Abstract
Background
Sanger sequencing (SS) of PCR products is still the most frequent method to test colorectal cancer for KRAS mutations in routine practice.
Methods
An audit of SS on 1720 routine cases was carried out, taking into account age, gender, specimen type (resection vs biopsies), tumour site (primary vs metastasis), tumour stage, neoplastic cells abundance (>30% vs <30%) and fixation type (buffered formalin vs simple formalin). In a subset of 50 wild-type (WT) patients correlations between SS findings and response rate (RR), progression-free survival (PFS) and overall survival (OS) were also evaluated.
Results
The tests were informative in 1691 cases (98.3%). Mutations were detected in 671 cases (39.6%). No significant differences in mutation rates were observed with respect to age (p=0.2), gender (p=0.2), specimen type (p=0.3) and formalin fixation (p=0.08). Conversely, KRAS mutant rate was higher in metastatic tissue (50% vs 39%, p=0.02), in samples with over 30% of neoplastic cells (43.4% vs 26.6%, p=0.02) and in tumours tested in stage IV (p=0.05). The RR of SS KRAS WT patients was 26% (one complete and 12 partial responses). The disease control rate (objective responses plus stable disease) was 56%. Median PFS was 4.4 months and median OS was 10.4 months.
Conclusions
Pathological criteria that make SS a more robust method for KRAS testing and treatment response prediction are neoplastic cell abundance, metastatic tissue sample and stage IV primary tumour.
Keywords: COLORECTAL CANCER, AUDIT, HISTOPATHOLOGY, LABORATORY TESTS, MOLECULAR PATHOLOGY
Introduction
The surgical pathologist has a key function in the multidisciplinary management of colorectal cancer (CRC) including disease diagnosis, prognostication and targeted therapy.1 It is well known that in metastatic CRC, the epidermal growth receptor (EGFR) is a major target.2 Monoclonal antibodies (moAb) targeting EGFR reduce the risk of disease progression, increase the chance of response to chemotherapy, and in some cases may even improve overall survival (OS).3 4 However, these effects have only been observed in patients with KRAS wild-type (WT) disease.1 Conversely in patients with a KRAS mutant tumour, approximately 40% of the target population, this treatment lacks efficacy and might be harmful.1
Community KRAS testing was introduced in 2008 following the guidelines issued by the European Society of Pathology (ESP)1 and the College of American Pathologists.5 The recommendations to the pathologist were very detailed. However, no advice was given on which assay should be performed. As a matter of fact, each testing institution decided either to validate a laboratory-based assay or to adopt commercial kits.6 This choice was usually based on the equipment, experience and personnel available in each molecular laboratory.7 However, regardless of the chosen method, the pathologist's assessment of sample quality and consistency before testing strongly influences the final result.
Although many different molecular methods are available,8–12 Sanger sequencing (SS) of PCR products is still very often used. In the most recent scheme of the ESP external quality assurance, SS was adopted by 28/76 (36.8%) laboratories (http://kras.eqascheme.org). SS is less accurate in samples with less than 30% of neoplastic cells, and different reports suggest that SS of PCR products should not be used when the specimen contains a low amount of tumour cells.13 Instead, whether its performance differs between surgical resections and biopsies, primary tumour and metastatic tissues, buffered and simple formalin-fixed samples needs further investigation. The tumour stage may also matter. These issues were investigated here on the largest series published to date. The final goal was to provide the practising pathologist with additional information on which sample parameters make SS efficient for KRAS mutational status assessment.
Methods
Patients and samples
Our molecular laboratory is an accredited Italian reference centre for KRAS testing.14 From February 2009 to January 2012, 1720 KRAS tests were performed to assess the eligibility to moAb targeting EGFR therapy of 960 men and 760 women harbouring CRC. Mean patient age was 67 years (range 31–96 years). One single tumour sample of a given location (primary tumour n=1609; metastases n=111) was tested for each patient. Only in a subset of 30 patients, for which paired samples were available in our institution, were both primary and metastatic sites processed.
