Abstract
Intrinsic stress response pathways are frequently mobilized within tumor cells. The mediators of these adaptive mechanisms and how they contribute to carcinogenesis remain poorly understood. A striking example is heat shock factor 1 (HSF1), master transcriptional regulator of the heat shock response. Surprisingly, we found that loss of the tumor suppressor gene neurofibromatosis type 1 (Nf1) increased HSF1 levels and triggered its activation in mouse embryonic fibroblasts. As a consequence, Nf1–/– cells acquired tolerance to proteotoxic stress. This activation of HSF1 depended on dysregulated MAPK signaling. HSF1, in turn, supported MAPK signaling. In mice, Hsf1 deficiency impeded NF1-associated carcinogenesis by attenuating oncogenic RAS/MAPK signaling. In cell lines from human malignant peripheral nerve sheath tumors (MPNSTs) driven by NF1 loss, HSF1 was overexpressed and activated, which was required for tumor cell viability. In surgical resections of human MPNSTs, HSF1 was overexpressed, translocated to the nucleus, and phosphorylated. These findings reveal a surprising biological consequence of NF1 deficiency: activation of HSF1 and ensuing addiction to this master regulator of the heat shock response. The loss of NF1 function engages an evolutionarily conserved cellular survival mechanism that ultimately impairs survival of the whole organism by facilitating carcinogenesis.
Introduction
Evolutionarily conserved from yeasts to humans, the heat shock transcription factor heat shock factor 1 (HSF1) is activated by a broad range of stressors that extend far beyond heat, including heavy metals, UV radiation, hypoxia, desiccation, and acidosis (1). During activation, HSF1 undergoes phosphorylation and other posttranslational modifications, trimerization, and nuclear translocation. This results in rapid, high-affinity binding of HSF1 to consensus heat shock elements (HSEs) within the promoters of target genes (2). Such binding drives the induction or suppression of hundreds of genes in mammalian cells (3).
The adaptive response unleashed by HSF1 activation is critical for maintaining homeostasis of the cell’s proteome, mediated in large part by increased expression of classical heat shock proteins such as HSP27, HSP70, and HSP90 (4). However, the effect of HSF1 activation goes far beyond these chaperones. It helps coordinate a range of fundamental cellular processes that are important to the fitness of malignant cells, including cell cycle control, ribosome biogenesis, protein translation, and glucose metabolism (5, 6). As a result, HSF1 both facilitates initial oncogenic transformation and maintains the malignant phenotype of established cancer cell lines driven by a wide range of mutations. In mice and in cell culture, genetic ablation of Hsf1 expression potently impairs tumorigenesis and cellular transformation driven by oncogene activation or tumor suppressor loss (5). The importance of HSF1 in enabling malignancy has been demonstrated by other recent work as well (7–9). Given the importance of HSF1 in diverse survival mechanisms and cancer, we sought here to investigate factors that might increase its transcriptional activity during tumorigenesis.
Neurofibromatosis type 1 (NF1) is a common hereditary cancer syndrome associated with dysregulation of RAS activity (10). Tumor predisposition results from loss-of-function mutations in NF1, which encodes neurofibromin, a 280-kDa protein possessing a central domain with Ras GTPase activating protein (GAP) activity (11). Consistent with neurofibromin functioning as a negative regulator of RAS, NF1-deficient cells are hyperresponsive to several growth factors that signal via the RAS/MAPK pathway (12). Indeed, when overexpressed, the GAP-related domain (NF1-GRD) can downregulate RAS-mediated mitogenic signaling (13).
Unexpectedly, we found that the tumor suppressor Nf1 was a strong regulator of HSF1 levels and activity. Indeed, genetic compromise of Nf1 was alone sufficient to cause immediate activation of HSF1 in cell culture. HSF1 levels were also increased in tumor samples removed directly from NF1 patients. Having found that Nf1 regulated HSF1 function, we investigated the underlying mechanisms of this control as well as its functional consequences. We demonstrate that HSF1 was embedded in the dysregulated MAPK/RAS signaling characteristic of NF1 and participated in a feed-forward loop that facilitated tumorigenesis in mice and humans. These findings suggest HSF1 as a potentially clinically relevant modifier of NF1-associated tumorigenesis.
Results
Nf1 deficiency upregulates HSF1 and activates the heat shock response.
Given the importance of HSF1 in supporting tumorigenesis, we sought to identify genes that might modulate HSF1 function and the transcriptional response that it regulates. We screened a focused library of lentiviral inhibitory shRNAs targeting 175 genes that were selected based on (a) Gene Ontology or Panther annotation under the categories of heat shock, stress response, sensitivity to stress, protein folding, or protein degradation and (b) iHOP interactome data indicating direct interaction with HSF1 (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI62727DS1). NF1 was added to the list because a deletion mutation of its yeast homolog reduces thermotolerance in that organism (14).
HSF1 activation was monitored using NIH3T3 cells stably engineered to express an eGFP–firefly luciferase fusion controlled by a highly HSF1-dependent promoter (15). Luminescence per well was recorded 5 days after lentiviral transduction. As expected, shRNAs targeting Hsp27/Hspb2, Hsp47/Serpinh1, α(B)-crystallin/Cryab, and several Hsp40 family members (Dnajb6 and Dnajc17) strongly activated the heat shock reporter (Figure 1A). shRNAs targeting individual Hsp90 and Hsp70 isoforms resulted in only weak reporter induction, most likely because shRNA targeting of individual isoforms may be insufficient to impair function of the many isoforms comprising the HSP70 and HSP90 class of chaperones. Surprisingly, 2 independent Nf1-targeted shRNAs were among the strongest activators.
Figure 1. Genetic compromise of NF1 induces the heat shock response.
(A) Whole cell–based screen of a heat shock–oriented shRNA library. Heat shock reporter NIH3T3 cells arrayed in a 384-well format were infected with lentiviral constructs designed to target 175 candidate genes. 5 days later, reporter activation was measured by luciferase assay. The mean Z score calculated from 2 biological replicates per construct is plotted as a measure of activation relative to the entire population of wells assayed. 2 independent Nf1-targeted shRNAs that strongly activate the heat shock reporter are highlighted in bold. (B) Nf1 knockdown increased HSP72 expression in an immortalized MEF cell line. MEFs were stably transduced with shRNA39 and shRNA42, which target different regions of the Nf1 mRNA, and a scrambled control shRNA (Scr). NF1 and HSP72 protein levels were examined by immunoblotting. (C) Nf1 knockdown transcriptionally activates the heat shock response. Transcript levels of chaperones in Nf1-knockdown MEFs were measured by 2-step real-time quantitative RT-PCR technique. Transcript levels relative to cells transduced with scrambled control shRNA for each gene are expressed as fold changes (mean ± SD, n = 3 or 4). *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t test.
Next, we examined the effect of stable Nf1 knockdown on the activation of HSF1 in a mouse embryonic fibroblast (MEF) cell line immortalized with the HPV16 E6/E7 oncoproteins (7). Compared to a scrambled control shRNA, 2 independent shRNAs (referred to herein as shRNA39 and shRNA42) efficiently suppressed neurofibromin protein levels and induced an increase in the level of the heat shock protein HSP72, a classic HSF1 target (Figure 1B). The shRNA that suppressed neurofibromin expression most efficiently, shRNA42, also induced the strongest expression of HSP72. We further examined transcript levels of HSF1 target genes by real-time quantitative RT-PCR analysis in these Nf1-knockdown cells. Compared with the control, knockdown of Nf1 by both shRNA39 and shRNA42 increased transcript levels of a collection of Hsp genes, including Hsp70s (Hsp72/Hspa1a and Hspa4l), Hsp40s (Dnajb1 and Dnajb2), Hsp90 (Hsp90aa1), and a small Hsp family member (Hspb8) (Figure 1C). shRNA42 induced the strongest expression of these Hsp genes. These results demonstrate that reducing Nf1 expression can activate the heat shock response in premalignant cells.
To measure the effect of Nf1-targeted shRNAs on HSF1 protein levels, we immunoblotted lysates from the same stably transduced Nf1-knockdown cells (Figure 2, A and B). HSF1 normally shuttles between the nucleus and cytoplasm, but differential residence time in these compartments results in predominantly cytoplasmic localization under basal conditions. Upon activation, however, HSF1 accumulated in the nucleus, where a portion became tightly bound to DNA (Supplemental Figure 2A). Both shRNA39 and shRNA42 increased HSF1 levels and translocation of HSF1 to the nucleus compared with the control shRNA (Figure 2, A and B). They also induced phosphorylation of HSF1 at Ser326 (referred to herein as p-Ser326; Figure 2, A and B), a modification strongly associated with HSF1 activation in vivo by thermal and chemical stressors (16). The kinases or signaling pathways responsible for this key modification has yet to be defined, but consistent with activation of HSF1 by stable Nf1 knockdown, HSP72 protein levels were elevated (Figure 2B). The transcript level for Hsf1 was not altered by Nf1 knockdown (Supplemental Figure 2B), suggestive of posttranscriptional regulation of HSF1 levels.
Figure 2. NF1 loss activates HSF1 via elevated MAPK signaling.
