of the TIR-NB-LRR class (i.e. BS4) and the extracellular LRR class (i.e. Cf-4 and Cf-9) was also not affected by coexpression of SPRYSEC-19 (Fig. 4). The P. infestans secreted elicitin INF1 has features of pathogen-associated molecular patterns and autonomously elicits a strong cell death response in leaves of N. benthamiana (Heese et al., 2007). The expression of SPRYSEC-19 did not suppress INF1-induced cell death in agroinfiltrated leaves of N. benthamiana (Fig. 4). The CC-NB-LRR protein NRC1 likely operates in signaling pathways downstream of different types of resistance proteins (e.g. Rx1, Mi-1.2, Cf4, and Cf-9; Gabriëls et al., 2007). To investigate whether SPRYSEC-19 modulates immune signaling downstream of resistance proteins, we coexpressed SPRYSEC-19 with an autoactive mutant of NRC1(D481V) by agroinfiltration in leaves of N. benthamiana. Expression of NRC1(D481V) caused a strong cell death response within 24 h after agroinfiltration in N. benthamiana leaves, which was not suppressed by SPRYSEC-19 (Fig. 4). Altogether, our data demonstrated that SPRYSEC-19 suppresses the programmed cell death mediated by a group of closely related CC-NB-LRR resistance proteins.
SPRYSEC-19 Suppresses Disease Resistance Mediated by Rx1
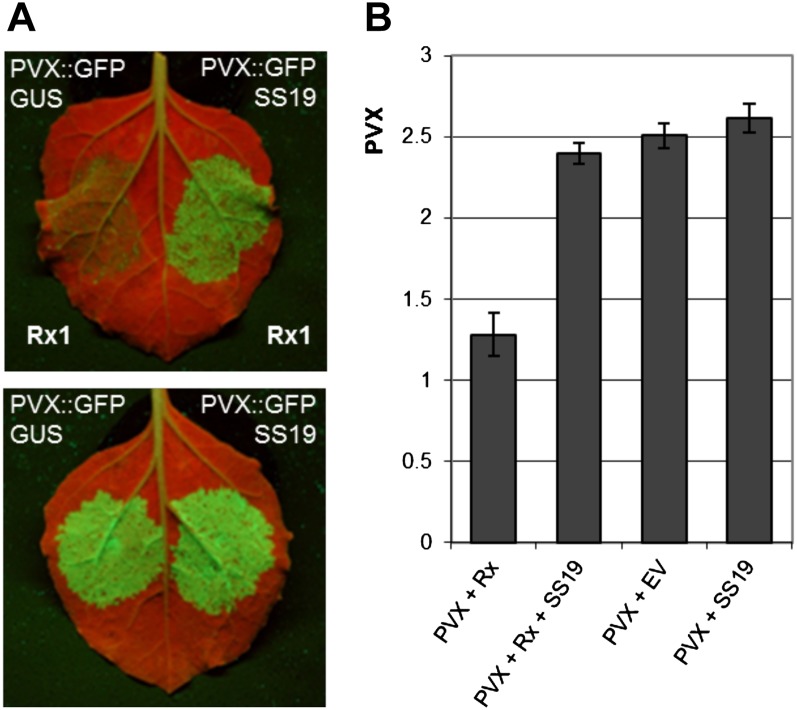
The local cell death mediated by resistance proteins may be a consequence rather than a prerequisite of disease resistance in plants (Coll et al., 2011). To determine if SPRYSEC-19 also suppresses disease resistance mediated by a CC-NB-LRR protein, we assessed the replication of the avirulent PVX strain UK106 in the presence of both the resistance protein Rx1 and SPRYSEC-19, and in the presence of Rx1 alone. To this purpose, PVX was introduced into N. benthamiana leaves by agroinfiltrating the complete viral amplicon including GFP (PVX::GFP). Virus replication was first deduced from the accumulation of GFP in mesophyll cells in infiltrated leaf areas (Fig. 5A). As expected, the coexpression of Rx1, PVX::GFP, and GUS resulted in poor accumulation of GFP in agroinfiltrated areas. However, replacing GUS with 4MYC-SPRYSEC-19 in the agroinfiltration mix led to a strong GFP signal. We also coexpressed PVX::GFP and 4MYC-SPRYSEC-19 alone in N. benthamiana mesophyll cells to demonstrate that 4MYC-SPRYSEC-19 targeted the action of Rx1 and not the replication of PVX directly (Fig. 5A). To confirm that the accumulation of GFP reflects PVX replication in mesophyll cells, we also quantified the accumulation of PVX coat protein by using a specific antibody in an ELISA on total protein extracts isolated from agroinfiltrated leaf areas (Fig. 5B). We concluded that the suppression of Rx1-mediated immune signaling by SPRYSEC-19 also results in loss of disease resistance.
Figure 5.
SPRYSEC-19 suppresses resistance to PVX mediated by the CC-NB-LRR protein Rx1. A, Transient expression of GFP-labeled PVX (PVX:GFP) in leaves of N. benthamiana after agroinfiltration together with the GUS gene or SPRYSEC-19 (SS19), with (top) or without (bottom) the resistance gene Rx1 under control of a leaky scan 35S CaMV promoter. GFP expression was visualized under a UV lamp 4 d post infiltration. Agroinfiltrations with the empty binary expression vector (EV) were included as a control. B, Quantification of PVX replication by ELISA directed against the PVX coat protein. Bars represent ELISA signal intensity; error bars represent se of the mean.
