Abstract
The formation of a long-lasting memory requires a transcription-dependent consolidation period that converts a short-term memory into a long-term memory. Nuclear receptors compose a class of transcription factors that regulate diverse biological processes, and several nuclear receptors have been implicated in memory formation. Here, we examined the potential contribution of nuclear receptors to memory consolidation by measuring the expression of all 49 murine nuclear receptors after learning. We identified 13 nuclear receptors with increased expression after learning, including all 3 members of the Nr4a subfamily. These CREB-regulated Nr4a genes encode ligand-independent “orphan” nuclear receptors. We found that blocking NR4A activity in memory-supporting brain regions impaired long-term memory but did not impact short-term memory in mice. Further, expression of Nr4a genes increased following the memory-enhancing effects of histone deacetylase (HDAC) inhibitors. Blocking NR4A signaling interfered with the ability of HDAC inhibitors to enhance memory. These results demonstrate that the Nr4a gene family contributes to memory formation and is a promising target for improving cognitive function.
Introduction
Memories are initially stored in a fragile form that can be disrupted by new information, but in the hours following learning a transcription-dependent process known as memory consolidation converts these short-term memories into stable long-term memories. The cellular mechanisms governing memory consolidation have been the subject of intense study over the past 30 years. The molecular underpinnings of memory consolidation have been most thoroughly studied in a region of the brain known as the hippocampus during spatial and contextual memory formation (1). Hippocampus-dependent memory formation requires 2 waves of protein synthesis (2), cAMP-dependent kinase (PKA) activity (2), and de novo transcription in the hippocampus (3) in the hours following learning.
Nuclear receptors (NRs) compose the largest class of transcription factors found in metazoans (4). Generally, NRs are regulated by lipophilic ligands, allowing rapid, ligand-dependent control of various developmental and metabolic processes. This family includes receptors for fat-soluble vitamins, endocrine hormones, thyroid hormones, fatty acids, bile acids, oxysterols, and dietary xenobiotic lipids. Additionally, “orphan” NRs either have no ligand or a ligand that has yet to be identified. Several NRs have been implicated in the formation of memory. For instance, agonists for glucocorticoid receptors, estrogen receptors (ERs), PPARs, and retinoic acid receptors (RARs) can improve long-term memory formation under certain conditions (5–8). Additionally, mice with mutations in the Erb (9), Rarb (10), or the orphan NR Nr4a2 have deficits in long-term memory (11).
Despite the importance of NRs to diverse physiological processes and data supporting a role of select NRs in memory formation, a systematic analysis of NR expression after learning has not been previously performed. Therefore, we surveyed the expression of all 49 NR genes after learning in the single-trial contextual fear-conditioning task. This training protocol produces a robust memory that requires the hippocampus, a site of increased gene expression after learning (12). We examined time points spanning the entire 24-hour period after learning and found that 13 NRs have increased hippocampal expression in the first 2 hours after training. Among these 13 learning-induced NRs were all 3 members of the Nr4a orphan NR family.
Interestingly, Nr4a family gene expression is activated by many of the same signaling cascades that are required for long-term memory formation, including cAMP, PKA, and cAMP-response element–binding protein (CREB) (reviewed in ref. 1). Further, a class of drugs that improves long-term memory formation through inhibition of histone deacetylases (HDACs) increases the expression of Nr4a genes (13). Therefore, we used a dominant-negative strategy to ascertain whether NR4A signaling contributes to long-term memory formation and the enhancement in memory caused by HDAC inhibitors. We found that transgenic expression of a dominant-negative form of NR4A in forebrain neurons impairs long-term contextual memory consolidation and blocks memory enhancement by intrahippocampal infusion of HDAC inhibitors after training. Further, we identify Bdnf and Fosl2 as targets of NR4A signaling that are also enhanced by HDAC inhibitor treatment. These results demonstrate a role for NR4A signaling in long-term memory formation and the enhancement in memory by HDAC inhibitors.
Results
NR gene expression in the hippocampus is regulated by contextual learning.
To address whether NR gene expression might be associated with memory consolidation, we examined hippocampal gene expression after contextual fear conditioning, a form of hippocampus-dependent memory (14). We chose this task because the anatomical circuitry and molecular signaling cascades underlying this form of memory are well established. Additionally, the timings of these molecular signaling events are directly measurable relative to a single training episode. Contextual fear conditioning is associated with 2 waves of CREB phosphorylation after training (15), and long-term contextual fear memory is sensitive to inhibitors of translation or PKA during 2 time windows that coincide with these 2 peaks of CREB phosphorylation (2). The first of these windows occurs within the first hour after learning, and the second occurs between the third and sixth hour after learning (2, 15). Newly expressed genes, such as Bdnf, appear to contribute to memory formation even as late as 12 hours after acquisition of fear memories (16). For these reasons, we analyzed gene expression in the hippocampus at multiple time points after learning (0.5, 1, 2, 4, 6, 8, 12, and 24 hours) in comparison to circadian controls (Figure 1A).
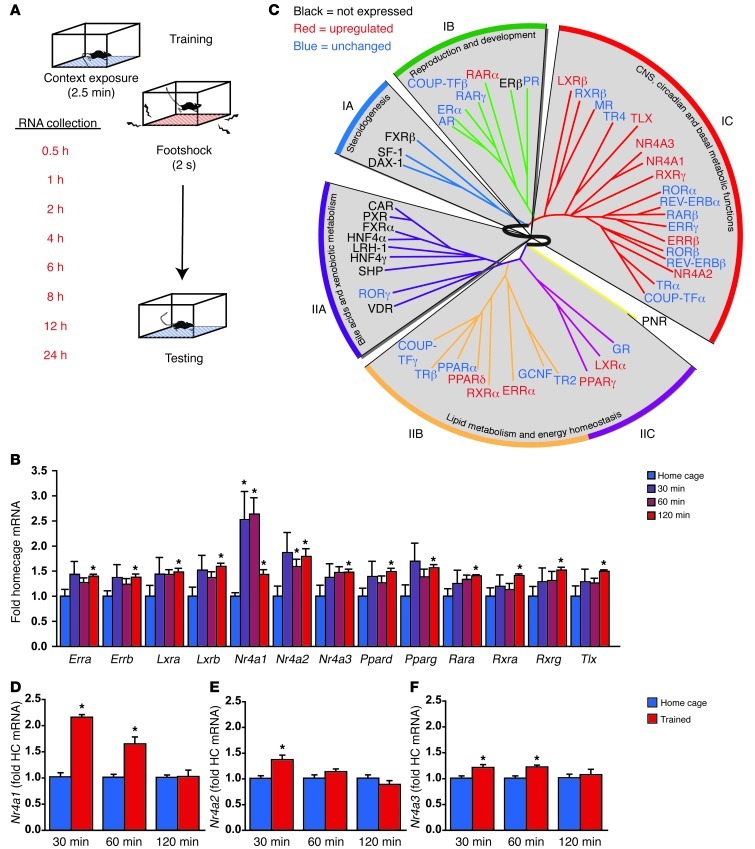
Figure 1. The formation of contextual fear memories induces expression of NR genes in the hippocampus.
