Abstract
Gadd45a, the first well-defined p53 downstream gene, can be induced by multiple DNA-damaging agents, which plays important roles in the control of cell cycle checkpoint, DNA repair process and signaling transduction. Our previous findings suggested that Gadd45a maintains cell-cell adhesion and cell contact inhibition. However, little is known about how Gadd45a participates in the suppression of malignancy in human cancer cells. To examine the functions of Gadd45a in cell invasion and metastasis, we performed the adhesion, wound-healing and transwell assays in Gadd45a+/+ and Gadd45a−/− MEF cell lines. We found the adhesion, migration and invasive abilities were much higher in Gadd45a deficient cells. We furthermore applied high-throughput cDNA microarray analysis and bioinformatics analysis to analyze the mechanisms of Gadd45a gene in invasion and metastasis. Compared with the Gadd45a wild type cells, the Gadd45a deficient cells showed a wide range of transcripts alterations. The altered gene pathways were predicted by the MAS software, which indicated focal adhesion,cell communication,ECM-receptor interaction as the three main pathways. Real-time PCR was employed to validate the differentially expressed genes. Interestingly, we figured out that the deregulations of these genes are caused neither by genomic aberrations nor methylation status. These findings provided a novel insight that Gadd45a may involve in tumor progression by regulating related genes expressions.
Keywords: Gadd45a, adhesion, migration, invasion, microarray, methylation
Introduction
Gadd45a, the first well-defined p53 downstream gene, is induced by multiple DNA-damaging agents and growth arrest signals, such as methylmethane sulfonate (MMS), hypoxia, ionizing radiation (IR), UV radiation (UVR), cisplatin, growth factor withdrawal and medium depletion.1-3 It is reported that high frequency point mutations are found in exon 4 of Gadd45a in human pancreatic cancer and the expression level of Gadd45a, combined with p53 status, significantly affects the survival of patients.4 There is also evidence to show Gadd45a as an abnormally methylated gene in breast cancer.5 The pivotal roles of Gadd45a have been well-demonstrated in various cellular processes. By physically interacting with Cdc2 kinase, Gadd45a can dissociate Cdc2/cyclinB1 complex and mediate G2/M cell cycle arrest.6,7 By interacting with proliferating cell nuclear antigen (PCNA), Gadd45a is involved in DNA repair process.8 In addition, it can induce apoptosis by promoting Bim translocation to mitochondria.9 Most recently, controversial roles of Gadd45a in the control of DNA demethylation have been reported.10,11 As a tumor-suppressor gene, Gadd45a negatively regulates cell malignancy. Gadd45a null mice are much more susceptible to DNA damage-induced tumors and mouse embryonic fibroblasts (MEFs) derived from Gadd45a-null mice exhibit genomic instability, single oncogene transformation, centrosome amplification and loss of normal cellular senescence.12
Tumor progression is considered to be a complex process, in which metastasis is the main cause of death in patients with malignancy.13 Several classes of proteins are involved in the cell metastatic process, including cell adhesion molecules (CAMs), integrins, extracellular matrix (ECM) and matrix metalloproteinases (MMPs).14,15 Gadd45a may not only play important roles in anti-tumorigenesis but also contribute to inhibiting tumor progression. It has been noted that Gadd45a could regulate matrix metalloproteinase through p38 MAP kinase and APC complex activation.16 Meanwhile, our previous study has revealed that Gadd45a maintained cell-cell adhesion and cell contact inhibition by regulating β-catenin subcellular distribution.17 However, little is known about how Gadd45a participates in the suppression of cell malignancy.
Here we report that Gadd45a regulates adhesion, migration and invasion of MEF cells in vitro. Additionally, Gadd45a affects the expression of various genes involved in ECM, cell communication, cell adhesion. However, deregulations of these genes are caused neither by genomic aberrations nor methylation status. Taken together, Gadd45a may be involved in tumor progression by regulating related genes expressions.
Results
Gadd45a inhibits adhesion ability of MEF cells in vitro
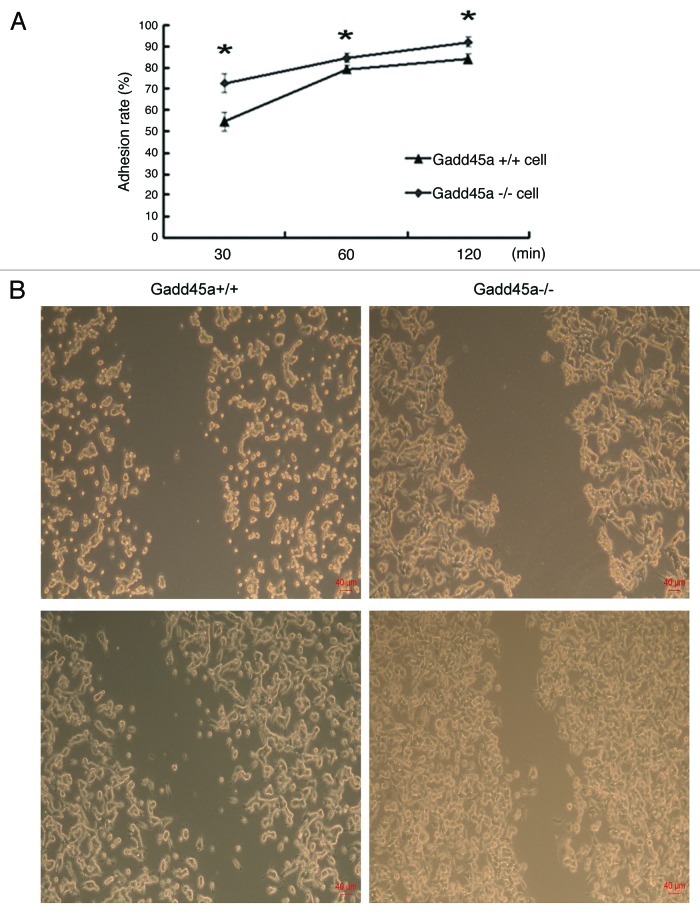
Cell adhesion to the extracellular matrix and molecules on the cell surface is a key step during cancer metastasis in vivo. To investigate the influence of Gadd45a on cell adhesive ability, we employed the adhesion assay. The results are shown in Figure 1A. The adhesion rates of Gadd45a+/+ MEF cells were 54.70% ± 4.03%, 79.25 ± 1.71% and 84.17% ± 2.20% at 30min, 60min and 120min after cell plating, respectively. While the adhesion rates of Gadd45a−/− MEF cells were 72.80% ± 4.39%, 84.79% ± 2.05%, 92.18% ± 2.34% at the same time points, respectively (p < 0.05). The Gadd45a-null mouse embryonic fibroblast cells (Gadd45a−/− MEFs) showed significantly increased attachment to fibronectin-coated surface, compared with wild type MEF cells (Gadd45a+/+ MEFs).
