Abstract
The PI3K/Akt pathway is activated in many cancers; therefore, we investigated NVP-BEZ235, a dual PI3K/mTOR inhibitor. BEZ235 was more potent than either the mTOR inhibitor rapamycin or the PI3K inhibitor LY294002 in blocking HIF-1α induction. BEZ235 decreases protein translation, and 7-methyl GTP chromatography showed that the drug induced robust recruitment of 4E-BP1 to eIF4E and a near absence of binding of eIF4G. BEZ235 also decreased expression of other proteins known to be regulated by eIF4E including cyclin B1 and D1 and vascular endothelial growth factor (VEGF). BEZ235 also decreased the level of eIF4G but not eIF4E. As HIF-1α has been associated with adaptation to hypoxic stress, we examined the effect of the drug on cell survival in low pO2. BEZ235 increased killing of cells under hypoxia, measured by short-term (MTT) and long-term (clonogenic) assays. To understand the underlying mechanism, we examined BEZ235’s effect on the expression of factors associated with cell survival. Under normoxia, Akt Ser473 phosphorylation decreased within an hour of BEZ235 treatment, but then increased by 24 h. In contrast, under hypoxia, BEZ235 caused prolonged suppression of Akt Ser473 phosphorylation. Furthermore, there was greater PARP cleavage in hypoxic cells than in normoxic cells, consistent with increased apoptosis. BEZ235 increased autophagy as measured by LC3-I to LC3-II conversion under both normoxic and hypoxic conditions, but our data indicate that this is actually a pro-survival mechanism. In conclusion, we have found that BEZ235 blocks HIF-1α induction by decreasing protein translation and increases cell killing under hypoxia, likely by increasing apoptosis.
Keywords: NVP-BEZ235, hypoxia, PI3K, mTOR, autophagy
Introduction
The PI3K/Akt pathway is activated in a large percentage of human cancers.1 This can occur through a variety of mechanisms including Ras mutation, loss of PTEN which encodes the phosphatase that opposes the action of PI3K, activation of growth factor receptors such as EGFR, and mutations in PIK3CA, the gene that encodes the alpha isoform of the p110 catalytic subunit of PI3K. Because of its central role in cancer development, the PI3K/Akt pathway is an attractive target for anti-cancer therapy.2 Two of the more commonly used inhibitors of the PI3K pathway are wortmannin and LY294002. However, due to their unfavorable pharmacologic properties and toxicity, these drugs are not suitable for clinical use.3,4 For this reason, new agents that can inhibit multiple targets in this pathway are being developed. We have focused our attention on the Novartis compound NVP-BEZ235, which is being used clinically and found to be relatively non-toxic.5,6 BEZ235 is a dual inhibitor, physically interacting with the ATP-binding clefts of both mammalian target of rapamycin (mTOR) and class I PI3 kinases. BEZ235 inhibits the α, γ and δ isoforms of the p110 subunits with an IC50 ranging from 4 - 7 nM and the β isoform with an IC50 of 75 nM.5 The IC50 for mTOR kinase is 20 nM; however, the IC50 for other kinases such as VEGFR1, HER1, cMet and Akt1 is orders of magnitude higher (>10,000 nM).
The PI3K/Akt pathway has been implicated in the regulation of cell growth, proliferation, survival and metabolism as well as protein translation. Specifically mutations in the PI3K/PTEN pathway contribute to increased rate of translation, even under hypoxic conditions.7 A key player in this pathway downstream of Akt is mTOR, a serine/threonine kinase that integrates mitogenic and nutrient signaling to regulate protein translation.8 mTOR can bind to Raptor to form the TORC1 complex9 or to Rictor to form the TORC2 complex.10 TORC1 phosphorylates major downstream proteins involved in translation including p70SK kinase (p70S6K) and eIF4G.11 eIF4G, eIF4A and the cap-binding protein eIF4E comprise the eukaryotic translation initiation factor 4F (eIF4F). During translation, mRNAs with a 7-methylguanosine cap are bound to eIF4E, while eIF4G serves as a scaffold for eIF4A, poly(A)-binding proteins (PABPs), and eIF3. TORC1 also phosphorylates the translational repressor 4E-BP1. Under non-proliferative conditions, 4EBP1 binds to eIF4E and prevents the latter from associating with eIF4G, thereby blocking cap-dependent translation.12 However, when it is phosphorylated, 4E-BP1 releases eIF-4E, so that the latter can bind eIF4G, allowing for cap-dependent protein translation to proceed.
The PI3K/Akt pathway has also been implicated in the modulation of hypoxia-inducible factors by many groups including our own.13-17 HIF-1 is a master transcription factor consisting of two subunits, the α subunit, which is induced by hypoxia, and the β subunit, which is expressed constitutively. HIF-1α binds to HIF-1β to transactivate target genes including VEGF, Glut1 and various glycolytic enzymes that help cells adapt to hypoxia.18 Hypoxia is a potent inducer of HIF-1α expression, but this induction can be augmented by PI3K/Akt activation.
We initiated the current study to investigate the effects of BEZ235 on HIF-1α expression under hypoxia. Because we found that BEZ235 interferes with HIF-1α synthesis and because it inhibits mTOR, we then examined the effect of the drug on the protein translation machinery. As HIF-1 is an important transcription factor under hypoxia, we also investigated the effects of BEZ235 on signaling under hypoxia. Our results indicate that the drug decreases cell survival under hypoxia and offers insight into how this happens. Our findings may have implications for the use of BEZ235 to treat tumors that are hypoxic, which are often resistant to radiation and chemotherapy.