(A) Contextual fear conditioning produces a long-lasting memory for the training context and the association of this context with a mild foot-shock. RNA was collected from whole hippocampi at multiple time points after training to survey the impact of training on NR gene expression using a standard ΔΔCT approach. (B) High-throughput qPCR data are illustrated for the 13 NR genes with statistically significant changes in gene expression during the first 2 hours after training, the window in which the majority of changes were observed. (C) The data from this screen indicate that 13 NRs have increased expression in the hippocampus within the first 2 hours after training (red), whereas 13 NRs are not appreciably expressed in the hippocampus (black). The remaining 23 NRs (blue) show no evidence of altered hippocampal expression in the first 2 hours after training. Expression changes are illustrated within clusters defined by anatomical expression profiling (IA, IB, IC, IIA, IIB, IIC) in a diagram modified with permission from Cell; ref. 17. (D) Nr4a1 expression is potently induced in the first hour after learning (P < 0.001). (E) Nr4a2 expression increases after fear conditioning (P = 0.033). (F) Nr4a3 expression increases after fear conditioning (P = 0.004). HC, home cage. Error bars represent SEM. *P < 0.05. See also Supplemental Figure 1.
Out of the 49 NRs, 13 were not expressed at appreciable levels in the hippocampus (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI64145DS1). These 13 genes cluster into the steroidogenesis and bile acid and xenobiotic metabolism functional categories (Figure 1C) identified previously by anatomical profiling of NRs (17). Of those genes expressed in the hippocampus, very few showed any evidence of altered gene expression in the late time points, more than 2 hours after learning (Supplemental Figure 1B). For this reason, we have focused on changes in the first 2 hours after training (Figure 1C).
Another 13 NRs showed increased expression in the first 2 hours after training (Figure 1B, red in Figure 1C) These 13 genes encode 6 orphan NRs (Tlx, also known as Nr2e1; NGFI-B, also known as Nr4a1; Nurr1, also known as Nr4a2; Nor1, also known as Nr4a3; Erra, also known as Nr3b1; Errb, also known as Nr3b2) and 7 receptors with known ligands (Lxra, also known as Nr1h3; LXRB, also known as Nr1h2; Ppard, also known as Nr1c2; Pparg, also known as Nr1c3; Rara, also known as Nr1b1; Rxra, also known as Nr2b1; Rxrg, also known as Nr2b3). Another 23 of the NRs were expressed in the hippocampus, but mRNA levels of these genes were not altered in the 2-hour window after training (blue in Figure 1C).
NR4A subfamily gene expression is regulated by contextual learning.
NRs can be grouped into 6 functionally — and evolutionarily — related subfamilies (NR1–NR6) and another “catch-all” subfamily (NR0) (18). Four of these seven subfamilies are represented with the group of training-induced genes found and shown in Figure 1, B and C; however, only the NR4 subfamily shows evidence for the entire family being induced by learning. The Nr4 gene family encodes 3 NR4A NRs: NR4A1 (also known NGFI-B, NUR77, and TR3), NR4A2 (also known as NURR1, HZF-3, and RNR1), and NR4A3 (also known as NOR1, MINOR, and TEC). Consistent with our results, in situ analysis has previously shown that Nr4a1 expression increases in hippocampal area CA1 (19) and that Nr4a2 expression increases in hippocampal areas CA1 and CA3 (20) following hippocampus-dependent learning. Additionally, Nr4a2 heterozygous null mice have impaired hippocampus-dependent passive avoidance memory (11), and Nr4a2 knockdown using antisense oligodeoxynucleotide injection into the hippocampus impairs long-term memory in a spatial discrimination task (21). Further, improved long-term spatial memory performance observed by deletion of HDAC3 in the hippocampus is attenuated by treatment with siRNA targeting Nr4a2 (22). Therefore, we chose to pursue the Nr4a gene family for follow-up studies by first confirming that expression of each of the 3 Nr4a genes increases after fear conditioning using low-throughput qPCR methods. Contextual fear conditioning had a significant overall effect on gene expression for each of the 3 Nr4a genes (Nr4a1, P < 0.001, Figure 1D; Nr4a2, P = 0.033 Figure 1E; Nr4a3, P = 0.004, Figure 1F). We found that Nr4a1 expression increases substantially at 30 minutes after contextual fear conditioning (P = 0.009), with transcript levels remaining elevated at 60 minutes after training (P = 0.009) but returning to baseline by 120 minutes after training. A similar pattern of expression was observed for Nr4a3 (30 minutes, P = 0.028; 60 minutes, P = 0.016). Nr4a2 expression was significantly elevated at 30 minutes after training (P = 0.009). Thus, Nr4a family gene expression increases within the hippocampus after training in a hippocampus-dependent task.
Generation of a NR4A dominant-negative transgenic mouse line.
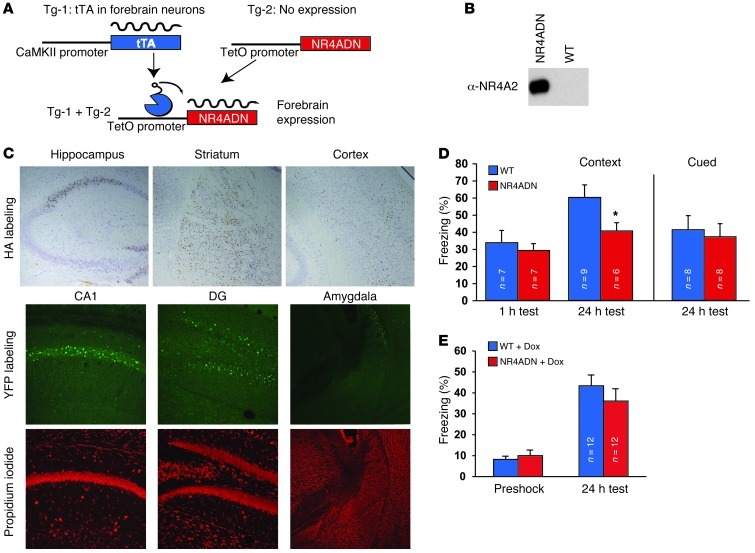
Our data demonstrate that learning induces de novo gene expression for all 3 Nr4a family genes during a window in which new gene synthesis is required for long-term memory formation (1). Because NR4A proteins are ligand-independent NRs (23), the level of gene expression is a major factor determining NR4A activity. In the hippocampus, Nr4a1 and Nr4a2 are 2 out of only 19 immediate early genes for which induction was blocked in CREB conditional mutant mice after seizure activity (24). Thus, Nr4a expression may be an important part of a CREB-initiated cascade of gene expression that contributes to the formation of long-term memory. This hypothesis is supported by existing data suggesting a role for Nr4a2 in long-term memory formation (11, 21). In other biological contexts, Nr4a family members have been observed to have redundant functions (25, 26). In these situations, a truncated version of NR4A1 that acts as a dominant-negative protein to silence all 3 family members has been an invaluable tool in deciphering the physiological roles of NR4A signaling (25, 27). Further, the involvement of NR4A signaling in distinct processes in different cell types (28, 29) and in different anatomical regions (29) calls for a cell-type and regionally restricted approach to examine the role of NR4A signaling in memory formation. For these reasons, we adapted the NR4A dominant-negative approach to allow us to block NR4A signaling in hippocampal neurons. The dominant-negative form of NR4A1 (NR4ADN) contains the DNA-binding and dimerization domains but lacks the transactivation domain (Supplemental Figure 2A), allowing it to form nonproductive dimers with all 3 NR4A proteins (25). As previously reported (25, 27), this truncated form of NR4A1 efficiently blocks NR4A-mediated transcriptional activity (Supplemental Figure 2B). We generated a transgenic mouse line expressing the NR4ADN construct under control of the tetracycline operator (tetO), which we combined with the CaMKII–tetracycline transactivator (CaMKII-tTA) transgene to achieve expression selectively within postnatal excitatory forebrain neurons (ref. 30 and Figure 2A). Endogenous NR4A2 protein was immunoprecipitated from hippocampal extracts with antibodies for the yellow fluorescent protein (YFP) tag of the NR4ADN protein (Figure 2B), showing that the dominant-negative protein interacts with NR4A proteins in vivo. Transgene expression was restricted in the forebrain to the striatum, sparse cortical areas, and subregions of the hippocampal formation (CA1 and the dentate gyrus) (Figure 2C). Transgene expression was not observed in the amygdala or in area CA3 of the hippocampus (Figure 2C).