All samples were formalin-fixed paraffin embedded. Overall, 406 samples (23.6%) were received from the gastrointestinal pathology unit and 1314 (76.4%) cases were received from 18 external pathology departments. Through a dedicated website, on obtaining patient consent, the oncologist and the primary pathologist submitted clinical and pathological data, respectively. Then the corresponding tissue sample was express mailed.
Information relative to tumour stage, from the Unio Internationale Contra Cancrum, were available in 1666 instances. In 899 (53.9%) cases the tumour was in stage IV; less frequently (n=767; 46.1%) the tumour was either in stages I–II (n=496) or III (n=271). Information on specimen type was available in 1427 cases (surgical specimens n=1263, 88.5%; biopsies n=164, 11.5%).
Sample fixation
Information on the fixation protocol was available on 1095 colectomy specimens. In 905 (82.6%) cases 10% buffered formalin was used, carefully monitoring the fixation time, which had not usually taken more than 24 h. In 190 (17.4%) cases fixation had been accomplished in simple formalin.
KRAS exon 2 SS validation
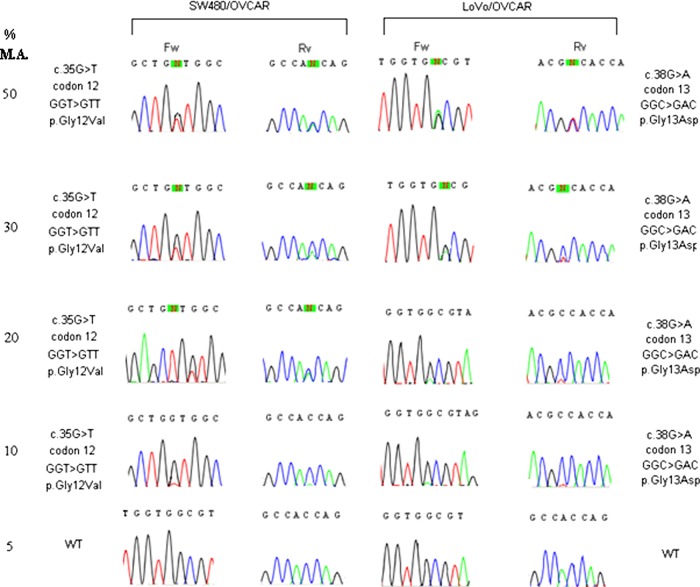
Before clinical implementation the SW480 (harbouring the G12V mutation), LoVo (harbouring the G13D mutation) and the OVOCAR (KRAS WT) cell lines were employed to standardise the KRAS mutation testing procedures.15 In addition, eight clinical samples (four mutated and four WT) previously tested by an accredited laboratory were also genotyped. All reactions were carried out in duplicate. Any single experiment gave the expected result. To test the smallest level of detectable mutant allele, we serially mixed mutated (SW480 and LoVo) and WT (OVOCAR) KRAS cell lines at 50%, 30%, 20%, 10% and 5% proportions. As show in figure 1, the detection limit of our method was 10% of mutated KRAS allele. The lowest level of neoplastic cellularity acceptable for our technique was thus 20%. Our laboratory conformed to the ESP external quality assurance 2010 and 2011 audits with a genotyping score of 100% (http://kras.eqascheme.org).
Figure 1.
Serial dilution experiments of DNA obtained from KRAS mutant tumour and wild-type (WT) cell lines. Serial dilution (50%, 30%, 20%, 10% and 5%) were tested. KRAS mutation was detected up to the 10% dilution. This figure is only reproduced in colour in the online version.