Results
BEZ235 suppresses HIF-1α induction under hypoxia
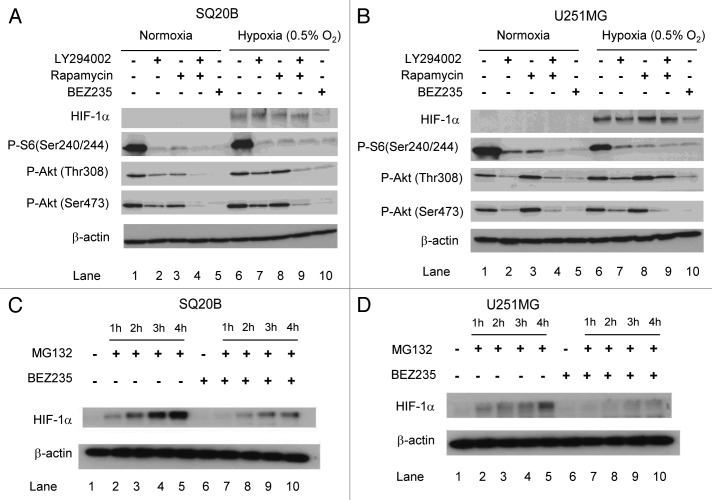
We compared the effects of the dual PI3K/mTOR inhibitor BEZ235 with LY294002 (PI3K inhibitor) and rapamycin (mTOR inhibitor) in the SQ20B head and neck squamous cell carcinoma cell line. BEZ235 almost completely abolished Akt phosphorylation on both the Thr308 and Ser473 residues whereas LY294002 had only a partial effect (Fig. 1A). If anything, rapamycin seemed to increase Akt phosphorylation (compare lanes 3 versus 1 and 8 versus 6). The three drugs, rapamycin, LY294002 and BEZ235, almost completely abolished phosphorylation of S6 at Ser240/244. We tested the effect of these drugs on HIF-1α induction. Figure 1A shows that the drug attenuated HIF-1α induction under hypoxia in SQ20B cells (compare lanes 6 and 10) whereas LY294002 and rapamycin had no effect (compare lane 6 with 7 or 8). Similar results were seen with U251MG glioblastoma cells (Fig. 1B).
Figure 1. Greater suppression of HIF-1α with BEZ235 than LY294002 or rapamycin. (A and B) Cells were treated with BEZ235, LY294002 or rapamycin for 3 h, and thereafter cells were exposed to 0.5% oxygen (lanes 6-10) for 3 h before cells were harvested. (C and D) Cells were treated with 100 nM BEZ235 for 24 h, followed by addition of MG132 for 1, 2, 3 and 4 h. After MG132 treatment, protein was harvested. For panels A–D, proteins samples were analyzed by SDS-PAGE and immunoblotting using antibodies as indicated, including HIF-1α and β-actin (loading control).
To determine whether this effect on HIF-1α protein might be due to an effect on the mRNA, we performed real-time quantitative RT PCR. Figure S1 shows that BEZ235 treatment did not reduce the level of HIF-1α mRNA. Hence, the effect of BEZ235 on decreasing HIF-1α protein expression was not at the level of mRNA but rather at the level of protein expression. To ascertain whether this decrease in HIF-1α expression was due to reduced stability or decreased translation of HIF-1α, we treated cells with BEZ235 for 24 h, followed by the addition of MG132 (a proteasomal inhibitor) for 1, 2, 3 and 4 h. We observed an accumulation of HIF-1α with time in the control SQ20B (Fig. 1C) and U251 cells (Fig. 1D). In contrast, SQ20B and U251 cells treated with BEZ235 showed considerably lower accumulation of HIF-1α, indicating that the drug interferes with protein synthesis rather than degradation.
BEZ235 decreases global protein translation and decreases eIF4G binding to eIF4E
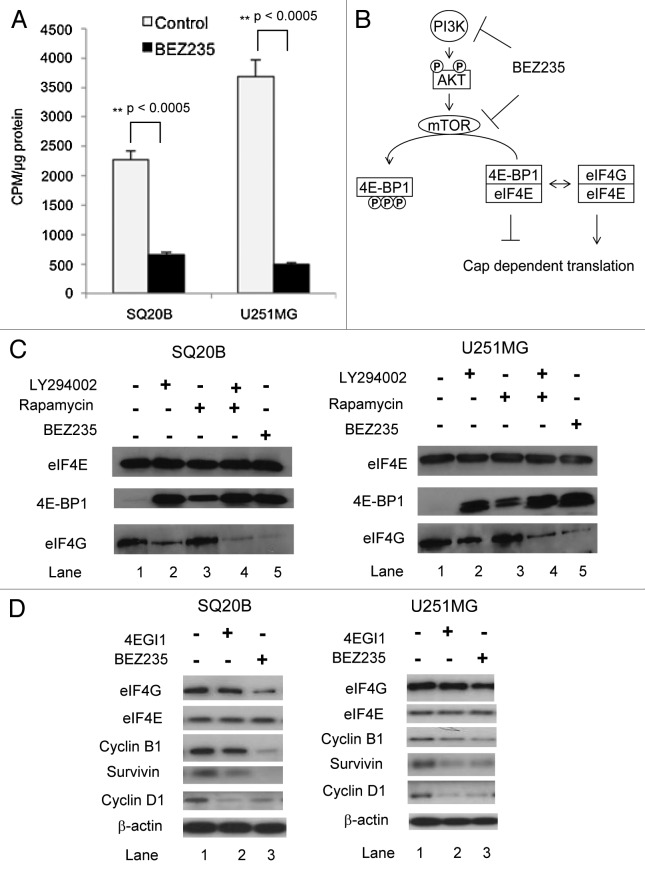
The results above led us to assess the effect of BEZ235 on global protein translation. To do this, we labeled cells with [35S] methionine, precipitated total protein, then measured radioactivity. Treatment with BEZ235 for 24 h led to a 70% inhibition of [35S] incorporation in SQ20B cells and 86% in U251MG cells (Fig. 2A).
Figure 2. BEZ235 treatment results in decreased global protein translation. (A) SQ20B or U251MG cells were treated with BEZ235 for 24 h. 30 minutes before the end of treatment, cells were labeled with [35S]methionine (50 µCi/ml). At completion of assay, cells were lysed and assessed for total protein content and radioactivity. Results are represented as the counts per minute (cpm) normalized to total protein concentration in the drug-treated cells compared to the untreated control cells. (B) The effect of PI3K/Akt/mTOR inhibition on cap dependent translation. The figure shows the interactions between eIF4E, 4E-BP1 and eIF4G. (C) SQ20B or U251MG cells were treated with drugs as indicated for 3 h. Whole cell extracts were prepared and eIF4E and its associated proteins purified by 7-methyl-GTP-Sepharose affinity chromatography. (D) SQ20B or U251MG cells were treated with 4EGI1 or BEZ235 for 24 h. After the drug treatment cells were exposed to hypoxia for 3 h. Thereafter, protein was harvested. For panels C and D, protein samples were analyzed by SDS-PAGE and immunoblotting for proteins using antibodies as shown.
