Abstract
This study reports on the use of a fibrinogen-derived peptide for the specific targeting and delivery of vancomycin to Staphylococcus epidermidis biofilms. One method by which S. epidermidis initially adheres to biomaterials uses the plasma protein fibrinogen as an intermediary, where the S. epidermidis surface protein SdrG binds to a short amino acid sequence near the amino terminus of the Bβ chain of fibrinogen. We mimicked this binding interaction and demonstrated the use of a synthetic fibrinogen-based β6-20 peptide to target and deliver vancomycin to S. epidermidis in vitro. The β6-20 peptide was synthesized and labeled with a nanogold probe, and its targeting capabilities were examined through the use of scanning electron microscopy. The Nanogold component was then replaced by vancomycin, utilizing a flexible, variable length poly(ethylene glycol) linker between the peptide and antibiotic to create the targeted vancomycin products, β6-20-PEGx-VAN. Initial binding to surface adherent S. epidermidis was increased in a concentration-dependent manner relative to vancomycin for all equivalent concentrations ≥4 μg/ml, with targeted vancomcyin content up to 22.9 times that of vancomycin alone. Retention of the targeted antibiotics was measured after an additional 24 hour incubation period, revealing levels 1.3 times that of vancomycin. The results demonstrate the improved targeting and retention of vancomycin within a biofilm due to the incorporation of a specific targeting motif.
Keywords: Staphylococcus epidermidis, SdrG, β6-20 peptide, Targeted Delivery, Vancomycin
INTRODUCTION
There are approximately 40 million surgical procedures involving artificial devices performed each year in the United States1. Complications arising from nosocomial infections pose a significant health risk to patients with synthetic implants. Staphylococci, enterococci, enterobacteriaceae, and Candida spp. are the common pathogens associated with infections of indwelling medical devices2, with the likelihood of infection, as well as the organism implicated in the infection, greatly dependent upon the type and location of the implant2. In the case of intravascular implants, coagulase-negative staphylococci (CoNS), particularly Staphylococcus epidermidis, are the most common cause of infection1,3.
The initial stage of S. epidermidis infection of intravascular devices involves bacterial adhesion through interactions with host plasma proteins that adsorb onto the biomaterial surface immediately following implantation3–12. Specific binding between S. epidermidis and host proteins is commonly mediated by microbial surface components recognizing adhesive matrix molecules (MSCRAMMs)13,14, with SdrG responsible for S. epidermidis binding to amino acids 6–20 of the fibrinogen Bβ chain9,11,15 with a KD of 0.9×10−7 M15. A synthetic peptide sequence representing the first 25 amino acid residues of the Bβ chain of fibrinogen (β1–25) was used by Davis et al15 to mimic the SdrG binding region, and this peptide bound to SdrG with a KD of 1.4×10−7 M. Thus, the significant amino acids necessary for the SdrG-fibrinogen binding interaction are present in the β1–25 peptide, and this linear sequence is in a nearly optimal conformational state. The fibrinogen binding domain was further localized to the β6–20 peptide, consisting of the amino acid sequence NEEGFFSARGHRPLD15, with amino acids β10–15 contributing most significantly to the SdrG-fibrinogen binding16. The affinity of the β6-20 peptide towards SdrG can be significantly reduced by creating substitution peptides, such as β6-20-R14A and β6-20-G15A, in which individual amino acid residues within the β10–15 binding region are replaced with an alanine residue16.
Targeted therapy of bacterial infections has been studied in various forms. Lytic bacteriophages represent a class of naturally occurring targeted antibacterial treatments, with phages identified that can infect various organisms, dissolve the exopolysaccharide matrix of existing biofilms, and in some cases even reduce the need for clinical use of antibiotics17. Another promising concept is the use of antibodies as opsonizing agents for colonizing bacteria. Opsonized S. epidermidis experience increased phagocytosis, resulting in a significantly lower incidence of infections in vivo18–21. However, targeted delivery of existing antibiotics remains a relatively unexplored area.
Vancomycin is a glycopeptide antibiotic that acts against gram positive bacteria by preventing the incorporation of N-acetylglucosamine and N-acetylmuramic acid into the growing peptidoglycan matrix22. Vancomycin forms a series of five hydrogen bonds with the terminal -D-Alanine-D-Alanine residues of the peptidoglycan intermediates, thereby interfering with the biosynthetic pathway used to build a stable cell wall23. The most common reason for Vancomycin resistance in a microbial population is a mutation that results in the peptidoglycan precursors terminating with -D-Alanine-D-Lactate, which eliminates one of the hydrogen bonds with vancomycin and lowers binding affinity by a factor on the order of 100024. In order to overcome this resistance, glycopeptide antibiotics form non-covalent dimers that are thought to improve antibiotic effectiveness by preferentially locating the antibiotic at the site of action. After the first vancomycin molecule binds its target, the second binding event can be considered an intramolecular event that is more difficult to antagonize than a solution-phase bimolecular binding event25. Thus, facilitating the binding interaction between vancomycin and its ligand can lead to improved antibiotic treatment by recovering binding efficiency lost due to the -D-Ala-D-Lac mutation.
Given the importance and prevalence of the SdrG-fibrinogen binding scheme in the initial stages of S. epidermidis infections, this binding behavior should be explored as a mechanism for specifically delivering therapeutic agents to S. epidermidis infections. The experiments presented here were designed to mimic the benefits obtained by dimerization of glycopeptide antibiotics, while adding a specific targeting and delivery aspect to the system through use of the synthetic β6-20 peptide. By covalently linking vancomycin to the β6-20 peptide, we have developed a system to specifically deliver vancomycin to the S. epidermidis biofilm.
MATERIALS AND METHODS
Peptide Synthesis
Two targeting peptides, NEEGFFSARGHRPLD (β6-20) and (Ac)-CNEEGFFSARGHRPLD (Cys-β6-20), as well as two substitution peptides used as non-binding controls, NEEGFFSAAGHRPLD (β6-20-R14A) and NEEGFFSARAHRPLD (β6-20-G15A), were synthesized by solid phase peptide synthesis on a 433a ABI peptide synthesizer (Applied Biosystems), using 9-fluoronylmethoxycarbonyl (fmoc) amino acids on a Knorr resin26. The Cys-β6-20 amino terminus was acetylated using acetic acid in a final automated coupling step on the synthesizer. The peptides were cleaved and deprotected using trifluoroacetic acid (TFA). Reverse phase high performance liquid chromatography (RP-HPLC) was used to purify the cleaved peptides, using water (w/ 0.1% TFA) and acetonitrile (w/ 0.082% TFA) as the aqueous and organic components of the mobile phase. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry was used to confirm the purified peptide products were of the correct molecular weight.