DNA extraction and SS analysis of KRAS exon 2
The whole series reported here was analysed by the following procedure. In any single case, DNA was extracted using a DNA minikit (Qiagen, Milan, Italy), following the manufacturer's instructions. A 220-bp PCR fragment of the exon 2 KRAS gene, including codons 12 and 13, was amplified by laboratory-developed primer pairs (KRAS F:5–GGTGGAGTATTTGATAGTGTATTAACC–3 and KRAS R: 5–AGAATGGTCCTGCACCAGTAA–3). Primers were designed on the GenBank reference sequence (accession NM_004985.3) with OligoCalc Software (http://www.basic.northwestern.edu/biotools/oligocalc.html). The concentration of the reagents was optimised using 200 ng of DNA, 0.4 mM of each primer and 0.5 U 5 PRIME Taq DNA Polymerase (5 PRIME; Eppendorf, Milan, Italy) in a total volume of 25 μl.
PCR conditions were as follows: initial denaturation for 5 min at 95°C, cyclic denaturation at 94°C for 30 s, annealing at 57°C for 30 s, elongation at 65°C for 30 s for 35 cycles and final extension at 72°C for 2 min. Following PCR the fragments were purified by the QiaQuick DNA purification kit (Qiagen, Crawley, West Sussex, UK) according to the manufacturer's instructions. Sequencing reactions were performed for both DNA strands by the Big Dye Terminator v1.1 (Applied Biosystems, Monza, Italy) on a total of 10 ng of purified PCR products. Dye purification was carried out by alcohol/sodium acetate precipitation. Sequence analysis was performed on an Applied Biosystems 310 genetic analyser. The files obtained were aligned and examined for mutations in codons 12 and 13 of the KRAS gene by CodonCode software.
Percentage of neoplastic cells
The relationship between the percentage of neoplastic cells and the rate of KRAS mutation by SS was carefully assessed in 578 cases. To this end, the area that had been marked for neoplastic cell enrichment on the original H&E stained slide was eyeballed adopting 30% of neoplastic cells as a threshold. This cut-off was selected as it was slightly higher than the lower limit of detection of our method. Samples with a tumour cell count less than 30% were also re-analysed using a mutation-specific PCR method (TheraScreen K-RAS mutation kit, DxS Ltd, Manchester, England) following manufacturer's instructions.
Treatment response evaluation
The predictive value of SS on the response to EGFR targeting moAb treatment was evaluated. From the 406 samples received from the gastrointestinal pathology unit, we selected 50 patients by the following criteria: (1) unresectable metastasis; (2) codon 12 and 13 KRAS gene WT status; and (3) at least one previous chemotherapy regimen for metastatic disease. A total of 50 patients met these inclusion criteria, 35 men and 15 women, with a median age of 61 years (range 29–77 years). Cetuximab was administered at the initial dose of 400 mg/m2 followed by weekly infusions of 250 mg/m2 along with chemotherapy, until unacceptable toxicity or progression of disease. Disease status was evaluated in all patients by total body CT scan before treatment onset and every 2 months thereafter.
The response rate (RR) was evaluated according to RECIST criteria (V.2.0). On the basis of the RR we classified patients as responders (complete or partial response) and non-responders (stable and progressive disease). Moreover, we considered the disease control rate (complete response, partial response and stable disease). Progression-free survival (PFS) was defined as the time from the first administration of cetuximab to the first evidence of disease progression or death from any cause. OS was considered the time from the first administration of cetuximab to death from any cause.
Statistical analysis
All the analyses were performed using Fisher's exact test on IBM SPSS Statistics 18 package software. A p value of 0.05 or less was considered to indicate statistical significance.
Results
Overall mutation distribution
Codon 12 and 13 KRAS mutational status was successfully assessed in 1691 (98.3%) tumour samples. In 29 (1.7%) cases insufficient starting material did not allow for the mutational analysis. Mutations within codons 12 and 13 were detected in 671 (39.6%) of the analysed samples. Whole cohort analysis did not underscore significant correlation regarding age (p=0.2) or gender, with mutations in 40.9% of women and in 38.6% of men (p=0.2).
Of the total number of mutations, 78% (523/671) were observed in codon 12 (GGT) and 22% (148/671) in codon 13 (GGC). In particular, for codon 12 the GAT codon (29.8%) leading to aspartic acid and the GTT codon (25.5%) leading to valine were the most frequently observed. In codon 13 the GAC codon (21.3%), leading to substitution of a glycine by aspartic acid, was by far the predominant mutation. The frequencies of all mutations, also including the less common genetic aberrations, are reported in table 1.