Figure 2. Blocking function of NR4A family NRs in the hippocampus impairs long-term memory formation.
(A) To impede NR4A signaling in forebrain neurons, a tTA transgene expressed selectively in the forebrain was used to activate a dominant-negative Nr4a transgene (NR4ADN) under control of the tetO. (B) An antibody to the YFP tag on the transgenic NR4ADN protein coimmunoprecipitates endogenous NR4A2 protein from hippocampal protein extracts, confirming the ability of the dominant-negative transgenic protein to heterodimerize with NR4A protein. (C) In the top row, immunolabeling for the NR4ADN hemagglutinin (HA) tag (brown) with cresyl violet counterstain (purple) shows expression in the hippocampus as well as in cortex and striatum (original magnification, ×100). Fluorescent immunolabeling for the YFP tag (middle row) and propidium iodide counterstaining (bottom row) illustrates transgene expression in hippocampal subregions CA1 (original magnification, ×250) and the dentate gyrus (DG) (original magnification, ×250) but not the amygdala (original magnification, ×62.5). (D) NR4ADN mice have selective deficits in long-term contextual fear memory, whereas neither short-term contextual nor long-term cued fear conditioning are impaired. (E) No difference in 24-hour contextual fear memory performance was detected between wild-type and NR4ADN mice after 4 weeks of doxycycline (dox) treatment (P = 0.87, n = 12 mice/group). All error bars denote SEM. *P < 0.05. See also Supplemental Figure 2.
NR4A dominant-negative transgenic mice display impaired long-term hippocampus-dependent memory.
To investigate the role of NR4A signaling in memory formation, we examined associative memory in NR4ADN mice using the fear-conditioning paradigm (Figure 2D). As discussed previously, the contextual version of this task requires hippocampal function. In contrast, long-term associative memory performance for a cue-induced fear response requires the same behavioral output, freezing, without requiring hippocampal function (14). NR4ADN mice had reduced freezing in a 24-hour test of long-term contextual memory (P = 0.03). In contrast, cued fear memory was unaffected in NR4ADN mice (P = 0.81), suggesting that the deficit in long-term contextual memory performance is likely due to defects in hippocampal function. Long-term memory deficits could result from either impaired learning or impaired memory consolidation, but performance in short-term memory tests requires learning without requiring transcription-dependent memory consolidation processes (1). Therefore, we examined short-term contextual fear memory in NR4ADN mice to test whether the Nr4a family of transcription factors contributes to learning or memory consolidation (Figure 2D). NR4ADN mice displayed levels of freezing in a 1-hour memory test that were similar to those of wild-type littermates (P = 0.71), demonstrating that NR4ADN mice are capable of learning the fear-conditioning task but have a reduced ability to retain the memory. Thus, it appears that Nr4a family function is involved in the consolidation of long-term contextual memory.
Memory deficit in NR4A mutant mice requires adult transgene expression.
Regulation of the NR4ADN transgene by CaMKII-tTA led to transgene expression selectively in postnatal neurons (Figure 2), but it is possible that the defect in long-term memory observed in these mice is due to a developmental requirement for Nr4a family function or is a consequence of transgene insertion. To address these possibilities, we reared NR4ADN mice and wild-type mice in the absence of doxycycline, as before, and then placed the mice on a doxycycline diet from weaning until 2 months of age. A 1-month-long treatment with doxycycline was sufficient to suppress transgene expression (Supplemental Figure 2E). After transgene suppression, 24-hour contextual fear memory performance in NR4ADN mice was equivalent to that in wild-type littermates that were also fed an identical doxycycline diet (P = 0.68, Figure 2E). These data are consistent with a requirement for Nr4a family function in the adult mouse brain, rather than a memory defect as a result of a developmental role of Nr4a family members or an effect caused by transgene insertion.
Intrahippocampal HDAC inhibitor treatment increases Nr4a gene expression.
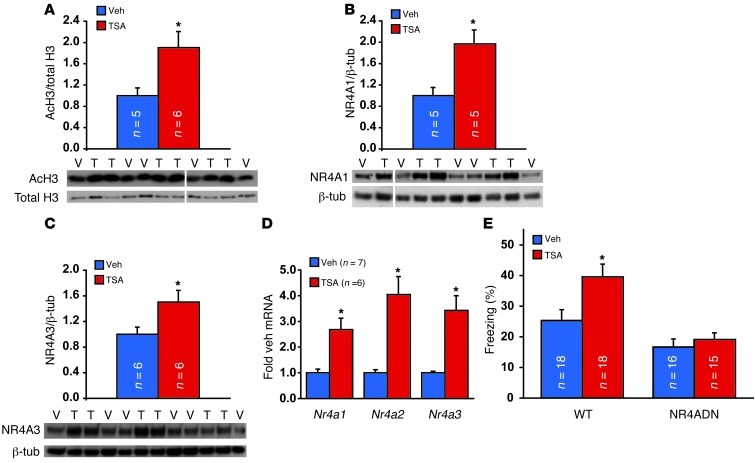
Memory enhancement by pharmacologically increasing histone acetylation with HDAC inhibitors requires CREB-mediated gene expression, and HDAC inhibitor treatment was observed to increase expression of only 2 out of 13 CREB target genes, Nr4a1 and Nr4a2 (13). This increase in Nr4a gene expression was accompanied by increased histone acetylation at the promoters of these genes (13). Additionally, intrahippocampal injection of siRNA targeting Nr4a2 attenuates the enhancement in memory observed with deletion of HDAC3 (22). These data suggest that Nr4a family gene expression may be an important component of the enhancement in memory by HDAC inhibitors. We tested this hypothesis by examining whether the HDAC inhibitor trichostatin A (TSA) is capable of increasing memory in NR4ADN mice. Injection of the HDAC inhibitor TSA into the dorsal hippocampus caused increased acetylation of histone H3 (Figure 3A). This increase in histone acetylation was accompanied by a similar rise in the protein level for NR4A1 (Figure 3B) using a specific antibody against NR4A1. Furthermore, these changes at the protein level were accompanied by increased transcript levels for all 3 Nr4a genes (Figure 3D). Thus, our results agree with previous findings that HDAC inhibitor treatment enhances Nr4a gene expression (13).
Figure 3. The ability of an HDAC inhibitor to enhance memory is blocked by the Nr4a dominant-negative transgene.
