Abstract
Background & objectives:
Though several viruses are responsible for conjunctivitis, but human adenovirus (HAdV) is by far the most common cause. Epidemic conjunctivitis causes morbidity and early detection of aetiological agent is essential in preventing spread of disease as some of serotypes of adenoviruses cause a severe form of conjunctivitis. This study was undertaken to identify the causative agent of conjunctivitis outbreak in Chennai in 2010.
Methods:
Conjunctival samples collected from 17 patients with conjunctivitis were subjected to virological investigations. Culture and PCR for detection of adenovirus and enterovirus were carried out. PCR positive products were further subjected for DNA sequencing. The nucleotide sequences of the hexons of isolates were analyzed by comparison with all 51 human adenovirus strains. Phylogenetic tree was constructed using DAMBE software.
Results:
Among 17 patients, seven were positive for adenovirus by PCR on the direct specimen, none was positive for enterovirus. Eleven of 30 conjunctival swabs showed cytopathic effect in HEp-2 cell line and were confirmed as HAdV by PCR. The DNA sequence data of the 11 isolates had equal percentage of homology with HAdV 6 and 2 on blast analysis. On phylogenetic analysis with GeneBank data of 51 adenovirus strains, 11 isolates from patients during the outbreak of conjunctivitis formed a separate clade indicating a new variant strain.
Interpretation & conclusions:
Based on phylogenetic analysis it was concluded that the recent conjunctivitis outbreak that occurred in Chennai was caused by a variant adenovirus strain.
Keywords: Adenovirus, conjunctivitis, PCR-based DNA sequencing, variant adenovirus
Conjunctivitis caused by adenoviruses may manifest as pharyngoconjunctival fever (serotypes 3, 7), epidemic keratoconjunctivitis (serotypes 4, 8, 9, 19a, 37)1 or acute haemorragic conjunctivitis (serotypes 7, 11, 21, 35). Though the diseases are self-limiting, these cause significant amount of morbidity in preventing people attending work and are spread rapidly to the susceptible populations resulting in outbreaks and epidemics in a given geographical area2,3. Genotyping the viruses is very important to understand the type of virus causing conjunctivitis in a particular region and a phylogenetic analysis carried out with the data provides the likely origin of the virus.
Outbreak of epidemic conjunctivitis is encountered every year in Chennai, Tamil Nadu, India, during the rainy season i.e. from August-November. Our earlier investigations during some of these outbreaks identified adenovirus serotype 4 in 1991, type 3 in 1992-19931. The identification on the causative agent was possible by virus isolation using tissue culture facility and serotyped using anti-adenoviral antiserum types 1, 2, 3, 4, 5, 6, 7a and 14 (NIAID Antisera, ATCC, Rockville, MD, USA). Later in 1996, viruses from clinical specimens were isolated in HEp-2 cell line, and PCR-RFLP technique confirmed these to be type 7a4. The outbreak of acute conjunctivitis in 1998 continued to December - January 1999 and was due to Coxsackie A24 virus5. The present study reports on detection, isolation and genotyping of virus responsible for the recent outbreak of conjunctivitis in Chennai (commenced in August with the peak reaching in September with a slow decline in the number in October 2010). Cultures of conjunctival secretions from the patients during the peak period were collected for isolation of viruses and were identified as adenoviruses. The phylogenetic analysis was carried out with the DNA sequences obtained from the isolates along with the well established 51 genotypes (obtained from GenBank) of adenoviruses.
Material & Methods
Patients and clinical specimens: Thirty conjunctival swabs were collected from 17 randomly selected patients (15 men, 2 women, age 30 to 76 yr) with acute conjunctivitis attending the out patient department of the ophthalmic hospital Sankara Nethralaya, Chennai during October 2010. Swabs were collected from both eyes in 11 patients and in eight others from one eye only as a routine diagnostic procedure which is followed in the hospital. The conjunctival material collected in sterile absorbable cotton swab was transported in viral transport medium [minimum essential medium (MEM) with 3% foetal calf serum (FCS) and antibiotics]. The specimens were collected from the patients between 2 - 5 days after onset of the symptoms and were processed within 20 - 30 min in the laboratory.