SPRYSEC-19 Overexpression Renders a Fungal Resistant Potato Genotype Susceptible to Verticillium dahliae
To investigate whether stable overexpression of SPRYSEC-19 enhances the susceptibility of plants to plant pathogens, we first inoculated transgenic potato plants (line V) overexpressing untagged or 4MYC-tagged SPRYSEC-19 with G. rostochiensis. Four weeks post inoculation, the number of adult females per plant was not significantly higher in at least twelve independent transgenic potato lines overexpressing SPRYSEC-19 as compared with transgenic plants harboring the corresponding empty binary expression vector (Supplemental Fig. S5). The draft genome sequence of the sister species G. pallida suggests that potato cyst nematodes carry over 200 different SPRYSEC genes (http://www.sanger.ac.uk/cgi-bin/blast/submitblast/g_pallida). We therefore reasoned that the overexpression of one specific SPRYSEC gene family member in a host plant might have little impact on the virulence of G. rostochiensis. However, the potato line V is resistant to V. dahliae, and the V. dahliae genome does not harbor homologs of nematode SPRYSEC effectors. We therefore challenged the transgenic potato lines overexpressing either SPRYSEC-19 or 4MYC-SPRYSEC-19 with V. dahliae strain 5361, to test whether SPRYSEC-19 alters the resistance of potato plants to this fungus. Four weeks post inoculation with V. dahliae, the SPRYSEC-19 overexpressing plants showed a strong reduction in shoot growth as compared with mock-inoculated plants, and as compared with the empty vector plants inoculated with V. dahliae (Fig. 6A). To further quantify the level of resistance to V. dahliae in the transgenic potato lines, we measured the accumulation of fungal biomass in plants harboring either SPRYSEC-19, 4MYC-SPRYSEC-19, or the empty expression vector by specifically amplifying the internal transcribed spacer (ITS) region of V. dahliae with PCR (Fradin et al., 2011). The ITS region of V. dahliae was amplified from plants overexpressing either SPRYSEC-19 or 4MYC-SPRYSEC-19 three weeks post inoculation with fungal spores (Fig. 6B). As expected, no amplification product of the ITS region in V. dahliae was observed in the empty vector plants three weeks post inoculation with fungal spores. These data suggest that SPRYSEC-19 suppresses a yet unidentified fungal resistance in potato, rendering these plants susceptible to an otherwise avirulent strain of V. dahliae.
Figure 6.
SPRYSEC-19 overexpression renders a resistant potato genotype susceptible to V. dahliae. Stable transgenic potato line V overexpressing SPRYSEC-19 (19.3) or 4MYC-tagged SPRYSEC-19 (M19.1 and M19.7) under control of the 35S CaMV promoter infected with V. dahliae strain 5361. A, Shoot growth of V. dahliae (V) and mock (M)-inoculated transgenic potato lines 4 weeks post inoculation. EV is a transgenic potato line harboring the corresponding empty binary expression vector. B, Quantification of V. dahliae biomass by PCR-amplification of the internal transcribed spacer region of V. dahliae in total DNA extracts of V. dahliae and mock-inoculated transgenic potato lines (top). SPRYSEC-19 and actin genes were PCR-amplified as internal controls (middle and bottom). 1kb+, DNA size marker. [See online article for color version of this figure.]
Suppression of Disease Resistance Responses by SPRYSEC-19 Does Not Require a Direct Interaction with R-Proteins
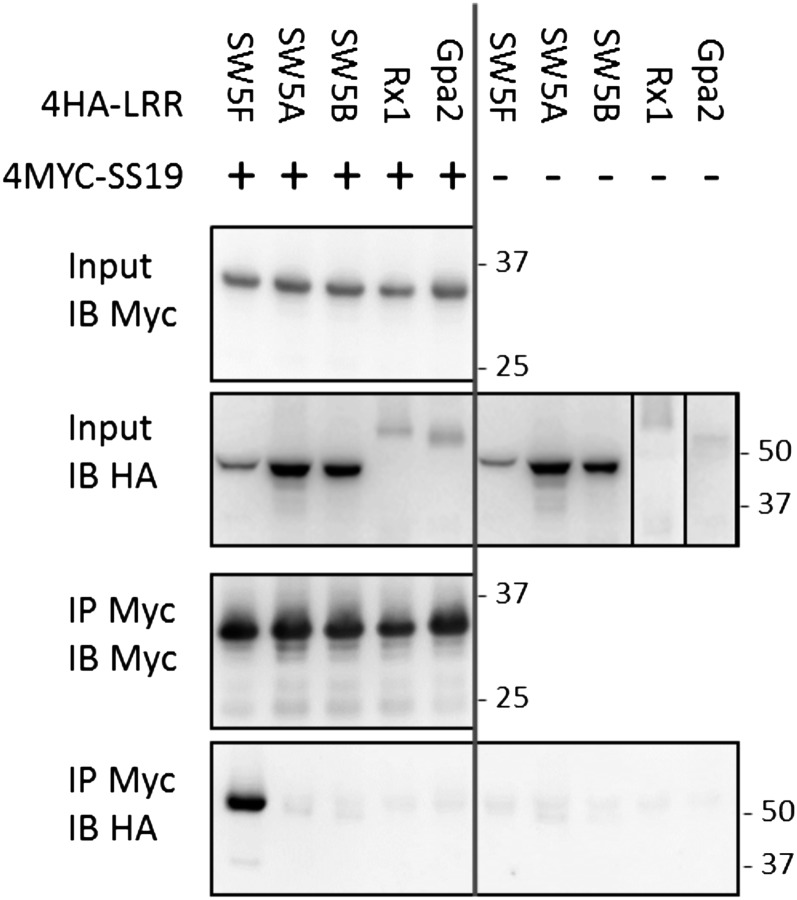
To investigate whether the suppression of Gpa2, Rx1, and autoactive SW5B requires a physical interaction with SPRYSEC-19, we coexpressed 4MYC-SPRYSEC-19 and the LRR domains of these proteins fused to a 4HA tag in leaves of N. benthamiana for Co-IP. Capturing 4MYC-SPRYSEC-19 in total protein extracts of agroinfiltrated leaf areas with anti-MYC beads did not result in the Co-IP of the LRR domains of Sw5B, Rx1, and Gpa2 (Fig. 7). We therefore concluded that SPRYSEC-19-mediated suppression of CC-NB-LRR-mediated programmed cell death and resistance does not require a physical interaction of SPRYSEC-19 with the LRR domains of these resistance proteins.
Figure 7.