Figure 1. Gadd45a decreases adhesion, migration abilities of MEF cells in vitro. (A) Gadd45a+/+ and Gadd45a−/− MEFs were seeded onto the 96-well plates precoated with fibronectin. After 30 min, 60 min and 120 min, the remaining cells per well were measured by MTT assay. (B) The migration ability of Gadd45a+/+ and Gadd45a−/− MEFs was compared by scratch wound assay. Representative images shown here were from three independent experiments. Original magnification, × 40.
Gadd45a decreases migration and invasion abilities of MEF cells in vitro
To elucidate the possible effects of Gadd45a on cell migration, wound-healing assay was performed. Gadd45a+/+ MEFs spread significantly slower than Gadd45a−/− MEFs along the wound edges (Fig. 1B).
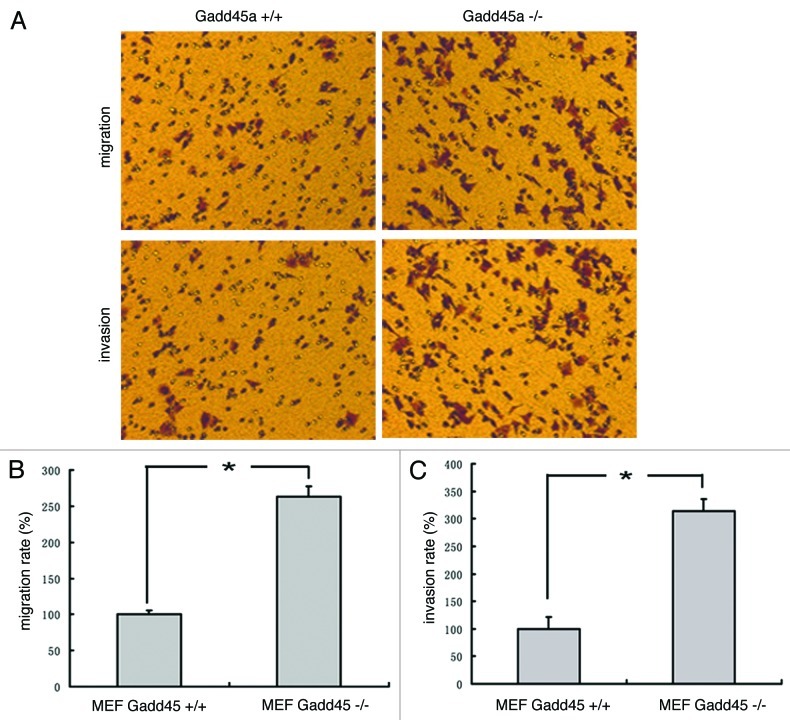
In addition, migration and invasion assays were performed in transwell chamber. Compared with the Gadd45a+/+ MEFs, the migration rate of Gadd45a−/− MEFs increased to 263 ± 5.64% and the invasion rate increased to 314 ± 6.37%. The results implied that the migration and invasive abilities of Gadd45a−/− MEFs were much higher than that of Gadd45a+/+ MEFs (p < 0.05) (Fig. 2A–C).
Figure 2. Gadd45a decrease migration and invasion abilities of MEF cells in vitro. (A)Migration of cells through a layer of polycarbonate was determinedaccording to the method above and incubated for 10 h. Invasion of cells are the same as the migration assay described above except that the filter was coated with 250 μl/ml Matrigel and the incubation time was 10 h. (B-C)Ten fields were counted for each filter and results were representative of at least three separate experiments and were expressed as mean ± SD.
Gadd45a decreases migration and invasion abilities of HepG2 cells in vitro
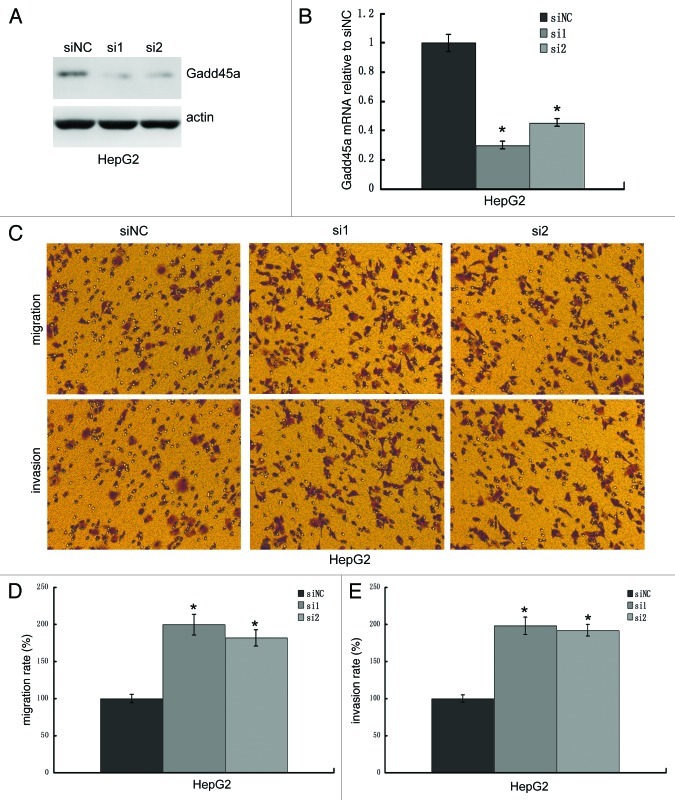
To clarify whether it is a cell line-specific phenomenon, we knocked down Gadd45a expression by siRNA and performed migration and invasion assays in HepG2 cells. The siRNA knockdown efficiency was examined by western-blot and realtime PCR. Gadd45a siRNA transfected cells dramatically decreased both Gadd45a mRNA and protein expression compared with the negative control siRNA transfected cells (Fig. 3A–B). As we expected, the migration rate of Gadd45a knockdown cells increased to 200 ± 2.57% (siRNA1) and 178 ± 6.33% (siRNA2) and the invasion rate of Gadd45a knockdown cells increased to 198 ± 4.67% (siRNA1) and 192 ± 4.37%(siRNA2), compared with negative control transfected cell, respectively (Fig. 3C–E).
