Abstract
Serotonin is a critical regulator of bone mass, fulfilling different functions depending on its site of synthesis. Brain-derived serotonin promotes osteoblast proliferation, whereas duodenal-derived serotonin suppresses it. To understand the molecular mechanisms of duodenal-derived serotonin action on osteoblasts, we explored its transcriptional mediation in mice. We found that the transcription factor FOXO1 is a crucial determinant of the effects of duodenum-derived serotonin on bone formation We identified two key FOXO1 complexes in osteoblasts, one with the transcription factor cAMP-responsive element–binding protein 1 (CREB) and another with activating transcription factor 4 (ATF4). Under normal levels of circulating serotonin, the proliferative activity of FOXO1 was promoted by a balance between its interaction with CREB and ATF4. However, high circulating serotonin levels prevented the association of FOXO1 with CREB, resulting in suppressed osteoblast proliferation. These observations identify FOXO1 as the molecular node of an intricate transcriptional machinery that confers the signal of duodenal-derived serotonin to inhibit bone formation.
Introduction
The serotonergic system is an evolutionary conserved system that is the target of stress stimuli (1, 2). In mammals, serotonin is made in the CNS, in which it acts as a neurotransmitter, as well as in peripheral tissues, in which it acts as a hormone via serotonin receptors present in these tissues (3–5). Serotonin does not cross the blood-brain barrier; hence, the 2 pools of serotonin may act independently of each other. Fluctuations in serotonin signaling in the brain or in peripheral tissues affect a wide spectrum of physiological processes, such as satiety, energy expenditure, thermoregulation, and immunity (6). Serotonin is also a potent regulator of bone mass. Consistent with the existence of 2 independent pools, serotonin exerts an opposite influence on bone mass accrual depending on its site of synthesis. In the brain, it suppresses sympathetic tone activity and promotes osteoblast proliferation (7). In contrast, serotonin produced by the gut is released in the circulation and acts as a hormone inhibiting osteoblast proliferation (8).
The suppressing function of gut-derived serotonin in osteoblast proliferation is central to bone homeostasis, because it directly applies to human diseases. Indeed, gut-derived serotonin is suppressed by low-density lipoprotein receptor-related protein 5 (LRP5), a potent regulator of bone mass that controls bone formation by promoting osteoblast proliferation (9). That gain-of-function mutations in LRP5 cause high bone mass (10–12) by increasing bone formation led to the hypothesis that inhibiting gut-derived serotonin synthesis may become a treatment for low bone mass diseases such as osteoporosis (13). Hence, it is imperative to decipher as precisely as possible the mechanism of action of gut-derived serotonin in osteoblasts.
Since serotonin is conserved from C. elegans to humans, knowledge gathered in model organisms can be used to address this question. In C. elegans daf-16, the worm homolog of FOXO transcription factors mediates serotonin signaling in response to adverse environmental conditions (14). Serotonin promotes FOXO1 nuclear accumulation to induce stress resistance via genes involved in detoxification or metabolism and to maintain survival. As daf-16 does in worms, FOXO1 in mammals regulates a balance between cell proliferation and survival. In health, FOXO1 promotes cell proliferation; however, in stress, it suppresses cell proliferation to promote cell survival by inducing cell cycle arrest and quiescence (reviewed in refs. 15, 16). Small alterations in the transcriptional activity of FOXO1 influence this balance and can profoundly change the biological effect of FOXO1 actions and convert activating to repressing functions (17).