Table 1.
Mutation type distribution in relation to clinical characteristics
| Patients | Clinical characteristics | Mutations |
Mutations | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G12D | G12V | G12C | G12A | G12S | G12R | G12dup | G13D | G13C | G13V | G13A | ||||
| 1691 | Gender | |||||||||||||
| Male | 112 | 92 | 41 | 23 | 20 | 20 | 1 | 68 | 0 | 0 | 1 | 365 | 671 | |
| Female | 88 | 79 | 23 | 19 | 16 | 2 | 0 | 75 | 3 | 1 | 0 | 306 | ||
| 1691 | Age (years) | |||||||||||||
| <59 | 46 | 41 | 16 | 10 | 7 | 2 | 0 | 20 | 0 | 0 | 0 | 142 | 671 | |
| ≥60 | 154 | 130 | 48 | 32 | 29 | 7 | 1 | 123 | 3 | 1 | 1 | 529 | ||
| 1691 | Site* | |||||||||||||
| Primary | 185 | 159 | 60 | 39 | 35 | 8 | 1 | 128 | 1 | 0 | 1 | 616 | 671 | |
| Metastasis | 15 | 14 | 4 | 3 | 0 | 1 | 0 | 15 | 2 | 1 | 0 | 55 | ||
| 543 | Neoplastic cells percentage* | |||||||||||||
| <30% | 3 | 5 | 1 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 12 | 240 | |
| ≥30% | 69 | 56 | 24 | 19 | 9 | 3 | 1 | 45 | 1 | 0 | 1 | 228 | ||
| 1406 | Specimens | |||||||||||||
| Resection | 143 | 139 | 51 | 28 | 24 | 7 | 1 | 105 | 2 | 1 | 1 | 502 | 567 | |
| Biopsy | 19 | 13 | 5 | 6 | 3 | 1 | 0 | 17 | 1 | 0 | 0 | 65 | ||
| 1095 | Fixation modalities (formalin) | |||||||||||||
| Buffered | 112 | 96 | 37 | 19 | 23 | 3 | 0 | 71 | 1 | 0 | 0 | 362 | 428 | |
| Simple | 25 | 12 | 5 | 3 | 2 | 1 | 0 | 17 | 1 | 0 | 0 | 66 | ||
| 1639 | Stage* | |||||||||||||
| I–III | 97 | 85 | 28 | 14 | 18 | 3 | 1 | 61 | 0 | 0 | 0 | 307 | 671 | |
| IV | 103 | 171 | 67 | 42 | 36 | 9 | 1 | 143 | 0 | 1 | 1 | 364 | ||
*Correlation between KRAS mutation rate and clinical characteristics show a statistical significance in the case of site (primary vs metastasis, p=0.02), neoplastic cell percentage (≥30% vs <30%, p=0.02) and tumour stage (I–III vs IV, p=0.05).
Sample types and mutant rates
The rates of mutant samples were similar between resection specimens and endoscopic biopsies (40% vs 43%; p=0.3). A similar mutation distribution was also observed, as reported in table 1. Noteworthy is the fact that biopsies were more often inadequate than resection specimens for insufficient material (8.0% vs 0.6%).
Mutation rates in samples from primary tumour and from distant metastases
In our series (n=1720) one single tumour sample of a given location was tested for each patient. The mutation distribution did not differ between the two sample groups, as reported in table 1. However, in samples from primary tumours (n=1609) the mutant rate was 39%; in metastatic samples (n=111) it was 50%. This difference was statistically significant (p=0.02). In a subset of 30 patients, for which paired samples were available in our institution, both primary and metastatic sites were processed. Discordant results were found in four patients (13.4%). In all cases the mutation (n=1 G12C, n=1 G12D, n=1 G12S and n=1 G12R) was restricted to the metastasis, whereas the primary tumour disclosed a WT genotype.