(A) Intrahippocampal injection of the HDAC inhibitor TSA (T) enhances acetylation of histone H3 (AcH3), as illustrated at 1 hour after TSA injection after training. Veh/V, vehicle. (B and C) Intrahippocampal TSA also increases expression of the (B) NR4A1 and (C) NR4A3 protein at the same time point. (D) Intrahippocampal TSA increases mRNA levels for each of the 3 Nr4a genes at 1 hour after injection. (E) Injection of TSA into hippocampi after training enhances 24-hour contextual fear memory of wild-type mice but fails to enhance memory of NR4ADN littermates. Error bars represent SEM. *P < 0.05. See also Supplemental Figure 3. Lanes were run on the same gel but were noncontiguous (white lines).
Memory enhancement by HDAC inhibitors is blocked in NR4A mutant mice.
The observation that Nr4a family members play a role in memory consolidation suggests that increased Nr4a gene expression after HDAC inhibitor treatment (Figure 3, B and C, and ref. 13) may contribute to memory enhancement produced by these drugs. Because NR4ADN mice retained some degree of long-term memory (Figure 2D), we were able to ask whether HDAC inhibitors are capable of enhancing memory in these mutant mice (Figure 3D). We injected the HDAC inhibitor TSA directly into the hippocampus immediately after contextual fear conditioning. Consistent with previous findings (13), intrahippocampal TSA injection after training increased long-term contextual fear memory in wild-type littermates of NR4ADN mice (Figure 3E, P = 0.038). However, TSA did not enhance memory in NR4ADN mutant mice (P = 0.99). Thus, the function of Nr4a family transcription factors is required for memory enhancement caused by HDAC inhibitors, suggesting that NR4A NRs contribute to this enhancement.
NR4ADN transgenic mice have reduced expression of NR4A target genes.
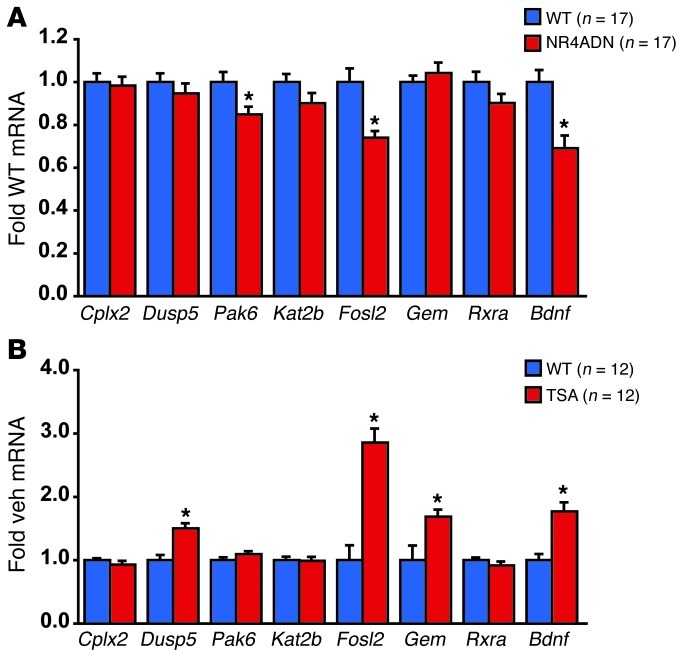
To identify targets of NR4A signaling that could contribute to the memory deficit observed in NR4ADN mutant mice, we surveyed gene expression using a high-throughput qPCR approach. A 224-member gene panel was selected based on previous microarray studies after learning and neuronal activity. Using this approach, we identified 8 potential NR4A targets (Supplemental Figure 3) that we then pursued by low-throughput qPCR. We found 3 genes that were clearly impacted by NR4ADN expression (Figure 4A). Each of these genes (Bdnf, Fosl2, and Pak6) has potential NR4A-binding sites in the proximal promoter region, and Bdnf has previously been identified as a direct target of NR4A2 (31). Thus, these genes may be part of the functionally relevant output of NR4A signaling during memory formation.
Figure 4. Common gene targets are impaired by NR4ADN expression and increased by TSA treatment.
(A) NR4ADN transgenic mice have reduced expression of several putative Nr4a target genes, including Bdnf, Fosl2, and Pak6. (B) Intrahippocampal TSA treatment increases expression of several genes, including Fosl2 and Bdnf. Error bars represent SEM. *P < 0.05. See also Supplemental Figure 4.
Because our pharmacological studies in NR4ADN mice suggest that it is likely that TSA enhances memory by potentiating expression of NR4A target genes, we chose to further refine the list of potentially relevant targets of NR4A signaling in memory storage by examining potential NR4A target genes after TSA treatment (Figure 4B). TSA treatment increased expression of 4 out of the 8 examined genes. Both Fosl2 and Bdnf were common targets of NR4A signaling and TSA treatment, suggesting that these genes might be downstream effectors for NR4A signaling that contribute to the function of NR4A signaling in memory formation (Figure 4C).
Discussion
In this work, we found that the NR gene expression increases in the hippocampus in the hours after learning a hippocampus-dependent long-term memory task. Although these NRs span multiple subfamilies, all 3 of the NR4A orphan NRs show increased expression after fear conditioning. Our results are consistent with previous reports suggesting Nr4a1 and Nr4a2 expression is induced by spatial and contextual exploration (19, 20). Furthermore, we found that blocking NR4A signaling in forebrain neurons, including in the hippocampus, selectively impairs long-term hippocampus-dependent contextual fear memory, without impacting short-term contextual fear memory or hippocampus-independent cued fear memory. These findings support previous suggestions that Nr4a function may contribute to memory formation. Further, we have confirmed that memory enhancement by HDAC inhibitors is accompanied by increased expression of Nr4a genes and found that blocking NR4A signaling prevents the HDAC inhibitor TSA from improving long-term memory. Finally, we identified Fosl2 and Bdnf as candidate genes at the critical junction between NR4A signaling and HDAC inhibitors, as these genes have reduced expression when NR4A signaling is blocked and increased expression when an HDAC inhibitor is infused into the hippocampus (Figure 5).
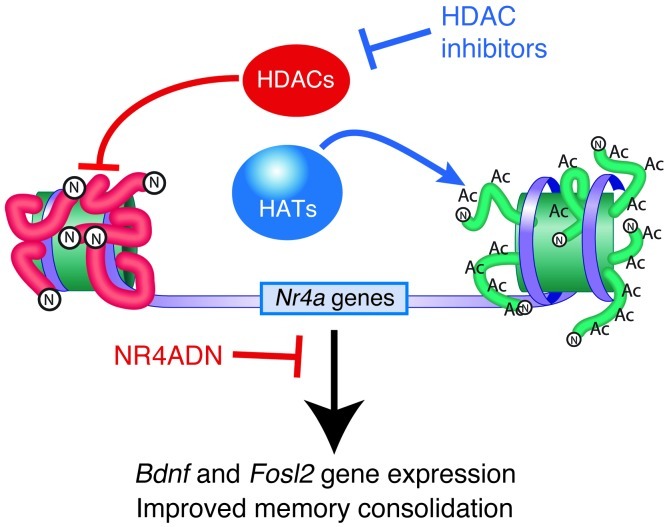
Figure 5. NR4A signaling contributes to memory formation and enhancement by HDAC inhibitors.