In this study, we asked whether, as it is the case in C. elegans, serotonin also recruits FOXO1 to mediate its functions in osteoblasts. Here, we provide evidence that gut-derived serotonin inhibits osteoblast proliferation by a mechanism that finely tunes the levels on FOXO1 transcriptional activity by controlling a functional balance between 2 FOXO1 complexes, one with cAMP-responsive element–binding protein 1 (CREB) and another with activating transcription factor 4 (ATF4).
Results
Foxo1 haploinsufficiency rescues the low bone mass phenotype caused by high circulating serotonin levels.
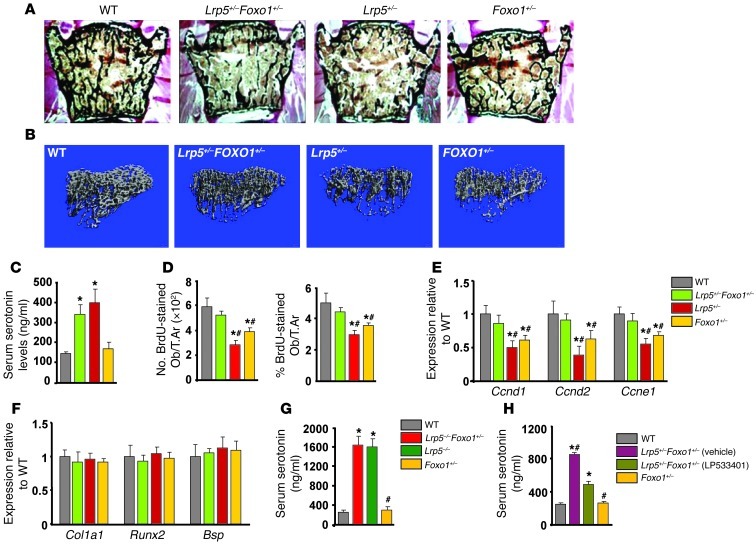
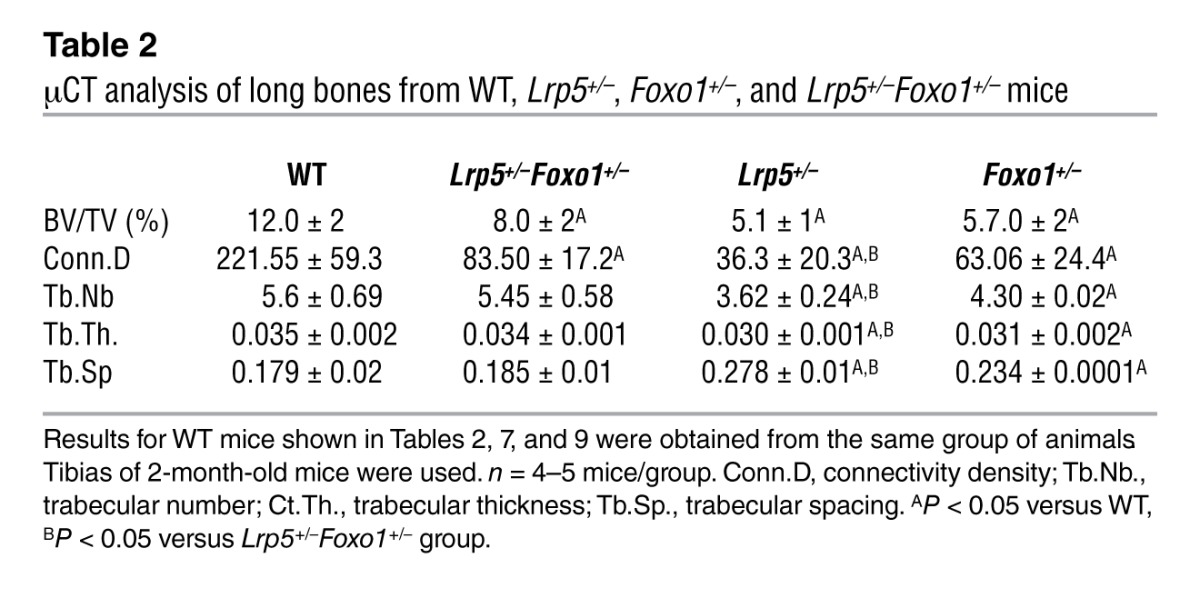
To determine whether FOXO1 is involved in mediating the antiproliferative effects of increased circulating serotonin levels, we used Lrp5-deficient mice in which serotonin levels are high as a tool (8). Specifically, we generated compound heterozygote Lrp5+/–Foxo1+/– animals and compared their bone phenotype with that of WT, Lrp5+/–, and Foxo1+/– littermates. As expected Lrp5+/– and Foxo1+/– mice showed decreased bone volume, bone formation rate, and osteoblast numbers as compared with those of WT controls (Figure 1, A and B, and Tables 1 and 2). Circulating serotonin levels increased in Lrp5+/– and Lrp5+/–Foxo1+/– mice (Figure 1C), whereas osteoblast proliferation was suppressed (Figure 1D). Surprisingly, removing one allele of Foxo1 reversed the decrease in osteoblast numbers and the low bone formation phenotype of Lrp5+/– mice by reversing the decrease in osteoblast proliferation (Figure 1, A–C, and Tables 1 and 2). Explaining the bone mass observations, Foxo1 haploinsufficiency prevented the decrease in cyclin D1 (Ccnd1), cyclin D2 (Ccnd2), and cyclin E1 (Ccne1) expression in Lrp5+/–Foxo1+/– bones (Figure 1E). Osteoblast differentiation was not affected in Lrp5+/–Foxo1+/– mice, as evidenced by the lack of changes in the expression of the osteoblast-differentiation markers Runx2, collagen 1 α 1(Col1a1), and bone sialoprotein (Bsp) (Figure 1F). Osteoclastogenesis was also not affected (Table 1).
Figure 1. Foxo1 haploinsufficiency rescues the low bone formation phenotype of Lrp5+/– mice.
(A) Representative vertebral section images from 2-month-old mice. Mineralized bone matrix is stained in black by Von Kossa reagent. Original magnification, ×4. (B) Representative 3D images of transverse CT slices of proximal tibia obtained by high-resolution μCT analysis. (C) Serum serotonin levels (n = 4 mice/group). (D) Osteoblast proliferation as the number of BrdU-stained osteoblasts (Obs) per trabecular area (T.Ar.) or as the percentage of total osteoblasts per trabecular area (n = 6 mice/group) in sections of femurs. (C and D) *P < 0.05 versus WT, #P < 0.05 versus Lrp5+/–Foxo1+/– group. (E and F) Real-Time PCR analysis of indicated genes in bone (n = 6 mice/group). *P < 0.05 versus WT, #P < 0.05 versus Lrp5+/–Foxo1+/– group. (G) Serum serotonin levels (n = 4 mice/group). *P < 0.05 versus WT, #P < 0.05 versus Lrp5–/–Foxo1+/– group. (H) Serum serotonin levels in WT, Lrp5+/–Foxo1+/–, and LP533401-treated Lrp5+/–Foxo1+/– mice. The gut-derived serotonin synthesis inhibitor LP533401 was administered as indicated at 200 mg/kg/d by oral gavage for 4 weeks. Mice were 2 months old (n = 6 mice/group). *P < 0.05 versus WT, #P < 0.05 versus LP533401-treated Lrp5+/–Foxo1+/– group.
Table 1.
Foxo1 haploinsufficiency rescues the low bone formation phenotype of Lrp5+/– mice
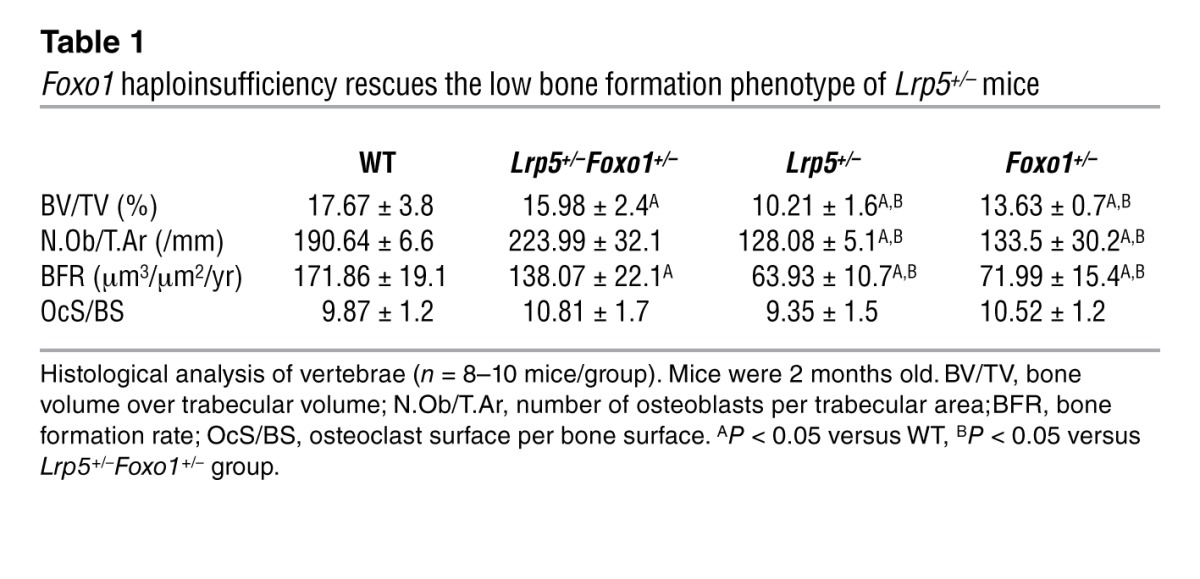
Table 2.