SPRYSEC-19 does not bind the LRR domains of suppressed R-proteins. Co-IP of different 4HA-tagged LRR domains of SW5F, -A, -B, Rx1, and Gpa2 with 4MYC-SPRYSEC-19 (SS19) transiently coexpressed in N. benthamiana. The LRR domains pulled-down (IP MYC/IB HA) by 4MYC-SPRYSEC-19 on anti-MYC agarose beads (IP MYC/IB MYC) were detected on western blots with anti-HA serum.
DISCUSSION
We have shown that the resistance protein SW5F of tomato interacts specifically with the effector SPRYSEC-19 of G. rostochiensis in planta. Surprisingly, this interaction did not lead to the effector-triggered activation of SW5F-mediated programmed cell death and nematode resistance. Instead, SPRYSEC-19 is the first nematode effector to demonstrate suppression of defense-related programmed cell death by some, but not all, CC-NB-LRR resistance proteins (i.e. SW5B, Rx1, Gpa2, and RGH10). The suppression of CC-NB-LRR-mediated signaling does not require a physical association between SPRYSEC-19 and these resistance proteins. Furthermore, the suppression of programmed cell death mediated by autoactive mutant CC-NB-LRR proteins suggested that SPRYSEC-19 most likely disturbs receptor-mediated immune signaling rather than effector recognition. In addition to abrogating the programmed cell death mediated by Rx1, the nematode effector SPRYSEC-19 also repressed virus resistance mediated by this CC-NB-LRR protein. Altogether, our data demonstrates that SPRYSEC-19 of G. rostochiensis functions as a suppressor of CC-NB-LRR-mediated programmed cell death and disease resistance.
SPRYSEC-19 physically associates with SW5F in planta through its interaction with seven C-terminal Leu-rich repeats of the LRR domain of SW5F. There are only a few other plant resistance proteins for which a physical interaction with a pathogen effector in planta has been demonstrated. These interactions agree with the model of effector-triggered immunity following direct recognition of effectors by plant immune receptors. Like ATR1/PPR1 and IPI-O1/RB, we expected that the physical association of SPRYSEC-19 and SW5F would also activate effector-triggered immunity to G. rostochiensis. However, the absence of SPRYSEC-19-dependent SW5F-mediated programmed cell death in N. benthamiana and SW5F-mediated resistance to G. rostochiensis in tomato and potato led us to reject this hypothesis.
We have demonstrated with four different experimental designs that the physical association between SPRYSEC19 and the LRR domain of SW5F is robust. That this association does not activate effector-triggered programmed cell death and resistance may indicate that SW5F is an inactive gene duplicate of a paralogous functional CC-NB-LRR resistance protein to G. rostochiensis. In this scenario, the lack of functional constraints on the SW5F gene may have rendered its activation domains (i.e. CC-NB) dysfunctional, while binding to the sensor (i.e. LRR) domain is still intact (Takken and Goverse, 2012). We tried to make SW5F, along with SW5B, constitutively active by introducing mutations at positions that switch several other CC-NB-LRR resistance proteins into a permanent “on” state. However, these mutations only induced autoactivity in SW5B, which is thus far the only member of the SW5 cluster linked to a known resistance (Spassova et al., 2001). The lack of autoactivity in SW5F mutants therefore favors the hypothesis that SW5F is a dysfunctional paralog of a functional nematode resistance gene.
As SPRYSEC-19 lacked any evident avirulence activity on the three SW5F homologs isolated in this study, we also reasoned that SPRYSEC-19 might interact with the LRR domain of SW5F to suppress the activation of the CC-NB-LRR-mediated immune signaling. Using agroinfiltration assays, we have demonstrated that SPRYSEC-19 suppresses programmed cell death mediated by some, but not all, CC-NB-LRR resistance proteins in N. benthamiana. Moreover, SPRYSEC-19 suppressed none of the members of the TIR-NB-LRR and extracellular LRR classes of resistance proteins tested in this study. We found no evidence in our Co-IPs that suppression of CC-NB-LRR-mediated programmed cell death requires the binding of SPRYSEC-19 to these receptor proteins. However, it should be noted that most high affinity interactions between proteins can be demonstrated with Co-IPs. We therefore cannot exclude the possibility that SPRYSEC-19 more transiently interacts with the LRR domains of the resistance proteins it suppresses.
As the suppression of autoactive mutant CC-NB-LRR proteins demonstrated, SPRYSEC19 most likely does not disturb the recognition of specific cognate pathogen effectors that activates these resistance proteins. It is nonetheless conceivable that SPRYSEC-19 is able to outcompete other SPRYSEC effectors of G. rostochiensis that trigger the activation of a functional homolog of SW5F. Such a mechanism seems to determine the virulence of P. infestans strains on potato plants harboring the RB resistance protein (Chen et al., 2012). Alternatively, as discussed earlier SPRYSEC-19 may also suppress CC-NB-LRR resistance proteins by targeting the immune receptors to the proteasome for degradation (Rehman et al., 2009). However, western blots of total protein extracts of agroinfiltrated leaf areas revealed no enhanced breakdown of CC-NB-LRR proteins or parts thereof in the presence of SPRYSEC-19. We therefore conclude that our current data does not support a model in which SPRYSEC-19 interacts with CC-NB-LRR resistance proteins to alter their turnover rate.
Programmed cell death in the site of pathogen infections is often associated with effector-triggered immunity in plants, but may not be required for disease resistance (Coll et al., 2011). It could therefore be argued that the suppression of programmed cell death by SPRYSEC-19 in agroinfiltration assays bears little biological significance with regard to disease resistance. Using an avirulent PVX strain that was modified to express GFP but that was still recognized and restrained by the resistance protein Rx1, we have demonstrated that SPRYSEC-19 also suppresses CC-NB-LRR-mediated disease resistance. Furthermore, our observation that the overexpression of SPRYSEC-19 in potato plants abrogated the resistance of this potato genotype to V. dahliae further supports that this effector functions as a suppressor of disease resistance.