(A and B) Cytoplasmic and nuclear fractions were prepared from immortalized MEFs after overnight treatment with DMSO or 10 μM U0126. Blotting for cytoplasmic lactate dehydrogenase (LDH) and nuclear lamins A/C confirmed appropriate fractionation. (C and D) HSF1 was activated in primary Nf1-knockout MEFs. (E) MEK inhibition impaired HSF1 transcriptional activity. EGFP or HSF1 plasmids were transfected with pHSE–firefly luciferase reporter plasmid and pCMV–renilla luciferase plasmid into HEK293T cells. After 1 day, cells were treated with DMSO or 20 μM U0126 overnight. Firefly luciferase signals were normalized against renilla luciferase signals (mean ± SD; n = 6). (F) Dominant-negative MEK1 impaired p-Ser326. EGFP or MEK1AA plasmid was transfected into HEK293T cells; after 3 days, cells were harvested for immunoblotting. (G) Dominant-negative MEK1 impaired HSF1 transcriptional activity. In HEK293T cells, EGFP or MEK1AA plasmid was transfected with the luciferase reporter plasmids. HSF1 plasmid was further cotransfected with either EGFP or MEK1AA plasmid. 3 days after transfection, luciferase signals were measured (mean ± SD; n = 4). (H) Proteasomal inhibition caused HSF1 protein accumulation. MEFs were treated with DMSO or MG132 overnight. (I) HSF1 polyubiquitination was suppressed in Nf1-knockdown cells and reestablished after MEK inhibition. MEFs were treated with either DMSO or 20 μM U0126 overnight, and whole cell lysates were immunoprecipitated for HSF1. Normal rat IgG served as the control. Precipitates were immunoblotted for polyubiquitinated conjugates and HSF1. LC, light chain; WLC, whole cell lysates. P values were determined by Student’s t test.
To confirm the effects demonstrated by shRNA knockdown experiments, we further tested the effect of Nf1 deficiency using primary MEFs in which the Nf1 locus was disrupted by gene targeting. Primary cultures derived from both Nf1+/– and Nf1–/– embryos had increased HSF1 protein levels compared with their Nf1+/+ counterparts (Figure 2, C and D). Nf1–/– cells displayed even higher levels of HSF1 protein and greater induction of HSP72 than Nf1+/– cells (Figure 2, C and D). Nf1–/– cells also had an increase in both nuclear HSF1 and the activating p-Ser326 modification than Nf1+/+ cells (Figure 2D).
Dysregulated MAPK signaling mediates HSF1 activation.
How might Nf1 loss increase p-Ser326, nuclear accumulation of HSF1, and total HSF1 levels? A hallmark of the NF1 phenotype is dysregulation of RAS activation with downstream effects on the MAPK/ERK signaling cascade. Hence, Nf1 loss could activate HSF1 through the MAPK pathway. We tested this possibility using U0126, a potent and selective inhibitor of MEK1 and MEK2, the immediate upstream activators of ERK1 and ERK2. U0126 reduced activation-associated p-HSF1 as well as nuclear accumulation of HSF1, leading to a decline in HSP72 levels in both control and Nf1-knockdown cells (Figure 2B). Interestingly, U0126 had a greater effect on Nf1-knockdown cells than on control cells (Supplemental Figure 2C). The effects of U0126 on primary Nf1–/– MEFs were similar to those seen on knockdown cells (Figure 2D and Supplemental Figure 2D). The compound also caused an increase in p-HSF1 and HSP72 levels in primary Nf1+/+ MEFs (Figure 2D). This likely reflects a biological difference between immortalized and primary cells. Either blocking this trophic signaling pathway is stressful to primary Nf1+/+ cells, or basal MAPK activities actually repress HSF1 activation in these cells. In any case, the data in Nf1-deficient cells indicate that hyperactivated MAPK signaling contributes to HSF1 Ser326 phosphorylation, nuclear translocation, and activation.
Next, we used a heat shock reporter assay to examine the effect of dysregulated MAPK signaling on HSF1 transcriptional activity in HEK293T cells. In the absence of environmental insults, the transfected reporter demonstrated a modest level of activation that was further enhanced by overexpression of HSF1 (Figure 2E). Inhibition of endogenous MEK activity by U0126 significantly suppressed both basal and HSF1-driven reporter activity (Figure 2E). To further validate the role of hyperactive MAPK signaling in HSF1 phosphorylation and activation, we transiently transfected HEK293T cells with a dominant-negative MEK1 mutant in which 2 key serine residues — Ser218 and Ser222 — are substituted with alanine (referred to herein as MEK1AA; ref. 17). In agreement with our U0126 data, expression of MEK1AA impaired p-Ser326 as well as HSF1-mediated reporter activation (Figure 2, F and G). Although significant, mutant MEK1 reduced reporter activity less than U0126. This is likely due to both the limited efficiency of HSF1 and MEK1AA cotransfection and the potential for MEK2 compensatory effects. Together, these data indicate that MEK hyperactivation is important for p-Ser326 and HSF1 Ser326 activation.
Does MAPK signaling also underlie the increase in total HSF1 protein? Because HSF1 mRNA is not increased in NF1-deficient cells, we investigated posttranscriptional mechanisms that might contribute to increasing HSF1 protein after NF1 loss. Interestingly, HSF1 levels in scrambled shRNA control cells increased more than 2-fold after proteasome inhibition with MG132, but there was only subtle change in HSF1 levels in cells with shRNA42-mediated Nf1 knockdown (Figure 2H and Supplemental Figure 2E). This finding suggested that HSF1 is a substrate for the proteasome, but that its degradation is impaired in Nf1-deficient cells.
To further explore the impaired degradation of HSF1 in Nf1-deficient cells, and to establish a link to MAPK signaling, we immunoprecipitated polyubiquitinated HSF1 from control and Nf1-knockdown cells (18). While polyubiquitinated HSF1 was readily detected in control cells, HSF1 polyubiquitination was markedly diminished in cells with shRNA42-mediated Nf1 knockdown (Figure 2I). Strikingly, inhibiting MEK with U0126 reestablished polyubiquitination of HSF1 (Figure 2I). Thus, MEK hyperactivation also regulated HSF1 polyubiquitination and proteasomal degradation.
HSF1 activation by Nf1 loss induces stress tolerance.
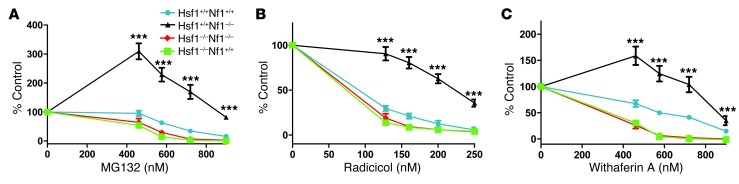
Next we asked whether loss of Nf1 alone, and subsequent activation of HSF1, is sufficient to provide cells with a survival advantage in the face of increased levels of proteotoxic stress. To test this, we generated primary MEFs from mice carrying targeted Hsf1 or Nf1 knockouts. Because the Nf1–/– genotype is embryonic lethal (19) and Hsf1–/– mice have reproductive defects (20), it was necessary to screen more than 120 embryos in order to isolate MEFs with 4 distinct genotypes: Hsf1+/+Nf1+/+, Hsf1+/+Nf1–/–, Hsf1–/–Nf1+/+, and Hsf1–/–Nf1–/–. We then measured the changes in cell number in the presence of 3 chemically and mechanistically distinct cellular stressors: the proteasome inhibitor MG132 (Figure 3A), the HSP90 inhibitor radicicol (Figure 3B), and the thiol-reactive compound withaferin A (Figure 3C and refs. 21, 22). Unlike the situation in yeast, in which deletion of the Nf1 homolog IRA2 reduces thermotolerance, Hsf1+/+Nf1–/– MEFs were significantly more resistant to all 3 proteotoxic stressors than were Hsf1+/+Nf1+/+ MEFs (Figure 3, A–C). In fact, Hsf1+/+Nf1–/– MEFs showed an increase in cell number at low concentrations of MG132 and withaferin A. In contrast, disrupting Nf1 in Hsf1–/– cells did not confer resistance to the stressors (Figure 3, A–C). Thus, the tolerance of Nf1-deficient cells to proteotoxic stress was mediated by Hsf1.
Figure 3. HSF1 activation by Nf1 knockout renders cells resistant to proteotoxic stress.
Primary MEFs of the indicated genotypes were plated at low density (2,000 cells/well) and exposed for 5 days to the indicated concentrations of (A) MG132, (B) radicicol, and (C) withaferin A. Resazurin dye reduction was assayed as a measure of relative viable cell number. The mean of triplicate determinations repeated in 2 separate experiments is presented (mean ± SD). ***P < 0.01, 2-way ANOVA.
Hsf1 compromise suppresses tumorigenesis in mice.
Because tumorigenesis imposes diverse stresses on the physiology of cells, we wondered whether HSF1 activation and the ensuing acquisition of tolerance to proteotoxic stress could facilitate NF1-associated tumorigenesis in vivo. To investigate this, we used NPcis mice. The tumor predisposition syndrome caused by NF1 loss in humans has proven difficult to faithfully model in mice. Homozygous germline disruption of Nf1 is embryonic lethal, predominantly due to defects in cardiovascular system development (23). Heterozygous mice display modest tumor predisposition, but they do not develop the classic spectrum of lesions characteristic of the human disease, especially neurofibroma and malignant peripheral nerve sheath tumors (MPNSTs; ref. 24). The observation that the tumor suppressor TP53 is frequently lost in human MPNSTs prompted development of mouse models in which alleles of the closely linked Trp53 and Nf1 genes are disrupted on the same chromosome (25, 26). The chromosome bearing deletion of both Nf1 and Trp53 in cis is designated NPcis. NPcis+/– heterozygotes develop soft tissue sarcomas resembling human MPNSTs with high frequency. Due to the close proximity of the Trp53 and Nf1 genes (in humans as well as in mice), such tumors often demonstrate loss of heterozygosity for the remaining wild-type alleles of both genes.