μCT analysis of long bones from WT, Lrp5+/–, Foxo1+/–, and Lrp5+/–Foxo1+/– mice
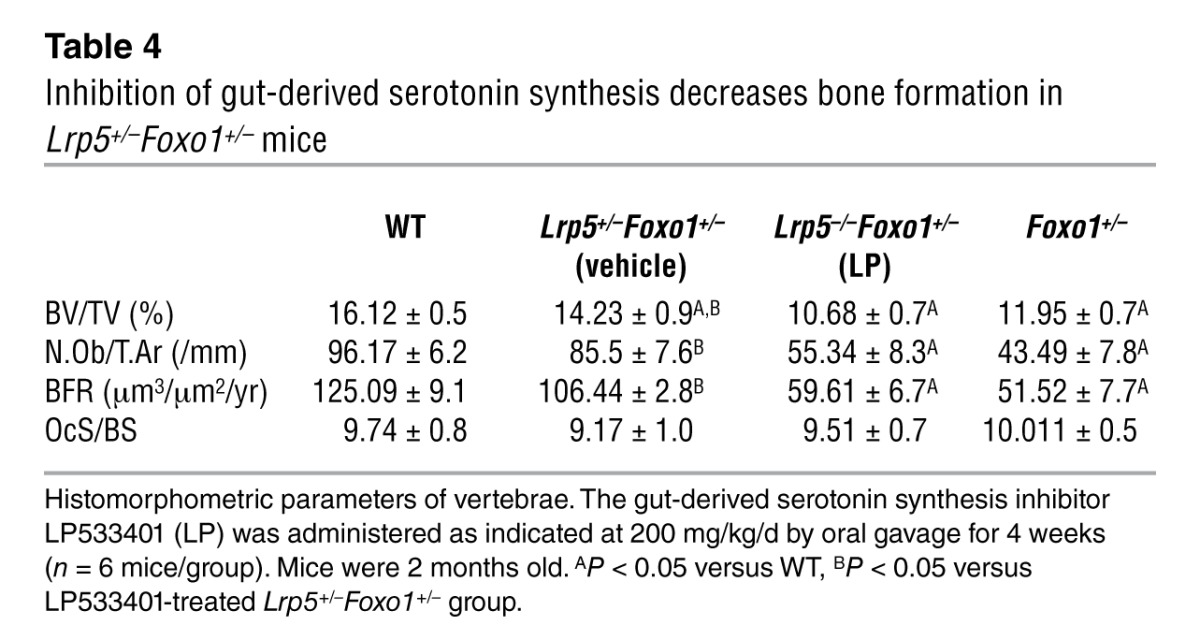
Similar to the observations in mice with double haploinsufficiency, removal of one Foxo1 allele corrected the low osteoblast number and low bone formation phenotype of Lrp5–/– mice (Table 3). Lrp5–/–Foxo1+/– mice showed increased circulating serotonin levels (Figure 1G). Collectively, these observations indicated that when circulating serotonin levels are high, FOXO1 has a role opposite to the one seen in WT mice. It is suppressing, rather than promoting, osteoblast proliferation. To confirm this hypothesis, we examined whether normalization of gut-derived serotonin levels in Lrp5+/– mice would reestablish the role of FOXO1 in promoting osteoblast proliferation. In support of our conclusion, normalization of duodenal-derived serotonin levels in Lrp5+/–Foxo1+/– mice by chemical inhibition of its synthesis reverted the rescue of low bone formation in Lrp5+/– mice and led to a low bone mass phenotype, characteristic of Foxo1+/– mice (Table 4 and Figure 1H). Our observations suggest that, in the presence of high serotonin levels in Lrp5+/– or Lrp5–/– mice, FOXO1 mediates the antiproliferative actions of serotonin. Removal of one Foxo1 allele in Foxo1+/–Lrp5+/– or Foxo1+/–Lrp5–/– mice, which also show high serotonin levels, rescues the suppressive effect of serotonin in osteoblast proliferation.
Table 3.
Foxo1 haploinsufficiency rescues the low bone formation phenotype of Lrp5–/– mice
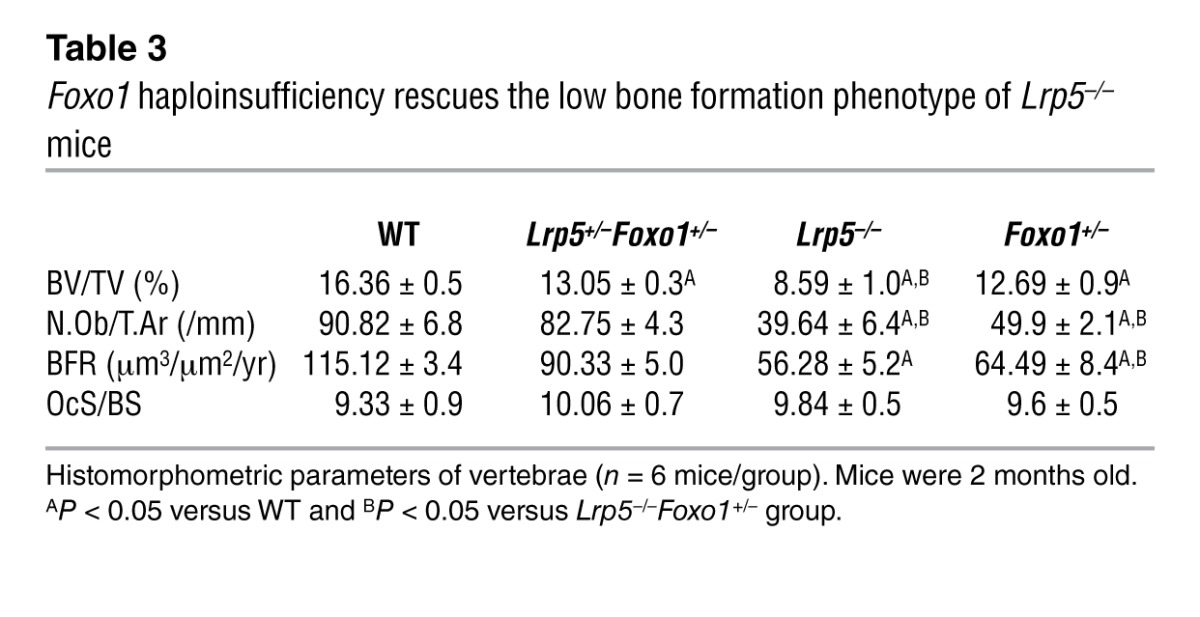
Table 4.