Next to the ability to induce and maintain feeding cells, the survival and reproduction of sedentary plant-parasitic nematodes is most likely determined by their ability to suppress host defenses. The molecular mechanisms underlying the suppression of host defense responses by plant-parasitic nematodes are not known. All known plant immune receptors conferring resistance to G. rostochiensis belong to the CC-NB-LRR class of resistance proteins (Molinari, 2011). Here we showed that G. rostochiensis has evolved several SPRYSEC effectors that selectively suppress CC-NB-LRR-mediated programmed cell death and disease resistance. The SPRYSECs in the potato cyst nematodes G. rostochiensis and G. pallida constitute the largest effector family found in a plant parasitic nematode to date. If the SPRYSEC effector family functions as suppressors of effector-triggered immunity, the expansion of this effector family may reflect adaptations to functional diversifications in plant immune receptors. As the SPRYSEC effector GpRBP1 of G. pallida suggests, on their turn, plants may have evolved novel NB-LRR plant immune receptors (e.g. Gpa2) that recognize and neutralize SPRYSEC effectors again. It will be highly interesting to investigate if GpRBP1 also suppresses CC-NB-LRR resistance proteins, and if the activation of Gpa2-mediated resistance also involves a physical association between the LRR domain of Gpa2 and GpRBP1.
MATERIALS AND METHODS
Plant Material
For nematode infection assays, explants of in vitro cultured tomato (Lycopersicon esculentum ‘GCR-161’; Kroon and Elgersma, 1993) or potato (Solanum tuberosum, line V, genotype 6487-9; Schouten et al., 1997) were grown on B5 medium (3.29 g/L Gamborg B5, 20 g/L Suc, 15 g/L bacto agar, pH 6.2) at 24°C and 16-/8-h photoperiod for 3 weeks prior to inoculation. All other experiments were performed year round on 3-week-old tomato ‘MoneyMaker’ or Nicotiana benthamiana plants that were grown in a greenhouse in 15-cm-diameter pots with potting soil.
Cloning and Plasmid Construction
SPRYSEC-19 was subcloned from pGBKT7-A18-2 (Rehman et al., 2009) as a BspMI-BamHI fragment and inserted jointly with the complementary oligo pair A18For + A18Rev (Supplemental Table S1) into pRAP digested with NheI-BglII. The coding regions of the mature peptides of other SPRYSECs without their native signal peptides for secretion were PCR-amplified from Globodera rostochiensis cDNA. The full-length SW5F genes of tomato ‘GCR161’ were PCR-amplified as described before (Rehman et al., 2009). The regions of R-genes coding for the LRR domain were subcloned from existing plasmids: SW5A and -B (Spassova et al., 2001), SW5F (Rehman et al., 2009), Gpa2 (Rairdan and Moffett, 2006), and Rx1 (Slootweg et al., 2010). PCR-amplification products were cloned into vector pRAP using specific restriction sites and confirmed by DNA sequencing. The fragments cloned into the pRAP vector were cloned in frame with the Cauliflower mosaic virus (CaMV) 35S promoter and N- or C-terminal 4HA or 4MYC affinity tags, or no additional tags. Primers and restriction sites used for the cloning of novel genes are listed in Supplemental Table S2. Expression cassettes of pRAP, including promoter, affinity tags, and the gene of interest, were subcloned into binary vector pBINPLUS (van Engelen et al., 1995) using AscI and PacI restriction sites. All SW5F genes were cloned with the 3′ untranslated region (polyadenylation signal and terminator) of the SW5F gene isolated from cv MoneyMaker (Rehman et al., 2009). Autoactive SW5 mutants were made by inserting the annealed oligo pair D879V-1 and D879V-2 (Supplemental Table S3) between the BspHI and XbaI restriction sites of the SW5 genes in pRAP. All of the constructs described above were mobilized to Agrobacterium tumefaciens strain MOG101 (Hood et al., 1993), which was selectively grown on 50 mg/L kanamycin and 20 mg/L rifampicin. For the expression of SPRYSEC-19 in tomato, the coding region for the mature peptide of SPRYSEC-19 without its signal peptide was PCR-amplified from G. rostochiensis cDNA using primers listed in Supplemental Table S1 and cloned into pENTR/D-TOPO (Invitrogen). After confirmation of the sequence by DNA sequencing, SPRYSEC-19 was subcloned to the expression vector SOL2085 (kindly provided by Patrick Smit, Laboratory of Phytopathology, Wageningen University) using LR clonase (Invitrogen), resulting in vector SOL2085:SS19. For agroinfiltrations in tomato leaves, the constructs were mobilized to A. tumefaciens strain 1D1249 (Wroblewski et al., 2005), which was selectively grown on 100 mg/L kanamycin, 100 mg/L spectinomycin, and 1 mg/L tetracyclin.
Agroinfiltrations
A. tumefaciens harboring the individual binary vectors was grown at 28°C in yeast extract peptone medium (per liter: 10 g peptone, 10 g yeast [Saccharomyces cerevisiae] extract, 5 g NaCl) with appropriate antibiotics. The bacteria were spun down and resuspended in infiltration medium (per liter: 5 g Murashige and Skoog salts, 1.95 g 2-(N-morpholino)ethanesulfonic acid, 20 g Suc). The bacterial solution was diluted to an optical density at 600 nm (OD600) of 0.5 (for infiltration in N. benthamiana) or 0.1 (for infiltration in tomato) in infiltration medium and infiltrated in the abaxial side of the leaves using a syringe. Coinfiltration of different constructs was performed by mixing equal volumes of the bacterial suspensions to a final OD600 as described above.