HDAC inhibitors increase Nr4a gene expression, and blocking NR4A signaling prevents memory enhancement by HDAC inhibitors, suggesting a model in which NR4A target genes contribute to memory enhancement by HDAC inhibition. In this figure, arrows represent processes that stimulate gene expression and enhance memory formation. In contrast, blunt ends signify pathways that repress gene expression and limit memory formation. Nucleosomes are indicated by the green barrels that are encircled by the gray ribbon, which illustrates promoter DNA. Acetylation (ac) of the histone proteins that constitute the nucleosome (N) is dictated by a dynamic equilibrium between HDAC and HAT activity. Impairing HAT activity would be predicted to reduce Nr4a gene expression and impair memory formation. As illustrated in this study, blocking HDAC activity increases Nr4a gene expression and enhances memory formation. Also, inhibiting the function of NR4A proteins using a dominant-negative protein blocks memory enhancement by HDAC inhibition and impedes expression of several putative NR4A target genes. The increase in Nr4a gene expression observed after TSA injection after training is accompanied by increased expression of the putative NR4A target genes, Bdnf and Fosl2, two memory-associated genes that may contribute to the molecular mechanism of memory enhancement by HDAC inhibitors.
We found that 13 different NR genes have increased expression after contextual fear conditioning. These NRs can be broadly divided into the group: NR4A, TLX, ERR, LXR, PPAR, RAR, and RXR. As discussed earlier, previous studies provided the suggestion that the NR4A group may contribute to memory storage (5–11), and we have provided compelling evidence supporting this possibility in this article. Yet, there is also evidence that the other NR groups identified in this study may also contribute to memory storage. For instance, TLX regulates the proliferation of adult neural stem cells, and conditional knockout of Tlx in the adult brain causes a defect in neurogenesis and spatial memory (32). Perhaps surprisingly, contextual fear memory was not impacted by Tlx deletion. ERRα and ERRβ both show increased expression after fear conditioning. Because ERRs bind to many of the same targets as the ER and estrogen signaling contributes to memory formation (6, 9), the relationship between this increase in ERR expression and estrogen signaling may be an interesting area of future study. Alternatively, the role played by ERRs in mitochondrial function and energy metabolism (33) may suggest that these processes contribute to learning and memory as has been suggested elsewhere (34). LXR (35) and PPAR agonists (36) have been reported to improve learning and memory deficits in Alzheimer disease mouse models, suggesting that defects in signaling for these 2 classes of NRs may contribute to pathological memory deficits. RXR and RAR both respond to retinoid acid signaling, which has been previously linked to learning and memory. Depletion of vitamin A, the dietary source of retinoids, leads to defects in synaptic plasticity (37) and hippocampus-dependent memory (8) that are acutely reversible by supplementing retinoids. Additionally, knockout mice for RARβ and RXRγ show abnormalities in hippocampus-dependent memory and synaptic plasticity (10). Therefore, it is possible that the changes in RXR and RAR expression observed in this study could contribute to memory by altering retinoid sensitivity after learning.
Increased expression of Nr4a genes after learning is consistent with these genes being important activity-dependent targets of CREB (24), a transcription factor involved in long-term memory formation (38). Additionally, memory enhancement by HDAC inhibitors requires the interaction between CREB and the histone acetyltransferase (HAT) CREB-binding protein (CBP) as well as being accompanied by increased gene expression for Nr4a1 and Nr4a2 (13). In other systems, NR4A signaling is mediated by redundant roles of these 3 transcription factors (see ref. 39 for review). For these reasons, we undertook an analysis of NR4A function in memory formation using a well-tested dominant-negative strategy that was developed for the study of NR4A function in other physiological contexts (25, 27). Using this approach, we found that impeding NR4A function impairs long-term memory formation. We have now found that impeding NR4A signaling blocks the ability to enhance memory by HDAC inhibitors, suggesting that this family of NRs may be a useful target for modulating memory function.
Previous studies have found that Nr4a1 gene expression increases in hippocampal area CA1 after contextual fear conditioning (19). Additionally, Nr4a2 gene expression increases within both CA1 and CA3 after spatial exploration (20). In this study, transgenic NR4A dominant-negative protein was expressed under control of the CaMKII promoter, limiting expression to forebrain excitatory neurons, including those in the hippocampus. Behavioral deficit in this transgenic mouse line supports a role for NR4A signaling in hippocampal memory consolidation. Transgene expression in this mouse line was fortuitously restricted to neurons within hippocampal area CA1 and the dentate gyrus. The HDAC inhibitor injection protocol used in this study targets hippocampal area CA1 (13, 40, 41). Together, our data suggest that NR4A proteins act within excitatory pyramidal neurons in area CA1 to support memory consolidation and the enhancement in memory achieved with HDAC inhibitor treatment.
An important future direction is the identification of the mechanism by which NR4A signaling supports memory formation, which is likely to be through the activation of downstream target genes. We found that mice expressing the NR4ADN transgene have impaired expression of genes Pak6, Fosl2, and Bdnf. Both Fosl2 and Bdnf are also potentiated by HDAC inhibitor treatment. Fosl2, also known as Fra2, is a member of the AP-1 family of transcription factors, a family that is known to be important for memory storage (42). FOSL2 is a long-lasting FOS-related antigen that, like ΔFosB, can be retained long after induction (43), suggesting that the persistence of this protein during memory storage may be an interesting subject of future inquiry. Bdnf is a known Nr4a target gene (31) that contributes to memory formation (44). The specific Bdnf promoter 1 that is impaired in NR4ADN mice is activated in a second, late response to neuronal stimulation (45). Thus, regulation of this promoter by NR4A signaling may represent an important mechanism governing Bdnf expression in later waves of transcription after learning, an intriguing idea in light of the growing appreciation that Bdnf contributes to memory at late time points after learning (44). The impact of the NR4ADN transgene on Bdnf expression provides a direct link between NR4A signaling and an effector gene known to be involved in memory formation (44). Additionally, Bdnf expression is potentiated by TSA treatment, suggesting that this gene might contribute to the enhancement in memory observed with TSA administration.
The present findings are especially exciting because Nr4a polymorphisms have been identified in patients with schizophrenia (46), and Nr4a gene expression is reduced in patients with schizophrenia (47). Thus, impaired Nr4a function may contribute to the cognitive impairments that accompany this psychiatric disorder. Agonists for other NRs, such as PPAR and LXR, have shown promise for the treatment of Alzheimer disease (48). Over the last several years, small molecules that increase the activity of one or more NR4A proteins have been identified (49, 50). Our data suggest that the therapeutic value of these NR4A agonists is worthy of further investigation. Future approaches to ameliorate the cognitive impairment associated with neuropsychiatric disorders will greatly benefit from the knowledge that Nr4a family function is required for memory enhancement by HDAC inhibitors. The specific requirement for Nr4a family function in memory enhancement by HDAC inhibitors promises to provide more refined targets for memory improvement than would be possible with even the most specific HDAC inhibitors.
Methods
Subjects.
Mice were maintained under standard conditions, with food and water available ad libitum. Adult mice (2 to 6 months of age) were kept on a 12-hour-light/12-hour-dark cycle, with lights on at 7 AM. All behavioral and biochemical experiments were performed during the light cycle, with training and testing at approximately 10 AM.