Figure 3. Gadd45a decrease migration and invasion abilities of HepG2 cells in vitro. (A and B) The knockdown efficiency of Gadd45a siRNA was examined by Western-blot and Realtime-PCR. (C) Migration of cells through a layer of polycarbonate was determined according to the method above and incubated for 10 h. Invasion of cells are the same as the migration assay described above except that the filter was coated with 250 μl/ml Matrigel. (D-E)Ten fields were counted for each filter and results were representative of at least three separate experiments and were expressed as mean ± SD.
Gadd45 alters the global RNA expression in MEFs
To investigate the genome-wide influence of Gadd45a, microarray analysis was performed for Gadd45a+/+ and Gadd45a−/− MEFs cells. Compared with the Gadd45a wild type cells, the Gadd45a deficient cells showed a wide range of transcripts alterations. There were 818 genes changed (more than 2-fold) in the expression profile with 473 genes upregulated and 345 genes downregulated.
Distinct changes of genes in certain pathways
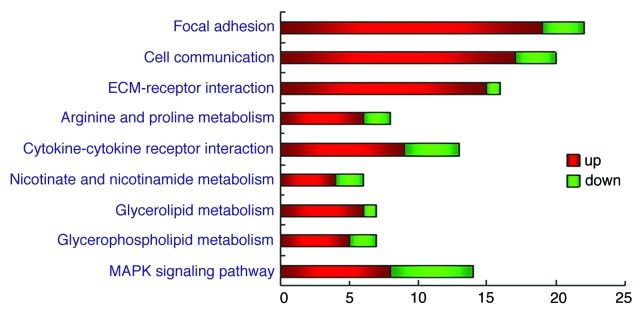
The pathways were predicted by the MAS software, which indicated focal adhesion,cell communication,ECM-receptor interaction as the three mainly altered pathways. These pathways were detected at the significance p-value of < 10−4, respectively. (Table 1 and Fig. 4)
Table 1. Distinct altered genes in certain pathways.
| Pathway Name | Gene symbol | Fold Change | RefSeq. | Gene Description |
|---|---|---|---|---|
|
Focal adhesion |
Itgb1 |
0.41 |
NM_010578 |
integrin β 1 |
| |
Col2a1 |
2.49 |
NM_031163 |
procollagen, type II, α 1 |
| |
Col1a2 |
7.39 |
NM_007743 |
procollagen, type I, α 2 |
| |
Col6a1 |
6.64 |
NM_009933 |
procollagen, type VI, α 1 |
| |
Col3a1 |
5.96 |
NM_009930 |
procollagen, type III, α 1 |
| |
Col6a2 |
5.14 |
NM_146007 |
procollagen, type VI, α 2 |
| |
Thbs2 |
6.94 |
NM_011581 |
thrombospondin 2 |
| |
Prkcb1 |
0.18 |
NM_008855 |
protein kinase C, β 1 |
| |
Fn1 |
2.30 |
NM_010233 |
fibronectin 1 |
| |
Cav2 |
2.03 |
NM_016900 |
caveolin 2 |
| |
Lama1 |
2.09 |
NM_008480 |
laminin, α 1 |
| |
Pdgfra |
4.87 |
NM_011058 |
platelet derived growth factor receptor, α polypeptide |
| |
Pik3ca |
0.45 |
NM_008839 |
phosphatidylinositol 3-kinase, catalytic, α polypeptide |
| |
Cav1 |
2.21 |
NM_007616 |
caveolin, caveolae protein 1 |
| |
Capn7 |
0.48 |
NM_009796 |
calpain 7 |
| |
Lama4 |
3.48 |
NM_010681 |
laminin, α 4 |
| |
Col5a3 |
5.84 |
NM_016919 |
procollagen, type V, α 3 |
| |
Col5a1 |
2.39 |
NM_015734 |
procollagen, type V, α 1 |
| |
Col5a2 |
3.63 |
NM_007737 |
procollagen, type V, α 2 |
| |
Tnc |
3.76 |
NM_011607 |
tenascin C |
| |
Col1a1 |
16.8 |
NM_007742 |
procollagen, type I, α 1 |
|
Cell Communication |
Krt2–4 |
0.34 |
NM_008475 |
keratin complex 2, basic, gene 4 |
| |
Des |
3.26 |
NM_010043 |
desmin |
| |
Col2a1 |
2.49 |
NM_031163 |
procollagen, type II, α 1 |
| |
Krt2–8 |
3.76 |
AK077597 |
Keratin, type II cytoskeletal 8 |
| |
Col1a2 |
7.39 |
NM_007743 |
procollagen, type I, α 2 |
| |
Col6a1 |
6.64 |
NM_009933 |
procollagen, type VI, α 1 |
| |
Col3a1 |
5.96 |
NM_009930 |
procollagen, type III, α 1 |
| |
Krt1–19 |
0.11 |
NM_008471 |
keratin complex 1, acidic, gene 19 |
| |
Col6a2 |
5.14 |
NM_146007 |
procollagen, type VI, α 2 |
| |
Thbs2 |
6.94 |
NM_011581 |
thrombospondin 2 |
| |
Fn1 |
2.30 |
NM_010233 |
fibronectin 1 |
| |
Lama1 |
2.09 |
NM_008480 |
laminin, α 1 |
| |
Lama4 |
3.48 |
NM_010681 |
laminin, α 4 |
| |
Col5a3 |
5.84 |
NM_016919 |
procollagen, type V, α 3 |
| |
Col5a1 |
2.39 |
NM_015734 |
procollagen, type V, α 1 |
| |
Col5a2 |
3.63 |
NM_007737 |
procollagen, type V, α 2 |
| |
Tnc |
3.76 |
NM_011607 |
tenascin C |
| |
Krt1–18 |
0.11 |
NM_010664 |
keratin complex 1, acidic, gene 18 |
| |
Col1a1 |
16.80 |
NM_007742 |
procollagen, type I, α 1 |
|
ECM-receptor interaction |
Col1a1 |
16.86 |
NM_007742 |
procollagen, type I, α 1 |
| |
Tnc |
3.76 |
NM_011607 |
tenascin C |
| |
Col5a2 |
3.63 |
NM_007737 |
procollagen, type V, α 2 |
| |
Col5a1 |
2.39 |
NM_015734 |
procollagen, type V, α 1 |
| |
Col5a3 |
5.84 |
NM_016919 |
procollagen, type V, α 3 |
| |
Lama4 |
3.48 |
NM_010681 |
laminin, α 4 |
| |
Lama1 |
2.09 |
NM_008480 |
laminin, α 1 |
| |
Fn1 |
2.30 |
NM_010233 |
fibronectin 1 |
| |
Sdc2 |
2.07 |
NM_008304 |
syndecan 2 |
| |
Thbs2 |
6.94 |
NM_011581 |
thrombospondin 2 |
| |
Col6a2 |
5.14 |
NM_146007 |
procollagen, type VI, α 2 |
| |
Col3a1 |
5.96 |
NM_009930 |
procollagen, type III, α 1 |
| |
Col6a1 |
6.64 |
NM_009933 |
procollagen, type VI, α 1 |
| |
Col1a2 |
7.39 |
NM_007743 |
procollagen, type I, α 2 |
| |
Col2a1 |
2.49 |
NM_031163 |
procollagen, type II, α 1 |
| Itgb1 | 0.41 | NM_010578 | integrin β 1 (fibronectin receptor β) |
Figure 4. Distinct altered genes in certain pathways. Differentially expressed genes are classified into pathways dealing with molecular function, biological processes and cellular components. Each bar indicates the number of the genes involved in the certain pathway. Changes of the expression levels are illustrated using color coding, the key to the color code is shown on the right. The p-value < 10−4 was used as a cut-off.