Peptide Labeling with Nanogold
The β6-20 peptide was labeled for imaging using sulfo-N-hydroxy succinimide Nanogold (Nanoprobes, Inc; Yaphank, NY). Briefly Nanogold (6 nmol) was dissolved in Millipore water (0.2 ml). A 20-fold excess of the β6-20 peptide was dissolved in HEPES buffer (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 0.02 M, pH 7.5, 1 ml). The two solutions were then mixed and incubated at room temperature for 1 hour while gently shaking. The labeled peptide was separated from the unlabeled peptide on the basis of molecular weight using a Millipore Centricon YM-10 centrifuge filter with a molecular weight cutoff of 10kDa. The peptide/Nanogold mixture was pipetted into the filter and spun at 4000× g. After 2 hours, the filtrate was removed and discarded, while the filter was inverted and spun for 3 minutes at 1000×g to recover the labeled peptide (β6-20-NG) into a new vial. This procedure was repeated twice to enhance the purification.
Peptide-Vancomycin Conjugation
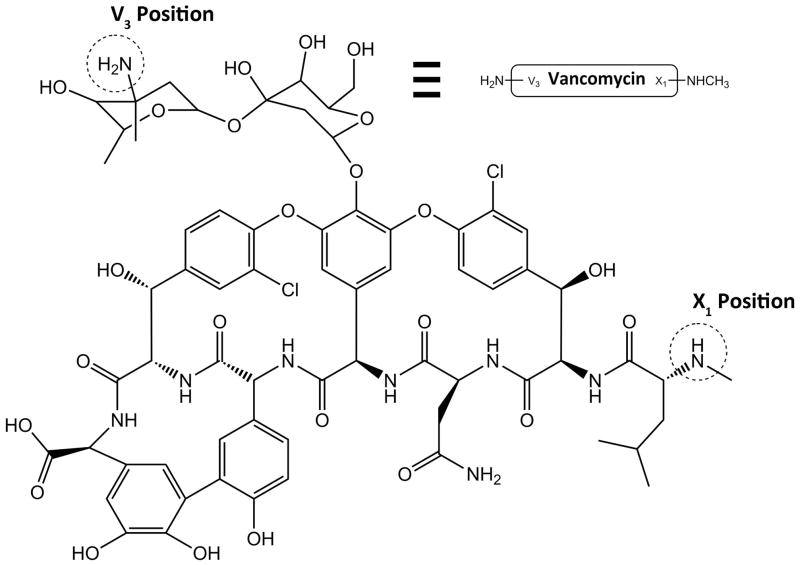
The β6-20-PEGx-Vancomycin (β6-20-PEGx-VAN) conjugate was synthesized using a Maleimide-PEGx-Succinimide heterofunctional crosslinker (x=3400 or x=5000 MW). Vancomycin contains both a primary amine and a secondary methylamine suitable for conjugation reactions, commonly described as the V3 and X1 positions, respectively (figure 1). In order to ensure that the PEG chain attaches to vancomycin at the desired V3 position as opposed to the X1 position, a two-step reaction was used (figure 2). The first step of the conjugation was carried out as described by Greenwald et al27. Briefly, MAL-PEG3400-SCM or MAL-PEG5000-SCM (Creative PEGWorks, Winston-Salem, NC) was added to a two-fold excess of vancomycin hydrochloride (Sigma) in anhydrous dimethylformamide (DMF) supplemented with a twenty-fold molar excess of triethylamine (TEA). The reaction was carried out under nitrogen, while stirring at room temperature for 12 hours, after which the mixture was added drop-wise to cold diethyl ether to precipitate the product. The product was pelleted by centrifugation, and rinsed an additional two times with fresh diethyl ether. Excess vancomycin was removed from the crude MAL-PEGx-VAN product by dialysis (3500 MWCO) against water adjusted to pH 5.0 with 1N HCl (in order to prevent maleimide hydrolysis). The intermediate product was dried and residual vancomycin was quantified by RP-HPLC (C18 column). Residual, unconjugated MAL-PEGx-SCM was estimated using 1H NMR.
Figure 1.
Chemical structure of vancomycin. Dashed circles indicate the potential locations for amine-targeted modifications. Reaction conditions can be tailored to favor the primary V3 amine over the secondary X1 methylamine.
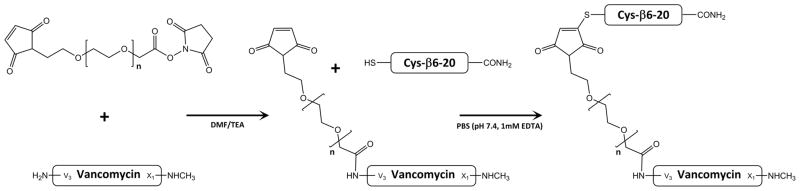
Figure 2.
Two-step reaction scheme for synthesis of β6-20-PEGx-Vancomycin. Step 1 Combines MAL-PEGx-SCM with vancomycin in DMF/TEA, favoring formation of an amide bond at the vancomycin V3 amine. Step 2 combines MAL-PEGx-Vancomycin with Cys-β6-20 in PBS to create a stable thioether bond between the targeting peptide and the PEG linker.
To prepare the final β6-20-PEGx-VAN product, a 1.5 molar excess of purified Cys-β6-20 peptide was added to MAL-PEGx-VAN in phosphate buffered saline (pH 7.4, 1 mM EDTA) and stirred for 2 hours at room temperature. Ellman’s assay was used to assess the extent of thiol disappearance in the reaction mixture, corresponding to formation of the desired thioether bond. Dialysis against water (3500 MWCO) was then used to remove excess peptide. Residual peptide and vancomycin were quantified by RP-HPLC (C18 column). Peptide:PEG:Vancomycin ratios were estimated using 1H NMR.
Determination of Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC)
The broth dilution method was used to determine the MIC and MBC of the newly synthesized targeted antibiotics. Tryptic soy broth (TSB, 1 ml) containing S. epidermidis (1×105 cfu/ml) and vancomycin, β6-20-PEG3400-VAN, or β6-20-PEG3400-VAN (0–128 μg/ml equivalent concentrations) was prepared. The samples (n=3) were then incubated for 24 hours at37°C with gentle shaking, after which the turbidity of the vials was observed. The MIC was taken as the lowest antibiotic concentration at which the TSB was no longer clear. In order to determine the MBC, 10 μl of the media from each tube was plated on a trypticase soy agar plate and incubated for an additional 24 hours at 37°C. The MBC was taken as the lowest antibiotic concentration at which no colonies were observed.