Isolation of human adenovirus: Fifty microlitres of specimen was inoculated on to a 24 h old monolayer of HEp2 (NCCS, India) cell culture grown over tissue culture plate after aspirating out the growth medium. The inoculated culture was kept in a rocker for 30 min. At the end of 30 min Dulbecco's minimal essential medium supplemented with 1 per cent foetal calf serum was added. The cultures were incubated at 37°C carbondioxide incubator (Thermo Scientifics, USA). The cultures were observed daily under phase contrast microscope (Nikon, Japan) for the presence of cytopathic effect. The isolation of adenovirus was confirmed by indirect immunofluorescence technique using antibodies raised against hexon protein of human adenovirus (Chemicon, UK) and anti-mouse antibody tagged with FITC (Dako, Germany).
DNA and RNA extraction: Part of the clinical specimen was used for RNA extraction using QIAGEN mini RNA extraction kit (Germany). DNA was extracted using QIAGEN DNA extraction kit (Hilden, Germany). The kit reagents were reconstituted before use as per the protocol of the manufacturers’ instructions.
Polymerase chain reaction (PCR) for human adenovirus: The extracted DNA was amplified for HAdV using primers specific for hexon region. The PCR protocol and primers used were as described earlier4. The primers were custom synthesized by Bangalore Genei, India. The forward primer Ad TU 7-5’ GCC ACC TTC TTC CCC ATG GC 3’ and the reverse primer Ad TU 4’-5’ GTA GCG TTG CCG GCC GAG AA 3’ were used to amplify a 1004 bp (base pair) product in the first round. For the second round of the nested PCR (nPCR) the forward primer US’-5’ TTC CCC ATG GCc CAC AAC AC 3’ and the reverse primer UA-5’ GCC TCG ATG ACG CCG CGC TG 3’ were used to amplify a 956 bp product.
Reverse transcriptase polymerase chain reaction (RT-PCR) for group specific human enterovirus genome (Human EV): The nRT-PCR was standardized using the standard strain of Coxsackie A24 (VR 583, Chemicon, USA), Enterovirus 70 (ATCC (R) 836, Chemicon, USA). RNA was extracted and enterovirus group specific PCR was carried out with the RNA. In brief, 20 μl RNA was subjected to amplification using one step RT-PCR kit (QIAGEN, Germany). The primers were within the 5’ non coding region of EV. The primers sequences used for the nested round of amplification were as follows - forward primer: S1 5’ CAA GCA CTT CTG TTT CCC CGG 3’, reverse primer: S2 5’ TCC TCC GGC CCC TGA ATG CG 3’. Second round primers were: forward primer: AS1 5’ ATT GTC ACC ATA AGC AGC CA 3’, reverse primer: AS2 5’ AAA CAC GGA CAC CCA AA GTA 3’6. Using 10 μl of the extracted RNA the first step of cDNA formation was carried out and then PCR for amplification was done using a single step RT-PCR kit (Qiagen, Germany).