Quantitative real-time PCR confirmation of the differential expression of candidate genes
In order to further confirm the reliability of our mircoarray data, we picked 10 candidate genes, which were involved in cell migration and invasion. We used the same RNA samples that had served for the cDNA microarray analysis and confirmed the expression of 5 upregulated genes and 5 downregulated genes by quantitative real-time PCR, which were MMP3 (269-folds), Col3a1 (45-folds), Col1a2 (523-folds), Lama4 (3.74-folds), Vcam1 (10.91-folds), Krt18 (0.001 -folds), Krt19 (0.06-folds), Alcam (0.16 -folds), Igsf4a (0.03 -folds) and Itgb1 (0.4-folds). (Fig. 5)
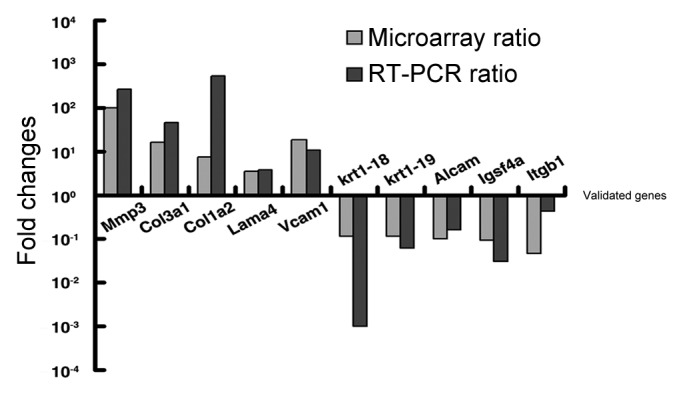
Figure 5. Validation of 10 candidate genes by real time PCR. Fold induction or repression of 10 genes associated with cell migration and invasion of Gadd45a+/+ cell, as compared with Gadd45a−/− cell. The gray bars indicate the fold changes in microarray analysis; the black bars indicate the fold changes in real-time PCR validation. The ratios are normalized by logarithm.
CGH array analysis of the chromosomal aberrations in MEF cells
To identify whether genomic aberrations were the main cause of deregulated expression, we applied comparative genomic hybridization (CGH) array on these two cell lines. As expected, very few chromosomal aberrations took place in MEF Gadd45a−/− cells, mainly at chromosome 4 and Y (Table S1). However, the deregulated genes in cDNA microarray scattered among the whole genome and especially, the locations of highly-changed genes could not match the abnormal regions in CGH array.
DMH array analysis of the aberrant methylation in MEF cells
To verify whether the deregulated expression is caused by methylation, we also applied Differential methylation hybridization (DMH) on these two MEF cell lines (Table S2). Our result showed that there were less aberrant methylation in MEF Gadd45a−/− cells compare with Gadd45a+/+ cells, which indicated that dysregulated expression may not be caused by aberrant methylation in MEF cells.
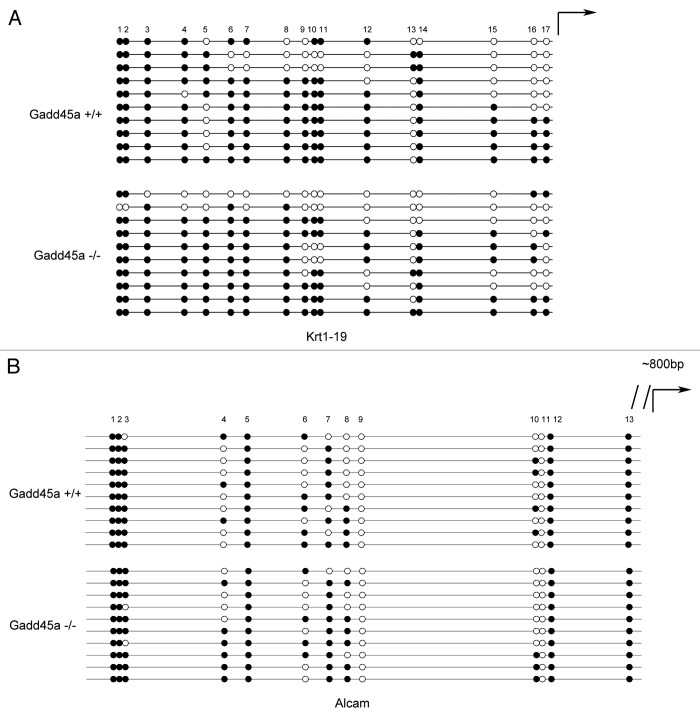
Analysis of Krt19 and Alcam promoter methylation in MEF cells
In order to further confirm that the differentially expressed genes are not caused by methylation alteration, we analyzed CpG islands of the 10 validated gene’s promoters and discovered that half of them didn't have CpG islands. Furthermore, we randomly picked two of the rest five genes, which were Krt19 and Alcam. The Krt19 gene contained two CpG islands and we analyzed the methylation patterns of the second one. Scattered methylation was found both in Gadd45a+/+ and Gadd45a−/− MEF cells (Fig. 6A). The Alcam gene also contained two CpG islands and we analyzed the methylation patterns of the first CpG island. In accordance with our previous results, these two cell lines didn't display too much different methylation patterns (Fig. 6B).
Figure 6. Analysis of Krt19 and Alcam promoter methylation by bisulfite sequencing. (A and B)Methylation status of Krt19 and Alcam gene in MEFs was analyzed, ten colonies of cloned BGS PCR products from each bisulfite-treated DNA sample were sequenced and each was shown as an individual row, representing a single allele of the promoter. Open circles denoted unmethylated CpG sites and filled circles represented methylated CpG sites, curved arrow represented transcription start site.