Seeding of Sample Surfaces
S. epidermidis strain RP62A was used for all procedures as a representative biofilm-positive strain. An inoculating loop was used to retrieve a small sample of S. epidermidis from a refrigerated culture plate not more than two weeks old. The bacteria were incubated in tryptic soy broth (TSB) for 18 hours at 37°C while shaking at 120 rpm, then pelleted and washed with fresh phosphate buffered saline (PBS, pH 7.4) before being resuspended in TSB to a final concentration of 0.5×108 cfu/ml, as determined by optical density (OD). Bacteria solution was then added to 96-well plates (150 μL) or 24-well plates (200 μl) and incubated at 37°C for 2 or 24 hours. The 24-well plates contained poly(ethylene terephthalate) (PET) surfaces (15mm diameter). Prior to seeding, the PET surfaces were cleaned with ethanol and secured in place using sterilized silicone tubing.
Nanogold Targeting
In order to test peptide targeting and delivery of Nanogold particles, 24-well plates were seeded and incubated for 2 hours, after which the sample surfaces were gently rinsed with fresh PBS to remove non-adherent bacteria. Unlabeled peptide (β6-20, β6-20-G15A, or β6-20-R14A) was added to the bacteria in 1% bovine serum albumin (BSA) for 1 hour at room temperature, followed by a gentle rinse with fresh 1% BSA and the subsequent addition of the β6-20-NG peptide for 1 hour. After binding, all wells were rinsed with 1% BSA. The 1.4nm Nanogold particles were enlarged to 20–50 nm using GoldEnhance (Nanoprobes, Inc; Yaphank, NY) in order to make them clearly visible in the scanning electron microscope (SEM). Finally, samples were covered with a 2.5% buffered glutaraldehyde solution and bacteria were fixed at 4°C overnight. Substrates were dehydrated for imaging using a water/ethanol gradient of 50%, 70%, 95%, and 100% ethanol. Samples were then covered 2×15min with hexamethyldisilazane (HMDS) and allowed to air dry. When dry, the samples were mounted onto SEM stubs, sputter coated with a 50 Ǻ layer of palladium and imaged using a Hitachi S-4500 SEM.
Vancomycin Targeting
In order to examine targeted delivery of vancomycin, 96-well plates were seeded and incubated for 2 or 24 hours, after which the sample surfaces were gently rinsed with PBS to remove non-adherent bacteria. β6-20-PEGx-VAN or free vancomycin (150 μl, 0–64 μg/ml in TSB) was added to the wells and incubated at 37°C for 1 or 24 hours. All wells were then rinsed twice with fresh PBS, followed by an additional incubation period in TSB at 37°C (0 or 24 hours). The vancomycin-targeted samples were then analyzed for vancomycin retention.
Vancomycin Retention Assay
Vancomycin-targeted samples were rinsed with 0.5% BSA, followed by the addition of an anti-vancomycin primary antibody (100 μl in 3% BSA, 5 μg/ml, rabbit IgG, Abcam) for 60 minutes at room temperature. All samples were then rinsed 3 times with fresh 0.5% BSA, followed by the addition of a secondary antibody conjugated to horseradish peroxidase (HRP, 100 μl, 1:1000 dilution, goat IgG anti-rabbit IgG, Molecular Probes) for an additional 60 minutes at room temperature, and then a final rinse with PBS. ABTS (2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid diammonium salt) was then added as an HRP substrate (200 μl, 0.1 mg/ml ABTS, 1 μl/ml hydrogen peroxide, 50 mM phosphate-citrate buffer, pH 5.0). After 20 minutes, 100 μl was transferred from each well into a new 96-well plate, and the wells were examined quantitatively for vancomycin content using a microplate reader (412 nm).
Statistics
Statistical analysis was done using Minitab 16. Comparisons between individual samples were made using an un-paired student’s t-test, while comparisons within groups were made using one-way ANOVA and Dunnett’s post-hoc test. α < 0.05 was considered significant.
RESULTS
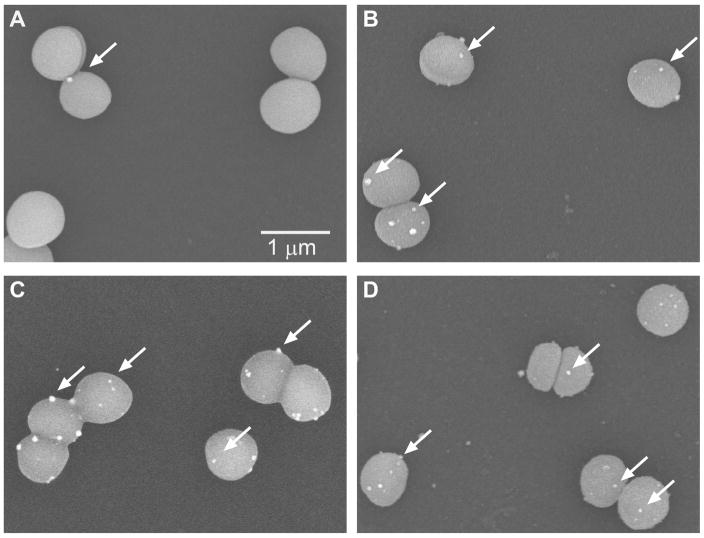
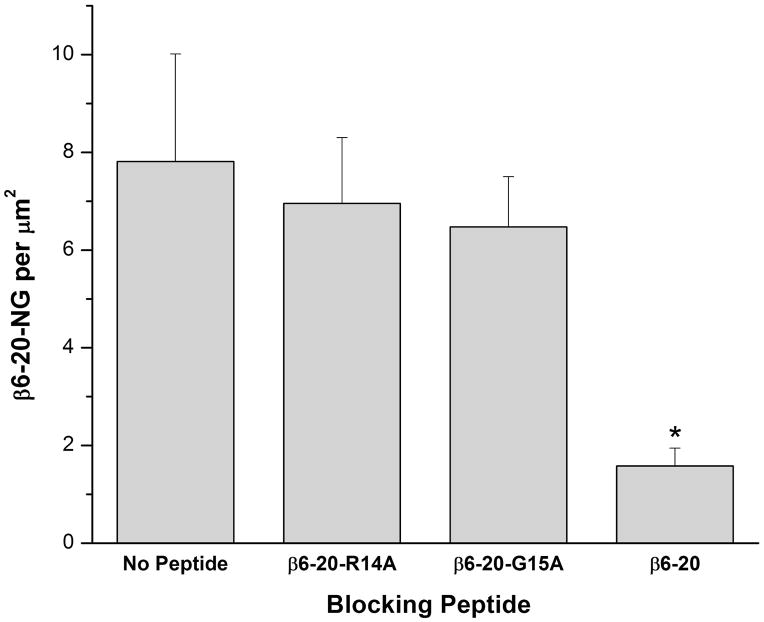
In order to test the targeting and delivery ability of the β6-20 peptide, blocking studies were carried out, with representative SEM images shown in Figure 3. The samples blocked with theβ6-20 peptide (A) show minimal Nanogold labels bound to the surface of the bacteria, while the unblocked samples (B) and samples blocked with the non-binding β6-20-R14A and β6-20-G15A peptides (C and D, respectively) show Nanogold labels specifically binding to the bacteria. Figure 4 quantifies the results obtained from the peptide blocking experiments, as determined by analysis with ImageJ28. The control data (no peptide) represents the labeled β6-20-NG peptide binding directly to the bacteria without an initial blocking step. When sample surfaces are incubated with the non-bindingβ6-20-R14A or β6-20-G15A peptide prior to adding the β6-20-NG peptide, there are no significant differences in binding when compared to the control (p< 0.74 and p< 0.59, respectively). However, when samples are incubated with the unlabeled β6-20 peptide prior to adding the labeled peptide, there is a significant decrease in specific binding relative to the control (p< 0.02).