The following cocktail was set up for standardiza-tion using the extracted RNA- in brief, the reaction mixture consisted of 400 μM of each dNTP, 0.6μM of each forward and reverse primers and the enzyme mix (Merck, India). The enzyme mix consisted of both the Omniscript and Sensiscript reverse transcriptase and Hot start Taq DNA polymerase (Synergy Scientifics, India). These enzymes are recombinant heterodimeric enzymes expressed in Escherichia coli and both exhibit higher affinity for RNA, facilitating transcription through secondary structures that inhibit other reverse transcriptases. The enzyme mix facilitated both reverse transcription and polymerase chain reaction. The reaction mix was incubated in the thermal cycler as follows: 50°C for 30 min for reverse transcription followed by the activation of Hot star Taq DNA polymerase enzyme and inactivation of Omniscript and Sensiscript reverse transcriptases at 95°C for 15 min. The PCR amplification was proceeded for 40 cycles, by denaturing the cDNA template at 94°C, annealing at 62°C for 30 sec and extension at 72°C for 1 min. The final extension was carried out at 72°C for 5 min. For the second round of the nested amplification, 2 μl of the first round product was added to 50 μl of the PCR mix consisting of 200 μM of each dNTP, 10 mM Tris-Cl, 0.6 μM of R3 and R4 primers and 2.5 U of Taq DNA polymerase. The PCR amplification was carried out for 25 cycles, denaturation at 94°C for 30 sec, annealing at 60°C for 30 sec and extension at 72°C for 1 min.
DNA sequencing: The amplified products in the samples positive by nPCR were subjected to DNA sequencing. Cycle sequencing of the amplified products was performed in a 10 μl reaction volume, containing 0.5 μl of ready reaction mix, 3.5 μl of sequencing buffer, 0.5 μl of forward primer (1:100 diluted), 3.5 μl MilliQ water and 0.5 μl of amplified product. Amplification was carried out in Perkin-Elmer thermocycler using 25 cycles at 96˚C for 10 sec, 50˚C for 5 sec, 60˚C for 4 min with initial denaturation at 96°C for 1 min. The cycle-sequenced products were then purified and sequenced using ABI Prism 3130 AVANT (Applied Biosystems, USA) genetic analyzer with polymer POP 6. The sequences were then analysed by Bio Edit sequence alignment software (www.softpedia.com/progDownload/BioEdit-Download-174716), and BLAST analysis (www.ncbi.nlm.nih.gov/BLAST) was done to confirm the sequenced data with the standard strains and to determine the per cent homology.
Phylogenetic analysis: The nucleotide sequences of the hexons were analyzed by comparison with all 51 HAdV7,8 and with novel HAdV serotype that caused epidemic keratoconjunctivitis at Japan9. The nucleotide sequences of the hexons were analyzed using DAMBE software (http://en.biosoft.net/format/DAMBE.html). Evolutionary distances were estimated using Kimura's two-parameter method and unrooted phylogenetic trees were performed by 1,000 resamplings of the data sets10.
Results
Eight samples collected from both eyes of four patients and three from one eye of three patients showed cytopathic effect in HEp 2 cell line and these were confirmed as HAdV by PCR. Seven were positive for HAdV by PCR from the direct specimen (Fig. 1). The seven direct PCR positive conjunctival specimens were also culture positive. All the specimens positive for culture were collected with in 2-3 days after onset of diseases symptoms and none of those with onset of disease beyond 3 days were positive either by PCR or culture. None of the clinical specimens was positive for human enterovirus genome by nRT-PCR.
Fig. 1.
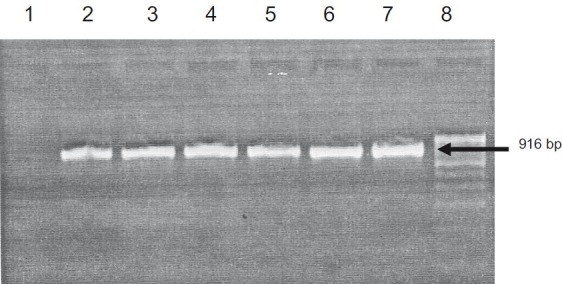
Agarose gel electrophoretogram showing HAdV PCR results of isolates from patients during the epidemic outbreak of acute keratoconjunctivitis. Lane 1: Negative control; Lane 2-6: Isolates; Lane 7: Adenovirus serotype 2 (ATCC (r) 846); Lane 8: Molecular weight marker
The DNA sequence data of the 11 isolates had 92-98 per cent homology with HAdV 6 and 2 on blast analysis. On multiple alignment with the prototype sequences of adenovirus 2 and 6, the isolates showed nucleotide variations in many regions. With these blast results, we could not definitely confirm the genotypes. Therefore, a phylogenetic analysis was carried out with DNA sequences of all known 51 HAdV strains obtained from GeneBank along with our 11 isolates and the novel HAdV strain described in Japan9. All the 11 Chennai outbreak HAdV strains formed a separate clade indicating a variant HAdV type as the cause of this outbreak (Fig. 2). The nucleotide sequences were 98 per cent identical. Therefore these did not form a monophyletic cluster but branched out as a separate clade from other genotypes. Based on Multain results of the sequences, the isolates had shown sequence similarity with group C at 1056, 1609 and 1791 nucleotide positions and the isolates had similarity with group A at 1038, 1057 and 1573 nucleotide positions.