In order to determine how BEZ235 decreased protein translation, we examined the drug’s effect on interactions between eIF4E, 4E-BP1 and eIF4G, all components of the eIF4F protein translation complex. We found that BEZ235 dramatically decreases 4E-BP1 phosphorylation (Fig. S2), which should increase 4E-BP1 binding to eIF4E (Fig. 2B). To assess this, we used 7 methyl GTP chromatography19 to recover cap-binding eIF4E and proteins bound to it. In SQ20B cells, treatment with LY294002 or BEZ235 led to a robust recruitment of 4E-BP1 to eIF4E, greater than that seen in rapamycin-treated cells (Fig. 2C; compare lanes 2, 3 and 5 with 1). This figure also shows that rapamycin did not alter the binding of eIF4G to eIF4E compared with control cells whereas BEZ235 resulted in a near absence of binding of eIF4G. LY24002 reduced the binding of eIF4G to eIF4E although not as robustly as BEZ235. The same qualitative result in terms of eIF4G binding was seen in U251MG with the effect of BEZ235>LY294002>rapamycin (Fig. 2C). Given that BEZ235 increases eIF4E binding to 4E-BP1 and decreases it’s binding to eIF4G, we decided to examine the effects of the drug on specific proteins whose expression is known to be regulated by eIF4E. Many of these are associated with cell proliferation or survival including cyclin D1, cyclin B1, survivin and vascular endothelial growth factor (VEGF).11,20,21 We compared the effects of BEZ235 to a known inhibitor of the eIF4E-eIF4G interaction, 4EGI1.22 Using a concentration shown by others to disrupt eIF4G/eIF4E interaction (50 µM), 4EGI1 decreased survivin and cyclin D1 levels in both SQ20B and U251MG cells, with little or no effect on cyclin B1, eIF4G and eIF4E levels (Fig. 2D). Treatment with BEZ235 caused a greater decrease in eIF4G, cyclin B1 and survivin protein levels. The level of eIF4E was not affected with BEZ235 treatment. BEZ235 also reduced the levels of VEGF secreted into the media of both the cell lines (Fig. S3).
However, BEZ235 did not reduce the levels of all proteins that we examined. For example, the level of β-actin was unaltered (Figs. 1 and 2), although it could be argued that β-actin is so stable that to see a decrease in its level would take prolonged treatment with the drug. Hence, we examined proteins that are known to have short half-lives, specifically c-Myc (half-life of 20- 30 minutes)23 and ornithine decarboxylase (ODC) (half-life of ~3 h).24Figure S4 shows that BEZ235 treatment of U251 cells clearly led to a clear decrease in cyclin D1 and survivin expression but not c-Myc or ODC.
Effect of BEZ235 on cell survival under hypoxia
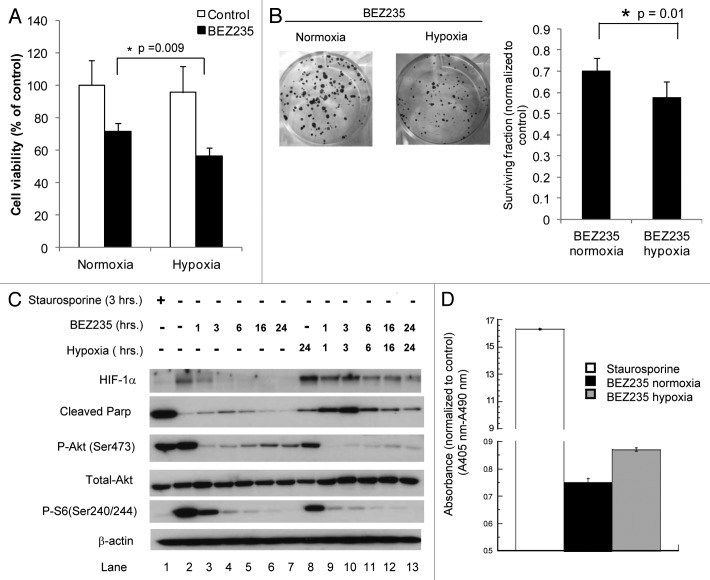
As we found that BEZ235 decreased HIF-1α, which has been associated with adaptation under hypoxia, we examined cell survival with the drug under different pO2 levels. Figure 3A shows that 24 hour treatment of hypoxic cells with the drug decreases cell viability (based on the MTT assay) to a greater extent than in normoxic cells (p = 0.009). The MTT assay measures the ability of mitochondrial enzymes to reduce the MTT substrate, resulting in a change in color of the media. Although it is not a direct measure of cell viability, this assay is often used as a surrogate for this. For a long-term measure, we performed clonogenic survival assays in which cells were exposed to hypoxia and/or normoxia and treated with ± BEZ235. Figure 3B shows visually that the combination of hypoxia (0.5% O2) and BEZ235 treatment led to fewer colonies than seen with drug under normoxia. Furthermore, the colonies in the dishes treated with BEZ235 under hypoxia showed smaller colonies than drug-treated dishes in normoxia. Under normoxia, the cloning efficiency was 30.4% with BEZ235 versus 43% without drug (ratio of 0.70 for drug: no drug). However, under hypoxia, the respective cloning efficiencies were 18% versus 33% (ratio of 0.58 for drug: no drug). The difference in the surviving fraction ratios for normoxia versus hypoxia was statistically significant (0.70 vs. 0.58; p = 0.01).
Figure 3. Effects of BEZ235 on survival under hypoxia. (A) SQ20B cells were exposed to 0.5% O2 for 24 h, and then treated with BEZ235 for 24 h. The cell viability was measured by the MTT assay. (B) SQ20B cells were exposed to 0.5% O2 for 24 h, and then treated with BEZ235 for 24 h. After 10 days the colonies were stained and counted. The graph underneath shows the surviving fraction ratios under hypoxia and normoxia (plating efficiency in the presence of BEZ235 divided by plating efficiency in the controls without the drug). (C) SQ20B cells were exposed to 0.5% O2 for 24 h, then treated with BEZ235 for 1, 3, 6, 16 and 24 h prior to protein harvesting. Proteins were analyzed by SDS-PAGE and immunoblotting with antibodies as shown. (D) SQ20B cells were exposed to 0.5% O2 for 24 h, and then treated with BEZ235 for 24 h. Cell death detection ELISA was used to quantitate the cytoplasmic oligonucleosomes as a measure of apoptosis.