Suppression of Programmed Cell Death
The suppression of programmed cell death in leaves of N. benthamiana was assessed using the pBINPLUS construct with MYC-tagged SPRYSEC-19 described above. The 4MYC:GFP construct was used as a negative control for suppression. The following pairs of resistance genes and cognate elicitors were used to induce programmed cell death in leaves: Gpa2 / RBP1 (Sacco et al., 2009), Rx1 / cp106 (Slootweg et al., 2010), Cf4 / Avr4 (Thomas et al., 2000), Cf9 / Avr9 (Thomas et al., 2000), R3a / AvrR3a (Huang et al., 2005), Rpi-blb2 / AvrBlb2 (van der Vossen et al., 2005), and BS4 (Schornack et al., 2005) / AvrBS4 (Ballvora et al., 2001). The following constructs of mutant CC-NB-LRR proteins were used to trigger an elicitor-independent programmed cell death: GG-GRR (Rairdan and Moffett, 2006), Mi-1.2(T557S) (Gabriëls et al., 2007), RGH10 (Finkers-Tomczak, 2011), NRC1(D481V; Gabriëls et al., 2007), and INF1 (Kamoun et al., 2003). Agroinfiltrated leaves were monitored up to 10 d for visual assessment of cell death.
SPRYSEC-19 in Tomato
SOL2085:GFP (kindly provided by Patrick Smit), SOL2085:SS19 (see above), pBIN61:Rx (D460V) (Bendahmane et al., 2002) in A. tumefaciens strain 1D1249 were agroinfiltrated in leaves of tomato ‘MoneyMaker.’ Agroinfiltrated leaves were monitored up to 10 d for visual assessment of cell death.
Bimolecular Fluorescence Complementation
The coding regions of SPRYSEC-18 and -19 without signal peptide and the coding regions of LRR7-13 of SW5B and SW5F were PCR-amplified from the pRAP vectors described above using the primers listed in Supplemental Table S4. The amplification products were cloned into vector pENTR/D-TOPO (Invitrogen) according to the manufacturer’s protocol and verified by DNA sequencing. Using the Gateway LR clonase reaction (Invitrogen), the amplification products were subcloned into vectors pGREENII:35S:YFPc and pGREENII:35S:YFPn (Zhong et al., 2008), and confirmed by restriction digestion. pGREEN vectors were mobilized to A. tumefaciens strain GV3101 (Holsters et al., 1980), which was selectively grown on 50 mg/L kanamycin, 20 mg/L rifampicin, and 50 mg/L carbenicillin. Two days after agroinfiltration in leaves of N. benthamiana, fluorescence analysis was performed on a Zeiss 510 confocal laser scanning microscope setup. Yellow fluorescent protein fluorescence was assessed at 514 nm (excitation) using an argon laser with an emission band of 535 to 590 nm and 650 nm (chlorophyll autofluorescence).
Co-IP
Total protein extracts of transient transformed N. benthamiana leaves were made by grinding leaf material in protein extraction buffer (50 mm Tris-HCl pH 7.5, 10% (v/v) glycerol, 150 mm NaCl, 1 mm EDTA, 2% (w/v) polyclar-AT polyvinylpolypyrrolidone (Serva), 0.4 mg/ml Pefabloc SC plus (Roche), 5 mm dithiothreitol) on ice. For Co-IP, the total protein extract was first passed over a Sephadex G-25 column (GE Healthcare). The protein extract was treated with rabbit-IgG agarose (40 μL slurry per mL protein extract). After preclearing, the protein extract was mixed with 25 μL anti-MYC agarose beads (Sigma) or anti-hemagglutinin (HA) agarose beads (Roche) and incubated for 2 h at 4°C. After washing six times with washing buffer (protein extraction buffer with 0.15% (v/v) Igepal CA-630; Sigma), the beads were resuspended in Laemmli buffer (Sambrook et al., 1989), and the bound protein was separated by SDS-PAGE and blotted on polyvinylidene difluoride membrane. For immunodetection, we used antibodies goat anti-MYC (Abcam) and horseradish peroxidase-conjugated donkey anti-goat (Jackson) or horseradish peroxidase-conjugated rat anti-HA (Roche). Peroxidase activity was visualized using Thermo Scientific SuperSignal West Femto or Dura substrate and imaging of the luminescence with G:BOX gel documentation system (Syngene).
Plant Transformation
Potato line V (genotype 6487-9) was transformed as described by (van Engelen et al., 1994) using A. tumefaciens strain MOG101 with vector pBINPLUS containing SPRYSEC-19, 4MYC:SPRYSEC-19, SW5F, or 4HA:SW5F under the control of a 35S promoter (described above). Genomic DNA was extracted from plant leaves by grinding tissues in liquid nitrogen and purifying DNA with the DNeasy Plant Mini Kit (Qiagen). For every construct, at least four independent transformation lines were tested, and for each line, 10 biological replicates were used.
Nematode Resistance Assay
Dried cysts of G. rostochiensis pathotype Ro1-Mierenbos were soaked on a 100-μm sieve in potato root diffusate to collect hatched ppJ2s (De Boer et al., 1992). Freshly hatched preparasitic second-stage juveniles in suspension were mixed with an equal volume of 70% (w/v) Suc in a centrifuge tube and covered with a layer of sterile tap water. Following centrifugation for 5 min at 1,000g, juveniles were collected from the Suc-water interface using a Pasteur pipette and washed three times with sterile tap water. The nematodes were surface sterilized by incubation for 20 min in 0.5% (w/v) streptomycin/penicillin solution, for 20 min in 0.1% (w/v) ampicillin/gentamycin solution, for 5 min in sterile tap water, and for 3 min in 0.1% (v/v) chlorhexidine solution. The nematodes were subsequently washed three times in sterile tap water, resuspended in sterile 0.7% (w/v) solution of Gelrite (Duchefa), and pipetted along the roots of 3-week-old in vitro-grown plants. Routinely, we used between 150 and 200 ppJ2s per plate containing one plant. Adult females per plate were counted 6 to 8 weeks after inoculation. For each transformant tested, at least 15 independent lines were used.