Inhibition of gut-derived serotonin synthesis decreases bone formation in Lrp5+/–Foxo1+/– mice
Circulating serotonin suppresses osteoblast proliferation through FOXO1 and FOXO1 target genes.
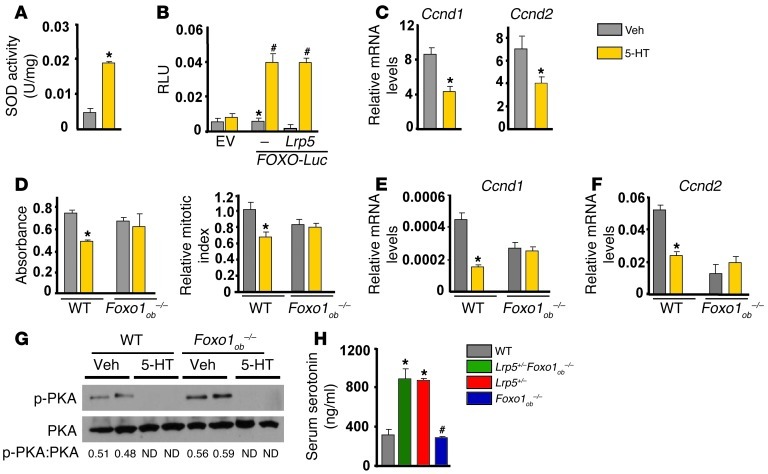
The rescue of the low bone mass phenotype of Lrp5+/– mice caused by Foxo1 haploinsufficiency in osteoblasts was unexpected, since osteoblast-specific deletion of Foxo1 also leads to a decrease in osteoblast numbers (18). To explain this discrepancy, we asked whether FOXO1 activity is regulated in the case of Lrp5 deficiency, by increased serotonin, a hormone specifically produced under this condition. Indeed, similar to Lrp5+/– mice, Lrp5+/–Foxo1+/– animals showed increased circulating serotonin levels (Figure 1C). Serotonin upregulated the activity of the FOXO1 target gene Sod2 in osteoblasts (Figure 2A). It also upregulated FOXO-mediated transcription on a FOXO reporter carrying a FOXO target sequence (19) in the Igf-1 promoter in osteoblasts (Figure 2B). This effect of serotonin was independent of LRP5, since transfection of an Lrp5 expression construct had no effect on FOXO1 activity unless serotonin was added to the medium (Figure 2B).
Figure 2. Serotonin suppresses osteoblast proliferation through FOXO1.
(A) SOD2 activity in osteoblasts treated with serotonin (5-HT; 100 μM) for 6 hours (n = 3). *P < 0.05 versus vehicle. (B) Calvaria cells cotransfected with Lrp5 and FOXO-Luc plasmids and treated with vehicle or 100 μM serotonin for 24 hours (n = 3). EV, empty vector. *P < 0.05 versus empty vector, #P < 0.05 versus FOXO-Luc vehicle. (C) Real-time PCR analysis of the expression of Ccnd1 and Ccnd2 in calvaria cells treated with serotonin (100 μM) for 6 hours (n = 3). *P < 0.05 versus vehicle. (D) Calvaria cells from WT and Foxo1ob–/– mice were treated with serotonin (100 μM) for 48 hours, and proliferation was assessed by Cell Titer One (left) or BrdU incorporation (right) (n = 3). *P < 0.05 versus vehicle. (E and F) Calvaria cells from WT and Foxo1ob–/– mice were treated with serotonin (100 μM) for 48 hours. Gene expression of (E) Ccnd1 and (F) Ccnd2 (n = 3). *P < 0.05 versus vehicle. (G) Western blot analysis showing phosphorylation of PKA in WT and Foxo1-deficient osteoblasts treated with serotonin (100 μM) for 30 minutes. (H) Serum serotonin levels (n = 4 mice/group). *P < 0.05 versus WT, #P < 0.05 versus Lrp5+/–Foxo1ob–/– group.
Next, we examined whether FOXO1 is required for the antiproliferative effects of gut-derived serotonin in osteoblasts. To this end, we first examined its effect on the expression of FOXO1 transcriptional targets that control cell cycle. Treatment of osteoblasts with serotonin suppressed expression of Ccnd1 and Ccnd2 (Figure 2C). However, whereas serotonin suppressed proliferation of WT osteoblasts, it had no effect on Foxo1–/– osteoblasts (Figure 2D). Consistent with these results, serotonin failed to suppress expression of Ccnd1 and Ccnd2 in Foxo1–/– osteoblasts (Figure 2, E and F). However, it suppressed PKA phosphorylation in Foxo1–/– osteoblasts as efficiently as in WT cells, indicating that serotonin signaling upstream of FOXO1 is normal in the absence of FOXO1 (Figure 2G). Confirming the in vitro observations in Foxo1-deficient osteoblasts, complete inactivation of Foxo1 in osteoblasts of Lrp5+/–Foxo1ob–/– mice resulted in a low bone formation phenotype that was similar to that of Foxo1ob–/– animals (Table 5). High levels of circulating serotonin in Lrp5+/–Foxo1ob–/– mice (Figure 2H), caused by inactivation of one Lrp5 allele, were not able to further suppress osteoblast numbers. These results indicate that gut-derived serotonin requires FOXO1 to exert its antiproliferative effects and that changes in FOXO1 activity caused by Lrp5 deficiency result from the increase in circulating serotonin levels.
Table 5.
Serotonin suppresses osteoblast proliferation through osteoblast-expressed FOXO1
Serotonin regulates FOXO1 activity via the Htr1b receptor.