Figure 3.
Scanning electron microscope images of peptide blocking studies. Bacteria were exposed to A)β6-20 peptide, B) no peptide (control), C) β6-20-R14A peptide, or D)β6-20-G15A peptide prior to the addition of the Nanogold-labeled β6-20-NG peptide. Nanogold labels (some are identified by white arrows) are bound to S. epidermidis in the control,β6-20-R14A, and β6-20-G15A blocking samples (B,C,D) while there are minimal labels bound to the bacteria in the β6-20 blocking sample (A).
Figure 4.
Peptide blocking study. There was no significant difference in specific binding between the positive control and the β6-20-R14A or β6-20-G15A negative control blocking peptides (p< 0.74 and p< 0.59, respectively). Using the β6-20 peptide to block specific binding, however, resulted in a significant decrease relative to the positive control (p< 0.02). Data represents mean plus standard error.
Synthesis of the β6-20-PEG3400-VAN and β6-20-PEG5000-VAN products was monitored for purity by RP-HPLC and 1H NMR in D2O (data not shown). RP-HPLC showed that the intermediate MAL-PEG3400-VAN and MAL-PEG5000-VAN products contained 1.27% (w/w) and 0.71% (w/w) residual vancomycin, respectively. In addition, 1H NMR characterization of the maleimide hydrogens (δ=6.8 ppm, 2H) and vancomycin methyl hydrogens (δ=0.9 ppm, 6H) indicated that the intermediate products were approximately 70 mol% MAL-PEGx-VAN and 30 mol% unreacted MAL-PEGx-COOH. After the Cys-β6-20 + MAL-PEGx-VAN conjugation step, RP-HPLC of the final β6-20-PEG3400-VAN and β6-20-PEG5000-VAN products indicated no residual peptide, and showed 0.13% (w/w) and 0.08% (w/w) residual vancomycin, respectively. The amounts of residual vancomycin and unreacted PEG were taken in to account when weighing out the final products for retention studies, and as such, the term equivalent vancomycin concentration is used when referring to these products.
Minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) were measured for the new targeted vancomycin derivatives (table 1). Vancomycin alone showed an MIC of 2 μg/ml and an MBC of 8 μg/ml. However, increases in MIC and MBC were seen for both β6-20-PEG3400-VAN and β6-20-PEG5000-VAN. The β6-20-PEG3400-VAN product showed MIC and MBC values of 16 μg/ml and 32 μg/ml, respectively. The β6-20-PEG5000-VAN product showed MIC and MBC values double those seen with the β6-20-PEG3400-VAN product, at 32 μg/ml and 64 μg/ml, respectively.
Table 1.
Minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) values for vancomycin, β6-20-PEG3400-VAN, and β6-20-PEG5000-VAN.
| Antibiotic | MIC (μg/ml) | MBC (μg/ml) |
|---|---|---|
| Vancomycin | 2 | 8 |
| β–20-PEG3400-VAN | 16 | 32 |
| β–20-PEG5000-VAN | 32 | 64 |
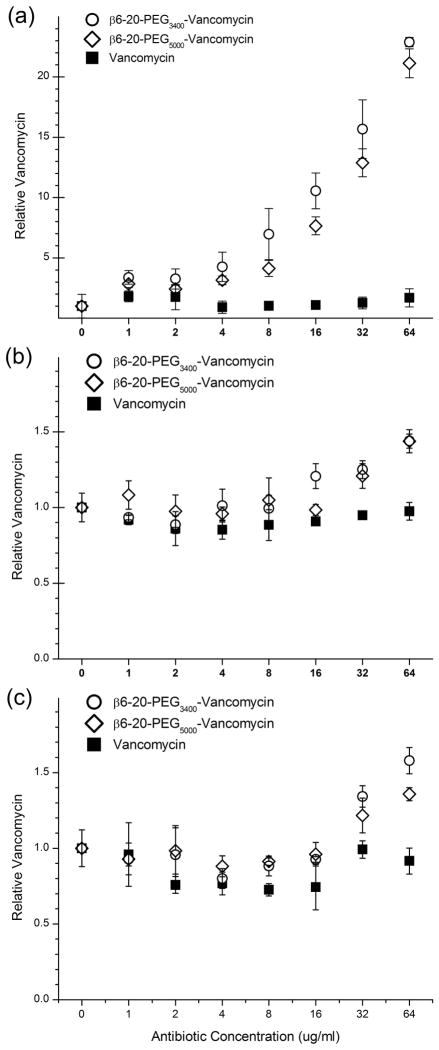
The β6-20-PEGx-VAN products were tested for their ability to target S. epidermidis and improve vancomycin retention. Retention was first measured on S. epidermidis in the early stages of biofilm formation by allowing the bacteria to adhere to 96-well plates for 2 hours prior to exposure to antibiotics (1 hour exposure, 0μg/ml to 64 μg/ml vancomycin equivalents). All data (n=3) was normalized to the 0 μg/ml control within each plate. Vancomycin showed no sigificant retention (p=0.48) at any concentration tested when compared to the 0 μg/ml control. Both β6-20-PEG3400-VAN and β6-20-PEG5000-VAN showed significantly increased retention (p<0.001) relative to vancomycin for all equivalent concentrations ≥4 μg/ml (Figure 5a), with retention increasing in a concentration-dependent manner. Maximum retention was observed at 64 μg/ml, with a 22.9-fold increase for β6-20-PEG3400-VAN and a 21.1-fold increase for β6-20-PEG5000-VAN. For equivalent concentrations <4 μg/ml, the targeted antibiotics showed no difference in retention when compared to vancomycin. Following the initial 1 hour exposure, all samples were incubated for an additional 24 hours before re-measuring the amount of antibiotics retained in the biofilms. Samples that had been treated with 32 μg/ml and 64 μg/ml vancomycin equivalents continued to show significant improvements, with retention of 1.3- and 1.8-fold for β6-20-PEG3400-VAN and 1.3- and 1.6-fold for β6-20-PEG5000-VAN (p<0.001, data not shown). For equivalent vancomycin concentrations ≤16 μg/ml, there were no differences in retained vancomycin between any of the samples.