Discussion
Gadd45a gene encodes a conserved 165 amino acid protein, which was first isolated from Chinese Hamster Ovarian cells (CHO) treated with UV radiation. Subsequently, it was found to be induced by a wide spectrum of DNA-damaging agents and growth arrest treatments. As a p53 and BRCA1 downstream gene, Gadd45a participates in various cellular responses to DNA damage agents, including cell cycle arrest, apoptosis and DNA repair. Most strikingly, the defect of Gadd45a is closely associated with genomic instability and tumorigenesis, which has been clearly proved in Gadd45a null mice.18 Our previous work has found that Gadd45a could induce β-catenin translocation to cell membrane for maintaining cell-cell adhesion/contact inhibition, which might contribute to its tumor suppressive function.17 However, the detailed mechanism is still poorly understood.
The mortality of patients with cancer is mainly caused by distant metastasis. The capability of invasion and metastasis enables cancer cells to escape the primary tumor mass and colonize new terrain in the body.14 In this study, we found that Gadd45a suppressed the migration and invasion abilities of MEF cells and also adhesion ability. As adhesion of cells to the ECM is a key step to the regulation of cellular migration and invasion,19 we hypothesized that pro-adhesive and pro-migration/invasive effects of Gadd45 might coordinately contribute to tumor metastasis. To better understand the mechanism of Gadd45a gene in invasion and metastasis, we applied high-throughput cDNA microarray analysis in both Gadd45a+/+ and Gadd45a−/− MEF cells. Among the deregulated genes in the Gadd45a−/− cells, many genes are classified in focal adhesion (FA), cell communication and ECM-receptor interaction pathways, which are highly concordant with the previous results in adhesion, migration and invasion assays. Changes in expression of CAMs, integrins and extracellular proteases are evident in invasive and metastatic cells.14 Functions of the validated genes will be discussed below, respectively.
Matrix Metalloproteinases (MMPs) are zinc-dependent endopeptidases, which play critical roles in the degradation of extracellular matrix (ECM) during the invasion process. Matrix Metalloproteinase-3 (MMP3, also known as stromelysin-1), which has been reported overexpressed in colorectal tumors and desmoid tumors,20,21 is highly upregulated in Gadd45a−/− cells both in microarray and real-time PCR for over a hundred folds. Basement membrane (BM), which plays an essential role in promoting the motility of tumor cells, is an important component of ECM and laminins are the major constituents of BM. Laminin-4 (LAMA4) has been reported to be highly upregulated in hepatocellular carcinoma and the downregulation of LAMA4 could inhibit glioma invasion.22,23 Interestingly, in our study, we found LAMA4 was overexpressed in Gadd45a−/− MEF cells. As another important component of BM, collagens also play curial roles in tumorigenesis. Several collagens were upregulated in MEF Gadd45a−/− cells according to microarray data. Since collagen type III α 1 and typeIalpha 2 have been reported to be overexpressed in ovarian and gastric carcinomas,24,25 we verified the expressions of them by RT-PCR. The cell adhesion molecules (CAMs) (including immunoglobulin and calcium-dependent cadherin families and integrins), which mediate the interactions of cell to cell and cells to extracellular matrix substrates, also display distinct changes in Gadd45a−/− MEF cells. Vascular cell adhesion molecule-1 (VCAM-1) expression increased over 10-folds in Gadd45a defect cells. It has been well-characterized that VCAM-1 takes a part in T-cell immunity escape. As tumor immune escape is a very important trail of tumorigenesis, the overexpression of VCAM-1 may play an important role in tumor progression. Among the downregulated genes, the altered expression of activated leukocyte cell adhesion molecule (ALCAM) has been reported in several human tumors. ALCAM can interact with CD6 and the interaction is required for optimal activation of T cells.26 It is also reported that the reduced expression of ALCAM correlated with poor prognosis and skeletal metastases in breast cancer.27,28 Another downregulated gene is immunoglobulin superfamily 4 (IGSF4), also named tumor suppressor in lung cancer (TSLC1), which was reported to be markedly reduced in many kinds of tumors, such as lung cancer, breast cancer, prostate cancer and gastric cancer.29 TSLC1 gene locates at chromosome 11q23, which was a frequently deleted region in diverse kinds of cancers, including breast cancer, lung cancer and Esophageal Squamous Cell Carcinoma.30-32 In addition to the aberration, it is also reported that the promoter methylation plays a role in the reduction of TSLC1.33,34 Integrins, which link cell to extracellular matrix substrates, also appear to play critical roles in the processes of metastasis. Integrin β1 (ITGB1), which has been reported downregulated in cervical squamous cell carcinoma,35 was downregulated in Gadd45a−/− cells. Keratin 18 (K18) and 19 (K19), whose reduction has been reported to associate with the malignant phenotype,36,37 were significantly decreased in RT-PCR validation. By and large, the expressions of MMP, ECM, CAMs genes largely altered in Gadd45 defective cells, which might result in increasing migration and invasion abilities and thus promoting the processes of metastasis.
Gadd45a has a substantial impact on the maintenance of genomic integrity, furthermore, disruption of this gene is associated with genomic instabilities, such as chromosomal aberrations, gene amplifications and abnormal mitosis.12 Considering the distinct influence of Gadd45a on multiple gene expression at mRNA levels, while it has not been reported as a transcriptional factor, we applied CGH array on these two cell lines to identify whether genomic aberrations were the main cause of deregulated expression. However, there were few aberrations in Gadd45a defective cells and the locations of highly-changed genes in cDNA array could not match the abnormal regions in CGH array. Herein, the mRNA expression alterations were not caused by genomic instabilities directly.
As we all know, acquired epigenetic abnormalities participate in genetic alterations and cause the dysregulation of gene expression, resulting in activation of oncogenes and inactivation of tumor suppressor genes.38 Recent reports have raised some questions concerning whether Gadd45a gene is involved in DNA demethylation. Guillermo Barreto reported that reduced Gadd45a may lead to hypermethylation and inactivation of tumor suppressor genes. In addition, Rai, K also reported Gadd45a participated in demethylation in zebrafish; meanwhile, Seung-Gi Jin insisted that Gadd45a wouldn’t promote DNA demethylation.10,11,39 To verify whether the deregulated expression was caused by methylation, we applied DMH array on these two MEF cell lines. Our result showed that there were few aberrant methylation in Gadd45a defective cells. As DMH array could not provide sufficient resolution at a single gene level, we analyzed the promoter CpG islands of the 10 validated genes and discovered that half of them didn't have CpG islands. We further analyzed the methylation status of Krt19 and Alcam genes by bisulfite DNA sequencing. However, scattered methylation was found in CpG islands of both Gadd45a+/+ and Gadd45a−/− MEF cells, It implied that dysregulated expression did not caused by aberrant methylation and Gadd45a could not promote demethylation at least in MEF cells. Taken together, the possibility that genomic aberrations or epigenetic changes are the main reasons for gene expression alterations in Gadd45a−/− cells has been excluded by the results above. Gadd45a may influence the expression of invasive related genes indirectly thus inhibit the migration and invasion ability of MEF cells.