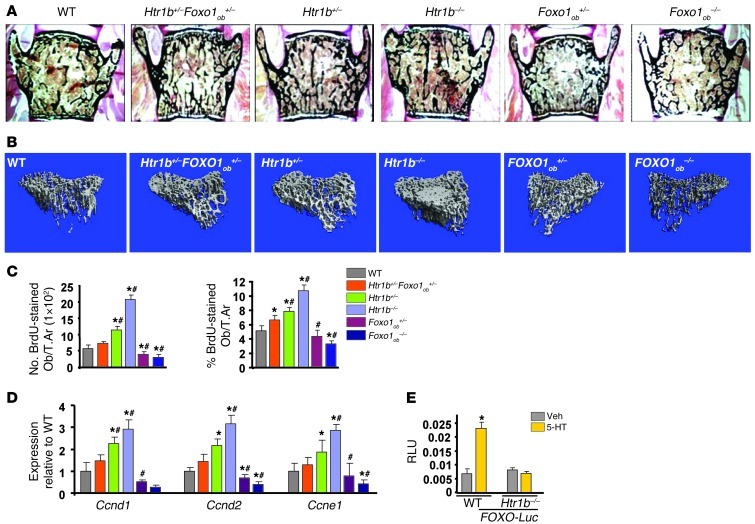
Serotonin exerts its direct antiproliferative action on osteoblasts through its 5-hydroxytryptamine receptor 1B (HTR1B) receptor (8). Thus, we examined whether Htr1b requires Foxo1 to confer the antiproliferative signal of serotonin in osteoblasts. To that end, we generated mice haploinsufficient for Htr1b and Foxo1 in osteoblasts (Htr1b+/–Foxo1ob+/– mice). Htr1b deletion and to a lesser extend Htr1b haploinsufficiency led to an increase in bone volume, osteoblast numbers, and bone formation rate due to a lack of serotonin signaling. Removal of one allele of Foxo1 from Htr1b+/– mice in the Htr1b+/–Foxo1ob+/– animals corrected the increased bone formation and osteoblast numbers caused by Htr1b haploinsufficiency (Figure 3, A and B, and Tables 6 and 7). The increase in osteoblast proliferation and expression of cyclins observed in Htr1b+/– mice was also restored to WT levels in Htr1b+/–Foxo1ob+/– animals (Figure 3, C and D). Osteoblast differentiation (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI64906DS1) and serotonin serum levels (Supplemental Figure 1B) were not altered in any of these models. Moreover, serotonin upregulated the activity of the FOXO-Luc reporter in WT osteoblasts but not in Htr1b-deficient osteoblasts (Figure 3E). These results indicate that Htr1b is the receptor used by serotonin to regulate FOXO1 activity.
Figure 3. Serotonin regulates FOXO1 transcriptional activity via its HTR1B receptor in osteoblasts.
(A) Representative vertebral section images from 2-month-old mice. Mineralized bone matrix is stained in black by Von Kossa reagent. Original magnification, ×4. (B) Representative 3D images of transverse CT slices of proximal tibia obtained by high-resolution μCT analysis. (C) In vivo osteoblast proliferation assessed in femoral sections expressed as the number of BrdU-stained osteoblasts per trabecular area or as the percentage of total osteoblasts per trabecular area (n = 3 mice/group). (D) Real-time PCR analysis of indicated genes in bones (n = 4 mice/group). (C and D) *P < 0.05 versus WT, #P < 0.05 versus Htr1b+/–Foxo1ob+/– group. (E) Calvaria cells from WT or Htr1b–/– mice transfected with FOXO-Luc and treated with vehicle or serotonin (100 μM) (n = 3). Mice were 2 months old. *P < 0.05 versus respective vehicle.
Table 6.
HTR1B requires FOXO1 to mediate the anti-osteoblastogenic signal of duodenal-derived serotonin
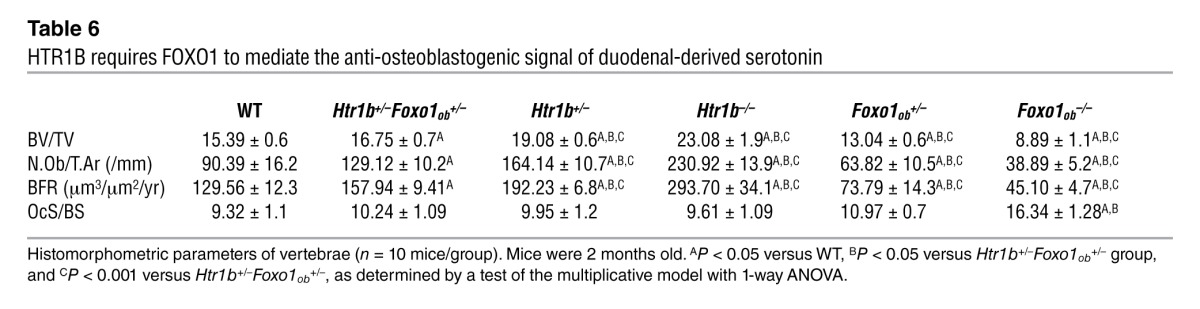
Table 7.
μCT analysis of long bones from WT, Htr1b+/–, Htr1b–/–, Htr1b+/–Foxo1ob+/–, Foxo1ob+/–, and Foxo1ob–/– mice
Serotonin promotes FOXO1 activity by inhibiting its association with CREB.
If serotonin acts through the HTR1B receptor to affect osteoblast activity, then serotonin-activated FOXO1 may interact in this cell with a set of proteins distinct from those used under physiological conditions. To look for FOXO1 transcriptional partners that differentially regulate its activity in the presence and absence of high serotonin levels, we focused on ATF4 and CREB, because HTR1B is a G protein–coupled receptor acting either via the PI3K/AKT/ATF4 or the cAMP/PKA/CREB signaling cascades (20, 21).
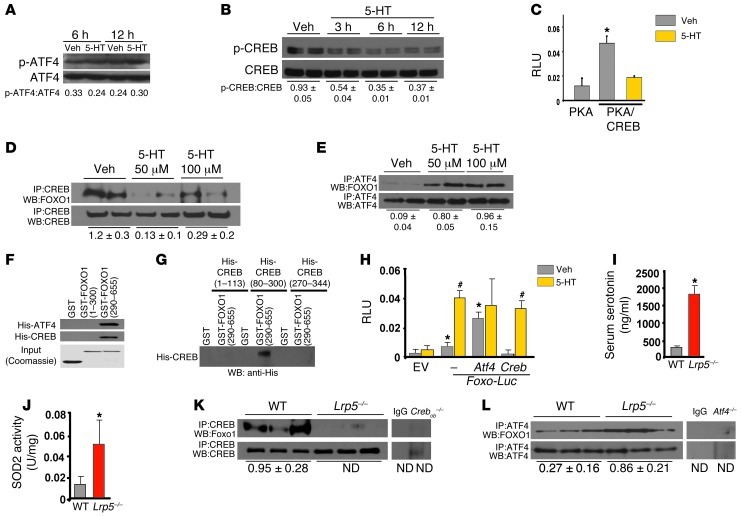
Treatment of osteoblasts with serotonin did not affect phosphorylation and activation of ATF4 but markedly suppressed CREB phosphorylation and activity (Figure 4, A–C). Moreover, under basal conditions, FOXO1 physically associated with both CREB and ATF4 in osteoblasts (Figure 4, D and E). However, whereas serotonin inhibited the interaction of FOXO1 with CREB, it increased its interaction with ATF4. We recently showed that the DNA-binding domain of ATF4 interacts with the transactivation domain of FOXO1 (22). Here, we used 2 FOXO1 GST-tagged fragments that carry either the N-terminal domain (aa 1–300 containing the Forkhead domain) or the C-terminal domain (aa 290–655 containing the transactivation domain of FOXO1) (23, 24) and 3 His-tagged fragments that carry the transactivation domain 1 (aa 1–113) or the kinase-inducible domain (aa 80–300) or the DNA-binding and leucine zipper domains (aa 270–341) of CREB. We found that CREB interacted with the C-terminal transactivation domain of FOXO1 (Figure 4F) through its kinase domain (Figure 4G).