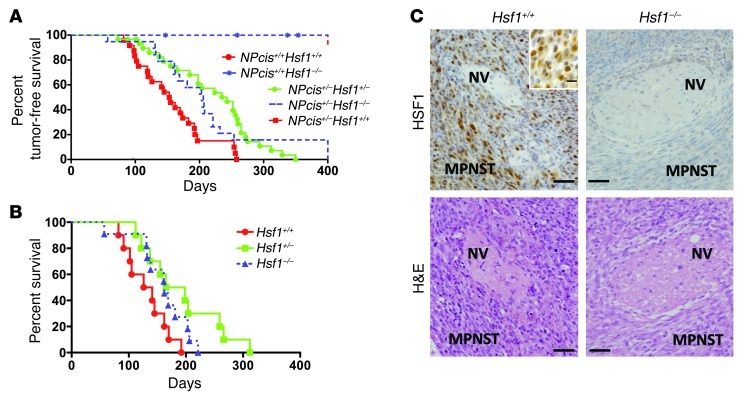
To examine the effect of HSF1 in this model, we crossed NPcis+/– mice with Hsf1+/– mice. NPcis+/–Hsf1+/– offspring was intercrossed to produce NPcis+/+Hsf1+/+, NPcis+/+Hsf1–/–, NPcis+/–Hsf1+/–, NPcis+/–Hsf1–/–, and NPcis+/–Hsf1+/+ genotypes. The NPcis+/+Hsf1+/+ and NPcis+/+Hsf1–/– genotypes served as controls for the effect of Hsf1 knockout on otherwise wild-type mice. As expected, these control mice had long-term tumor-free survival (Figure 4A). Genotypes NPcis+/–Hsf1+/–, NPcis+/–Hsf1–/–, and NPcis+/–Hsf1+/+ were tested for the effect of Hsf1 dosage on tumorigenesis in the NPcis background. All NPcis mice with wild-type Hsf1 developed tumors by about 250 days, with median tumor-free survival of 154 days (Figure 4A). In contrast, NPcis mice with hemizygous or homozygous Hsf1 knockout showed significantly prolonged tumor-free survival, with medians of 235 and 206 days, respectively.
Figure 4. Genetic compromise of Hsf1 prolongs survival in a mouse model of NF1.
(A) Tumor occurrence was suppressed in NPcis mice when Hsf1 was disrupted. Tumor-free survival in relationship to Hsf1 genotype is plotted for NPcis+/+ and NPcis+/– mice. Median survival for NPcis mice was as follows: Hsf1+/+, 22 weeks (n = 24); Hsf1+/–, 34 weeks (n = 30; P = 0.0002 vs. Hsf1+/+, log-rank test); Hsf1–/–, 29 weeks (n = 19; P = 0.028 vs. Hsf1+/+, log-rank test). (B) Representative micrographs of serial sections from MPNSTs arising in NPcis mice were immunostained for HSF1, showing increased levels in tumor, but not adjacent normal nerve (NV). The inset shows nuclear HSF1 staining in tumors. H&E staining is shown to demonstrate histology and provide orientation. Scale bars: 100 μm; 10 μm (inset). (C) Survival time of NPcis mice that developed MPNSTs only was prolonged by Hsf1 compromise. Median survival was as follows: Hsf1+/+, 19 weeks; Hsf1+/–, 23.5 weeks (P = 0.0152 vs. Hsf1+/+, log-rank test); Hsf1–/–, 23 weeks (P = 0.0869 vs. Hsf1+/+, log-rank test).
Conversely, the difference in tumor-free survival between Hsf1–/– and Hsf1+/– knockouts was not statistically significant, in contrast to our previous results in mice carrying a hotspot mutation in Trp53. In that case, Hsf1+/– mice also had improved outcome compared with Hsf1+/+ mice, but Hsf1–/– mice survived even longer. While being a prominent longevity factor in nematodes (27, 28), the influence of HSF1 on mammalian lifespan has been little explored. In some strains, Hsf1 compromise has no effect on lifespan (5, 29). However, we found that in the mixed background used for our NPcis experiments, Hsf1 knockouts had a shorter lifespan than Hsf1+/+ controls. This effect was evident in the depiction of overall survival shown in Supplemental Figure 3A, which includes deaths from all causes, not solely tumor-related deaths, as used for analysis of tumor-free survival (Figure 4A). Thus, the additional survival advantage that might have been conferred on tumor-associated mortality by loss of the second Hsf1 allele was likely masked by general effects of Hsf1 on lifespan in the NPcis strain.
To characterize the tumors arising in NPcis mice, we first examined their HSF1 status. In the same sections, MPNST cells had much stronger HSF1 immunoreactivity than did adjacent normal tissues, such as nerve (Figure 4B). The absence of reactivity in tumors arising in Hsf1–/– mice confirmed the specificity of the HSF1 staining.
NF1 patients can develop a broad range of tumor types, most of which also occur in NPcis mice (Supplemental Figure 4). To capture all tumor types arising in this model, we performed a full necropsy on each euthanized animal, and all major tissues were harvested for histological examination. In addition to prolonging tumor-free survival, Hsf1 deficiency altered the distribution of tumor types arising in this model. Compared with Hsf1+/+ mice, the incidence of lymphoma, neuroblastoma, angiosarcoma, osteosarcoma, pheochromocytoma, and histocytic sarcoma were all reduced in Hsf1–/– mice (Supplemental Figure 3B). Surprisingly, the overall incidence of MPNSTs and gliomas was not significantly different. Excluding gliomas and MPNSTs, however, tumor incidence was significantly lower in Hsf1–/– (5.9%) and Hsf1+/– (26.9%) mice compared with Hsf1+/+ mice (60.9%). The MPNSTs arising in Hsf1+/+, Hsf1+/–, and Hsf1–/– tumors appeared histologically similar.
Although MPNST incidence did not differ, Hsf1 deficiency did prolong survival time in mice that only developed MPNSTs (Figure 4C). This effect was statistically significant in Hsf1 hemizygous mice (P = 0.0152), and there was a trend toward significance in Hsf1–/– mice (P = 0.0869). Given the late-occurring, slowly progressing behavior of glioma in this NPcis model, the apparent increase in glioma incidence in Hsf1-deficient mice is likely due simply to their prolonged survival. Indeed, there was a strong positive correlation between glioma incidence and median survival across Hsf1 genotypes (Supplemental Figure 3C).
To circumvent placental defects associated with the Hsf1-null state in highly inbred genetic backgrounds (20), Hsf1 mice were maintained on a mixed genetic background (129/SvJ and Balb/cJ). Previously, 2 genetic loci mapping to chromosomes 15 and 19, as well as sex-specific effects, have been reported to modify tumorigenesis in the NPcis model (30, 31). To address potential confounding effects of these modifiers, we examined the correlation of mapped genetic loci or sex with animal survival (Supplemental Tables 1 and 2). Neither the states of these loci nor sex demonstrated a significant effect on survival (Supplemental Figure 5). Taken together, our data established Hsf1 as a modifier of NF1-associated tumorigenesis in this mouse model.
HSF1 supports MAPK/ERK signaling in Nf1-deficient cells.
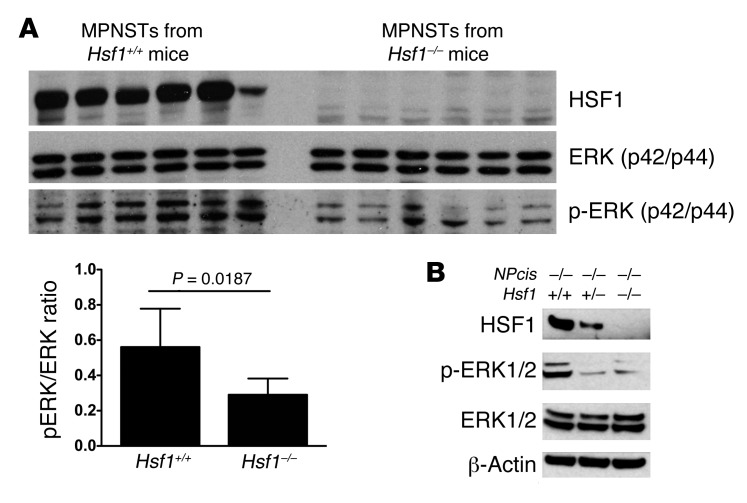
In light of the importance of dysregulated MAPK signaling we observed in cell culture, we next investigated the effect of Hsf1 loss on the MAPK/ERK pathway in tumors arising spontaneously in NPcis mice. All MPNSTs demonstrated comparable levels of total ERK protein, but those arising in Hsf1–/– mice had significantly lower levels of ERK phosphorylation (p-ERK) compared with MPNSTs in Hsf1+/+ mice (Figure 5A). This finding is consistent with our previous observations, in other contexts, that HSF1 supports transformation and tumorigenesis by oncogenic RAS (7).
Figure 5. Hsf1 deficiency attenuates MAPK signaling in NPcis mice.
(A) Immunoblotting demonstrated equivalent levels of total ERK, but less activation-associated p-ERK, in MPNSTs arising in Hsf1–/– mice. Densitometric quantitation of immunoblots confirmed a statistically significant difference in mean ratio of p-ERK to total ERK (Student’s t test). (B) Basal p-ERK was diminished in NPcis–/– MEFs derived from Hsf1+/– and Hsf1–/– embryos.