PVX Resistance Assay
A. tumefaciens strain MOG101 carrying vector pBINPLUS with 35S:4MYC:SPRYSEC-19 (described above), 35SLS:Rx1:GFP (Slootweg et al., 2010), 35S:GFP:PVX (Peart et al., 2002), or GPA2:GUS (Koropacka, 2010) were used for agroinfiltration of N. benthamiana leaves. Three days post infiltration, GFP expression of GFP-tagged PVX was visualized under UV light. Virus concentration was determined using double antibody sandwich-ELISA (Mäki-Valkama et al., 2000). Plates were coated with a 1:1000 dilution of a polyclonal antibody against PVX to bind the antigen, and a second polyclonal antibody against PVX conjugated with alkaline phosphatase was used for detection via the phosphatase substrate p-nitrophenyl-P.
Verticillium dahliae Resistance Assay
V. dahliae isolate 5361 (kindly provided by Richard Cooper) was grown on 4% potato dextrose media (Duchefa) at 28°C for 2 weeks. Fungal spores were transferred to sterile deionized water to a concentration of 1 × 106 spores/mL. The roots of 3-week-old in vitro-grown transgenic potato plants were soaked in spore suspension for 5 min and transferred to pots with soil in a greenhouse. For each transformant, at least 10 independent lines were used with four biological controls. At 20 d post inoculation, pictures were taken, and to determine the fungal biomass in infected plants, stem pieces were cut from the potato plants just above ground level and flash frozen in liquid nitrogen. Total DNA was extracted from plant tissues using DNeasy Plant Mini Kit (Qiagen). A 200-bp fragment of the ITS gene of V. dahliae was PCR-amplified using primers ITS1-F (Gardes and Bruns, 1993) and ST-VE1 (Lievens et al., 2006) on DNA samples using FirePol polymerase (Solis BioDyne). As an internal control, potato actin was amplified from the same templates using primers StActinF and StActinR (Nicot et al., 2005).
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: JX026913-JX026918 (SRYSEC-4 – SPRYSEC-15); JX026924 (SPRYSEC-16); JX026920 (SPRYSEC-19); JX026925 (SW5F GCR161-1.1); JX026926 (SW5F GCR161-1.2); JX026927 (SW5F cv MoneyMaker).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. BiFC and Co-IPs showing specificity of SPRYSEC-19-SW5F interaction.
Supplemental Figure S2. Photograph of an agroinfiltrated N. benthamiana leaf coexpressing SPRYSECs and SW5F.
Supplemental Figure S3. Protein alignment of SW5F isoforms identified in tomato.
Supplemental Figure S4. Amino acid identity matrix of R-proteins compared in this study.
Supplemental Figure S5. Susceptibility of transgenic potato lines overexpressing SPRYSEC-19 to G. rostochiensis.
Supplemental Table S1. Oligonucleotides used to subclone SPRYSEC-19.
Supplemental Table S2. Primers and restriction sites used to clone SPRYSECs, SW5F, and R-gene LRR regions.
Supplemental Table S3. Oligonucleotides used to construct autoactive SW5 mutants.
Supplemental Table S4. Primers used to clone SPRYSECs and LRRs into BiFC vectors.
Acknowledgments
We thank Sillin Zhong for providing the BiFC vectors, Richard Kormelink for providing SW5A and -B constructs, and Koste Yadeta for helping with the Verticillium spp. assay. Bert Essenstam and Henk Smid are acknowledged for outstanding plant care.
Glossary
- cDNA
complementary DNA
- BiFC
bimolecular fluorescence complementation
- Co-IP
coimmunoprecipitation
- PVX
Potato virus X
- ITS
internal transcribed spacer
- CC
coiled-coiled
- NB
nucleotide-binding
- LRR
Leu-rich repeat
- TIR
Toll/IL-1 receptor like
- OD600
optical density at 600 nm
- CaMV
Cauliflower mosaic virus
- HA
hemagglutinin
- CC
coiled-coil
References
- Ballvora A, Pierre M, van den Ackerveken G, Schornack S, Rossier O, Ganal M, Lahaye T, Bonas U. (2001) Genetic mapping and functional analysis of the tomato Bs4 locus governing recognition of the Xanthomonas campestris pv. vesicatoria AvrBs4 protein. Mol Plant Microbe Interact 14: 629–638 [DOI] [PubMed] [Google Scholar]
- Bendahmane A, Farnham G, Moffett P, Baulcombe DC. (2002) Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J 32: 195–204 [DOI] [PubMed] [Google Scholar]
- Bendahmane A, Kanyuka K, Baulcombe DC. (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11: 781–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane A, Köhn BA, Dedi C, Baulcombe DC. (1995) The coat protein of potato virus X is a strain-specific elicitor of Rx1-mediated virus resistance in potato. Plant J 8: 933–941 [DOI] [PubMed] [Google Scholar]
- Boiteux LS, de Giordano LB. (1993) Genetic basis of resistance against two Tospovirus species in tomato (Lycopersicon esculentum). Euphytica 71: 151–154 [Google Scholar]
- Bulgarelli D, Biselli C, Collins NC, Consonni G, Stanca AM, Schulze-Lefert P, Valè G. (2010) The CC-NB-LRR-type Rdg2a resistance gene confers immunity to the seed-borne barley leaf stripe pathogen in the absence of hypersensitive cell death. PLoS ONE 5: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Liu Z, Halterman DA. (2012) Molecular determinants of resistance activation and suppression by Phytophthora infestans effector IPI-O. PLoS Pathog 8: e1002595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung EH, da Cunha L, Wu AJ, Gao Z, Cherkis K, Afzal AJ, Mackey D, Dangl JL. (2011) Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe 9: 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll NS, Epple P, Dangl JL. (2011) Programmed cell death in the plant immune system. Cell Death Differ 18: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier SM, Moffett P. (2009) NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci 14: 521–529 [DOI] [PubMed] [Google Scholar]
- Davis EL, Hussey RS, Mitchum MG, Baum TJ. (2008) Parasitism proteins in nematode-plant interactions. Curr Opin Plant Biol 11: 360–366 [DOI] [PubMed] [Google Scholar]