Figure 4. Serotonin regulates a FOXO1-CREB interaction.
(A and B) Phosphorylation status of (A) ATF4 and (B) CREB in OB-6 cells treated with serotonin (100 μM). (C) OB-6 cells transfected with expression plasmids for PKA, the active domain of CREB fused to GAL4, and a construct with GAL4-binding sites upstream of the luciferase gene and treated with 100 μM serotonin for 24 hours (n = 3). *P < 0.05 versus PKA plasmid vehicle. (D and E) Immunoprecipitation and immunoblotting in OB-6 cells treated with serotonin (100 μM) for 1 hour. Blots were representative of 3 experiments. ND, not detected. (F) Binding of GST-FOXO1 fragments and His-CREB for mapping the FOXO1 interaction domain. (G) Binding of GST-FOXO1 (aa 290–655) and His-CREB fragments for mapping the CREB interaction domain. (H) Calvaria cells transfected with indicated plasmids and treated with vehicle or serotonin (100 μM) for 24 hours (n = 3). *P < 0.05 versus EV vehicle; #P < 0.05 versus respective vehicle. (I) Serum serotonin levels, (J) SOD2 activity in tibial lysates, and (K and L) immunoprecipitation and immunoblotting in femoral lysates from WT and Lrp5–/– mice (n = 3–4 mice/group). *P < 0.05 versus WT.
Subsequently, we examined the effect of CREB and ATF4 on the activity of FOXO1 in the absence and presence of serotonin. CREB suppressed FOXO1 transcriptional activity, and serotonin attenuated this inhibitory action of CREB (Figure 4H). In contrast, ATF4 upregulated FOXO1 activity. In agreement with the in vitro observations, high levels of serotonin in Lrp5-deficient mice (Figure 4I) increased FOXO1 transcriptional activity, as documented by the increase in SOD2 activity in Lrp5-deficient bone (Figure 4J). Furthermore, while FOXO1 associated with both ATF4 and CREB in WT mice, the interaction of FOXO1 with CREB was abolished in Lrp5–/– bone (Figure 4K), while the one with ATF4 was increased (Figure 4L). Collectively these observations suggest that serotonin promotes FOXO1 transcriptional activity by disrupting its interaction with CREB.
Differential regulation of FOXO1 activity by serotonin.
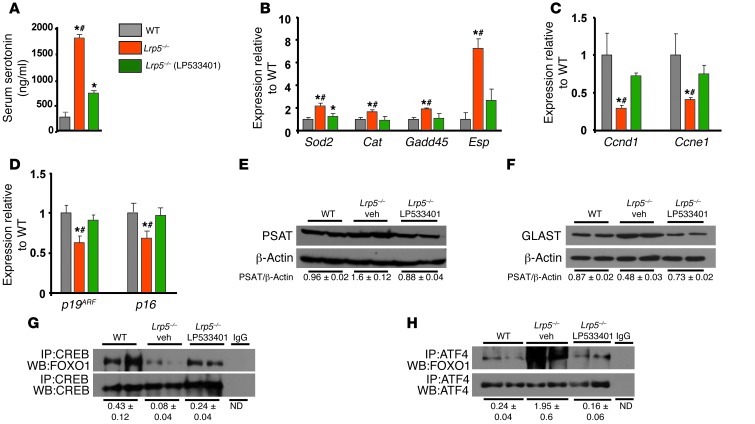
FOXO1 is required for normal osteoblast proliferation (18), yet the serotonin-triggered increase in FOXO1 activity suppresses osteoblast proliferation. Decreased osteoblast proliferation in Foxo1+/– mice is due to ATF4-dependent compromise in protein synthesis (18). This event leads to increased levels of oxidative stress in osteoblasts, suppression of p19ARF and p16, and subsequent activation of stress-dependent p53 signaling (18). We reasoned that if FOXO1 activity is regulated by ATF4 and CREB, serotonin may be preferentially eliciting one interaction instead of the other. To test this hypothesis, we examined ATF4- and CREB-dependent FOXO1 responses in the presence and absence of serotonin in vivo. We found that, in Lrp5–/– mice with high levels of circulating serotonin (Figure 5A), the decreased association with CREB unleashed FOXO1 transcriptional activity, as evidenced by upregulation of Sod2, catalase (Cat), Gadd45, and Esp expression and by suppression of cyclin genes expression (Figure 5, B and C). The upregulating effects are mediated by direct binding of FOXO1 to its cognate binding sites on the promoters of Sod2, catalase, Gadd45, and Esp (25–28). The suppressive effect of FOXO1 on cyclins, although widely reported, has no identified mechanism (17, 29, 30).
Figure 5. Differential regulation of FOXO1 interactions with ATF4 and CREB by serotonin.
(A) Serum serotonin levels, (B–D) real-time PCR analysis of indicated genes in bones, (E and F) immunoblotting in femoral lysates, and (G and H) immunoprecipitation and immunoblotting in femoral lysates of WT, Lrp5–/–, and LP533401-treated Lrp5–/– mice. The gut-derived serotonin synthesis inhibitor LP533401 was administered daily by gavage at 200 mg/kg/d for 7 days (n = 4 mice/group). Mice were 2 months old. *P < 0.05 versus WT, #P < 0.05 versus LP533401-treated Lrp5–/– group.