Figure 5.
Retention of β6-20-PEG3400-VAN, β6-20-PEG5000-VAN, and vancomycin by S. epidermidis after (a) 2 hour bacterial adhesion, 1 hour antibiotic exposure, (b) 24 hour biofilm growth, 1 hour antibiotic exposure, and (c) 24 hour biofilm growth, 24 hour antibiotic exposure. All data (n=3) are normalized to the 0 μg/ml concentration value.
Retention was also measured with more mature biofilms grown for 24 hours prior to exposure to antibiotics (1 hour exposure, 0μg/ml to 64 μg/ml vancomycin equivalents). Vancomycin showed no sigificant retention (p=0.16) in the biofilm at any concentration tested when compared to the 0 μg/ml control. β6-20-PEG3400-VAN showed improved retention (p<0.001) relative to vancomycin for equivalent concentrations ≥16μg/ml, while β6-20-PEG5000-VAN showed improved retention (p<0.001) for equivalent concentrations ≥32μg/ml (Figure 5b). Retention ranged from 1.2-1.4-fold increases for both β6-20-PEG3400-VAN and β6-20-PEG5000-VAN. Following the initial 1 hour exposure, all wells were incubated for an additional 24 hours before re-measuring the amount of antibiotics retained in the biofilms. There were no differences in retained vancomycin between any of the samples at this time point (p=0.79, data not shown).
Finally, retention was measured using 24 hour biofilms with a longer period of exposure to antibiotics (24 hour exposure, 0 μg/ml to 64 μg/ml vancomycin equivalents). Vancomycin showed no sigificant retention in the biofilm at any concentration tested when compared to the 0μg/ml control, with the exception of 4 μg/ml (p=0.01). Both β6-20-PEG3400-VAN and β6-20-PEG5000-VAN showed significant increases in retention (p<0.001) relative to vancomycin for equivalent concentrations ≥32μg/ml (Figure 5c). Retention ranged from 1.3–1.6-fold increases for β6-20-PEG3400-VAN and 1.2–1.4-fold increases for β6-20-PEG5000-VAN. Following the initial 24 hour exposure, all wells were incubated for an additional 24 hours before re-measuring the amount of antibiotics retained in the biofilms (data not shown). β6-20-PEG3400-VAN showed 1.3- and 1.4-fold increases in retention (p=0.01) for concentrations of 32 μg/ml and 64 μg/ml, while β6-20-PEG5000-VAN showed a 1.3-fold increase in retention at 32 μg/ml (p=0.01).
DISCUSSION
Staphylococcus epidermidis is part of the normal human flora and considered to be noninvasive and nonpathogenic under most circumstances. However, implantation of a medical device provides a foreign surface that can be colonized by bacteria that may be unintentionally introduced during the procedure. The SdrG-fibrinogen interaction has been shown to play a significant role in the initial stages of bacterial adhesion: recombinant SdrG is able to block the adherence of S. epidermidis to fibrinogen in a concentration dependent manner29, adding SdrG under saturating conditions prevents S. epidermidis from binding to fibrinogen11, and mutating the fibrinogen-binding region of the SdrG A-domain of S. epidermidis greatly impairs the ability to bind to immobilized fibrinogen.16,30. Furthermore, both the gene encoding for SdrG31 and the SdrG protein itself32 are present in 100% of the various clinical S. epidermidis isolates tested, and studies of infected and recovering patients reveal anti-SdrG antibodies in the serum, confirming that the bacteria express SdrG during the infectious stages20,32. An in-vivo intravascular catheter infection model confirmed the importance of SdrG: an SdrG-negative S. epidermidis mutant had a 20% infection rate, compared to a 100% rate for the SdrG-positive strain33.
Our results indicate that SdrG is a viable target for drug delivery using the β6-20 ligand. In order to confirm the peptide’s targeting and delivery abilities, the effects of the β6-20,β6-20-R14A, and β6-20-G15A peptides were studied in regards to their effectiveness at preventing β6-20-NG from binding to S. epidermidis. When the bacteria were exposed to the β6-20 peptide prior to adding β6-20-NG (Figure 3A), there were very few visible Nanogold-labeled peptides bound to the surface of the bacteria, indicating that the unlabeled peptide has occupied the available binding sites. When the labeled peptide was subsequently introduced, there were no SdrG proteins available for binding, and as a result the labeled peptide was washed away during the rinsing step. The β6-20-R14A and β6-20-G15A substitution peptides were used as negative controls in these studies, having a greatly reduced binding affinity for SdrG16. Results in Figure 3C and 3D demonstrate the inability of the β6-20-R14A and β6-20-G15A peptides to prevent subsequent binding of the β6-20-NG peptide, with results similar to those shown in Figure 3B where no blocking peptide was used. Quantitatively (Figure 4), the β6-20 blocking peptide significantly reduced targeted nanogold delivery relative to the control, while the negative control peptides were unable to significantly reduce this specific binding. These results indicate that the β6-20 peptide is able to specifically target and deliver nanogold particles to S. epidermidis in vitro, and additional studies are warranted wherein the nanogold particle is replaced by vancomycin.