To explore this further, we performed a time course experiment in which hypoxic cells were exposed to BEZ235 for varying lengths of time. Figure 3C shows that there is a greater increase in cleaved PARP seen in hypoxic cells treated with the drug for 1–16 h than in normoxic cells, consistent with increased apoptosis. This was confirmed using an ELISA assay that measures histone associated DNA fragments (mono- and oligonucleosomes) after apoptosis in cytoplasmic fractions of cell lysates. The assay showed greater enrichment of oligonucleosomes in hypoxic vs. normoxic cells treated with the drug (Fig. 3D), consistent with increased PARP cleavage (p = 0.175).
In Figure 3C we also noted that under normoxic conditions, Akt phosphorylation decreased within an hour of BEZ235 treatment, as expected, but then increased by 24 h. In contrast, under hypoxia the drug had a prolonged effect on suppressing Akt phosphorylation, even at 24 h. As active (phosphorylated) Akt has an anti-apoptotic effect, this could explain why the drug led to greater apoptosis in hypoxia. S6 phosphorylation was also decreased by the drug at 1 hour in normoxic cells but to an even greater extent in hypoxic cells. Therefore, we hypothesize that BEZ235 results in greater cell killing under hypoxia than normoxia by leading to sustained inhibition of Akt phosphorylation, thereby promoting apoptosis.
Effects of BEZ235 on autophagy
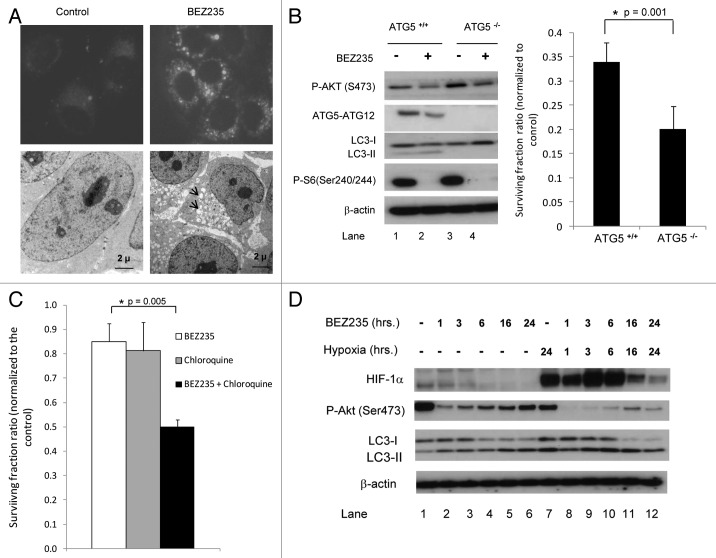
Since BEZ235 has been reported to induce autophagy in tumor cells25 we investigated this in SQ20B cells. We transfected cells with GFP-labeled LC3 and performed immunofluorescence analysis. LC3 is a protein that is lipidated during active autophagy and localizes to the autophagosomes; hence, GFP-LC3 is commonly used as a marker of this process. Figure 4A, upper panel shows that BEZ235 treatment led to the accumulation of punctate foci in the cytoplasm, consistent with localization of the fluorescent protein to the autophagosomes. Electron microscopy imaging analysis of SQ20B cells treated with the drug for 24 h did not show the classical double-membraned autophagosomes, however, there were numerous vacuoles in the cytoplasm (Fig. 4A, lower panel, arrowheads). We speculate that this represents cells at a later stage of autophagy during which the autophagosomes have merged with lysosomal vacuoles to form single-membraned autolysosomes containing degraded cytoplasmic contents as has been described previously.26
Figure 4. Effects of BEZ235 on autophagy. (A) Upper panel. SQ20B cells were stably transfected with GFP-LC3. The cells were then treated with or without BEZ235 for 24 h and examined by fluorescence microscopy. Lower panel. SQ20B cells were treated with BEZ235 for 24 h, fixed and analyzed by electron microscopy. There were numerous vacuoles in the cytoplasm of the drug treated cells (arrows). (B) ATG5 wild-type (ATG5+/+) and knockout (ATG5-/-) MEFs were treated with BEZ235 for 24 h. Left panel. After drug treatment cells were harvested for protein. Right panel. Cells were refed with drug free DMEM and after 10 days the colonies were stained and counted. The graph shows the surviving fraction ratios in ATG5+/+ and ATG5-/- cells (plating efficiency in the presence of BEZ235 divided by plating efficiency in the controls without the drug). (C) SQ20B cells were treated either with BEZ235 or chloroquine or a combination of these drugs for 6 h. After 10 days the colonies were stained and counted. Graph shows the surviving fraction ratios in SQ20B cells (plating efficiency in the presence of drug(s) divided by plating efficiency in the controls without drug). (D) SQ20B cells were exposed to 0.5% O2 for 24 h, then treated with BEZ235 for 1, 3, 6, 16 and 24 h prior to protein harvesting. Proteins were analyzed by SDS-PAGE and immunoblotting using antibodies as shown.
Autophagy has been implicated as a means of promoting cell survival in the face of adverse conditions; therefore, a critical question is whether the autophagy induced by BEZ235 leads to cell death. To address this question, we used fibroblasts from ATG5 knockout mice, which are defective for autophagy. Figure 4B (left panel) shows that ATG5/ATG12 is lacking in these cells. BEZ235 treatment (for 24 h) of the wildtype ATG5+/+ cells led to a slight decrease in P-Akt along with conversion of LC3-I to LC3-II. During autophagy LC3-I is converted to LC3-II, the lipidated form27; hence, this decrease in LC3-I is consistent with increased autophagic flux. In ATG5-/- cells, LC3-II was completely absent, consistent with a loss of autophagic capacity. We performed a clonogenic assay in order to assess long-term viability of the cells and their ability to propagate and form clones. The wildtype ATG5+/+ fibroblasts had a baseline plating efficiency of 32%, which was reduced to 11% with 24 hour BEZ235 treatment (surviving fraction ratio of 0.34 for drug: no drug). The ATG5-/- cells showed a baseline plating efficiency of 19%, which was reduced to 4% with 24 hour BEZ235 treatment (surviving fraction ratio of 0.20 for drug: no drug). This difference in the surviving fraction ratios for ATG5+/+ to ATG5-/- cells (0.34 vs. 0.20) was statistically significant (p = 0.001; Fig. 4B right panel). Hence, the inability to undergo autophagy in ATG5-/- cells actually decreased their clonogenic survival following BEZ235 treatment; therefore, autophagy appears to be playing a protective role in this setting. To further address the role of autophagy in survival after BEZ235 treatment, we inhibited active autophagy using chloroquine, a lysosomotropic agent that blocks the lysosome-autophagosome fusion.28 We found that the combination of BEZ235 and chloroquine was more cytotoxic than either of the agents alone (Fig. 4C), which has been noted in other cell lines.25,29 Therefore, chloroquine, which inhibits autophagy, appears to be promoting cell death.