CREB-released FOXO1 was used to increase its association with ATF4. The increased FOXO1/ATF4 interaction resulted in decreased expression of p19ARF and p16 and increased protein synthesis, measured by the ATF4 transcriptional targets phosphoserine aminotransferase (PSAT) and EAAT1/glutamate aspartate transporter (GLAST) (Figure 5, D–H). We have previously shown that p19ARF and p16 are transcriptional targets of the FOXO1/ATF4 synergism in osteoblasts (18). Normalization of serum serotonin levels in Lrp5–/– mice, by inhibition of gut-derived serotonin synthesis (Figure 5A), reestablished the association of FOXO1 with CREB (Figure 5G), decreased the association with ATF4 (Figure 5H), reversed the increase in PSAT and GLAST protein levels (Figure 5, E and F) and the upregulation in the expression of FOXO1 targets (Sod2, Cat, Gadd45, and Esp), and normalized Ccnd1 and Ccne1 expression and p19 and p16 expression (Figure 5, B–D). These data indicate that, in high serotonin levels, there is a shift in FOXO1 target genes from CREB- to ATF4-dependent responses.
PKA/JNK signaling confers the signal of serotonin to Foxo1.
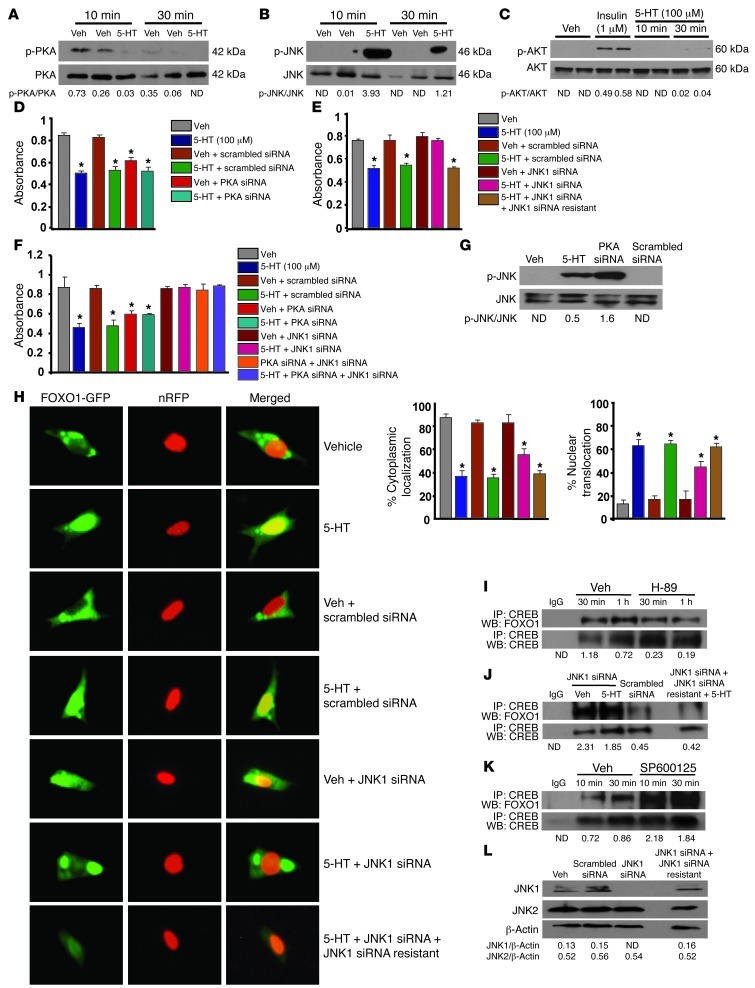
HTR1B is coupled to G(i)-type G proteins that can inhibit adenyl cyclase activity and suppress cAMP levels and PKA activity (31). To delineate the sequence of molecular events that lead from serotonin to CREB/FOXO1 association and transcriptional activation of FOXO1, we examined whether serotonin controls PKA activity. Serotonin treatment of osteoblasts suppressed PKA activity (Figure 6A). In addition, phosphorylation of JNK1 was increased by serotonin treatment (Figure 6B). On the other hand, serotonin had no effect on the phosphorylation of AKT, indicating that the PI3K/AKT pathway, which phosphorylates and inactivates FOXO1, was not induced by serotonin (Figure 6C). This is consistent with the fact that the HTR1B receptor mediates serotonin actions in osteoblasts (8) and does not activate PI3K or AKT. In agreement with the notion that antiproliferative effects of serotonin in osteoblasts involve PKA and JNK1, knockdown of PKA suppressed osteoblast proliferation, whereas silencing JNK1 suppressed the antiproliferative action of serotonin (Figure 6, D and E). Furthermore, inhibition of JNK expression in PKA-silenced osteoblasts blocked the siPKA-induced suppression of osteoblast proliferation, suggesting that PKA regulates JNK activity (Figure 6F). Consistent with this hypothesis, PKA knockdown by siRNA increased JNK1 phosphorylation in osteoblasts, demonstrating that JNK1 activity is regulated by PKA (Figure 6G).
Figure 6. PKA/JNK1-dependent signaling localizes FOXO1 to the nucleus.
Immunoblot showing (A) PKA, (B) JNK1, and (C) AKT phosphorylation in OB-6 cells treated with serotonin (100 μM). (D and E) Proliferation in primary osteoblasts transfected with (D) PKA siRNA oligos or control scrambled siRNA oligos or (E) JNK1 siRNA or control scrambled siRNA oligos, in the presence or absence of siRNA-resistant JNK1, followed by treatment with vehicle or serotonin (100 μM) for 48 hours (n = 3). (F) Proliferation in primary osteoblasts transfected with indicated siRNAs or control scrambled siRNA oligos, followed by treatment with vehicle or serotonin (100 μM) for 48 hours (n = 3). (D–F) *P < 0.05 versus vehicle. (G) Immunoblot of JNK1 phosphorylation in OB-6 cells transfected with PKA siRNA and control scrambled siRNA oligos. (H) Subcellular localization of Foxo1 in primary osteoblasts transfected with JNK1 siRNA oligos or control scrambled siRNA oligos, followed by cotransfection with FOXO1-GFP and nRFP plasmids, in the presence or absence of siRNA-resistant JNK1. Transfected cells were treated with serotonin (100 μM) for 6 hours. Subcellular localization of FOXO1 was expressed as the percentage cytoplasmic or nuclear localization (n = 3). Original magnification, ×4. *P < 0.05 versus vehicle. (I–K) Immunoprecipitation and immunoblotting of CREB and FOXO1 in OB-6 cells (I) treated with PKA inhibitor (H-89) or (J) transfected with JNK1 siRNA, with or without siRNA-resistant JNK1 or control scrambled siRNA oligos, or (K) treated with the JNK inhibitor SP600125 at 10 μM. (L) Immunoblotting for JNK1 and JNK2 protein levels in OB-6 cells. Blots were representative of 3 experiments.