- De Boer JM, Overmars HA, Bakker J, Gommers FJ. (1992) Analysis of two-dimensional protein patterns from developmental stages of the potato cyst-nematode, Globodera rostochiensis. Parasitology 105: 461–474 [Google Scholar]
- de la Fuente van Bentem S, Vossen JH, de Vries KJ, van Wees S, Tameling WIL, Dekker HL, de Koster CG, Haring MA, Takken FLW, Cornelissen BJC. (2005) Heat shock protein 90 and its co-chaperone protein phosphatase 5 interact with distinct regions of the tomato I-2 disease resistance protein. Plant J 43: 284–298 [DOI] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. (2003) Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA 100: 8024–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitas TK, Dangl JL. (2010) NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol 13: 472–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JG, Dodds PN, Lawrence GJ. (2007) Flax rust resistance gene specificity is based on direct resistance-avirulence protein interactions. Annu Rev Phytopathol 45: 289–306 [DOI] [PubMed] [Google Scholar]
- Farnham G, Baulcombe DC. (2006) Artificial evolution extends the spectrum of viruses that are targeted by a disease-resistance gene from potato. Proc Natl Acad Sci USA 103: 18828–18833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkers-Tomczak A, Bakker E, de Boer J, van der Vossen E, Achenbach U, Golas T, Suryaningrat S, Smant G, Bakker J, Goverse A. (2011) Comparative sequence analysis of the potato cyst nematode resistance locus H1 reveals a major lack of co-linearity between three haplotypes in potato (Solanum tuberosum ssp.). Theor Appl Genet 122: 595–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkers-Tomczak AM. (2011) Co-evolution between Globodera rostochiensis and potato driving sequence diversity of NB-LRR resistance loci and nematode suppressors of plant immunity. PhD thesis. Wageningen University, Wageningen, The Netherlands
- Fradin EF, Abd-El-Haliem A, Masini L, van den Berg GCM, Joosten MHAJ, Thomma BPHJ. (2011) Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol 156: 2255–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriëls SHEJ, Vossen JH, Ekengren SK, van Ooijen G, Abd-El-Haliem AM, van den Berg GC, Rainey DY, Martin GB, Takken FLW, de Wit PJ, et al. (2007) An NB-LRR protein required for HR signalling mediated by both extra- and intracellular resistance proteins. Plant J 50: 14–28 [DOI] [PubMed] [Google Scholar]
- Gardes M, Bruns TD. (1993) ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol 2: 113–118 [DOI] [PubMed] [Google Scholar]
- Haegeman A, Mantelin S, Jones JT, Gheysen G. (2012) Functional roles of effectors of plant-parasitic nematodes. Gene 492: 19–31 [DOI] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AME, He K, Li J, Schroeder JI, Peck SC, Rathjen JP. (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsters M, Silva B, Van Vliet F, Genetello C, De Block M, Dhaese P, Depicker A, Inzé D, Engler G, Villarroel R, et al. (1980) The functional organization of the nopaline A. tumefaciens plasmid pTiC58. Plasmid 3: 212–230 [DOI] [PubMed] [Google Scholar]
- Hood EE, Gelvin SB, Melchers LS, Hoekema A. (1993) New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res 2: 208–218 [Google Scholar]
- Huang S, van der Vossen EAG, Kuang H, Vleeshouwers VGAA, Zhang N, Borm TJA, van Eck HJ, Baker B, Jacobsen E, Visser RGF. (2005) Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. Plant J 42: 251–261 [DOI] [PubMed] [Google Scholar]
- Hussey RS. (1989) Disease-inducing secretions of plant-parasitic nematodes. Annu Rev Phytopathol 27: 123–141 [Google Scholar]
- Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B. (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J 19: 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MG, Northcote DH. (1972) Nematode-induced syncytium—a multinucleate transfer cell. J Cell Sci 10: 789–809 [DOI] [PubMed] [Google Scholar]
- Kamoun S, Hamada W, Huitema E. (2003) Agrosuppression: a bioassay for the hypersensitive response suited to high-throughput screening. Mol Plant Microbe Interact 16: 7–13 [DOI] [PubMed] [Google Scholar]
- Koropacka M. (2010) Molecular contest between potato and the potato cyst nematode Globodera pallida: modulation of Gpa2-mediated resistance. PhD thesis. Wageningen University, Wageningen, The Netherlands
- Krasileva KV, Dahlbeck D, Staskawicz BJ. (2010) Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell 22: 2444–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon BAM, Elgersma DM. (1993) Interactions between race 2 of Fusarium oxysporum f. sp. lycopersici and near-isogenic resistant and susceptible lines of intact plants or callus of tomato 58731 F71 137. J Phytopathol 137: 1–9 [Google Scholar]
- Lievens B, Brouwer M, Vanachter ACRC, Cammue BPA, Thomma BPHJ. (2006) Real-time PCR for detection and quantification of fungal and oomycete tomato pathogens in plant and soil samples. Plant Sci 171: 155–165 [Google Scholar]
- Lukasik E, Takken FL. (2009) STANDing strong, resistance proteins instigators of plant defence. Curr Opin Plant Biol 12: 427–436 [DOI] [PubMed] [Google Scholar]
- Mäki-Valkama T, Valkonen JPT, Kreuze JF, Pehu E. (2000) Transgenic resistance to PVY(O) associated with post-transcriptional silencing of P1 transgene is overcome by PVY(N) strains that carry highly homologous P1 sequences and recover transgene expression at infection. Mol Plant Microbe Interact 13: 366–373 [DOI] [PubMed] [Google Scholar]
- Molinari S. (2011) Natural genetic and induced plant resistance, as a control strategy to plant-parasitic nematodes alternative to pesticides. Plant Cell Rep 30: 311–323 [DOI] [PubMed] [Google Scholar]