Hypoxia itself has been shown to induce autophagy,30-33 therefore, we wish to investigate the effect of hypoxia combined with BEZ235 on autophagy. We performed Western blotting on protein lysates for LC3 (Fig. 4D). This panel shows that HIF-1α induction under hypoxia was blunted by BEZ235, especially at 16 and 24 h. As shown in Figure 3C, this panel also shows that the drug suppressed P-Akt to a greater extent in hypoxia than under normoxia. Furthermore, Figure 4D indicates that there is less LC3-I at 16 and 24 h after drug treatment in hypoxic cells than in normoxic cells (compare lanes 11 and 12 with 5 and 6), consistent with a greater effect on promotion of autophagy.
Discussion
The PI3K/Akt pathway has been implicated in the regulation of HIF-1α and VEGF expression; hence, initially our goal was to assess the effects of BEZ235 on the expression of HIF-1α.17 We compared its efficacy to the mTOR inhibitor rapamycin and the PI3K inhibitor, LY294002. Neither LY294002 nor rapamycin decreased HIF-1α induction appreciably whereas BEZ235 clearly did. The discordant effects of these drugs on HIF-1α induction might be explained by differences in their ability to inhibit components of the PI3K/Akt/mTOR pathway. Specifically, rapamycin is a more potent inhibitor of the mTORC1 complex compared to the mTORC2 complex,10,34 although this may be cell line-dependent.35 BEZ235 surpassed LY294002 in decreasing Akt phosphorylation, at both the Ser473 and Thr308 sites. Rapamycin actually seemed to increase Akt phosphorylation. This has been noted by other investigators and has been ascribed to a negative feedback loop that involves the insulin-like growth factor-1 (IGF-1).8,36,37 Specifically, it has been hypothesized that mTORC1 inhibition by rapamycin prevents serine phosphorylation of insulin receptor substrate-1 (IRS-1) and enhances IRS-1 association with IGF-I receptors. This leads to increased autocrine signaling through the IGF-1/IRFR pathway and results in downstream Akt phosphorylation. Consistent with our results, a dual mTOR/PI3K inhibitor such as BEZ235 would not be expected to lead to increased Akt phosphorylation because it would inhibit the PI3K pathway connecting IGF-1/IGFR to Akt.
The suppression of HIF-1α expression by BEZ235 was due to decreased translation, which led to our investigation of the drug’s effects on the eIF4F translation initiation complex, consisting of eIF4E, eIF4G and other proteins. These proteins participate in cap-dependent translation which is thought to be the major mode by which HIF-1α mRNA is translated.38 BEZ235 interferes with the eIF4F complex in at least two ways, first by decreasing phosphorylation of 4EBP1 (via its inhibitory effect on mTOR), leading to increased 4EBP1 binding to eIF4E and decreased binding of the latter to eIF4G. LY294002, which targets only PI3K and not mTOR, had only a modest effect on eIF4E/eIF4G binding. Rapamycin does target mTOR but has a much weaker effect on decreasing eIF4G binding to eIF4E. Therefore, our data indicate that the dual inhibitory action of BEZ235 results in increased potency in its ability to interfere with the eIF4F translation complex. A second means by which BEZ235 interferes with the eIF4F translation complex is by decreasing the level of the eIF4G protein. The fact that the level of eIF4E was not altered argues against this being a non-specific effect. The Akt/mTOR pathway has not previously been implicated in regulation of eIF4G protein expression.
We found that, in addition to HIF-1α, several other proteins were decreased by BEZ235 including cyclin D1 and survivin. We examined these proteins because they have been reported to be regulated by eIF4E.11,20,21 BEZ235 does not simply decrease the levels of all short-lived proteins, as neither c-Myc nor ODC levels were affected. One could speculate that the varying effect of BEZ235 on different proteins is due to differential effects on cap-dependent versus cap-independent translation. c-Myc is thought to have an internal ribosome entry site (IRES), allowing for translation in a cap-independent manner,39,40 and perhaps this is why its level was not decreased by BEZ235 treatment. The mechanism of HIF-1α translation is controversial with some groups suggesting that the translation of HIF-1α and other proteins under hypoxia may occur via a cap-independent (IRES-dependent) mechanism.41,42 although one group did not find that HIF-1α could be translated using an IRES.38
Our finding that BEZ235 interferes with HIF-1α expression led us to investigate its effects on cell survival under hypoxia. While BEZ235 was cytotoxic against normoxic cells, it had a greater effect on hypoxic cells. Whether the effect of BEZ235 in decreasing survival under hypoxia is actually due to HIF-1α inhibition is not clear. An alternate (or additional) possibility is that the effects of BEZ235 on survival under hypoxia is actually via it ability to inhibit Akt phosphorylation. Under normoxic conditions, this inhibition appears to be short-lived as Akt phosphorylation disappears within a few hours but then rises by 16-24 h. In contrast, under hypoxic conditions, there is more sustained suppression of Akt phosphorylation. Since Akt activation (phosphorylation) is associated with anti-apoptotic effect, prolonged suppression should lead to increased apoptosis. Consistent with this idea, under hypoxic conditions, there was greater PARP cleavage with the drug than under normoxic conditions. This was substantiated by a different assay that measures cytoplasmic mono- and oligonucleosomes. Therefore, we hypothesize that treatment of hypoxic cells with BEZ235 leads to increased cell death by induction of apoptosis.