Strikingly, HSF1 supported MAPK signaling not only in Nf1-deficient tumors, but also in primary MEFs. In the homozygous NPcis mutant background, Hsf1+/– or Hsf1–/– primary MEFs showed p-ERK that was much reduced compared with their Hsf1+/+ counterparts (Figure 5B). Reduced MAPK signaling in both tumors and primary cells that were Hsf1 deficient demonstrated a critical role for HSF1 in supporting activity of the RAS/MAPK pathway. Taken together with data from Figure 2, these findings indicate that loss of Nf1 activates HSF1, at least in part, via activation of the MAPK/ERK pathway, and that HSF1 in turn is necessary to support MAPK signaling.
HSF1 is activated in NF1-deficient human tumors.
Results in mouse models do not always reflect the pathophysiology of human disease. Does a pathobiology comparable to that seen in the NPcis mouse model operate in humans? To address this question, we first examined HSF1 activation status in a panel of 5 human cell lines derived from patients clinically diagnosed with NF1. These lines express Schwann cell markers, such as S100β and p75 neurotrophin receptor, consistent with Schwann cell origin (32). 3 cell lines (sNF96.2, 90-8TL, and S462) were devoid of neurofibromin, consistent with their reported NF1 mutation status (33–35). Despite their clinical derivation, however, lines sNF02.2 and sNF94.3 expressed full-length neurofibromin protein (Figure 6A and refs. 33, 36, 37). Although the specific NF1 mutation(s) in these 2 cell lines remain unknown, despite sequencing of multiple exons, their elevated RAS-GTP levels suggest some impairment of neurofibromin function (38). However, these 2 lines exhibited significantly lower levels of activating phosphorylation of key RAS downstream effectors, C-RAF and MEK1/2 (17, 39, 40), compared with the 3 cell lines devoid of neurofibromin (Figure 6A). This result supports only partial loss of neurofibromin function in these 2 lines.
Figure 6. HSF1 is overexpressed and activated in human MPNST cell lines lacking neurofibromin.
(A) Immunoblotting of cell lines with complete loss of neurofibromin showed elevated p–C-RAF, p-MEK1/2, total HSF1, p-Ser362, and HSP72 levels compared with lines that maintain full-length neurofibromin expression. (B) IF staining confirmed HSF1 overexpression in cells without neurofibromin. Insets show nuclear HSF1 staining. DAPI counterstaining is presented to visualize cell nuclei. All images were acquired at identical magnification. Scale bars: 25 μm; 5 μm (insets). (C) Immunoblotting of cross-linked cell lysates demonstrated increased activation-associated trimeric (Tri) versus inactive monomeric (Mon) forms of HSF1 in MPNST cells without NF1 compared with lysates from NF1-expressing cell lines. The trimeric/monomeric ratio in lysates, as measured by densitometry, is indicated. (D and E) MEK inhibition reduced p-Ser326 and HSF1 nuclear translocation in human MPNST cells. S462 cells were treated with 20 μM U0126 overnight. Total p-Ser326 and nuclear HSF1 were detected by immunoblotting.
In analyzing our panel of human MPNST cells, our distinct findings demonstrated that HSF1 is activated to a greater degree in those cell lines lacking neurofibromin than in those expressing it. First, levels of HSF1 and the highly HSF1-regulated heat shock protein HSP72 were increased (Figure 6A). Second, as shown by immunofluorescent (IF) staining, HSF1 was increased and localized to the nucleus (Figure 6B). Third, p-Ser326 was robust (Figure 6A). Fourth, HSF1 was trimerized, the characteristic state of the activated protein (Figure 6C). This activation was constitutive, occurring in the absence of environmental stressors.
Furthermore, consistent with the results in Nf1-deficient MEFs (Figure 2, B and D), U0126 reduced p-Ser326 and nuclear translocation of HSF1 in MPNST cells (Figure 6, D and E). Thus, in human NF1 tumor cells, aberrant MAPK signaling also critically regulated HSF1 activity.
Human MPNST cells are addicted to HSF1 and to MEK signaling.
Do these cell lines require HSF1 to maintain their malignant phenotype? To test this, we knocked down HSF1 expression with lentiviral shRNAs. First, we tested the efficacy of different shRNAs on HSF1 knockdown. As previously reported in other cell types (5), the hA6 shRNA was more effective than the hA9 shRNA in MPNST cells (Figure 7A). Suppression of HSF1 by both hA6 and hA9 markedly decreased cell number in lines lacking neurofibromin (Figure 7B). Moreover, transduction with hA6 caused a greater reduction than hA9, consistent with a dose-dependent effect of HSF1 on viability. As an additional control, we transduced one of the lines with low HSF1 levels that maintained neurofibromin expression with the same panel of lentiviral supernatants. The cell number was much less reduced, which not only ruled out nonspecific cytotoxicity as the basis for the activity of HSF1-targeted lentiviral preparations, but also supported the idea that NF1 function is only partially compromised in sNF02.2 cells.
Figure 7. HSF1 knockdown impairs human MPNST cell growth and attenuates MAPK signaling.
(A) Differential efficacy of HSF1-targeting shRNA constructs. S462 cells were transduced with GSP or scrambled control or with hA9 or hA6 HSF1-targeting lentiviral supernatants and harvested for immunoblotting. (B) HSF1 knockdown impaired net growth and survival of MPNST cells. After plating, cells were transduced with viral supernatants as indicated. Relative viable cell number in each well was measured 4 days after viral transduction by resazurin dye reduction assay. Raw fluorescence data were normalized to values obtained in wells that underwent mock transduction (mean ± SD, n = 5). #P < 0.001, 2-way ANOVA. (C) Decreased levels of KSR1, p-ERK, HSP90α, and HSP72 after HSF1 knockdown. S462 cells were stably transduced with the indicated viral supernatants and harvested for immunoblotting. (D) Decreased levels of KSR1 and p-ERK after HSP90 inhibition. S462 cells were treated overnight with 1 μM geldanamycin (GA) and harvested for immunoblotting. (E) KSR1 knockdown induced apoptosis in MPNST cells. S462 cells were transfected with a nontargeting siRNA (NC) or 3 independent KSR1-targeting siRNAs at 25 nM final concentration. 2 days after transfection, cells were harvested for immunoblotting. (F) KSR1 knockdown impaired net growth and survival of MPNSTs. S462 cells plated in a 96-well format were transiently transfected without or with 25 nM siRNAs. Relative viable cell number in each well was measured as described in B 3 days after transfection (mean ± SD, n = 5). ***P < 0.001, 1-way ANOVA.
To provide mechanistic insight into the interplay of HSF1 and MAPK signaling in human MPNST cells, we examined the effect of HSF1 knockdown on phosphorylation of ERK, the downstream target of RAS/MAPK signaling. This endpoint had been strongly HSF1-dependent in both mouse tumors and NPcis MEFs (Figure 5). Compared with cells transduced with scrambled control shRNAs, transduction with either hA6 or hA9 virus caused a marked decline in p-ERK, with hA6 again having the greatest effect (Figure 7C). RAS requires the scaffold protein KSR1 for efficient signaling of the MAPK/ERK pathway, and KSR1 deletion inhibits RAS-driven oncogenesis (41, 42). HSF1 knockdown sharply reduced KSR1 levels in a dose-dependent manner. Because KSR1 function depends on molecular chaperones (43), the decrease in HSP72 and HSP90 levels after HSF1 knockdown (Figure 7C) could contribute to KSR1 loss. Consistent with this hypothesis, inhibition of HSP90 by geldanamycin reduced KSR1 protein levels and p-ERK in both S462 and 90-8TL cells (Figure 7D and Supplemental Figure 6A). As expected, levels of AKT, a known HSP90 client protein, were also reduced by geldanamycin.
To examine the role of KSR1 in maintaining the malignant phenotype of human MPNST cells, we transiently transfected a nontargeting and 3 independent KSR1-targeting siRNAs into S462 cells. In these cells, knockdown of KSR1 induced apoptosis, as indicated by elevated levels of cleaved PARP and caspase 7 (Figure 7E), and reduced cell number (Figure 7F). Overall, the data support a model in which HSF1 enables the malignant phenotype of neurofibromin-deficient cells at least in part by stabilizing KSR1 and supporting dysregulated MAPK signaling.