The HA-tagged truncated Nr4a1 plasmid (27) (obtained from Jacques J. Tremblay, CRCHUQ, Quebec City, Quebec, Canada) was subcloned in-frame with the YFP tag in the pcDNA6.2 N-YFP-GW TOPO vector (Invitrogen, no. 45-1903). The YFP-HA-NR4ADN fragment was ligated into the EcoRV site of MM400 to place the YFP-NR4ADN into a hybrid intron structure under control of the tetO (30). MM400-YFP-HA-NR4ADN was purified by CsCl gradient centrifugation. The 2.7-kb tetO-YFP-HA-NR4ADN transgene fragment was injected into pronuclei of C57BL/6 zygotes (Transgenic and Chimeric Mouse Facility at the University of Pennsylvania). Founders were crossed to C57BL/6J mice bearing the CaMKII-tTA (line B) transgene (30). Genotyping was performed by Southern blotting using transgene-specific probes and/or PCR.
Behavior.
Fear conditioning was performed as previously described (13), with handling for 3 days prior to conditioning. Briefly, the conditioning protocol entailed a single 2-second, 1.5-mA foot shock, terminating at 2.5 minutes after placement of the mouse in the chamber. Testing was performed at 1 hour or 24 hours after training over a 5-minute interval. For experiments involving TSA injection, 0.75-mA shock intensity was used to avoid a ceiling effect. Cued fear conditioning was performed as described for contextual fear conditioning, except that a 30-second cue (white noise) co-terminated with a 0.75-mA foot shock. For cued testing, mice were placed into a new context (a distinct conditioning chamber with smooth flat flooring, altered chamber dimensions, and a different odorant) for 2-minutes in the absence of the conditioned stimulus (white noise), followed by a 3-minute presentation of the auditory conditioned stimulus. Conditioning was quantified by measuring freezing behavior, the absence of nonrespiratory movement (14), using automated scoring software (Clever Systems).
Intrahippocampal TSA injection.
Based on previous work (13, 40, 41), bilateral 22-gauge guide cannula were implanted 1 week prior to training at the following coordinates: anteroposterior, –1.7 mm; mediolateral, ± 1.5 mm; 1.5 mm dorsoventral. Injection cannula extended 0.7 mm below the guide cannula. TSA (16.5 mM, AG Scientific) or vehicle (50% ethanol) was injected at a rate of 0.5 μl/min for 1 minute immediately after training.
RNA preparation.
Hippocampal dissections were performed on ice after conditioning alternating between control and experimental groups. RNA was prepared using a modified TRIzol RNA extraction, followed by RNeasy (Qiagen) purification and DNA-free (Ambion) DNase treatment. RNA concentration was ascertained using a NanoDrop spectrophotometer (ThermoFisher Scientific).
cDNA synthesis.
Real-time PCR template cDNA was produced using the RETROscript Kit (Ambion). For each reaction, 1 μg total RNA was added to a 20 μl total reaction volume composed of 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 5 mM dithiothreitol, 500 μM each dNTP, 5 μM random decamer, 10 units RNase inhibitor, and 100 units MMLV-RT. Control reactions were performed lacking template or reverse transcriptase. Reactions were performed at 44°C for 1 hour, followed by heat inactivation at 100°C for 10 minutes. Reactions were diluted to 2 ng/μl in water to 500 μl final volume.
Real-time RT-PCR.
High-throughput qPCR was performed as described previously (17). Briefly, 10 ng cDNA was assayed using multiplexed TaqMan qPCR probes. Data were normalized to 18S rRNA, and t tests were performed against the appropriate circadian control group. Subsequent, low-throughput qPCR was performed on the ABI7500 Fast or Viia7 Real-Time PCR system using 3 separate housekeeper genes for normalization (Gapdh, Tuba4a, and Hprt). Relative quantification of gene expression was based on the ABI users’ bulletin using a ΔΔCt method and described previously (13). Fold difference in mean value for biological replicates is presented, and all samples are distinct biological replicates.
Immunolabeling.
For immunolabeling experiments, mice were transcardially perfused with 4% PFA, and 30-μM coronal cryostat sections were prepared. Sections were permeabilized in 0.1% Triton X-100 and then incubated in 1% H2O2 in PBS for 15 minutes at room temperature. Sections were given 3 washes for 5 minutes each before being blocked in PBS with 5% preimmune serum and 0.3% Triton X-100 for 50 minutes at room temperature. Sections were triple washed for 5 minutes each in PBS and then incubated overnight at 4°C in PBS with 2% preimmune serum (same species as that of secondary antibody; Jackson ImmunoResearch Inc.), 0.3% Triton X-100, and primary antibody (anti-GFP, 1:1,000, Invitrogen no. A11122; anti-HA, 1:1,000, Roche clone 3F10). Sections were triple washed for 5 minutes each in PBS, followed by incubation in biotinylated secondary antibody for 2 hours at room temperature in PBS. For HA immunohistochemistry, biotinylated goat anti-rat antibody (1:1,000, Jackson ImmunoResearch Inc.) was used as secondary antibody, and biotinylated donkey anti-rabbit antibody (1:1,000, Jackson ImmunoResearch Inc.) was used for the remaining labeling. Three 5-minute washes in PBS were followed with incubation for 1.5 hours at room temperature in Vectastain ABC solution (Vector Laboratories). Sections were again triple washed in PBS, incubated for 10 to 15 minutes in 0.2 mg/ml 3,3′-diaminobenzidine in PBS, and then triple washed in PBS with sodium azide (0.01%). Stained sections were mounted onto glass microscope slides in 0.7% gelatin. Sections were examined and digitally photographed through a light microscope. For immunofluorescence, Alexa Fluor 488–coupled chicken anti-rabbit IgG (1:5,000; Invitrogen) was used as secondary antibody. After incubation in fluorescent antibody, sections were washed 3 times in PBS for 5 minutes each and then mounted on to glass slides in 50% glycerol/PBS containing 20 μg/ml propidium iodide. Images were captured using a Leica (Wetzlar) TCS SP2 confocal microscope.
Coimmunoprecipitation.
Mice were cervically dislocated 30 minutes after placement in a novel context. Hippocampi were dissected and flash frozen on dry ice. Hippocampi were homogenized in 1 ml of hypotonic lysis buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 1 mM EDTA, plus protease and phosphatase inhibitors [Sigma-Aldrich]), followed by rocking at 4°C for 30 minutes, and then centrifugation at 1,000 × g for 15 minutes at 4°C to pellet nuclei. Nuclear pellets were resuspended in 300 μl E1A lysis buffer (250 mM NaCl, 0.1% Nonidet P-40, 50 mM HEPES [pH 7.0], 5 mM EDTA, plus protease and phosphatase inhibitors [Sigma-Aldrich]) and gently rocked at 4°C for 30 minutes. Nuclear lysates were centrifuged at 1,000 × g for 15 minutes and 4°C to pellet nuclear debris. Supernatants were diluted to 1 ml in E1A lysis buffer containing 1 μg biotinylated anti-GFP (Invitrogen) and gently rocked overnight at 4°C. For immunoprecipitation, 50 μl of Pierce streptavidin magnetic beads (Thermo) were then added to the protein solution on the following day. Beads were washed 3 times in E1A buffer, and antigens were eluted in 30 μl of 0.1 M glycine (pH 2.5).
Western blotting.