We also found that BEZ235 induced autophagy, so we explored the role of this process in cell death. Our results with chloroquine, which blocks autophagy, and with ATG5-/- cells suggest that the autophagy induced by BEZ235 actually plays a protective role. We also found that BEZ235 increased the conversion of LC3-I to LC3-II under hypoxia. However, the role of autophagy on survival under hypoxia is highly controversial. Early work suggested that hypoxia could induce autophagic-induced cell death via BNIP33; however, other reports suggest that hypoxia-mediated autophagy is part of a cell survival mechanism.30,31
Our finding that the drug BEZ235 is more effective in killing hypoxic cells than normoxic cells has obvious clinical implications. Hypoxia has long been recognized to be present in human cancers43,44 and is associated with resistance to both radiation and some chemotherapeutic agents.45 Therefore, we speculate that using the drug in combination with radiation could help control the hypoxic fractions in these tumors.
The fact that BEZ235 disrupts eIF4E could also be clinically important. A recent report found that the formation of eIF4F was a critical determinant of the response of anticancer drugs targeting EGFR or HER2.46 Overexpression of eIF4E in breast tumors was associated with resistance to the anti-HER2 agent trastuzumab. If eIF4F truly is an important mediator of resistance to tyrosine kinase receptor antagonists, then BEZ235, by its disruption of eIF4F function, might be able to overcome this resistance.
mTOR inhibitors have been tested extensively in the clinic in many human cancers.47 In particular in metastatic renal cell carcinoma, in which there can be constitutive activation of the HIF pathway due to loss of VHL, a number of trials have demonstrated that mTOR inhibitors such as rapamycin, everolimus and temsirolimus can improve progression-free survival and even overall survival.48,49 Given that BEZ235 more effectively targets TORC1 and TORC2 complexes as well as PI3K, the question arises whether this agent might be more effective than the standard mTOR inhibitors that have been tried to date.
Materials and Methods
Chemicals
NVP-BEZ235 was supplied by Novartis and used at a concentration of 100 nM in all experiments. Rapamycin (LC Laboratories) was used at 10 nM, LY294002 (Alexis Corporation) at 20 µM, MG132 (Calbiochem) at 10 μM and 4EG1I (Calbiochem) at 50 µM, chloroquine (Sigma) at 20 µM.
Tissue culture
SQ20B head and neck squamous cell carcinoma, U251MG glioblastoma cells, ATG5 wild-type (ATG5+/+) and ATG5 knockout (ATG5-/-) mouse embryonic □broblasts (MEFs) were cultured in DMEM (4,500 mg/L glucose; Invitrogen) containing 10% fetal bovine serum (Atlanta Biologicals) and ATG5+/+ and ATG5-/- MEFs were generated from ATG5 heterozygous mice generously provided by Dr. Noboru Mizushima (Tokyo Medical and Dental University).50 The identities of the all the cell lines were tested at the RADIL research lab. All cells were grown in an incubator containing 5% CO2 and 21% O2. Hypoxic conditions were established in the Invivo2 400 hypoxia workstation (Ruskinn Technology Ltd.) containing 5% CO2 and 0.5% O2.
Real Time RT-PCR
Total RNA was isolated using TRIzol reagent (Invitrogen). Single strand cDNA was reverse transcribed using First-Strand cDNA Synthesis kit (GE Healthcare). Real time PCR reactions were performed on ABI Prism 7300 Sequence Detection System (Applied Biosystems) using SYBR Green PCR Master Mix (Applied Biosystems). The PCR reactions were performed in triplicate for each gene. Relative HIF-1α transcript quantities were calculated with β-actin as the reference gene amplified from the samples.
Protein extraction and Western blot analysis
Protein isolation and quantitation and Western blotting were performed as described previously.17 The following antibodies were procured from Cell Signaling Technology: anti–phospho-Akt antibody (Ser473 and Thr308), anti-phospho-S6, anti-eIF4E, anti-eIF4G, anti-4E-BP1, anti-Cyclin B1, anti-Cyclin D1, anti-Survivin, LC3B, cleaved-PARP. Other antibodies were those directed against HIF-1α (BD Transduction Lab) and β-actin antibody (Sigma-Aldrich). The secondary antibody used for these blots was a goat anti-mouse and goat anti-rabbit antibody (Thermo Scientific). Antibody binding was detected by chemiluminescence using an enhanced chemiluminescence kit (GE Healthcare). All results shown in the Western blots in the Results section are based on a minimum of 2 independent experiments.
7 methyl GTP sepaharose chromatography
Cells were seeded and 24 h later the respective drugs were added for a period of 3 h. Sepharose 4B and 7-Methyl GTP-Sepharose 4B was obtained from Sigma and Amersham Pharmacia Biotech respectively. After washing with PBS, cells were lysed directly with 0.5 ml lysis buffer (50 mM HEPES at pH 7.4, 100 mM NaCl, 1.5 mM MgCl2, 2 mM EDTA, 2 mM Na3VO4, Complete Mini protease inhibitor cocktail (Roche Diagnostics) and 0.25% NP-40) and extracts clarified by centrifugation. Equal amount of supernatant was precleared with Sepharose 4B (30 μl of settled bed volume for 1 hour). After centrifugation the supernatants were next incubated with 7-Methyl GTP-Sepharose 4B (50 μl of settled bed volume) overnight at 4°C. Pelleted beads were washed four times with 0.5 ml of lysis buffer, and resuspended in 0.5 ml lysis buffer containing 1 mM GTP for 1 h at 4°C. After four washes with 0.5 ml of lysis buffer, the beads were suspended in 2x Laemmli sample buffer (Bio-Rad) and boiled. After pelleting the beads the supernatant was transferred to a fresh tube and the proteins were analyzed by SDS-PAGE and immunoblotted for eIF4E, eIF4G and 4EBP1.
MTT assay
Cells were plated in 96 well plates and the cell viability assay was performed using a commercial kit (Roche Diagnostics) as per the manufacturer’s instructions.
Clonogenic assay
Cells were seeded and allowed to attach overnight. Next day, the cells were treated with the drug. For hypoxia experiments, cells were either exposed to 0.5% O2 or kept in the incubator for 24 h, and then treated with BEZ235 for 24 h under normoxia or hypoxia. Following the treatment, the cells were re-fed with drug free DMEM. After 10 days the colonies were stained with crystal violet and counted.