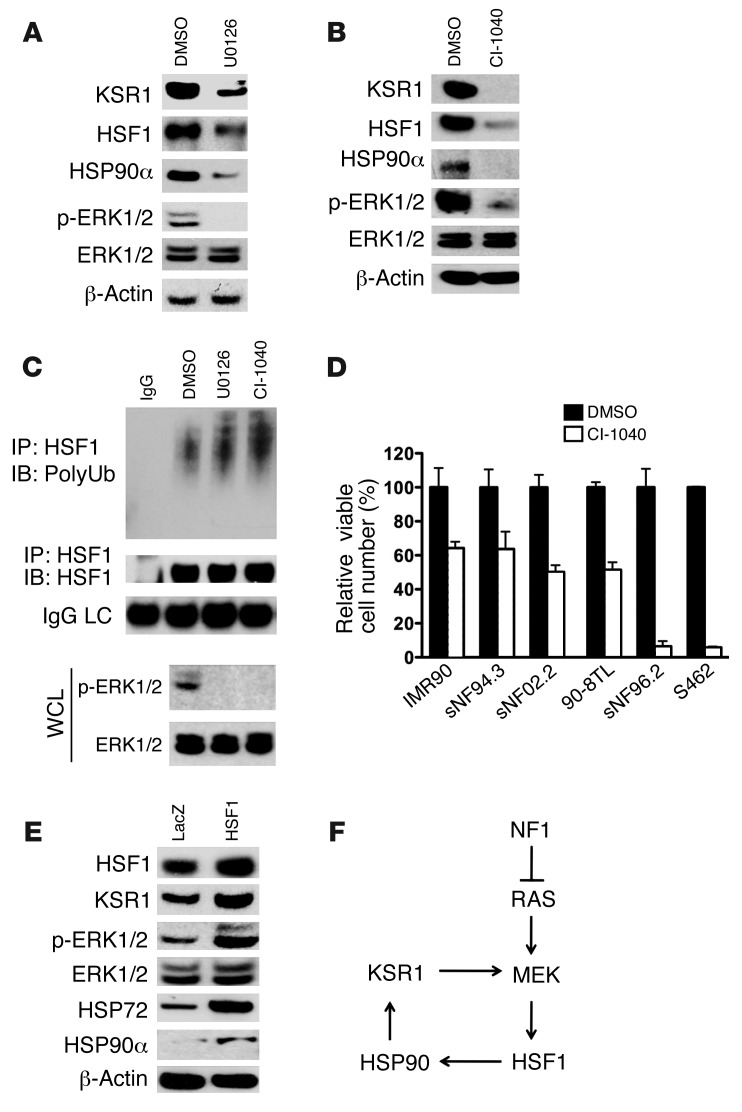
MEK provides an attractive therapeutic target for human cancers. To examine the effects of MEK inhibition on human MPNST cells, S462 cells were treated with either U0126 or CI-1040, the first MEK inhibitor to enter clinical trials (44). Prolonged MEK inhibition over the course of several days markedly reduced the protein levels of HSF1, HSP90α, and KSR1 (Figure 8, A and B). Consistent with our observation that inhibiting MEK reconstituted polyubiquitination of HSF1 (Figure 2I), enhanced HSF1 polyubiquitination was seen in S462 cells after overnight exposure to U0126 or CI-1040 (Figure 8C).
Figure 8. A feed-forward loop of MEK/HSF1.
(A and B) Prolonged MEK inhibition decreased HSF1, HSP90α, and KSR1 protein levels in MPNST cells. S462 cells were treated with 20 μM U0126 for 4 days or 20 μM CI-1040 for 3 days. Whole cell lysates were subjected to immunoblotting. (C) MEK inhibition enhanced HSF1 polyubiquitination. S462 cells were treated with 20 μM U0126 or CI-1040 overnight. HSF1 polyubiquitination was detected as described in Figure 2I. (D) MEK inhibition impaired net growth and survival of MPNST cells. MPNST cells were plated in a 96-well format (2,000 cells/well) and treated with DMSO or 20 μM CI-1040 for 4 days. Relative viable cell number in each well was measured (mean ± SD, n = 3). (E) HSF1 overexpression increased HSP90α and KSR1 protein levels and enhanced p-ERK. S462 cells were transduced with lentiviral LacZ or HSF1 particles. After stable selection with blasticidin, cells were harvested for immunoblotting. (F) Proposed feed-forward loop of MEK/HSF1.
We also examined whether MEK inhibition reduced the cell number of human MPNST cells. 2 neurofibromin-expressing MPNST cell lines and a normal diploid fibroblast control cell line were relatively resistant to MEK inhibition with CI-1040. However, 2 of the 3 neurofibromin-deficient cell lines displayed enhanced sensitivity to CI-1040 treatment (Figure 8D). Why the third line, 90-8TL, was relatively insensitive is unknown, but warrants further investigation. Previous reports suggest that a number of factors could be involved, including mutations in BRAF and PI3KCA (45, 46).
Next, we asked whether enhanced HSF1 function is sufficient to augment MAPK signaling. We generated isogenic S462 cells stably overexpressing HSF1 by viral transduction. Compared with cells stably transduced with LacZ, cells with HSF1 overexpression showed higher levels of KSR1 and p-ERK (Figure 8E). This result was also reproduced in HEK293 cells and immortalized MEFs (Supplemental Figure 6, B and C). As expected, levels of HSP90α and HSP72 were increased in these HSF1-overexpressing cells. These data, together with the role we have demonstrated for HSF1 in supporting MAPK signaling, establish HSF1 as an important modulator of the RAS/MAPK signaling axis. Such a feed-forward loop could enable a powerful shift in the survival landscape that facilitates tumorigenesis (Figure 8F).
HSF1 is overexpressed and activated in MPNSTs resected from patients.
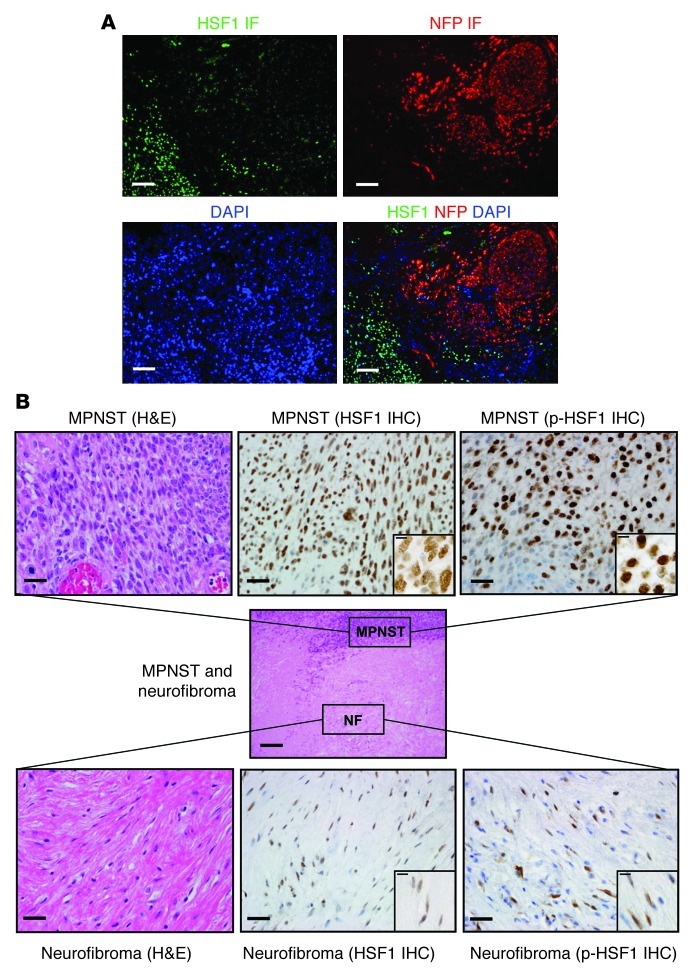
To move from laboratory models to clinical material, we used both IF and immunohistochemistry (IHC) to examine HSF1 levels and localization in a panel of 10 human MPNST surgical resections. High levels of HSF1 were found in the nuclei of tumor cells in all specimens examined. HSF1 levels were markedly higher in MPNST cells than in surrounding normal tissues, including nerve and perineurium (Supplemental Figure 7A). Benign neurofibroma demonstrated an intermediate level (Supplemental Figure 7B). Co-IF also clearly demonstrated high HSF1 levels in MPNST cells, but very low levels in the infiltrated nerve (Figure 9A). Consistent with our findings in mice (Figure 5) and cultured human tumor cells (Figure 6), we detected activation-associated p-ERK at much higher levels in HSF1-overexpressing MPNST cells than in nerve (Supplemental Figure 8). Intermediate levels of p-ERK were observed in benign neurofibroma (Supplemental Figure 8).
Figure 9. HSF1 is overexpressed and activated in NF1 patient tumor resections.
(A) Photomicrographs of dual IF staining for HSF1 (green signal) and neurofilament protein (red signal). DAPI counterstaining was used to visualize cell nuclei (blue signal). Scale bars: 25 μm. (B) Photomicrographs of IHC staining for HSF1 and p-HSF1 in a surgical specimen that shows an MPNST arising from adjacent neurofibroma. Brown signal depicts immunoreactivity. Pale blue counterstaining was provided by Mayer hematoxylin. Scale bars: 250 μm (center); 25 μm (higher magnification); 10 μm (insets).
In one specimen, we captured an MPNST developing from a neurofibroma; it is uncommon to observe a truly high-grade MPNST adjacent to its low-grade counterpart in an individual microscopic specimen. This opportune sample allowed us to visualize levels of HSF1 and assess its phosphorylation status during tumor progression. Captured side by side, HSF1 staining was indeed present in the nuclei of many cells in the neurofibroma, but there was more intense staining for HSF1 protein in the nuclei of MPNST cells (Figure 9B and Supplemental Figure 7, A and B). Whereas p-Ser326 levels were heterogeneous in benign neurofibroma cells, levels were markedly higher and uniformly expressed in the MPNST cells, consistent with increasing activation of HSF1 during tumor progression (Figure 9B).
Discussion
HSF1, the master regulator of the heat shock response in eukaryotes, is a powerful modifier of carcinogenesis (5, 7–9). How HSF1 is activated in tumors, however, has been largely unknown. Although heat shock proteins were known to be elevated in cancer, this was commonly ascribed to a classic heat shock response in reaction to the environmental and proteomic stresses that are inherent to the malignant state. Here, we report that HSF1 is activated by a simple genetic alteration that underlies loss of the Nf1 tumor suppressor, but precedes malignancy. Upon loss of Nf1 function, dysregulated MAPK signaling engages HSF1. In turn, HSF1 supports and further augments activation of the MAPK pathway to drive tumorigenesis. This feed-forward loop arises from intrinsic genetic alteration, not from environmental stress. Activation of HSF1 in this fashion allows cells to withstand a range of proteotoxic insults, even during very early stages of tumorigenesis, thereby enabling the process. The same pathobiology is at work in NF1-associated malignancies in human patients: activation of HSF1 arises in neurofibroma, and more robust and uniform HSF1 activation then occurs during progression to MPNST.