NuPage SDS loading buffer and 2-mercaptoethanol (Invitrogen) were added to samples prior to a 10-minute incubation at 100°C. Proteins were resolved by polyacrylamide electrophoresis on a NuPage Bis-Tris gels (Invitrogen) and transferred to a PVDF membrane (Invitrogen) for Western blotting. Blocking was performed with 5% milk. Washes and primary antibody incubations were performed in either TBS-0.1% Tween-20 (anti-AcH3k9/14, 1:5,000, Millipore 06-599; anti-H3, 1:5,000, Abcam ab1791; anti–β-tubulin, 1:20,000, Sigma-Aldrich T4026; anti-NR4A1, 1:1,000, eBiosciences 14-5965; anti-Nr4a3, 1:1,000, Abcam ab41918) or PBS-0.1% Tween-20 (anti-NR4A2, 1:2,000, Santa Cruz Biotechnology Inc. SC991). Three 5-minute washes were followed by incubation in PBST with HRP-conjugated goat anti-rabbit secondary antibody (1:1000, Santa Cruz Biotechnology Inc.). The blot was washed 3 times in PBST for 5 minutes each and treated with ECL Western Blotting Detection Reagent (GE Healthcare) for chemiluminescence detection. Membranes were then exposed to film for 1 to 10 minutes and developed for analysis.
Statistics.
Data are expressed as mean ± SEM. For behavioral experiments, ANOVAs were performed, followed by Student-Newman-Keuls post-hoc tests. Gene expression survey of NRs was analyzed with 2-tailed t tests. Follow-up real-time qPCR data were analyzed with ANOVAs to detect overall effects of training and nonparametric Kruskal-Wallis ANOVAs for planned comparisons to home cage controls. High-throughput qPCR data to detect NR4ADN target genes was performed using ANOVAs with training and genotype as factors. For all statistical tests, P < 0.05 was considered significant. Experimenters were blind to genotype, and genotypes were confirmed after experiments were completed.
Study approval.
All animal care and experiments were consistent with NIH guidelines and approved by the Institutional Care and Use Committee at the University of Pennsylvania.
Supplementary Material
Acknowledgments
We thank Jacques Tremblay for providing the truncated Nr4a1 clone and Jean Richa for transgene injections. Also, we thank Peter Giese, Julie Blendy, and Robbert Havekes for comments on the article and experimental design. This work was funded by NIH grants MH060244 (to T. Abel), U19DK62434 (to D.J. Manglesdorf), and GM007062 (to A.L. Bookout); Howard Hughes Medical Institute (to D.J. Manglesdorf); and the Robert A. Welch Foundation (grant I-1275 to D.J. Manglesdorf). Funding from the UPENN Provost Summer Program supported M.E. Sulewski. Further funding from NIH Predoctoral Training Program in Genetics (T32-GM-008216-20) provided support to J.D. Hawk.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(10):3593–3602. doi:10.1172/JCI64145.
References
- 1.Hawk JD, Abel T. Encyclopedia of Behavioral Neuroscience. 2010. Role of gene transcription in long–term memory storage. In: Koob GF, Le Moal M, Thompson RF, eds. pp. 161–179. Vol. 3. Oxford, United Kingdom: Academic Press; [Google Scholar]
- 2.Bourtchouladze R, Abel T, Berman N, Gordon R, Lapidus K, Kandel ER. Different training procedures recruit either one or two critical periods for contextual memory consolidation, each of which requires protein synthesis and PKA. Learn Mem. 1998;5(4–5):365–374. [PMC free article] [PubMed] [Google Scholar]
- 3.Igaz LM, Vianna MRM, Medina JH, Izquierdo I. Two time periods of hippocampal mRNA synthesis are required for memory consolidation of fear-motivated learning. J Neurosci. 2002;22(15):6781–6789. doi: 10.1523/JNEUROSCI.22-15-06781.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mangelsdorf DJ, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83(6):835–839. doi: 10.1016/0092-8674(95)90199-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roozendaal B, et al. Membrane-associated glucocorticoid activity is necessary for modulation of long-term memory via chromatin modification. J Neurosci. 2010;30(14):5037–5046. doi: 10.1523/JNEUROSCI.5717-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu F, et al. Activation of estrogen receptor-beta regulates hippocampal synaptic plasticity and improves memory. Nat Neurosci. 2008;11(3):334–343. doi: 10.1038/nn2057. [DOI] [PubMed] [Google Scholar]
- 7.Gemma C, et al. Rosiglitazone improves contextual fear conditioning in aged rats. Neuroreport. 2004;15(14):2255–2259. doi: 10.1097/00001756-200410050-00023. [DOI] [PubMed] [Google Scholar]
- 8.Cocco S, et al. Vitamin A deficiency produces spatial learning and memory impairment in rats. NeuroScience. 2002;115(2):475–482. doi: 10.1016/S0306-4522(02)00423-2. [DOI] [PubMed] [Google Scholar]
- 9.Day M, Sung A, Logue S, Bowlby M, Arias R. Beta estrogen receptor knockout (BERKO) mice present attenuated hippocampal CA1 long-term potentiation and related memory deficits in contextual fear conditioning. Behav Brain Res. 2005;164(1):128–131. doi: 10.1016/j.bbr.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 10.Chiang MY, et al. An essential role for retinoid receptors RARbeta and RXRgamma in long-term potentiation and depression. Neuron. 1998;21(6):1353–1361. doi: 10.1016/S0896-6273(00)80654-6. [DOI] [PubMed] [Google Scholar]
- 11.Rojas P, Joodmardi E, Hong Y, Perlmann T, Ogren SO. Adult mice with reduced Nurr1 expression: an animal model for schizophrenia. Mol Psychiatry. 2007;12(8):756–766. doi: 10.1038/sj.mp.4001993. [DOI] [PubMed] [Google Scholar]
- 12.Keeley MB, et al. Differential transcriptional response to nonassociative and associative components of classical fear conditioning in the amygdala and hippocampus. Learn Mem. 2006;13(2):135–142. doi: 10.1101/lm.86906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vecsey CG, et al. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J Neurosci. 2007;CREB(23):6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maren S, Quirk GJ. Neuronal signalling of fear memory. Nat Rev Neurosci. 2004;5(11):844–852. doi: 10.1038/nrn1535. [DOI] [PubMed] [Google Scholar]
- 15.Stanciu M, Radulovic J, Spiess J. Phosphorylated cAMP response element binding protein in the mouse brain after fear conditioning: relationship to Fos production. Brain Res Mol Brain Res. 2001;94(1–2):15–24. doi: 10.1016/S0169-328X(01)00174-7. [DOI] [PubMed] [Google Scholar]
- 16.Bekinschtein P, Cammarota M, Igaz LM, Bevilaqua LR, Izquierdo I, Medina JH. Persistence of long-term memory storage requires a late protein synthesis- and BDNF- dependent phase in the hippocampus. Neuron. 2007;53(2):261–277. doi: 10.1016/j.neuron.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 17.Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126(4):789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laudet V. Evolution of the nuclear receptor superfamily: early diversification from an ancestral orphan receptor. J Mol Endocrinol. 1997;19(3):207–226. doi: 10.1677/jme.0.0190207. [DOI] [PubMed] [Google Scholar]