Cell death detection ELISA
Cells were seeded in 96 well plates and treated with BEZ235 for 24 h. Then, the plates were centrifuged at 200g for 10 minutes, and apoptosis was measured using the Cell Death Detection ELISA-plus System (Roche Diagnostics), which detects cytoplasmic histone-complexed DNA fragments. The data is reported as enrichment factor, which was calculated by using the formula: mean absorbance of BEZ235 treated cells/mean absorbance of negative control.
[35S]Methionine incorporation assay
Cells were treated for 24 h with BE5235 and then the [35S] methionine incorporation assay was performed as described previously.51 Briefly, thirty minutes before the end of treatment, cells were labeled with [35S] methionine (50 µCi/ml). Results were normalized as the percent increase or decrease of [35S] methionine incorporation compared with untreated control cells.
GFP-LC3 immunofluorescence microscopy
DH5α competent cells were transformed with pGFP-LC3 plasmid and put under selection. Single colonies were selected and cultured further. Plasmid was isolated using Qiagen maxi prep kit. SQ20B cells were transfected with pCMV-GFP-LC3 plasmid and put under selection to get stable transfected colonies. The cells were treated with BEZ235 for 24 h. Following treatment, cells were fixed and subjected to immunofluorescence analysis.
Electron microscopy
SQ20B cells were treated with BEZ235 for 24 h and then fixed and given to the Electron Microscopy Resource facility for further processing.
Statistical analysis
A two tailed t-test was used to compare the mean values of the control and BEZ235 treated samples.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Financial Support
This study was supported by National Cancer Institute (NIH R01 CA093638; PI: Amit Maity).
Supplemental Material
Supplementary material can be found here:
Footnotes
† These authors contributed equally to this work.
Previously published online: www.landesbioscience.com/journals/cbt/article/21144
References
- 1.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–19. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 2.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–44. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, et al. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gharbi SI, Zvelebil MJ, Shuttleworth SJ, Hancox T, Saghir N, Timms JF, et al. Exploring the specificity of the PI3K family inhibitor LY294002. Biochem J. 2007;404:15–21. doi: 10.1042/BJ20061489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maira SM, Stauffer F, Brueggen J, Furet P, Schnell C, Fritsch C, et al. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol Cancer Ther. 2008;7:1851–63. doi: 10.1158/1535-7163.MCT-08-0017. [DOI] [PubMed] [Google Scholar]
- 6.Schnell CR, Stauffer F, Allegrini PR, O’Reilly T, McSheehy PM, Dartois C, et al. Effects of the dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor NVP-BEZ235 on the tumor vasculature: implications for clinical imaging. Cancer Res. 2008;68:6598–607. doi: 10.1158/0008-5472.CAN-08-1044. [DOI] [PubMed] [Google Scholar]
- 7.Kaper F, Dornhoefer N, Giaccia AJ. Mutations in the PI3K/PTEN/TSC2 pathway contribute to mammalian target of rapamycin activity and increased translation under hypoxic conditions. Cancer Res. 2006;66:1561–9. doi: 10.1158/0008-5472.CAN-05-3375. [DOI] [PubMed] [Google Scholar]
- 8.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–75. doi: 10.1016/S0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 10.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 11.Mamane Y, Petroulakis E, LeBacquer O, Sonenberg N. mTOR, translation initiation and cancer. Oncogene. 2006;25:6416–22. doi: 10.1038/sj.onc.1209888. [DOI] [PubMed] [Google Scholar]
- 12.Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15:807–26. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- 13.Zundel W, Schindler C, Haas-Kogan D, Koong A, Kaper F, Chen E, et al. Loss of PTEN facilitates HIF-1-mediated gene expression. Genes Dev. 2000;14:391–6. [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang BH, Jiang G, Zheng JZ, Lu Z, Hunter T, Vogt PK. Phosphatidylinositol 3-kinase signaling controls levels of hypoxia-inducible factor 1. Cell Growth Differ. 2001;12:363–9. [PubMed] [Google Scholar]
- 15.Zhong H, Chiles K, Feldser D, Laughner E, Hanrahan C, Georgescu MM, et al. Modulation of hypoxia-inducible factor 1alpha expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: implications for tumor angiogenesis and therapeutics. Cancer Res. 2000;60:1541–5. [PubMed] [Google Scholar]
- 16.Cho DC, Cohen MB, Panka DJ, Collins M, Ghebremichael M, Atkins MB, et al. The efficacy of the novel dual PI3-kinase/mTOR inhibitor NVP-BEZ235 compared with rapamycin in renal cell carcinoma. Clin Cancer Res. 2010;16:3628–38. doi: 10.1158/1078-0432.CCR-09-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pore N, Jiang Z, Shu HK, Bernhard E, Kao GD, Maity A. Akt1 activation can augment hypoxia-inducible factor-1alpha expression by increasing protein translation through a mammalian target of rapamycin-independent pathway. Mol Cancer Res. 2006;4:471–9. doi: 10.1158/1541-7786.MCR-05-0234. [DOI] [PubMed] [Google Scholar]
- 18.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–32. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 19.Walsh D, Arias C, Perez C, Halladin D, Escandon M, Ueda T, et al. Eukaryotic translation initiation factor 4F architectural alterations accompany translation initiation factor redistribution in poxvirus-infected cells. Mol Cell Biol. 2008;28:2648–58. doi: 10.1128/MCB.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J Cell Biol. 2006;175:415–26. doi: 10.1083/jcb.200607020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mamane Y, Petroulakis E, Martineau Y, Sato TA, Larsson O, Rajasekhar VK, et al. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLoS One. 2007;2:e242. doi: 10.1371/journal.pone.0000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–67. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 23.Gregory MA, Hann SR. c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt’s lymphoma cells. Mol Cell Biol. 2000;20:2423–35. doi: 10.1128/MCB.20.7.2423-2435.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanamoto R, Kameji T, Iwashita S, Igarashi K, Hayashi S. Spermidine-induced destabilization of ornithine decarboxylase (ODC) is mediated by accumulation of antizyme in ODC-overproducing variant cells. J Biol Chem. 1993;268:9393–9. [PubMed] [Google Scholar]