In summary, we proposed a novel function of Gadd45a that it alters various genes’ expression in mammalian cells; and furthermore, we found these genes are mainly classified in focal adhesion, cell communication and ECM-receptor interaction pathways. However, neither gene copy number nor epigenetic changes cause the alterations and the distinct changes of gene expressions might be responsible for the different invasiveness and metastatic ability of Gadd45a+/+ cells compare with Gadd45a−/− cells. Therefore, aberrant modulation and expression of Gadd45a could result in dysregulation of invasion related genes as well as increasing the invasion and metastatic potentials in tumor development and progression.
Materials and Methods
Cell culture and gene silencing
Gadd45a+/+ and Gadd45a−/− mouse embryonic fibroblasts (MEFs) cell lines were kindly provided by Professor Albert J. Fornace and M. Christine Hollander and cultured in Dulbecco’s modified Eagle’s medium (DMEM), while HepG2 cell line was cultured in .Eagle's minimum essential medium (EMEM), all supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin, maintained at 37°C humidified atmosphere of 5% carbon dioxide.
For Gadd45a knockdown, two validated Gadd45a siRNAs (siRNA1: sense: 5′- GAG CAG AAG ACC GAA AGG AUG GAU A-3′; anti-sense: 5′-UAU CCA UCC UUU CGG UCU UCU GCU C-3′; siRNA2: 5′-GGU GAC GAA UCC ACA UUC AUC UCA A-3′; anti-sense: 5′-UUG AGA UGA AUG UGG AUU CGU CAC C -3′) were used. In brief, 3 × 105 HepG2 cells per well in 6-well plates were transfected at a final concentration of 50nM of siRNA or negative control RNA using lipofectamine RNAiMAX (Invitrogen). After incubation for 36 to 48 h, cells were prepared for subsequent assays.
Adhesion assay
The adhesion assay was performed by MTT assay.40 The monolayers of Gadd45a+/+ and Gadd45a−/− MEF cells (1 × 105) were planted into the fibronectin-precoated (100 μg/ ml) 96-well plate in triplicate. The groups of cells were washed with phosphate buffered solution (PBS) at 30 min, 60 min and 120 min, respectively, to remove the non-adherent cells. After washing, the adhered cells were measured with MTT assay at 490-nm wavelength. The OD values of washed groups compared with non-washed groups reflect the proportion of cells which adhered to the fibronectin-coated 96-well plate.
Wound-healing assay
Gadd45a+/+ and Gadd45a−/− MEFs were assessed with a scratch wound-healing assay. The confluent cell monolayers were carefully wounded with sterile pipette tips and washed with PBS once to remove cellular debris. After acquired the first image of the wounded cell monolayers, incubated dishes for 4 h and photographed again.
Migration assay
In vitro migration assay was performed using a 24-well transwell unit with polycarbonate filters (Corning Costar). Lower compartment was filled with serum-free media containing 0.2% BSA. Cells (5 × 104) were placed in the upper part of the transwell plate, incubated for indicated hours in 37þC, 5% CO2 incubator. Cells that migrated to the bottom of the membrane were fixed with methanol and stained with crystal violet. The migration phenotypes were determined by counting the cells that migrated to the lower side of the filter with microscopy. Ten fields were counted for each filter and three independent experiments were performed for each set.
Invasion assay
In vitro invasion assay was performed using 24-well Transwell unit with polycarbonate filters. Experimental procedures are the same as the in vitro migration assay described above except that the filter was coated with 250 μl/ml Matrigel (BD Biosciences) for the invasion assay.
Microarray based gene expression analysis
The 22 K oligonucleotide microarrays were constructed by CapitalBio Corp. (CapitalBio) using the Human Genome Oligo Set Version 2.1 (Operon) and comprise 21,329 5′-amino-modified 70-mer probes. After hybridization, microarrays were scanned with a confocal scanner LuxScanTM 10KA (CapitalBio) and the data of obtained images were extracted with LuxScan 3.0Software (CapitalBio). The raw data was normalized using a space and intensity-dependent LOWESS program. For each test and control sample, two hybridizations were performed by using a dye-swap strategy.41
Bioinformatics analysis
After normalization, genes that showed ≥ 2.0- and ≤ 0.5-fold changes in their expression levels when compared with control were considered to be upregulated and downregulated, respectively. To determine the biological roles of the differentially expressed genes, functional classification was performed using MAS software (http://bioinfo.capitalbio.com/mas). It classifies the genes into relevant pathways dealing with molecular function, biological processes and cellular components. The p-value < 10−4 was used as a cut-off.
Real time quantitative RT-PCR (qRT-PCR)
Total RNA was isolated from GADD45+/+ and GADD45−/− MEFs cells with TRIzol reagent ((Invitrogen). 10 µg of total RNA was incubated with 10 U RNase-free DNase I (Invitrogen) at 37°C for 30 min. RNA was further purified with an RNeasy Mini kit (Qiagen) for subsequent use. The purified total RNA (2 µg) was reverse transcribed according to the manufacturer’s instructions (Invitrogen). Quantitative RT-PCR was performed by employing a DNA Master SYBR green I kit in ABI 7300 thermocycler (Applied Biosystems). Target gene and reference gene (GAPDH) were amplified in parallel. The primer sequences are shown inTable S1. The results were analyzed by using System SDS software (Applied Biosystems).
Array-based comparative genomic hybridization
Total DNA was isolated from GADD45+/+ and GADD45−/− MEFs cells with Genomic DNA Extraction Kit (Tiangen). Custom NimbleGen 3 × 720 K microarrays contain up to 4.2 million oligonucleotide probes designed and fabricated on a single slide, resulting in a median probe spacing of 3537 bp. Standard genomic DNA labeling (Cy3 for samples and Cy5 for references), hybridizations, array scanning, data normalization and segmentation were performed at CapitalBio Corporation.
Differential methylation hybridization
Total DNA was isolated from GADD45+/+ and GADD45−/− MEFs cells as described previously. Approximately 2ug genomic DNA was restricted to completion with MseI (New England Biolabs).The digests were purified with QIAquick column (QIAGEN) and then were ligated with linkers. Using the polymerase chain reaction (PCR) to amplify the unrestricted DNA fragments and then hybridizing them onto microarrays. Standard digestion, ligation with linkers, PCR, hybridizations, array scanning, data normalization and segmentation were performed at CapitalBio Corporation.