The success of S. epidermidis infections depends upon the ability of the bacteria to form a biofilm following initial adhesion to a material3,34. The biofilm environment offers the encapsulated bacteria increased resistance to antibiotics, often times able to survive antibiotic concentrations several orders of magnitude higher than the minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) measured in planktonic suspensions35–37. However, after dispersing the cells in a biofilm, the newly suspended bacteria are once again susceptible to the same MIC and MBC levels found in planktonic bacteria38. Furthermore, S. epidermidis biofilms formed on dialysis membranes allow vancomycin to penetrate the biofilm and cross the membrane39, while biofilms formed on stainless steel prostheses have been shown to uptake significant quantities of vancomycin38. It is likely that antibiotic resistance within a biofilm is not obtained through irreversible genotypic changes, and that biofilm penetration is not a limiting factor in antibiotic effectiveness. As such, it is possible that increasing the binding capabilities of vancomycin through use of a targeting ligand could prove beneficial in treating infections.
Covalently tethering vancomycin to a synthetic targeting ligand yields significant increases in antibiotic retention by the S. epidermidis biofilm. Interestingly, the amount of β6-20-PEG3400-VAN product retained by the S. epidermidis samples was slightly greater than the amount of β6-20-PEG5000-VAN retained under most conditions tested. One likely reason for this difference is the potential for the longer PEG chain to obstruct the binding interactions between peptide and bacteria. In addition, it is also possible that the higher molecular weight of the β6-20-PEG5000-VAN product decreases the speed at which the molecule can penetrate the growing biofilm. These findings correlate well with the MIC and MBC values observed for the new targeted vancomycin derivatives. While both β6-20-PEG3400-VAN and β6-20-PEG5000-VAN were shown to be active against S. epidermidis, the MIC and MBC values for β6-20-PEG5000-VAN product were twice as high as those observed for β6-20-PEG3400-VAN.
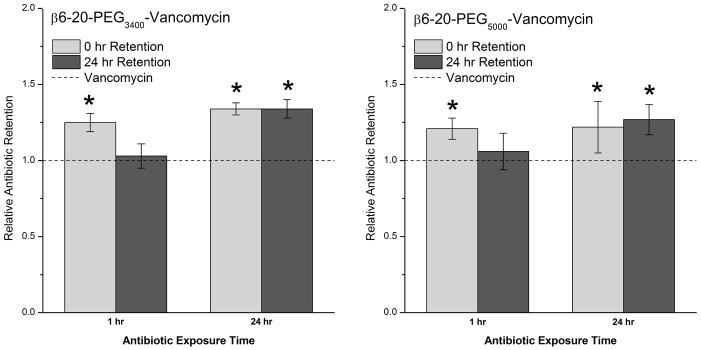
Within the typical clinical range for vancomycin concentrations (figure 6), initial retention of the targeted antibiotics after 1 hour or 24 hour exposure was 20–30% greater than retention of vancomycin alone. The amount of vancomycin retained by the mature biofilms after an additional 24 hour incubation period depended upon the initial duration of vancomycin exposure prior to the rinsing step. The biofilms that were treated for just 1 hour ended up with targeted vancomycin levels that were not significantly different from those of vancomycin alone. A potential explanation for this inability to retain the antibiotics is due to the larger size of the targeted vancomycin: 1 hour may not be enough time for such a large molecule to fully penetrate a mature biofilm, and as such, there was no real advantage over vancomycin. However, when the mature biofilms were exposed to the targeted vancomycin for a full 24 hours prior to the final incubation step, there were negligible changes in retained targeted vancomycin when comparing initial and final values, with final retention 27–34% higher than seen with vancomycin alone.
Figure 6.
Retention of targeted antibiotics by 24 hour S. epidermidis biofilms as determined by indirect ELISA. S. epidermidis was exposed to vancomycin, β6-20-PEG3400-VAN, or β6-20-PEG5000-VAN (32 μg/ml vancomycin equivalents) for 1 hour or 24 hours. Vancomycin levels were measured 0 and 24 hours post-treatment. All data (n=3) are reported as increases in retention relative to untargeted vancomycin, with (*) indicating a statistically significant increase (α<0.05).
When surface adherent bacteria are exposed to the targeted vancomycin prior to biofilm formation, the targeting peptide significantly improves the vancomycin retention, with 22.1–22.9-fold increases observed after just a 1 hour exposure. However, as the biofilm develops, it is likely that the size difference between vancomycin and the two targeted species becomes more of a factor, resulting in the observed decrease in retention of the targeted molecules relative to the untargeted vancomycin. This becomes most evident when the biofilm is allowed to mature prior to being exposed to the targeted vancomycin molecules. Extending the duration of exposure to the targeted vancomycin allows more time for the larger molecule to penetrate the biofilm, which in turn results in a higher relative retention.
It is expected that increased retention within the biofilm will provide benefits analogous to those seen with vancomycin dimerization. Dimerization of glycopeptide antibiotics such as Vancomycin occurs naturally, improving effectiveness of the antibiotic by preferentially locating the antibiotic at the site of action where subsequent binding interactions are thought to be more of an intramolecular event25. Dimerization has been shown to improve antibiotic effectiveness in a number of ways. A series of 40 covalently linked vancomycin dimers were synthesized, many of which displayed improved effectiveness against both vancomycin-susceptible organisms and vancomycin-resistant Enterococci (VRE)40. Meanwhile, a multi-valent polymer of vancomycin demonstrated enhanced antibacterial activity against VRE that was 8- to 60-fold more effective than traditional vancomycin. While vancomycin binds to -D-Ala-D-Ala with a KD of ~1×10−6 M, a trivalent system of vancomycin binds a trivalent -D-Ala-D-Ala ligand with a KD of ~4×10−17, which is 25 times tighter than biotin-avidin23. Finally, vancomycin covalently tethered to a titanium surface maintained its ability to bind -D-Ala-D-Ala as well as kill S. aureus, and indicated that a high surface density of antibiotic is equivalent to an enormous solution concentration of free vancomycin41. Similar results can be seen with the antibiotic teicoplanin, which does not form dimers but instead anchors to the bacterial membrane by way of its fatty acid chain. Teicoplanin, once anchored to a surface, functions intramolecularly and displays improved effectiveness when compared to binding events in the absence of a membrane25,42.