- 25.Fan QW, Cheng C, Hackett C, Feldman M, Houseman BT, Nicolaides T, et al. Akt and autophagy cooperate to promote survival of drug-resistant glioma. Sci Signal. 2010;3:ra81. doi: 10.1126/scisignal.2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eskelinen EL. Fine structure of the autophagosome. Methods Mol Biol. 2008;445:11–28. doi: 10.1007/978-1-59745-157-4_2. [DOI] [PubMed] [Google Scholar]
- 27.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carew JS, Nawrocki ST, Cleveland JL. Modulating autophagy for therapeutic benefit. Autophagy. 2007;3:464–7. doi: 10.4161/auto.4311. [DOI] [PubMed] [Google Scholar]
- 29.Xu CX, Zhao L, Yue P, Fang G, Tao H, Owonikoko TK, et al. Augmentation of NVP-BEZ235’s anticancer activity against human lung cancer cells by blockage of autophagy. Cancer Biol Ther. 2011;12:549–55. doi: 10.4161/cbt.12.6.16397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frezza C, Zheng L, Tennant DA, Papkovsky DB, Hedley BA, Kalna G, et al. Metabolic profiling of hypoxic cells revealed a catabolic signature required for cell survival. PLoS One. 2011;6:e24411. doi: 10.1371/journal.pone.0024411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaakkola PM, Pursiheimo JP. p62 degradation by autophagy: another way for cancer cells to survive under hypoxia. Autophagy. 2009;5:410–2. doi: 10.4161/auto.5.3.7823. [DOI] [PubMed] [Google Scholar]
- 32.Mazure NM, Pouysségur J. Hypoxia-induced autophagy: cell death or cell survival? Curr Opin Cell Biol. 2010;22:177–80. doi: 10.1016/j.ceb.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 33.Tracy K, Dibling BC, Spike BT, Knabb JR, Schumacker P, Macleod KF. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol Cell Biol. 2007;27:6229–42. doi: 10.1128/MCB.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soliman GA, Acosta-Jaquez HA, Dunlop EA, Ekim B, Maj NE, Tee AR, et al. mTOR Ser-2481 autophosphorylation monitors mTORC-specific catalytic activity and clarifies rapamycin mechanism of action. J Biol Chem. 2010;285:7866–79. doi: 10.1074/jbc.M109.096222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Copp J, Manning G, Hunter T. TORC-specific phosphorylation of mammalian target of rapamycin (mTOR): phospho-Ser2481 is a marker for intact mTOR signaling complex 2. Cancer Res. 2009;69:1821–7. doi: 10.1158/0008-5472.CAN-08-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamburini J, Chapuis N, Bardet V, Park S, Sujobert P, Willems L, et al. Mammalian target of rapamycin (mTOR) inhibition activates phosphatidylinositol 3-kinase/Akt by up-regulating insulin-like growth factor-1 receptor signaling in acute myeloid leukemia: rationale for therapeutic inhibition of both pathways. Blood. 2008;111:379–82. doi: 10.1182/blood-2007-03-080796. [DOI] [PubMed] [Google Scholar]
- 37.Shi Y, Yan H, Frost P, Gera J, Lichtenstein A. Mammalian target of rapamycin inhibitors activate the AKT kinase in multiple myeloma cells by up-regulating the insulin-like growth factor receptor/insulin receptor substrate-1/phosphatidylinositol 3-kinase cascade. Mol Cancer Ther. 2005;4:1533–40. doi: 10.1158/1535-7163.MCT-05-0068. [DOI] [PubMed] [Google Scholar]
- 38.Young RM, Wang SJ, Gordan JD, Ji X, Liebhaber SA, Simon MC. Hypoxia-mediated selective mRNA translation by an internal ribosome entry site-independent mechanism. J Biol Chem. 2008;283:16309–19. doi: 10.1074/jbc.M710079200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stoneley M, Paulin FE, Le Quesne JP, Chappell SA, Willis AE. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–8. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 40.Johannes G, Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA. 1998;4:1500–13. doi: 10.1017/S1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Braunstein S, Karpisheva K, Pola C, Goldberg J, Hochman T, Yee H, et al. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol Cell. 2007;28:501–12. doi: 10.1016/j.molcel.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 42.Lang KJ, Kappel A, Goodall GJ. Hypoxia-inducible factor-1alpha mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol Biol Cell. 2002;13:1792–801. doi: 10.1091/mbc.02-02-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomlinson RH, Gray LH. The histological structure of some human lung cancers and the possible implications for radiotherapy. Br J Cancer. 1955;9:539–49. doi: 10.1038/bjc.1955.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evans SM, Koch CJ. Prognostic significance of tumor oxygenation in humans. Cancer Lett. 2003;195:1–16. doi: 10.1016/S0304-3835(03)00012-0. [DOI] [PubMed] [Google Scholar]
- 45.Brown JM, Wilson WR. Exploiting tumour hypoxia in cancer treatment. Nat Rev Cancer. 2004;4:437–47. doi: 10.1038/nrc1367. [DOI] [PubMed] [Google Scholar]
- 46.Zindy P, Bergé Y, Allal B, Filleron T, Pierredon S, Cammas A, et al. Formation of the eIF4F translation-initiation complex determines sensitivity to anticancer drugs targeting the EGFR and HER2 receptors. Cancer Res. 2011;71:4068–73. doi: 10.1158/0008-5472.CAN-11-0420. [DOI] [PubMed] [Google Scholar]
- 47.Zaytseva YY, Valentino JD, Gulhati P, Evers BM. mTOR inhibitors in cancer therapy. Cancer Lett. 2012;319:1–7. doi: 10.1016/j.canlet.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 48.Posadas EM, Figlin RA. Systemic therapy in renal cell carcinoma: advancing paradigms. Oncology (Williston Park) 2012;26:290–301. [Williston Park] [PubMed] [Google Scholar]
- 49.Coppin C, Kollmannsberger C, Le L, Porzsolt F, Wilt TJ. Targeted therapy for advanced renal cell cancer (RCC): a Cochrane systematic review of published randomised trials. BJU Int. 2011;108:1556–63. doi: 10.1111/j.1464-410X.2011.10629.x. [DOI] [PubMed] [Google Scholar]
- 50.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 51.Gupta AK, Li B, Cerniglia GJ, Ahmed MS, Hahn SM, Maity A. The HIV protease inhibitor nelfinavir downregulates Akt phosphorylation by inhibiting proteasomal activity and inducing the unfolded protein response. Neoplasia. 2007;9:271–8. doi: 10.1593/neo.07124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.