Tumor suppressor loss as a mode of HSF1 activation.
During tumorigenesis, microenvironmental challenges, such as nutrient deprivation and hypoxia (47), and cell-autonomous challenges, such as genomic instability, altered metabolism, reactive oxygen species, and the burden of mutated and misfolded oncoproteins, must all be successfully managed by malignant cells to permit their survival (48, 49). These stresses, inherent to malignant transformation, progression, and metastasis, are made tolerable by a range of innate adaptive responses. The HSF1-mediated response is one of several critical systems that make adaptation not only possible, but efficient. Other responses mediated by regulators, such as HIF1, NF-κB, and stress-activated protein kinases (p38 MAPK and JNK), participate as well to enable cancer cells to adapt and prosper (50–52).
Understanding how stress response regulators are engaged during malignant transformation is of both basic interest and potential therapeutic value. In the case of HSF1, the drivers of activation, the point in tumorigenesis at which this occurs, and the functional consequences have remained poorly defined. The stimulus most widely assigned to drive HSF1 activation in cancer is increased substrate burden on the heat shock protein/chaperone machinery. Dysregulation of the protein translation machinery (53), imbalanced protein production due to aneuploidy (54, 55), and accumulation of mutated, highly chaperone-dependent oncoproteins all strain the chaperone machinery (56–58). Support for this model of HSF1 activation comes from observations that the HSP90-based chaperone machinery may be pressed to operate at its maximal capacity in cancer cells (59), thereby releasing HSF1 from a repressive complex with HSP90 (60).
Alongside this model of HSF1 activation, we now describe another driver for HSF1 activation: loss of the tumor suppressor Nf1. Not only did HSF1 accumulate in the nucleus of neurofibromin-deficient cells, but total levels of HSF1 protein and activating phosphorylation of HSF1 all increased. This indicates a role for the HSF1-adaptive response even in the earliest phases of tumorigenesis, when dysplasia and premalignant alterations begin to arise. By coordinating the diverse biological processes required for tumorigenesis, HSF1 can play a critical role in providing a permissive cellular environment that cultivates the very inception of a neoplasm. Interestingly, in a recent study in breast cancer, we observed that HSF1 is frequently activated in developing neoplasms prior to invasion while the tumor cells still remain in situ (61). Our present findings in Nf1–/– cells suggest that HSF1 can be at work even earlier in tumorigenesis, before atypical changes in cell morphology become visually evident.
Although NF1 was cloned almost 2 decades ago (62), our understanding of exactly how neurofibromin affects growth, metabolism, and survival to act as a key tumor suppressor remains incomplete. The findings reported here further expand our understanding of this tumor suppressor, revealing a function for NF1 in modulating the cellular response to proteotoxic stress that we believe to be previously unrecognized. The tumor suppressor NF1 prevents activation of the heat shock response under basal conditions by restraining improper activation of MAPK signaling. Upon NF1 loss, however, dysregulated MAPK signaling induces p-HSF1, nuclear translocation, and an increase in the level of HSF1 protein through impaired proteasomal degradation.
Dysregulated MAPK signaling is important for HSF1 activation during tumor development, which supports a relationship between tumorigenesis and HSF1 activation that is deeply rooted in the molecular biology and signaling aberrations of NF1-associated tumorigenesis, not solely in environmental and proteotoxic triggers. The importance of MAPK signaling in HSF1 recruitment suggests that other mitogenic pathways dysregulated by dominantly acting oncogenes may also affect HSF1 activity (56). The question therefore arises as to whether loss of other classical tumor suppressors, such as p53, RB, or PTEN, are alone sufficient to activate HSF1. It seems likely that many factors in combination evoke the robust activation of HSF1 observed in very high-grade tumors, compared with the more modest activation seen in the lower-grade counterparts from which they often develop.
HSF1 shapes the tumor landscape.
The functional consequences of HSF1 activation are only beginning to emerge. Here, we found that Nf1 loss led to an HSF1-dependent capacity to tolerate a broad range of proteotoxic perturbations. The chemical stressors we used likely phenocopy some of the internal proteomic imbalances as well as the environmental pressures that tumors must accommodate during their initiation and progression. By fostering an ability to thrive in unfavorable conditions, HSF1 ultimately can promote enhanced malignant behavior (63). Indeed, in a separate study of more than 1,800 women with invasive breast cancer from the Nurses’ Health Study, we recently found that HSF1 activation is associated with tumor size, histologic grade, and likelihood of metastasis (61). This work demonstrates that HSF1 is an independent predictor of poor outcome in breast cancer (61). These strong epidemiological associations implicating HSF1 in malignancy were supported here by knockdown experiments that demonstrated a direct and critical role for HSF1 in promoting the viability of MPNST cell lines derived from advanced cancers.
In addition to facilitating survival under stress, HSF1 also supported tumor growth through its integral role in a feed-forward loop supporting the proproliferative MAPK pathway. Not only was HSF1 activated by MAPK signaling, but its activation, in turn, robustly supported increased flux through this signaling axis, both in MEFs and in MPNST cells. By supporting this particular signaling pathway, which is central to the pathophysiology of NF1, HSF1 alters the NF1 phenotype, as suggested by our observation of a shift in tumor spectrum and latency. How it might affect other features of clinical NF1, such as neurodevelopmental abnormalities, would be fascinating to investigate. Finally, important as MAPK signaling may be, the effect of HSF1 on NF1-associated malignancies undoubtedly extends beyond effects on this one signaling pathway and is likely rooted in the compendium of diverse cellular processes that HSF1 uniquely regulates in tumors.
HSF1 as a therapeutic target in NF1?
The critical dependence of established MPNST cell lines on HSF1, the expression of HSF1 at high levels in surgical resection specimens, and the activation-associated phosphorylation of HSF1 firmly establish its importance in enabling NF1-associated tumorigenesis in humans. Targeting HSF1 could represent a new way of approaching anticancer therapy. Nongenotoxic strategies would be particularly important in NF1, because patients are at a markedly increased risk of developing treatment-related myeloid leukemias after exposure to conventional alkylating agents (64). Moreover, our previous finding that HSF1 expression and activation are associated with malignant potential and outcome in breast cancer (61) suggests that HSF1 status should be evaluated as a potential prognostic marker for human NF1 malignancies, particularly MPNSTs.
Is HSF1 a plausible therapeutic target? Genetic knockout of Hsf1 was sufficient to completely abrogate induction of the heat shock response after temperature shift. Normal basal expression of the major heat shock protein classes was preserved, however, which indicates HSF1 is dispensable for growth and survival of mammalian cells under physiological conditions. Indeed, knockout mice deficient for Hsf1 develop essentially normally and prosper under nonstress conditions (20, 65). This feature, and our finding that even haploinsufficiency of Hsf1 impaired tumorigenesis, suggests that a useful therapeutic index for selective inhibitors of HSF1 likely exists.
Unfortunately, no small, drug-like inhibitors of the heat shock response have yet been identified that target HSF1 in a convincingly specific manner. Most, if not all, appear to act on the cascade of posttranslational modifications mediated by kinases, phosphatases, acetylases, and conjugating enzymes involved in regulating HSF1 activation (66, 67). Discovery efforts are in their infancy, however, with great potential for further progress (66–69).
Many important questions remain, but the findings reported here provide a deeper basic understanding of how cellular physiology accommodates constitutive loss of NF1 function. The adaptive HSF1 program set in motion enhanced cell survival, but came at the cost of enabling tumor formation in this common cancer predisposition syndrome. A similar phenomenon likely occurs in sporadic human cancers driven by other tumor suppressor abnormalities or even mutant oncogenes.
Methods
Cells, tissues, and reagents.
See Supplemental Methods.
Real-time quantitative RT-PCR.
Total RNAs were extracted using a USB PrepEase RNA Spin Kit (Affymetrix). 100 ng RNAs were used for reverse transcription using a Maxima First Strand cDNA Synthesis kit (Fermentas). Equal amounts of cDNA were used for quantitative PCR reaction using Maxima SYBR Green/ROX qPCR Master Mix (Fermentas). Signals were detected by an ABI 7500 Real-Time PCR System (Applied Biosystems). See Supplemental Methods for sequences of individual primers for each gene.
Transfection and luciferase reporter assay.
Plasmids were transfected into HEK293T cells with Arrest-In transfection reagent (Open Biosystems). The following plasmids were used in transfection experiments: pBabe-EGFP (generated from pEGFP-N3 plasmid), pBabe–MEK1AA (generated by site-directed mutagenesis), pHSE–firefly luciferase (Clontech), pCMV–renilla luciferase (Promega), and pBabe-HSF1 (catalog no. 1948; Addgene). Luciferase activities were quantitated using a Promega Dual-Glo Luciferase Assay System using a VICTOR3 Multilabel plate reader (PerkinElmer).
Tumorigenesis studies in NPcis mice.
NPcis mice (129/Sv) were provided by K. Cichowski (Brigham and Women’s Hospital, Boston, Massachusetts, USA). C57BL/6 mice heterozygous for disruption of Nf1 were provided by K. Reilly (National Cancer Institute, Frederick, Maryland, USA). Mice were crossed to Hsf1-modified mice (mixed 129/SvJ and Balb/cJ), a gift from I. Benjamin (University of Utah, Salt Lake City, Utah, USA; ref. 29). Moribund mice or mice with severely compromised body condition were euthanized, a full necropsy was performed, and all major tissues were harvested and fixed in 10% formalin (soft tissues) or Bouin’s fixative (bone). Incidental tumors were documented in the spectrum of tumors depicted in Supplemental Figure 3B, but were not included in the assessment of tumor-free survival (Figure 4A). Tumors were identified and diagnosed in consultation with R. Bronson (Tufts University, Boston, Massachusetts, USA).