Fig. 2.
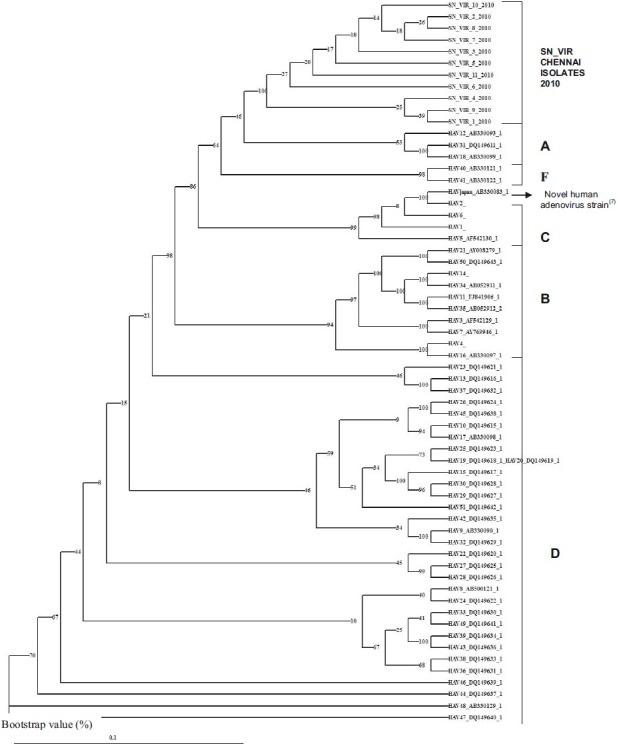
Phylogenetic analyses of the isolates from patients with keratoconjunctivitis in Chennai. The 916 bp sequence of a partial hexon gene of the representative isolates were analyzed together with all the 51 HAdV serotypes. The numbers at the nodes are percentages of 1,000 bootstrap pseudoreplicates containing the cluster distal to the node.
The sequences were submitted to Genbank and the 11 isolates were given the accession numbers from JN181144 to JN181154.
Sensitivity of the nRT-PCR for the detection of human enterovirus (EV): The sensitivity of the primers was determined by diluting the initial standard strain inoculated cell culture harvest 10-fold in DMEM, following which the RNA extraction was performed from each of the log10 dilutions. The standardized nRT-PCR was set up for each of the dilutions and the detection limit was 12 femtograms.
The optimized nRT-PCR for 5’ non-coding region was specific for enteroviruses since there was no amplification of RNAs from infective agents namely HSV, Chlamydia trachomatis, adenovirus and human RNA included in the reactions.