Mutation rate and percentage of neoplastic cells
The relationship between the percentage of neoplastic cells and the KRAS mutant rate was carefully reviewed in a subset of 578 cases. Overall, 528 (91.3%) cases had more than 30%, whereas 50 (8.7%) cases had less than 30% of neoplastic cells. The SS mutant rate was significantly (p=0.02) lower (26.7% vs 43.4%) in the group with less than 30%. This latter group re-analysis by the TheraScreen K-RAS mutation kit confirmed all mutations (n=13) detected by SS; in addition, six more mutated cases were found, leading to an overall rate of 38%.
Mutation rate and tumour stage
The rates of mutant samples were correlated to stage categories. The mutant rate was 35.9% in stages I–II, 38% in stage III and 41.8 in stage IV. We found that the mutant rate difference between non-metastatic (stages I–III) and metastatic (stage IV) patients was statistically significant (p=0.05).
Mutation rates and fixation modalities
The mutant rate in buffered formalin-fixed cases was 40.9%, whereas it was 34.9% in simple formalin-fixed samples. This difference was not significant (p=0.08).
RR evaluation
Twenty-five patients received irinotecan associated with cetuximab and the other 25 were administered a combination regimen (22 FOLFIRI and three FOLFOX). Twenty-eight patients were treated in second line and 22 in third line (table 2). In 13 out of 50 patients a response to treatment occurred (26%); in particular, one (2%) complete and 12 (24%) partial responses were observed. No response was observed in 27 patients (74%); 22 out of 50 had a progression as best response. Stable disease rates were 30%. The disease control rate (objective responses plus stable disease) was 56% (table 3). Concerning survival, the median PFS was 4.4 months and the median OS was 10.4 months.
Table 2.
Patients’ treatment regimens
| Cetuximab association | 2nd line CT | 3rd line CT |
|---|---|---|
| Irinotecan (25 patients) | 3 | 22 |
| FOLFIRI (22 patients) | 22 | – |
| FOLFOX (3 patients) | 3 | – |
| Total | 28 | 22 |
Table 3.
Patients’ RR
| KRAS WT | N (%) |
|---|---|
| CR | 1 (2) |
| PR | 12 (24) |
| SD | 15 (30) |
| PD | 22 (44) |
| DCR | 28 (56) |
| PFS (months) | 4.4 |
| OS (months) | 10.4 |
CR, complete response; DCR, disease control rate; OS, overall survival; PD, progression disease; PFS, progression-free survival; PR, partial response; RR, response rate; SD, stable disease; WT, wild type.
Discussion
It is widely held that SS has a limited sensitivity for detecting KRAS mutations.13 Therefore, the suitability of any sample should be determined by the pathologist on a case-by-case basis. To date, the only criteria is the percentage of neoplastic cells. Previous studies showed that SS is equivalent to more sensitive methods on tumour samples containing more than 30% of neoplastic cells.13 16 Above this cut-off more mutant (p=0.02) cases were also detected by our SS assay. Conversely, below 30% of neoplastic cells, the more sensitive TheraScreen kit increased the mutant rate by 12%. Therefore, the pathologist should be aware that SS can generate a not insignificant false negative results rate in cases with scant neoplastic cells.
In addition, we highlighted that the performance of SS may be influenced by two other criteria: the site and the stage of the tumour. The mutant rates, obtained from routinely testing one single tumour sample for each of the 1720 patients examined, differed between the group of 1609 primary tumours and that of 111 metastases. In particular, the group of cases from distant metastases featured more (50%) mutants than those (39%) from primary tumours (p=0.02). A similar discrepancy was observed from the analysis of paired primary and metastatic samples obtained from the same patients. In fact, in four of 30 (13.4%) patients, the mutation was restricted to the metastatic site. These differences may reflect heterogeneity between primary and metastatic tumours,17 but could also suggest that in samples taken from metastatic sites SS is more efficient from a technical point of view. Similarly, stage IV tumours yielded more mutants (p=0.05) than samples from the earlier stages. Therefore, samples taken from either advanced primary tumours or metastatic sites yielded a higher KRAS mutant rate by SS, probably reflecting the higher ratio of mutated and non-mutated cells featuring in late-stage CRC. Our data concur with the notion that KRAS intratumoral heterogeneity may occur only in earlier tumour stages.18 In fact, discrepancy between SS and more sensitive techniques has been reported only in stages I–II.18 As the ESP guidelines recommend upfront (reflex) testing of CRC including patients with stages II–III,1 our data challenge the suitability of SS in this setting. Further investigation is warranted to define the best upfront testing methodology.