CONCLUSION
The Fibrinogen-SdrG interaction is an important component of bacterial adhesion to blood-contacting biomaterials, and the specificity of the β6-20 peptide demonstrated in vitro indicates the potential for using this peptide to specifically deliver Nanogold particles to biomaterial-adherent bacteria. By replacing the Nanogold label with vancomycin, the covalently tethered antibiotic can be used for localized treatment with a higher retention rate within the biofilm. Significant improvements in initial binding of vancomycin to S. epidermidis were obtained utilizing this targeted vancomycin scheme, and 27–34% improvements in retention rates were achieved. If successfully developed, a similar technique could be applied to target and treat infections of other bacterial species.
Acknowledgments
The project was supported by Award Number 5R01EB000279 from the National Institute of Biomedical Imaging and Bioengineering.
References
- 1.Darouiche Rabih AO. Device-Associated Infections: A Macroproblem that Starts with Microadherence. Clinical Infectious Diseases. 2001;33(9):1567–1572. doi: 10.1086/323130. [DOI] [PubMed] [Google Scholar]
- 2.Schierholz JM, Beuth J. Implant infections: a haven for opportunistic bacteria. Journal of Hospital Infection. 2001;49(2):87–93. doi: 10.1053/jhin.2001.1052. [DOI] [PubMed] [Google Scholar]
- 3.Vuong C, Otto M. Staphylococcus epidermidis infections. Microbes Infect. 2002;4(4):481–9. doi: 10.1016/s1286-4579(02)01563-0. [DOI] [PubMed] [Google Scholar]
- 4.Bale MD, Wohlfahrt LA, Mosher DF, Tomasini B, Sutton RC. Identification of Vitronectin as a Major Plasma-Protein Adsorbed on Polymer Surfaces of Different Copolymer Composition. Blood. 1989;74(8):2698–2706. [PubMed] [Google Scholar]
- 5.Chugh TD, Burns GJ, Shuhaiber HJ, Bahr GM. Adherence Of Staphylococcus-Epidermidis To Fibrin-Platelet Clots Invitro Mediated By Lipoteichoic Acid. Infection And Immunity. 1990;58(2):315–319. doi: 10.1128/iai.58.2.315-319.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heilmann C, Hussain M, Peters G, Gotz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Molecular Microbiology. 1997;24(5):1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 7.Hussain M, Heilmann C, Peters G, Herrmann M. Teichoic acid enhances adhesion of Staphylococcus epidermidis to immobilized fibronectin. Microbial Pathogenesis. 2001;31(6):261–270. doi: 10.1006/mpat.2001.0469. [DOI] [PubMed] [Google Scholar]
- 8.Li DQ, Lundberg F, Ljungh A. Characterization of vitronectin-binding proteins of Staphylococcus epidermidis. Current Microbiology. 2001;42(5):361–367. doi: 10.1007/s002840010230. [DOI] [PubMed] [Google Scholar]
- 9.Nilsson M, Frykberg L, Flock JI, Pei L, Lindberg M, Guss B. A fibrinogen-binding protein of Staphylococcus epidermidis. Infection and Immunity. 1998;66(6):2666–2673. doi: 10.1128/iai.66.6.2666-2673.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pei L, Arvholm IL, Lonnies L, Flock JI. GST-F be can recognize beta-chains of fibrin(ogen) on explanted materials. Journal of Chromatography B-Analytical Technologies in the Biomedical and Life Sciences. 2003;786(1–2):319–325. doi: 10.1016/s1570-0232(02)00744-4. [DOI] [PubMed] [Google Scholar]
- 11.Pei L, Palma M, Nilsson M, Guss B, Flock JI. Functional studies of a fibrinogen binding protein from Staphylococcus epidermidis. Infection and Immunity. 1999;67(9):4525–4530. doi: 10.1128/iai.67.9.4525-4530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams RJ, Henderson B, Sharp LJ, Nair SP. Identification of a fibronectin-binding protein from Staphylococcus epidermidis. Infection And Immunity. 2002;70(12):6805–6810. doi: 10.1128/IAI.70.12.6805-6810.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patti JM, Allen BL, Mcgavin MJ, Hook M. MSCRAMM-Mediated Adherence of Microorganisms to Host Tissues. Annual Review of Microbiology. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 14.Patti JM, Hook M. Microbial Adhesins Recognizing Extracellular-Matrix Macromolecules. Current Opinion In Cell Biology. 1994;6(5):752–758. doi: 10.1016/0955-0674(94)90104-x. [DOI] [PubMed] [Google Scholar]
- 15.Davis SL, Gurusiddappa S, McCrea KW, Perkins S, Hook M. SdrG, a fibrinogen-binding bacterial adhesin of the microbial surface components recognizing adhesive matrix molecules subfamily from Staphylococcus epidermidis, targets the thrombin cleavage site in the B beta chain. Journal of Biological Chemistry. 2001;276(30):27799–27805. doi: 10.1074/jbc.M103873200. [DOI] [PubMed] [Google Scholar]
- 16.Ponnuraj K, Bowden MG, Davis S, Gurusiddappa S, Moore D, Choe D, Xu Y, Hook M, Narayana SVL. A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell. 2003;115(2):217–228. doi: 10.1016/s0092-8674(03)00809-2. [DOI] [PubMed] [Google Scholar]
- 17.Donlan RM. Preventing biofilms of clinically relevant organisms using bacteriophage. Trends in microbiology. 2009;17(2):66–72. doi: 10.1016/j.tim.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Bloom B, Schelonka R, Kueser T, Walker W, Jung E, Kaufman D, Kesler K, Roberson D, Patti J, Hetherington S. Multicenter study to assess safety and efficacy of INH-A21, a donor-selected human staphylococcal immunoglobulin, for prevention of nosocomial infections in very low birth weight infants. The Pediatric infectious disease journal. 2005;24(10):858. doi: 10.1097/01.inf.0000180504.66437.1f. [DOI] [PubMed] [Google Scholar]