- 19.von Hertzen LS, Giese KP. Memory reconsolidation engages only a subset of immediate-early genes induced during consolidation. J Neurosci. 2005;25(8):1935–1942. doi: 10.1523/JNEUROSCI.4707-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pena de Ortiz S, Maldonado-Vlaar CS, Carrasquillo Y. Hippocampal expression of the orphan nuclear receptor gene hzf-3/nurr1 during spatial discrimination learning. Neurobiol Learn Mem. 2000;74(2):161–178. doi: 10.1006/nlme.1999.3952. [DOI] [PubMed] [Google Scholar]
- 21.Colon-Cesario WI, et al. Knockdown of Nurr1 in the rat hippocampus: implications to spatial discrimination learning and memory. Learn Mem. 2006;13(6):734–744. doi: 10.1101/lm.407706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McQuown SC, et al. HDAC3 is a critical negative regulator of long-term memory formation. J Neurosci. 2011;31(2):764–774. doi: 10.1523/JNEUROSCI.5052-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, et al. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature. 2003;423(6939):555–560. doi: 10.1038/nature01645. [DOI] [PubMed] [Google Scholar]
- 24.Lemberger T, Parkitna JR, Chai M, Schutz G, Engblom D. CREB has a context-dependent role in activity-regulated transcription and maintains neuronal cholesterol homeostasis. FASEB J. 2008;22(8):2872–2879. doi: 10.1096/fj.08-107888. [DOI] [PubMed] [Google Scholar]
- 25.Cheng LE, Chan FK, Cado D, Winoto A. Functional redundancy of the Nur77 and Nor-1 orphan steroid receptors in T-cell apoptosis. EMBO J. 1997;16(8):1865–1875. doi: 10.1093/emboj/16.8.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ke N, et al. Nuclear hormone receptor NR4A2 is involved in cell transformation and apoptosis. Cancer Res. 2004;64(22):8208–8212. doi: 10.1158/0008-5472.CAN-04-2134. [DOI] [PubMed] [Google Scholar]
- 27.Robert NM, Martin LJ, Tremblay JJ. The orphan nuclear receptor NR4A1 regulates insulin-like 3 gene transcription in Leydig cells. Biol Reprod. 2006;74(2):322–330. doi: 10.1095/biolreprod.105.044560. [DOI] [PubMed] [Google Scholar]
- 28.Saijo K, et al. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell. 2009;137(1):47–59. doi: 10.1016/j.cell.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kadkhodaei B, et al. Nurr1 is required for maintenance of maturing and adult midbrain dopamine neurons. J Neurosci. 2009;29(50):15923–15932. doi: 10.1523/JNEUROSCI.3910-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, Kandel ER. Control of memory formation through regulated expression of a CaMKII transgene. Science. 1996;274(5293):1678–1683. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- 31.Volpicelli F, et al. Bdnf gene is a downstream target of Nurr1 transcription factor in rat midbrain neurons in vitro. J Neurochem. 2007;102(2):441–453. doi: 10.1111/j.1471-4159.2007.04494.x. [DOI] [PubMed] [Google Scholar]
- 32.Zhang CL, Zou Y, He W, Gage FH, Evans RM. A role for adult TLX-positive neural stem cells in learning and behaviour. Nature. 2008;451(7181):1004–1007. doi: 10.1038/nature06562. [DOI] [PubMed] [Google Scholar]
- 33.Giguere V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr Rev. 2008;29(6):677–696. doi: 10.1210/er.2008-0017. [DOI] [PubMed] [Google Scholar]
- 34.Weeber EJ, et al. The role of mitochondrial porins and the permeability transition pore in learning and synaptic plasticity. J Biol Chem. 2002;277(21):18891–18897. doi: 10.1074/jbc.M201649200. [DOI] [PubMed] [Google Scholar]
- 35.Riddell DR, et al. The LXR agonist TO901317 selectively lowers hippocampal Abeta42 and improves memory in the Tg2576 mouse model of Alzheimer’s disease. Mol Cell Neurosci. 2007;34(4):621–628. doi: 10.1016/j.mcn.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 36.Pedersen WA, McMillan PJ, Kulstad JJ, Leverenz JB, Craft S, Haynatzki GR. Rosiglitazone attenuates learning and memory deficits in Tg2576 Alzheimer mice. Exp Neurol. 2006;199(2):265–273. doi: 10.1016/j.expneurol.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 37.Misner DL, et al. Vitamin A deprivation results in reversible loss of hippocampal long-term synaptic plasticity. Proc Natl Acad Sci U S A. 2001;98(20):11714–11719. doi: 10.1073/pnas.191369798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Josselyn SA, Nguyen PV. CREB, synapses and memory disorders: past progress and future challenges. Curr Drug Targets CNS Neurol Disord. 2005;4(5):481–497. doi: 10.2174/156800705774322058. [DOI] [PubMed] [Google Scholar]
- 39.Hawk JD, Abel T. The role of NR4A transcription factors in memory formation. Brain Res Bull. 2011;85(1–2):21–29. doi: 10.1016/j.brainresbull.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oliveira AM, Hawk JD, Abel T, Havekes R. Post-training reversible inactivation of the hippocampus enhances novel object recognition memory. Learn Mem. 2010;17(3):155–160. doi: 10.1101/lm.1625310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hawk JD, Florian C, Abel T. Post-training intrahippocampal inhibition of class I histone deacetylases enhances long-term object-location memory. Learn Mem. 2011;18(6):367–370. doi: 10.1101/lm.2097411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fleischmann A, et al. Impaired long-term memory and NR2A-type NMDA receptor-dependent synaptic plasticity in mice lacking c-Fos in the CNS. J Neurosci. 2003;23(27):9116–9122. doi: 10.1523/JNEUROSCI.23-27-09116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nankova BB, Rivkin M, Kelz M, Nestler EJ, Sabban EL. Fos-related antigen 2: potential mediator of the transcriptional activation in rat adrenal medulla evoked by repeated immobilization stress. J Neurosci. 2000;20(15):5647–5653. doi: 10.1523/JNEUROSCI.20-15-05647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cunha C, Brambilla R, Thomas KL. A simple role for BDNF in learning and memory? Front Mol Neurosci. 2010;3:1. doi: 10.3389/neuro.02.001.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marmigere F, Rage F, Tapia-Arancibia L. Regulation of brain-derived neurotrophic factor transcripts by neuronal activation in rat hypothalamic neurons. J Neurosci Res. 2001;66(3):377–389. doi: 10.1002/jnr.1230. [DOI] [PubMed] [Google Scholar]
- 46.Buervenich S, et al. NURR1 mutations in cases of schizophrenia and manic-depressive disorder. Am J Med Genet. 2000;96(6):808–813. doi: 10.1002/1096-8628(20001204)96:6<808::AID-AJMG23>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 47.Xing G, Zhang L, Russell S, Post R. Reduction of dopamine-related transcription factors Nurr1 and NGFI-B in the prefrontal cortex in schizophrenia and bipolar disorders. Schizophr Res. 2006;84(1):36–56. doi: 10.1016/j.schres.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 48.Mandrekar-Colucci S, Landreth GE. Nuclear receptors as therapeutic targets for Alzheimer’s disease. Expert Opin Ther Targets. 2011;15(9):1085–1097. doi: 10.1517/14728222.2011.594043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dubois C, Hengerer B, Mattes H. Identification of a potent agonist of the orphan nuclear receptor Nurr1. ChemMedChem. 2006;1(9):955–958. doi: 10.1002/cmdc.200600078. [DOI] [PubMed] [Google Scholar]
- 50.Zhan Y, et al. Cytosporone B is an agonist for nuclear orphan receptor Nur77. Nat Chem Biol. 2008;4(9):548–556. doi: 10.1038/nchembio.106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.