Differences in mutant rates relative to patient mean age (p=0.2) and gender (p=0.2), between resection and biopsy specimens (p=0.3) and in relation to fixation modalities (p=0.08), were not significant. Simple formalin is thus suitable for molecular analysis, as long as care is taken to design primers that amplify short PCR products.19 Endoscopic samples may be useful in rectal adenocarcinoma specimens with low tumour cellularity, after neoadjuvant treatment. The reliability of endoscopic biopsy has also been proved by two very recent studies showing that the KRAS mutant rate is highly concordant in matched endoscopic and resection specimens.20 21 However, in our series in 8.0% of biopsies the residual material following standard histology and ancillary techniques was insufficient.
Previous studies reported that the RR to anti-EGFR moAb varies from 10% to 20% in patients with metastatic CRC.22 Our data showed a response to treatment in 13 out of 50 KRAS WT patients (26%). Our SS-based method thus predicted the response to cetuximab treatment efficiently. It is noteworthy that the microscopic review of the H&E sections corresponding to the selected cases showed a high (≥30%) amount of invasive neoplastic component in all instances. Further investigation is warranted to verify the RR of direct sequencing in KRAS WT patients whose tumours harbour less than 30% of neoplastic cells.
In conclusion, data from our large experience from treatment-predictive KRAS testing may be useful to the practising pathologist. Besides neoplastic cellularity assessment, the pathologist should evaluate two additional criteria, as we showed that SS is more efficient on tissue taken from a metastatic site or from an advanced stage primary tumour.
Take-home messages.
The suitability of any sample for KRAS mutation detection by SS should be determined by the pathologist on a case-by-case basis. Besides tumour cellularity, we highlight two other criteria: the site and the stage of the tumour.
Samples taken from either advanced primary tumours or metastatic sites yielded a high KRAS mutant rate by SS. Conversely, our data challenge the suitability of SS in stages II–III CRC patients.
Acknowledgments
The autrhors would like to thank Suzanne Kamel-Reid (Molecular Diagnostics Laboratory, Department of Pathology, The University Health Network, Toronto, Ontario, Canada) for providing clinical samples whose KRAS status was known for SS validation.
Footnotes
Contributors: UM and GT conceived the study. CB contributed as referral pathologist. UM, MS, CdL and ER contributed as molecular biologists. PR, LS, FMM, GV and ON contributed as primary pathologists. AdS and CM contributed as oncologists. GT supervised the study and wrote the paper with the contribution of the other authors. GT is the guarantor of the paper. All authors read and approved the final version.
Competing interest: None.
Patient consent: Obtained.