Proliferation and survival assays.
Relative cell growth and survival was measured in 96-well microplate format, using the fluorescent detection of resazurin dye reduction as an endpoint. Proliferation of primary MEFs was measured after a 5-day exposure to the indicated compounds.
Immunoblotting and immunoprecipitation.
Whole-cell protein extracts were prepared in cold lysis buffer (100 mM NaCl; 30 mM Tris-HCl, pH 7.6; 1% NP-40; 30 mM sodium fluoride; 1 mM EDTA; 1 mM sodium orthovanadate; protease inhibitor cocktail tablet from Roche Diagnostics). Proteins were separated on SDS-PAGE gels and transferred to nitrocellulose membranes. Primary antibodies were applied in wash buffer for 1 hour at room temperature or overnight at 4°C. Peroxidase-conjugated secondary antibodies were applied at room temperature for 1 hour, and signal was visualized by chemiluminescent substrate (Thermo-Fisher Scientific) followed by exposure to film.
For immunoprecipitation, 500 μg whole-cell lysates were incubated with either 4 μg rat monoclonal HSF1 antibodies (catalog no. RT-629-PABX; Thermo-Fisher Scientific) or normal rat IgG overnight at 4°C. Next, 50 μl protein G magnetic beads (GenScript) were added for 1 hour at room temperature. After 3 washes with PBS, beads were boiled in 50 μl sample loading buffer for 5 minutes before loading on SDS-PAGE.
Immunostaining of cells and tissues.
Archival clinical specimens and mouse tissues were processed as formalin-fixed, paraffin-embedded tissue blocks. Serial 4-μm sections were cut for H&E staining and immunostaining. Detection of NFP (diluted 1:900; clone 2F11; Monosan), p-ERK (diluted 1:500; catalog no. 4370; Cell Signaling Technology), and p-HSF1 (diluted 1:1,200; catalog no. 2092-1; Epitomics) was achieved by IHC. Deparaffinized sections were blocked with 3% H2O2, and antigen retrieval was achieved using a pressure cooker with Dako citrate buffer (pH 6.0) at 120°C ± 2°C, 15 ± 5 PSI. Primary antibodies were incubated on sections at room temperature for 40 minutes followed by 30 minutes of incubation with Dako-labeled Polymer-HRP IgG as a secondary antibody. Immunoreactivity was visualized with 3,3′-diaminobenzidine (Dako Envision+ System). For HSF1 immunostaining, rat monoclonal antibodies (diluted 1:1,000) were used with the ABC method. Slides were blocked with 3% normal rabbit serum. Biotinylated rabbit anti-rat (catalog no. BA-4000; Vector Labs) was applied as secondary antibody, and detection was achieved by 30 minutes of incubation with Vectastain Elite ABC reagent (catalog no. PK-6100; Vector Labs). Mayer hematoxylin was used for counterstaining. For IF staining, reactivity was visualized with secondary anti-IgG antibodies labeled with either Cy3 or Cy5. Primary antibody dilutions were 1:250 for anti-HSF1 and 1:300 for anti-NFP. DAPI (0.5 μg/ml) was applied to visualize cell nuclei.
Statistics.
All statistical analyses were performed using Prism 5.0 (GraphPad software). Significance of differences was determined using 2-tailed Student’s t test, unless otherwise indicated as ANOVA or log-rank test. A P value of 0.05 or less was considered statistically significant.
Study approval.
Paraffin blocks of human surgical resection specimens were obtained from an archive maintained by the Department of Pathology, Brigham and Women’s Hospital, under an excess tissue protocol approved by the Brigham and Women’s Hospital Partners Healthcare Human Research Committee. Ethics committee review specifically waived the need for informed consent. All mouse experiments were performed under a protocol approved by the MIT Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We thank S. Silver and D. Root (Broad Institute RNAi platform) for assisting in the design and execution of the shRNA screening experiments. We thank A.W. Cheng and E. Yeger-Lotem for assistance in selecting heat shock–related genes for shRNA targeting. M. Topolski and M. Koeva provided expert technical assistance, and members of the Lindquist laboratory provided many helpful comments. S. Lindquist is a senior investigator of the Howard Hughes Medical Institute. S. Santagata is supported by NIH grant K08NS064168 and grants from the Valvano Foundation, the American Brain Tumor Association, and the Brain Science Foundation. This work was supported in part by the USAMRMC Neurofibromatosis Research Program (to S. Lindquist), the Johnson & Johnson COSAT program (to L. Whitesell), the Marble Fund (to S. Santagata and S. Lindquist), the Children’s Tumor Foundation, and NIH grant 1DP2OD007070 (to C. Dai).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(10):3742–3754. doi:10.1172/JCI62727.
References
- 1.Ahn SG, Thiele DJ. Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev. 2003;17(4):516–528. doi: 10.1101/gad.1044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voellmy R. On mechanisms that control heat shock transcription factor activity in metazoan cells. Cell Stress Chaperones. 2004;9(2):122–133. doi: 10.1379/CSC-14R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trinklein ND, Murray JI, Hartman SJ, Botstein D, Myers RM. The role of heat shock transcription factor 1 in the genome-wide regulation of the mammalian heat shock response. Mol Biol Cell. 2004;15(3):1254–1261. doi: 10.1091/mbc.E03-10-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319(5865):916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 5.Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130(6):1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mendillo ML, et al. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150(3):549–562. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Min JN, Huang L, Zimonjic DB, Moskophidis D, Mivechi NF. Selective suppression of lymphomas by functional loss of Hsf1 in a p53-deficient mouse model for spontaneous tumors. Oncogene. 2007;26(35):5086–5097. doi: 10.1038/sj.onc.1210317. [DOI] [PubMed] [Google Scholar]
- 8.Jin X, Moskophidis D, Mivechi NF. Heat shock transcription factor 1 is a key determinant of HCC development by regulating hepatic steatosis and metabolic syndrome. Cell Metab. 2011;14(1):91–103. doi: 10.1016/j.cmet.2011.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott KL, et al. Proinvasion metastasis drivers in early-stage melanoma are oncogenes. Cancer Cell. 2011;20(1):92–103. doi: 10.1016/j.ccr.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bollag G, et al. Loss of NF1 results in activation of the Ras signaling pathway and leads to aberrant growth in haematopoietic cells. Nat Genet. 1996;12(2):144–148. doi: 10.1038/ng0296-144. [DOI] [PubMed] [Google Scholar]
- 11.Cichowski K, Jacks T. NF1 tumor suppressor gene function: narrowing the GAP. Cell. 2001;104(4):593–604. doi: 10.1016/S0092-8674(01)00245-8. [DOI] [PubMed] [Google Scholar]
- 12.Largaespada DA, Brannan CI, Jenkins NA, Copeland NG. Nf1 deficiency causes Ras-mediated granulocyte/macrophage colony stimulating factor hypersensitivity and chronic myeloid leukaemia. Nat Genet. 1996;12(2):137–143. doi: 10.1038/ng0296-137. [DOI] [PubMed] [Google Scholar]
- 13.Gutmann DH, Boguski M, Marchuk D, Wigler M, Collins FS, Ballester R. Analysis of the neurofibromatosis type 1 (NF1) GAP-related domain by site-directed mutagenesis. Oncogene. 1993;8(3):761–769. [PubMed] [Google Scholar]
- 14.Tanaka K, et al. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell. 1990;60(5):803–807. doi: 10.1016/0092-8674(90)90094-U. [DOI] [PubMed] [Google Scholar]
- 15.Turbyville TJ, et al. Search for Hsp90 inhibitors with potential anticancer activity: isolation and SAR studies of radicicol and monocillin I from two plant-associated fungi of the Sonoran desert. J Nat Prod. 2006;69(2):178–184. doi: 10.1021/np058095b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guettouche T, Boellmann F, Lane WS, Voellmy R. Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC Biochem. 2005;6:4. doi: 10.1186/1471-2091-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng CF, Guan KL. Activation of MEK family kinases requires phosphorylation of two conserved Ser/Thr residues. EMBO J. 1994;13(5):1123–1131. doi: 10.1002/j.1460-2075.1994.tb06361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitin-proteasome system. Nat Rev Drug Discov. 2006;5(7):596–613. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- 19.Gitler AD, Epstein JA. Regulating heart development: the role of Nf1. Cell Cycle. 2003;2(2):96–98. [PubMed] [Google Scholar]
- 20.Xiao X, et al. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J. 1999;18(21):5943–5952. doi: 10.1093/emboj/18.21.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu YM, et al. 2,3-Dihydrowithaferin A-3beta-O-sulfate, a new potential prodrug of withaferin A from aeroponically grown Withania somnifera. Bioorg Med Chem. 2009;17(6):2210–2214. doi: 10.1016/j.bmc.2008.10.091. [DOI] [PubMed] [Google Scholar]
- 22.Santagata S, et al. Using the heat-shock response to discover anticancer compounds that target protein homeostasis. ACS Chem Biol. 2012;7(2):340–349. doi: 10.1021/cb200353m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gitler AD, et al. Nf1 has an essential role in endothelial cells. Nat Genet. 2003;33(1):75–79. doi: 10.1038/ng1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacks T, Shih TS, Schmitt EM, Bronson RT, Bernards A, Weinberg RA. Tumour predisposition in mice heterozygous for a targeted mutation in Nf1. Nat Genet. 1994;7(3):353–361. doi: 10.1038/ng0794-353. [DOI] [PubMed] [Google Scholar]