- Nicot N, Hausman JF, Hoffmann L, Evers D. (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56: 2907–2914 [DOI] [PubMed] [Google Scholar]
- Oh SK, Young C, Lee M, Oliva R, Bozkurt TO, Cano LM, Win J, Bos JIB, Liu HY, van Damme M, et al. (2009) In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell 21: 2928–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart JR, Cook G, Feys BJ, Parker JE, Baulcombe DC. (2002) An EDS1 orthologue is required for N-mediated resistance against tobacco mosaic virus. Plant J 29: 569–579 [DOI] [PubMed] [Google Scholar]
- Rairdan GJ, Moffett P. (2006) Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell 18: 2082–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman S, Postma W, Tytgat T, Prins P, Qin L, Overmars H, Vossen J, Spiridon LN, Petrescu AJ, Goverse A, et al. (2009) A secreted SPRY domain-containing protein (SPRYSEC) from the plant-parasitic nematode Globodera rostochiensis interacts with a CC-NB-LRR protein from a susceptible tomato. Mol Plant Microbe Interact 22: 330–340 [DOI] [PubMed] [Google Scholar]
- Rhodes DA, de Bono B, Trowsdale J. (2005) Relationship between SPRY and B30.2 protein domains. Evolution of a component of immune defence? Immunology 116: 411–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco MA, Koropacka K, Grenier E, Jaubert MJ, Blanchard A, Goverse A, Smant G, Moffett P. (2009) The cyst nematode SPRYSEC protein RBP-1 elicits Gpa2- and RanGAP2-dependent plant cell death. PLoS Pathog 5: e1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis TA. (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schornack S, Peter K, Bonas U, Lahaye T. (2005) Expression levels of avrBs3-like genes affect recognition specificity in tomato Bs4- but not in pepper Bs3-mediated perception. Mol Plant Microbe Interact 18: 1215–1225 [DOI] [PubMed] [Google Scholar]
- Schouten A, Roosien J, de Boer JM, Wilmink A, Rosso MN, Bosch D, Stiekema WJ, Gommers FJ, Bakker J, Schots A. (1997) Improving scFv antibody expression levels in the plant cytosol. FEBS Lett 415: 235–241 [DOI] [PubMed] [Google Scholar]
- Slootweg E, Roosien J, Spiridon LN, Petrescu A-J, Tameling W, Joosten M, Pomp R, van Schaik C, Dees R, Borst JW, et al. (2010) Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell 22: 4195–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczak M, Avrova A, Jupowicz J, Phillips MS, Ernst K, Kumar A. (2005) Characterization of susceptibility and resistance responses to potato cyst nematode (Globodera spp.) infection of tomato lines in the absence and presence of the broad-spectrum nematode resistance Hero gene. Mol Plant Microbe Interact 18: 158–168 [DOI] [PubMed] [Google Scholar]
- Sobczak M, Golinowski W, Berg R, Taylor C. (2009) Structure of cyst nematode feeding sites. In Cell Biology of Plant Nematode Parasitism, Vol 15. Springer, Berlin, pp 153–187
- Spassova MI, Prins TW, Folkertsma RT, Klein-Lankhorst RM, Hille J, Goldbach RW, Prins M. (2001) The tomato gene Sw5 is a member of the coiled coil, nucleotide binding, leucine-rich repeat class of plant resistance genes and confers resistance to TSWV in tobacco. Mol Breed 7: 151–161 [Google Scholar]
- Tae H, Casarotto MG, Dulhunty AF. (2009) Ubiquitous SPRY domains and their role in the skeletal type ryanodine receptor. Eur Biophys J 39: 51–59 [DOI] [PubMed] [Google Scholar]
- Takken FL, Goverse A. (2012) How to build a pathogen detector: structural basis of NB-LRR function. Curr Opin Plant Biol 15: 375–384 [DOI] [PubMed] [Google Scholar]
- Tameling WIL, Vossen JH, Albrecht M, Lengauer T, Berden JA, Haring MA, Cornelissen BJC, Takken FLW. (2006) Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol 140: 1233–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasset C, Bernoux M, Jauneau A, Pouzet C, Briére C, Kieffer-Jacquinod S, Rivas S, Marco Y, Deslandes L. (2010) Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1-R-mediated immunity in Arabidopsis. PLoS Pathog 6: e1001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CM, Tang S, Hammond-Kosack K, Jones JDG. (2000) Comparison of the hypersensitive response induced by the tomato Cf-4 and Cf-9 genes in Nicotiana spp. Mol Plant Microbe Interact 13: 465–469 [DOI] [PubMed] [Google Scholar]
- Van der Biezen EA, Jones JDG. (1998) Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci 23: 454–456 [DOI] [PubMed] [Google Scholar]
- van der Vossen EAG, Gros J, Sikkema A, Muskens M, Wouters D, Wolters P, Pereira A, Allefs S. (2005) The Rpi-blb2 gene from Solanum bulbocastanum is an Mi-1 gene homolog conferring broad-spectrum late blight resistance in potato. Plant J 44: 208–222 [DOI] [PubMed] [Google Scholar]
- van Engelen FA, Molthoff JW, Conner AJ, Nap JP, Pereira A, Stiekema WJ. (1995) pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res 4: 288–290 [DOI] [PubMed] [Google Scholar]
- van Engelen FA, Schouten A, Molthoff JW, Roosien J, Salinas J, Dirkse WG, Schots A, Bakker J, Gommers FJ, Jongsma MA, et al. (1994) Coordinate expression of antibody subunit genes yields high levels of functional antibodies in roots of transgenic tobacco. Plant Mol Biol 26: 1701–1710 [DOI] [PubMed] [Google Scholar]
- van Ooijen G, Mayr G, Kasiem MMA, Albrecht M, Cornelissen BJC, Takken FLW. (2008) Structure-function analysis of the NB-ARC domain of plant disease resistance proteins. J Exp Bot 59: 1383–1397 [DOI] [PubMed] [Google Scholar]
- Wroblewski T, Tomczak A, Michelmore R. (2005) Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol J 3: 259–273 [DOI] [PubMed] [Google Scholar]
- Zhong S, Lin Z, Fray RG, Grierson D. (2008) Improved plant transformation vectors for fluorescent protein tagging. Transgenic Res 17: 985–989 [DOI] [PMC free article] [PubMed] [Google Scholar]