Ethics approval: Ethics approval was obtained from the Università degli Studi di Napoli Federico II, Comitato Etico per le attività biomediche ‘Carlo Romano’ n. study 46/12.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.van Krieken JH, Jung A, Kirchner T, et al. KRAS mutation testing for predicting response to anti-EGFR therapy for colorectal carcinoma: proposal for an European quality assurance program. Virchows Arch 2008;453:417–31 [DOI] [PubMed] [Google Scholar]
- 2.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med 2008;358:1160–74 [DOI] [PubMed] [Google Scholar]
- 3.Tol J, Punt CJ. Monoclonal antibodies in the treatment of metastatic colorectal cancer: a review. Clin Ther 2010;32:437–53 [DOI] [PubMed] [Google Scholar]
- 4.Van Cutsem E, Kohne CH, Hitre E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med 2009;360:1408–17 [DOI] [PubMed] [Google Scholar]
- 5.Allegra CJ, Jessup JM, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 2009;27:2091–6 [DOI] [PubMed] [Google Scholar]
- 6.Weichert W, Schewe C, Lehmann A, et al. KRAS genotyping of paraffin-embedded colorectal cancer tissue in routine diagnostics: comparison of methods and impact of histology. J Mol Diagn 2010;12:35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellon E, Ligtenberg MJ, Tejpar S, et al. External quality assessment for KRAS testing is needed: setup of a european program and report of the first joined regional quality assessment rounds. Oncologist 2011;16:467–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angulo B, Garcia-Garcia E, Martinez R, et al. A commercial real-time PCR kit provides greater sensitivity than direct sequencing to detect KRAS mutations. A morphology-based approach in colorectal carcinoma. J Mol Diagn 2010;12:292–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cross J. DxS Ltd. Pharmacogenomics 2008;9:463–7 [DOI] [PubMed] [Google Scholar]
- 10.Do H, Krypuy M, Mitchell PL, et al. High resolution melting analysis for rapid and sensitive EGFR and KRAS mutation detection in formalin fixed paraffin embedded biopsies. BMC Cancer 2008;8:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franklin WA, Haney J, Sugita M, et al. KRAS mutation: comparison of testing methods and tissue sampling techniques in colon cancer. J Mol Diagn 2010;12:43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon MJ, Lee SE, Kang SY, et al. Frequency of KRAS, BRAF, and PIK3CA mutations in advanced colorectal cancers: comparison of peptide nucleic acid-mediated PCR clamping and direct sequencing in formalin-fixed, paraffin-embedded tissue. Pathol Res Pract 2011;207:762–8 [DOI] [PubMed] [Google Scholar]
- 13.Tol J, Dijkstra JR, Vink-Borger ME, et al. High sensitivity of both sequencing and real-time PCR analysis of KRAS mutations in colorectal cancer tissue. J Cell Mol Med 2010;14:2122–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malapelle U, de Rosa N, Rocco D, et al. EGFR and KRAS mutations detection on lung cancer liquid-based cytology: a pilot study. J Clin Pathol 2012;65:87–91 [DOI] [PubMed] [Google Scholar]
- 15.Kamel-Reid S, Zhang T, Persons DL, et al. Validation of KRAS testing for anti-EGFR therapeutic decisions for patients with metastatic colorectal carcinoma. Arch Pathol Lab Med 2012;136:26–32 [DOI] [PubMed] [Google Scholar]
- 16.Kobunai T, Watanabe T, Yamamoto Y, et al. The frequency of KRAS mutation detection in human colon carcinoma is influenced by the sensitivity of assay methodology: a comparison between direct sequencing and real-time PCR. Biochem Biophys Res Commun 2010;395:158–62 [DOI] [PubMed] [Google Scholar]
- 17.Longo DL. Tumor heterogeneity and personalized medicine. N Engl J Med 2012;366:956–7 [DOI] [PubMed] [Google Scholar]
- 18.Baldus SE, Schaefer KL, Engers R, et al. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res 2010;16:790–9 [DOI] [PubMed] [Google Scholar]
- 19.Kotoula V, Charalambous E, Biesmans B, et al. Targeted KRAS mutation assessment on patient tumor histologic material in real time diagnostics. PLoS One 2009;4:e7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krol LC, t Hart NA, Methorst N, et al. Concordance in KRAS and BRAF mutations in endoscopic biopsy samples and resection specimens of colorectal adenocarcinoma. Eur J Cancer 2012;48:1108–15. [DOI] [PubMed] [Google Scholar]
- 21.Yang QH, Schmidt J, Soucy G, et al. KRAS mutational status of endoscopic biopsies matches resection specimens. J Clin Pathol 2012;65:604–7. [DOI] [PubMed] [Google Scholar]
- 22.Van Cutsem E, Kohne CH, Lang I, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol 2011;29:2011–19 [DOI] [PubMed] [Google Scholar]