- 19.Ohlsen K, Lorenz U. Immunotherapeutic strategies to combat staphylococcal infections. International Journal of Medical Microbiology. 2010;300(6):402–410. doi: 10.1016/j.ijmm.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 20.Rennermalm A, Nilsson M, Flock JI. Fibrinogen binding protein of Staphylococcus epidermidis is a target for opsonic antibodies. Infection and Immunity. 2004;72(5):3081–3083. doi: 10.1128/IAI.72.5.3081-3083.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vernachio JH, Bayer AS, Ames B, Bryant D, Prater BD, Syribeys PJ, Gorovits EL, Patti JM. Human immunoglobulin G recognizing fibrinogen-binding surface proteins is protective against both Staphylococcus aureus and Staphylococcus epidermidis infections in vivo. Antimicrobial Agents and Chemotherapy. 2006;50(2):511–518. doi: 10.1128/AAC.50.2.511-518.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watanakunakorn C. The antibacterial action of vancomycin. Rev Infect Dis. 1981;3 (suppl):S210–5. [PubMed] [Google Scholar]
- 23.Arimoto H, Nishimura K, Kiniumi T, Hayakawa I, Uemura D. Multi-valent polymer of vancomycin - enhanced antibacterial activity against VRE. Chem Commun. 1999;15:1361–1362. [Google Scholar]
- 24.Williams DH. The glycopeptide story--how to kill the deadly ‘superbugs’. Nat Prod Rep. 1996;13(6):469–77. doi: 10.1039/np9961300469. [DOI] [PubMed] [Google Scholar]
- 25.Beauregard DA, Williams DH, Gwynn MN, Knowles DJ. Dimerization and membrane anchors in extracellular targeting of vancomycin group antibiotics. Antimicrob Agents Chemother. 1995;39(3):781–785. doi: 10.1128/AAC.39.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fields G, Noble R. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int J Pept Protein Res. 1990;35(3):161–214. doi: 10.1111/j.1399-3011.1990.tb00939.x. [DOI] [PubMed] [Google Scholar]
- 27.Greenwald RB, Zhao H, Xia J, Martinez A. Poly (ethylene glycol) transport forms of vancomycin: a long-lived continuous release delivery system. Journal of medicinal chemistry. 2003;46(23):5021–5030. doi: 10.1021/jm030202g. [DOI] [PubMed] [Google Scholar]
- 28.Abramoff M, Magelhaes P, Ram S. Image Processing with ImageJ. Biophotonics International. 2004;11(7):36–42. [Google Scholar]
- 29.Hartford O, O’Brien L, Schofield K, Wells J, Foster TJ. The Fbe (SdrG) protein of Staphylococcus epidermidis HB promotes bacterial adherence to fibrinogen. Microbiology-Sgm. 2001;147:2545–2552. doi: 10.1099/00221287-147-9-2545. [DOI] [PubMed] [Google Scholar]
- 30.Pei L, Flock JI. Lack of fbe, the gene for a fibrinogen-binding protein from Staphylococcus epidermidis, reduces its adherence to fibrinogen coated surfaces. Microbial Pathogenesis. 2001;31(4):185–193. doi: 10.1006/mpat.2001.0462. [DOI] [PubMed] [Google Scholar]
- 31.Bowden MG, Chen W, Singvall J, Xu Y, Peacock SJ, Valtulina V, Speziale P, Hook M. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology-Sgm. 2005;151:1453–1464. doi: 10.1099/mic.0.27534-0. [DOI] [PubMed] [Google Scholar]
- 32.McCrea KW, Hartford O, Davis S, Eidhin DN, Lina G, Speziale P, Foster TJ, Hook M. The serine-aspartate repeat (Sdr) protein family in Staphylococcus epidermidis. Microbiology-Uk. 2000;146:1535–1546. doi: 10.1099/00221287-146-7-1535. [DOI] [PubMed] [Google Scholar]
- 33.Guo BN, Zhao X, Shi YG, Zhu DM, Zhang YY. Pathogenic implication of a fibrinogen-binding protein of Staphylococcus epidermidis in a rat model of intravascular-catheter-associated infection. Infection and Immunity. 2007;75(6):2991–2995. doi: 10.1128/IAI.01741-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Queck SY, Otto M. Staphylococcus epidermidis and other Coagulase-Negative Staphylococci. In: Lindsay JA, editor. Staphylococcus Molecular Genetics. Norfolk, UK: Caister Academic Press; 2008. [Google Scholar]
- 35.Cerca N, Jefferson KK, Maira-Litran T, Pier DB, Kelly-Quintos C, Goldmann DA, Azeredo J, Pier GB. Molecular basis for preferential protective efficacy of antibodies directed to the poorly acetylated form of staphylococcal poly-N-acetyl-beta-(1-6)-glucosaminev. Infection and Immunity. 2007;75(7):3406–3413. doi: 10.1128/IAI.00078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. The Calgary Biofilm Device: New Technology for Rapid Determination of Antibiotic Susceptibilities of Bacterial Biofilms. J Clin Microbiol. 1999;37(6):1771–1776. doi: 10.1128/jcm.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olson M, Ceri H, Morck D, Buret A, Read R. Biofilm bacteria: formation and comparative susceptibility to antibiotics. Can J Vet Res. 2002;66(2):86–92. [PMC free article] [PubMed] [Google Scholar]
- 38.Darouiche RO, Dhir A, Miller AJ, Landon GC, Raad II, Musher DM. Vancomycin penetration into biofilm covering infected prostheses and effect on bacteria. J Infect Dis. 1994;170(3):720–3. doi: 10.1093/infdis/170.3.720. [DOI] [PubMed] [Google Scholar]
- 39.Dunne WM, Mason E, Kaplan S. Diffusion of Rifampin and Vancomycin through a Staphylococcus epidermidis Biofilm. Antimicrobial Agents and Chemotherapy. 1993;37(12):2522–2526. doi: 10.1128/aac.37.12.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griffin JH, Linsell MS, Nodwell MB, Chen Q, Pace JL, Quast KL, Krause KM, Farrington L, Wu TX, Higgins DL, et al. Multivalent Drug Design. Synthesis and In Vitro Analysis of an Array of Vancomycin Dimers. Journal of the American Chemical Society. 2003;125(21):6517–6531. doi: 10.1021/ja021273s. [DOI] [PubMed] [Google Scholar]
- 41.Jose B, Antoci V, Jr, Zeiger AR, Wickstrom E, Hickok NJ. Vancomycin Covalently Bonded to Titanium Beads Kills Staphylococcus aureus. Chemistry & Biology. 2005;12(9):1041–1048. doi: 10.1016/j.chembiol.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 42.Westwell MS, Bardsley B, Dancer RJ, Try AC, Williams DH. Cooperativity in ligand binding expressed at a model cell membrane by the vancomycin group antibiotics. Chemical Communications. 1996;5:589–590. [Google Scholar]