- 25.Cichowski K, et al. Mouse models of tumor development in neurofibromatosis type 1. Science. 1999;286(5447):2172–2176. doi: 10.1126/science.286.5447.2172. [DOI] [PubMed] [Google Scholar]
- 26.Vogel KS, Klesse LJ, Velasco-Miguel S, Meyers K, Rushing EJ, Parada LF. Mouse tumor model for neurofibromatosis type 1. Science. 1999;286(5447):2176–2179. doi: 10.1126/science.286.5447.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300(5622):1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 28.Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15(2):657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steele AD, et al. Heat shock factor 1 regulates lifespan as distinct from disease onset in prion disease. Proc Natl Acad Sci U S A. 2008;105(36):13626–13631. doi: 10.1073/pnas.0806319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reilly KM, et al. An imprinted locus epistatically influences Nstr1 and Nstr2 to control resistance to nerve sheath tumors in a neurofibromatosis type 1 mouse model. Cancer Res. 2006;66(1):62–68. doi: 10.1158/0008-5472.CAN-05-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walrath JC, Fox K, Truffer E, Gregory Alvord W, Quinones OA, Reilly KM. Chr 19(A/J) modifies tumor resistance in a sex- and parent-of-origin-specific manner. Mamm Genome. 2009;20(4):214–223. doi: 10.1007/s00335-009-9179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Y, Ghosh P, Charnay P, Burns DK, Parada LF. Neurofibromas in NF1: Schwann cell origin and role of tumor environment. Science. 2002;296(5569):920–922. doi: 10.1126/science.1068452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fieber LA, Gonzalez DM, Wallace MR, Muir D. Delayed rectifier K currents in NF1 Schwann cells. Pharmacological block inhibits proliferation. Neurobiol Dis. 2003;13(2):136–146. doi: 10.1016/S0969-9961(03)00031-7. [DOI] [PubMed] [Google Scholar]
- 34.Frahm S, et al. Genetic and phenotypic characterization of tumor cells derived from malignant peripheral nerve sheath tumors of neurofibromatosis type 1 patients. Neurobiol Dis. 2004;16(1):85–91. doi: 10.1016/j.nbd.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 35.Barkan B, Starinsky S, Friedman E, Stein R, Kloog Y. The Ras inhibitor farnesylthiosalicylic acid as a potential therapy for neurofibromatosis type 1. Clin Cancer Res. 2006;12(18):5533–5542. doi: 10.1158/1078-0432.CCR-06-0792. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, et al. Notch and Schwann cell transformation. Oncogene. 2004;23(5):1146–1152. doi: 10.1038/sj.onc.1207068. [DOI] [PubMed] [Google Scholar]
- 37.Miller SJ, et al. Integrative genomic analyses of neurofibromatosis tumours identify SOX9 as a biomarker and survival gene. EMBO Mol Med. 2009;1(4):236–248. doi: 10.1002/emmm.200900027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perrin GQ, et al. Plexiform-like neurofibromas develop in the mouse by intraneural xenograft of an NF1 tumor-derived Schwann cell line. J Neurosci Res. 2007;85(6):1347–1357. doi: 10.1002/jnr.21226. [DOI] [PubMed] [Google Scholar]
- 39.Diaz B, Barnard D, Filson A, MacDonald S, King A, Marshall M. Phosphorylation of Raf-1 serine 338-serine 339 is an essential regulatory event for Ras-dependent activation and biological signaling. Mol Cell Biol. 1997;17(8):4509–4516. doi: 10.1128/mcb.17.8.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alessi DR, et al. Identification of the sites in MAP kinase kinase-1 phosphorylated by p74raf-1. EMBO J. 1994;13(7):1610–1619. doi: 10.1002/j.1460-2075.1994.tb06424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Therrien M, Michaud NR, Rubin GM, Morrison DK. KSR modulates signal propagation within the MAPK cascade. Genes Dev. 1996;10(21):2684–2695. doi: 10.1101/gad.10.21.2684. [DOI] [PubMed] [Google Scholar]
- 42.Kortum RL, Lewis RE. The molecular scaffold KSR1 regulates the proliferative and oncogenic potential of cells. Mol Cell Biol. 2004;24(10):4407–4416. doi: 10.1128/MCB.24.10.4407-4416.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart S, Sundaram M, Zhang Y, Lee J, Han M, Guan KL. Kinase suppressor of Ras forms a multiprotein signaling complex and modulates MEK localization. Mol Cell Biol. 1999;19(8):5523–5534. doi: 10.1128/mcb.19.8.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allen LF, Sebolt-Leopold J, Meyer MB. CI-1040 (PD184352), a targeted signal transduction inhibitor of MEK (MAPKK). Semin Oncol. 2003;30(5 Suppl 16):105–116. doi: 10.1053/j.seminoncol.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 45.Corcoran RB, Dias-Santagata D, Bergethon K, Iafrate AJ, Settleman J, Engelman JA. BRAF gene amplification can promote acquired resistance to MEK inhibitors in cancer cells harboring the BRAF V600E mutation. Sci Signal. 2010;3(149):ra84. doi: 10.1126/scisignal.2001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wee S, et al. PI3K pathway activation mediates resistance to MEK inhibitors in KRAS mutant cancers. Cancer Res. 2009;69(10):4286–4293. doi: 10.1158/0008-5472.CAN-08-4765. [DOI] [PubMed] [Google Scholar]
- 47.Fang JS, Gillies RD, Gatenby RA. Adaptation to hypoxia and acidosis in carcinogenesis and tumor progression. Semin Cancer Biol. 2008;18(5):330–337. doi: 10.1016/j.semcancer.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136(5):823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ono K, Han J. The p38 signal transduction pathway: activation and function. Cell Signal. 2000;12(1):1–13. doi: 10.1016/S0898-6568(99)00071-6. [DOI] [PubMed] [Google Scholar]
- 51.Karin M. NF-kappaB as a critical link between inflammation and cancer. Cold Spring Harb Perspect Biol. 2009;1(5):a000141. doi: 10.1101/cshperspect.a000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29(5):625–634. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cencic R, et al. Reversing chemoresistance by small molecule inhibition of the translation initiation complex eIF4F. Proc Natl Acad Sci U S A. 2011;108(3):1046–1051. doi: 10.1073/pnas.1011477108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Theriault JR, He H, Gong J, Calderwood SK. Expression of a dominant negative heat shock factor-1 construct inhibits aneuploidy in prostate carcinoma cells. J Biol Chem. 2004;279(31):32651–32659. doi: 10.1074/jbc.M401475200. [DOI] [PubMed] [Google Scholar]
- 55.Pavelka N, et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468(7321):321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khaleque MA, et al. Induction of heat shock proteins by heregulin beta1 leads to protection from apoptosis and anchorage-independent growth. Oncogene. 2005;24(43):6564–6573. doi: 10.1038/sj.onc.1208798. [DOI] [PubMed] [Google Scholar]
- 57.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5(10):761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 58.Stanhill A, et al. Ha-ras(val12) induces HSP70b transcription via the HSE/HSF1 system, but HSP70b expression is suppressed in Ha-ras(val12)-transformed cells. Oncogene. 2006;25(10):1485–1495. doi: 10.1038/sj.onc.1209193. [DOI] [PubMed] [Google Scholar]
- 59.Kamal A, et al. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature. 2003;425(6956):407–410. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- 60.Voellmy R, Boellmann F. Chaperone regulation of the heat shock protein response. Adv Exp Med Biol. 2007;594:89–99. doi: 10.1007/978-0-387-39975-1_9. [DOI] [PubMed] [Google Scholar]
- 61.Santagata S, et al. High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc Natl Acad Sci U S A. 2011;108(45):18378–18383. doi: 10.1073/pnas.1115031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Y, et al. Somatic mutations in the neurofibromatosis 1 gene in human tumors. Cell. 1992;69(2):275–281. doi: 10.1016/0092-8674(92)90408-5. [DOI] [PubMed] [Google Scholar]
- 63.Whitesell L, Santagata S, Lin NU. Inhibiting HSP90 to treat cancer: a strategy in evolution [published online ahead of print July 17, 2012]. Curr Mol Med. doi: 10.2174/156652412803306657. In press. [DOI] [PubMed] [Google Scholar]
- 64.Chao RC, et al. Therapy-induced malignant neoplasms in Nf1 mutant mice. Cancer Cell. 2005;8(4):337–348. doi: 10.1016/j.ccr.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 65.McMillan DR, Xiao X, Shao L, Graves K, Benjamin IJ. Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J Biol Chem. 1998;273(13):7523–7528. doi: 10.1074/jbc.273.13.7523. [DOI] [PubMed] [Google Scholar]
- 66.Whitesell L, Lindquist S. Inhibiting the transcription factor HSF1 as an anticancer strategy. Expert Opin Ther Targets. 2009;13(4):469–478. doi: 10.1517/14728220902832697. [DOI] [PubMed] [Google Scholar]
- 67.Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;11(8):545–555. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Au Q, Zhang Y, Barber JR, Ng SC, Zhang B. Identification of inhibitors of HSF1 functional activity by high-content target-based screening. J Biomol Screen. 2009;14(10):1165–1175. doi: 10.1177/1087057109347472. [DOI] [PubMed] [Google Scholar]
- 69.Yoon YJ, et al. KRIBB11 inhibits HSP70 synthesis through inhibition of heat shock factor 1 function by impairing the recruitment of positive transcription elongation factor b to the hsp70 promoter. J Biol Chem. 2011;286(3):1737–1747. doi: 10.1074/jbc.M110.179440. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.