Discussion
In the present study, all the 11 isolates of the 2010 Chennai outbreak of acute keratoconjunctivitis formed a separate phylogenetic clade indicating that a new HAdV type was the aetiological cause. To understand the genetic relationships among the HAdV DNA sequences, a phylogenetic tree based on the partial hexon nucleotide sequences was constructed. Though adenoviruses are classified into 51 genotypes, many variants belonging to each of the genotypes are known to cause conjunctivitis. Molecular methods are highly sensitive to identify these variants. Since DNA sequence data of the 11 isolates showed equal percentage of homology with adenovirus 6 and 2 on blast analysis and using multiple alignment with the prototype sequences of adenovirus 2 and 6, the isolates showed nucleotide variations in many regions. Therefore, with blast results the genotype to the present isolates could not be identified. So using DAMBE Software, a phylogenetic tree was developed with the DNA sequences of all known 51 adenoviruses and including the new strain isolated in Japan9. The results showed that all 11 isolates of the present epidemic formed a separate clade and possibly a new variant strain of HAdV. The isolates could be a putative recombinant as these had sequences belonging to both C and A group. We could not authentically conclude this as the region sequenced was partial.
The clinical disease was acute keratoconjunctivitis caused by these viruses and was not different from that caused by other HAdV listed in literature.
We could diagnose HAdV infections by PCR and type infectious HAdVs by phylogeny rapidly. Phylogeny-based system is considered highly useful when rapid diagnosis and typing are required. Compared to blast analysis data, phylogeny is more reliable to find the closer relationship of genomic origin.
In conclusion, results of our phylogenetic analysis suggested that the 2010 outbreak of keratoconjunctivitis at Chennai was probably caused by a variant adenovirus strain.
References
- 1.Madhavan HN. Laboratory investigations on viral and Chlamydia trachomatis infections of the eye: Sankara Nethralaya experiences. Indian J Ophthalmol. 1999;47:241–6. [PubMed] [Google Scholar]
- 2.Richmond S, Burman R, Crosdale E, Cropper L, Longson D, Enoch BE, et al. A large outbreak of keratoconjunctivitis due to adenovirus type 8. J Hyg (London) 1984;93:285–91. doi: 10.1017/s0022172400064810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World WSM, Howrwitz MS. Adenoviridae. In: Knipe DMH, Howley PM, editors. Fields virology. 5th ed. Vol. 2. Philadelphia PA: Lippincott Williams & Wilkins; 2007. pp. 2395–436. [Google Scholar]
- 4.Dalapathy S, Lily TK, Roy S, Madhavan HN. Development and use of nested polymerase chain reaction (PCR) for the detection of adenovirus from conjunctivitis specimens. J Clin Virol. 1998;11:77–84. doi: 10.1016/s0928-0197(98)00021-x. [DOI] [PubMed] [Google Scholar]
- 5.Madhavan HN, Malathy J, Priya K. An outbreak of acute conjunctivitis caused by Coxsackie virus A 24. Indian J Ophthalmol. 2000;48:159. [PubMed] [Google Scholar]
- 6.Shulman LM, Manor Y, Azar R, Handsher R, Vonsover A, Mendelson E, et al. Identification of a new strain of fastidious enterovirus 70 as the causative agent of an outbreak of hemorrhagic conjunctivitis. J Clin Microbiol. 1997;35:2145–9. doi: 10.1128/jcm.35.8.2145-2149.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebner K, Pinsker W, Lion T. Comparative sequence analysis of the hexon gene in the entire spectrum of human adenovirus serotypes: phylogenetic, taxonomic, and clinical implication. J Virol. 2005;79:12635–42. doi: 10.1128/JVI.79.20.12635-12642.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimada Y, Ariga T, Tagawa Y, Aoki K, Ohno S, Ishiko H. Molecular diagnosis of human adenoviruses D and E by a phylogeny-based classification method using a partial hexon sequence. J Clin Microbiol. 2004;42:1577–84. doi: 10.1128/JCM.42.4.1577-1584.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishiko H, Shimada Y, Konno T, Hayashi A, Ohguchi T, Tagawa Y, et al. Novel human adenovirus causing nosocomial epidemic keratoconjunctivitis. J Clin Microbiol. 2008;46:2002–8. doi: 10.1128/JCM.01835-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saitou N, Nei M. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]