Bisulfite treatment and sequencing
A total of 200 ng genomic DNA from each sample was bisulfite-treated with the Methylamp DNA Modification Kit (Epigentek). The number and distribution of CpG islands of Krt1-19 and Alcam gene have been analyzed and primers were designed by using the online program MethPrimer (available at www.urogene.org).
Bisulfite-treated DNA was subjected to PCR using primers as follows: 5′-GTT AGG GTT TTA TTA AAA TTT TTA T -3′ (Krt19-forward) and 5′-CCA AAC CTC AAC TAT ATC TTC CTT TAA-3′ (Krt19-reverse); 5′-GGT GAT TGT TTT TAG GGA TAG ATT T-3′ (Alcam-forward), 5′-ACA TCT AAA ATA TCC CCT TCT TAC C-3′ (Alcam-reverse). PCR cycle conditions were as follows: 5 min at 95°C, then 30 sec at 95°C, 30 sec at 57°C and 45 sec at 72°C for 35 cycles, followed by 5 min at 72°C. PCR products were gel purified and cloned into the vector pMD19-T (Takara) according to the manufacturer’s protocol. Colonies were grown on agar plates and 10 of them were randomly selected and sequenced with the M13 reverse primer via automated sequencing (Invitrogen).
Statistical analyses
The data were analyzed by ANOVA. The statistical analysis was performed using SPSS version 11.0 software and p < 0.05 was considered as significant.
Supplementary Material
Disclosure of Potential Conflicts of Interest
The authors declare there are no conflicts of interest.
Acknowledgments
The authors would like to thank Professor Albert J. Fornace and M. Christine Hollander for kindly giving us the Gadd45a+/+ and Gadd45a−/− (MEFs) cell lines.
This work is supported by the funding from the 973 National Key Fundamental Research Program of China (2009CB521801) and the National Natural Science Foundation of China (81021061).
Footnotes
Previously published online: www.landesbioscience.com/journals/cbt/article/21186
References
- 1.Fornace AJ, Jr., Alamo I, Jr., Hollander MC. DNA damage-inducible transcripts in mammalian cells. Proc Natl Acad Sci U S A. 1988;85:8800–4. doi: 10.1073/pnas.85.23.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papathanasiou MA, Fornace AJ., Jr. DNA-damage inducible genes. Cancer Treat Res. 1991;57:13–36. doi: 10.1007/978-1-4615-3872-1_2. [DOI] [PubMed] [Google Scholar]
- 3.Papathanasiou MA, Kerr NC, Robbins JH, McBride OW, Alamo I, Jr., Barrett SF, et al. Induction by ionizing radiation of the gadd45 gene in cultured human cells: lack of mediation by protein kinase C. Mol Cell Biol. 1991;11:1009–16. doi: 10.1128/mcb.11.2.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamasawa K, Nio Y, Dong M, Yamaguchi K, Itakura M. Clinicopathological significance of abnormalities in Gadd45 expression and its relationship to p53 in human pancreatic cancer. Clin Cancer Res. 2002;8:2563–9. [PubMed] [Google Scholar]
- 5.Wang W, Huper G, Guo Y, Murphy SK, Olson JA, Jr., Marks JR. Analysis of methylation-sensitive transcriptome identifies GADD45a as a frequently methylated gene in breast cancer. Oncogene. 2005;24:2705–14. doi: 10.1038/sj.onc.1208464. [DOI] [PubMed] [Google Scholar]
- 6.Zhan Q, Antinore MJ, Wang XW, Carrier F, Smith ML, Harris CC, et al. Association with Cdc2 and inhibition of Cdc2/Cyclin B1 kinase activity by the p53-regulated protein Gadd45. Oncogene. 1999;18:2892–900. doi: 10.1038/sj.onc.1202667. [DOI] [PubMed] [Google Scholar]
- 7.Jin S, Tong T, Fan W, Fan F, Antinore MJ, Zhu X, et al. GADD45-induced cell cycle G2-M arrest associates with altered subcellular distribution of cyclin B1 and is independent of p38 kinase activity. Oncogene. 2002;21:8696–704. doi: 10.1038/sj.onc.1206034. [DOI] [PubMed] [Google Scholar]
- 8.Smith ML, Chen IT, Zhan Q, Bae I, Chen CY, Gilmer TM, et al. Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science. 1994;266:1376–80. doi: 10.1126/science.7973727. [DOI] [PubMed] [Google Scholar]
- 9.Tong T, Ji J, Jin S, Li X, Fan W, Song Y, et al. Gadd45a expression induces Bim dissociation from the cytoskeleton and translocation to mitochondria. Mol Cell Biol. 2005;25:4488–500. doi: 10.1128/MCB.25.11.4488-4500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barreto G, Schäfer A, Marhold J, Stach D, Swaminathan SK, Handa V, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–5. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 11.Jin SG, Guo C, Pfeifer GP. GADD45A does not promote DNA demethylation. PLoS Genet. 2008;4:e1000013. doi: 10.1371/journal.pgen.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hollander MC, Sheikh MS, Bulavin DV, Lundgren K, Augeri-Henmueller L, Shehee R, et al. Genomic instability in Gadd45a-deficient mice. Nat Genet. 1999;23:176–84. doi: 10.1038/13802. [DOI] [PubMed] [Google Scholar]
- 13.Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 14.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 15.Chiang AC, Massagué J. Molecular basis of metastasis. N Engl J Med. 2008;359:2814–23. doi: 10.1056/NEJMra0805239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hildesheim J, Belova GI, Tyner SD, Zhou X, Vardanian L, Fornace AJ., Jr. Gadd45a regulates matrix metalloproteinases by suppressing DeltaNp63alpha and beta-catenin via p38 MAP kinase and APC complex activation. Oncogene. 2004;23:1829–37. doi: 10.1038/sj.onc.1207301. [DOI] [PubMed] [Google Scholar]
- 17.Ji J, Liu R, Tong T, Song Y, Jin S, Wu M, et al. Gadd45a regulates beta-catenin distribution and maintains cell-cell adhesion/contact. Oncogene. 2007;26:6396–405. doi: 10.1038/sj.onc.1210469. [DOI] [PubMed] [Google Scholar]
- 18.Zhan Q. Gadd45a, a p53- and BRCA1-regulated stress protein, in cellular response to DNA damage. Mutat Res. 2005;569:133–43. doi: 10.1016/j.mrfmmm.2004.06.055. [DOI] [PubMed] [Google Scholar]
- 19.Gumbiner BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–57. doi: 10.1016/S0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 20.Zucker S, Vacirca J. Role of matrix metalloproteinases (MMPs) in colorectal cancer. Cancer Metastasis Rev. 2004;23:101–17. doi: 10.1023/A:1025867130437. [DOI] [PubMed] [Google Scholar]
- 21.Denys H, De Wever O, Nusgens B, Kong Y, Sciot R, Le AT, et al. Invasion and MMP expression profile in desmoid tumours. Br J Cancer. 2004;90:1443–9. doi: 10.1038/sj.bjc.6601661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang X, Ji G, Wu Y, Wan B, Yu L. LAMA4, highly expressed in human hepatocellular carcinoma from Chinese patients, is a novel marker of tumor invasion and metastasis. J Cancer Res Clin Oncol. 2008;134:705–14. doi: 10.1007/s00432-007-0342-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagato S, Nakagawa K, Harada H, Kohno S, Fujiwara H, Sekiguchi K, et al. Downregulation of laminin alpha4 chain expression inhibits glioma invasion in vitro and in vivo. Int J Cancer. 2005;117:41–50. doi: 10.1002/ijc.21102. [DOI] [PubMed] [Google Scholar]
- 24.Tapper J, Kettunen E, El-Rifai W, Seppala M, Andersson LC, Knuutila S. Changes in gene expression during progression of ovarian carcinoma. Cancer Genet Cytogenet. 2001;128:1–6. doi: 10.1016/S0165-4608(01)00386-7. [DOI] [PubMed] [Google Scholar]
- 25.Oue N, Hamai Y, Mitani Y, Matsumura S, Oshimo Y, Aung PP, et al. Gene expression profile of gastric carcinoma: identification of genes and tags potentially involved in invasion, metastasis and carcinogenesis by serial analysis of gene expression. Cancer Res. 2004;64:2397–405. doi: 10.1158/0008-5472.CAN-03-3514. [DOI] [PubMed] [Google Scholar]
- 26.Hassan NJ, Barclay AN, Brown MH. Frontline: Optimal T cell activation requires the engagement of CD6 and CD166. Eur J Immunol. 2004;34:930–40. doi: 10.1002/eji.200424856. [DOI] [PubMed] [Google Scholar]
- 27.King JA, Ofori-Acquah SF, Stevens T, Al-Mehdi AB, Fodstad O, Jiang WG. Activated leukocyte cell adhesion molecule in breast cancer: prognostic indicator. Breast Cancer Res. 2004;6:R478–87. doi: 10.1186/bcr815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davies SR, Dent C, Watkins G, King JA, Mokbel K, Jiang WG. Expression of the cell to cell adhesion molecule, ALCAM, in breast cancer patients and the potential link with skeletal metastasis. Oncol Rep. 2008;19:555–61. [PubMed] [Google Scholar]
- 29.Mao X, Seidlitz E, Truant R, Hitt M, Ghosh HP. Re-expression of TSLC1 in a non-small-cell lung cancer cell line induces apoptosis and inhibits tumor growth. Oncogene. 2004;23:5632–42. doi: 10.1038/sj.onc.1207756. [DOI] [PubMed] [Google Scholar]
- 30.Koreth J, Bakkenist CJ, McGee JO. Allelic deletions at chromosome 11q22-q23.1 and 11q25-qterm are frequent in sporadic breast but not colorectal cancers. Oncogene. 1997;14:431–7. doi: 10.1038/sj.onc.1200847. [DOI] [PubMed] [Google Scholar]
- 31.Murakami Y, Nobukuni T, Tamura K, Maruyama T, Sekiya T, Arai Y, et al. Localization of tumor suppressor activity important in nonsmall cell lung carcinoma on chromosome 11q. Proc Natl Acad Sci U S A. 1998;95:8153–8. doi: 10.1073/pnas.95.14.8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yen CC, Chen YJ, Chen JT, Hsia JY, Chen PM, Liu JH, et al. Comparative genomic hybridization of esophageal squamous cell carcinoma: correlations between chromosomal aberrations and disease progression/prognosis. Cancer. 2001;92:2769–77. doi: 10.1002/1097-0142(20011201)92:11<2769::AID-CNCR10118>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 33.Ito T, Shimada Y, Hashimoto Y, Kaganoi J, Kan T, Watanabe G, et al. Involvement of TSLC1 in progression of esophageal squamous cell carcinoma. Cancer Res. 2003;63:6320–6. [PubMed] [Google Scholar]
- 34.Lindsey JC, Lusher ME, Anderton JA, Bailey S, Gilbertson RJ, Pearson AD, et al. Identification of tumour-specific epigenetic events in medulloblastoma development by hypermethylation profiling. Carcinogenesis. 2004;25:661–8. doi: 10.1093/carcin/bgh055. [DOI] [PubMed] [Google Scholar]
- 35.Manavi M, Hudelist G, Fink-Retter A, Gschwandtler-Kaulich D, Pischinger K, Czerwenka K. Gene profiling in Pap-cell smears of high-risk human papillomavirus-positive squamous cervical carcinoma. Gynecol Oncol. 2007;105:418–26. doi: 10.1016/j.ygyno.2006.12.032. [DOI] [PubMed] [Google Scholar]
- 36.Bühler H, Schaller G. Transfection of keratin 18 gene in human breast cancer cells causes induction of adhesion proteins and dramatic regression of malignancy in vitro and in vivo. Mol Cancer Res. 2005;3:365–71. doi: 10.1158/1541-7786.MCR-04-0117. [DOI] [PubMed] [Google Scholar]
- 37.Zhou C, Nitschke AM, Xiong W, Zhang Q, Tang Y, Bloch M, et al. Proteomic analysis of tumor necrosis factor-alpha resistant human breast cancer cells reveals a MEK5/Erk5-mediated epithelial-mesenchymal transition phenotype. Breast Cancer Res. 2008;10:R105. doi: 10.1186/bcr2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase and gadd45. Cell. 2008;135:1201–12. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu J, Qian H, Li Y, Wang Y, Zhang X, Liang X, et al. Arsenic trioxide (As2O3) reduces the invasive and metastatic properties of cervical cancer cells in vitro and in vivo. Gynecol Oncol. 2007;106:400–6. doi: 10.1016/j.ygyno.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 41.Yu J, Zhang L, Chen A, Xiang G, Wang Y, Wu J, et al. Identification of the gene transcription and apoptosis mediated by TGF-beta-Smad2/3-Smad4 signaling. J Cell Physiol. 2008;215:422–33. doi: 10.1002/jcp.21325. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.