Next, we examined whether JNK1 activation affected FOXO1 activity. Since the JNK1-dependent phosphorylation sites for FOXO1 are not defined, we measured the activation status of FOXO1 by nuclear translocation. Knocking down JNK1 by siRNA in osteoblastic cells inhibited nuclear translocation of FOXO1 and retained FOXO1 in the cytoplasm (Figure 6H). Add-back of siRNA-resistant JNK1 inhibited the reduction in FOXO1 translocation caused by JNK1 siRNA. Finally, confirming that the association of FOXO1 with CREB was PKA and JNK1 dependent, inhibition of PKA abrogated FOXO1/CREB association, whereas inhibition of JNK1 prevented the ability of serotonin to dissociate the FOXO1/CREB complex in osteoblasts (Figure 6, I–K). As a control, JNK2 protein levels were not affected by JNK1 silencing (Figure 6L). This line of experiments indicates that circulating serotonin acts in osteoblasts through Htr1b to suppress PKA activity. This, in turn, promotes JNK1 activity, and activated JNK1 promotes nuclear localization of FOXO1.
Serotonin suppresses bone formation by controlling Foxo1/CREB interactions.
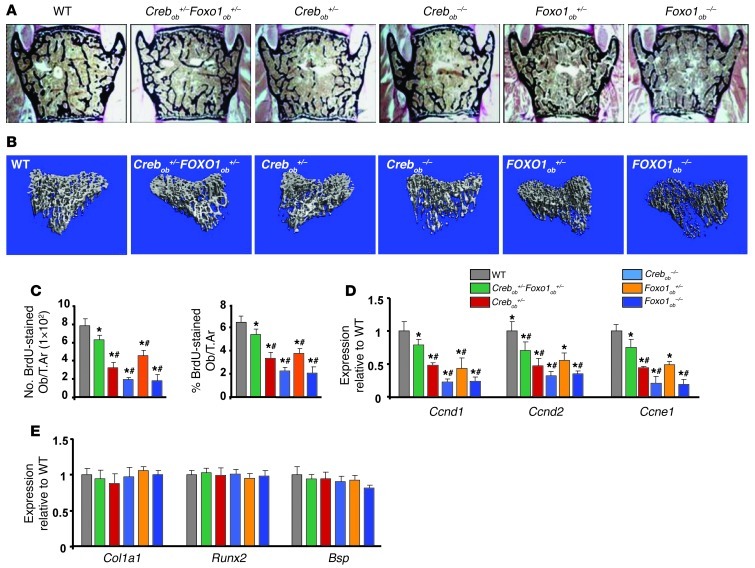
The importance of the interactions of FOXO1 with CREB and ATF4 and their effect in bone formation was verified genetically. Consistent with the notion that CREB is required for osteoblast function, its inactivation in osteoblasts led to low bone volume, due to a decrease in osteoblast proliferation (Figure 7, A–D, and Tables 8 and 9). FOXO1 haploinsufficiency reversed the low bone mass phenotype of Crebob+/– mice by increasing osteoblast numbers in Crebob+/–Foxo1ob+/– mice (Figure 7, A and B, and Tables 8 and 9). Partial rescue was observed in Crebob+/–Foxo1+/– mice as compared with that in WT animals, probably due to the fact that CREB is required for osteoblast proliferation through actions dependent and independent of FOXO1. Consistent with these data, decreases in osteoblast proliferation (Figure 7C) and in expression of cyclins (Figure 7D) were also reversed in Crebob+/–Foxo1ob+/– mice as compared with that in Crebob+/– mice. Osteoblast differentiation was not affected in any of these models (Figure 7E).
Figure 7. Gut-derived serotonin inhibits bone formation by regulating FOXO1-CREB interaction.
(A) Representative vertebral section images from 2-month-old mice. Mineralized bone matrix is stained in black by Von Kossa reagent. Original magnification, ×4. (B) Representative 3D images of transverse CT slices of proximal tibia obtained by high-resolution μCT analysis. (C) Osteoblast proliferation expressed as the number of BrdU-stained osteoblasts per trabecular area or as the percentage of total osteoblasts per trabecular area in sections of femurs (n = 6 mice/group). (D and E) Real-time PCR analysis of indicated genes in bones (n = 4 mice/group). Mice were 2 months old. *P < 0.05 versus WT, #P < 0.05 versus Crebob+/–Foxo1ob+/– group by ANOVA.
Table 8.
Gut-derived serotonin inhibits bone formation by regulating a FOXO1-CREB interaction
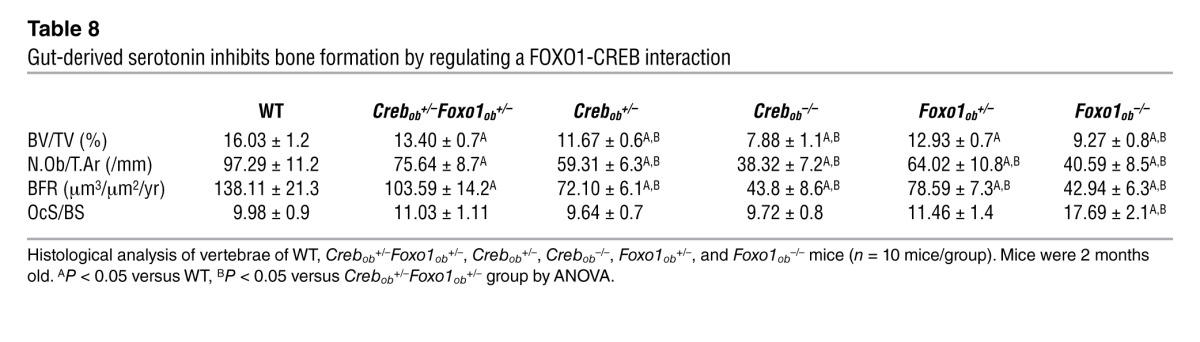
Table 9.
μCT analysis of long bones from WT, Crebob+/–, Crebob–/–, Crebob+/–Foxo1ob+/–, Foxo1ob+/–, and Foxo1ob–/– mice
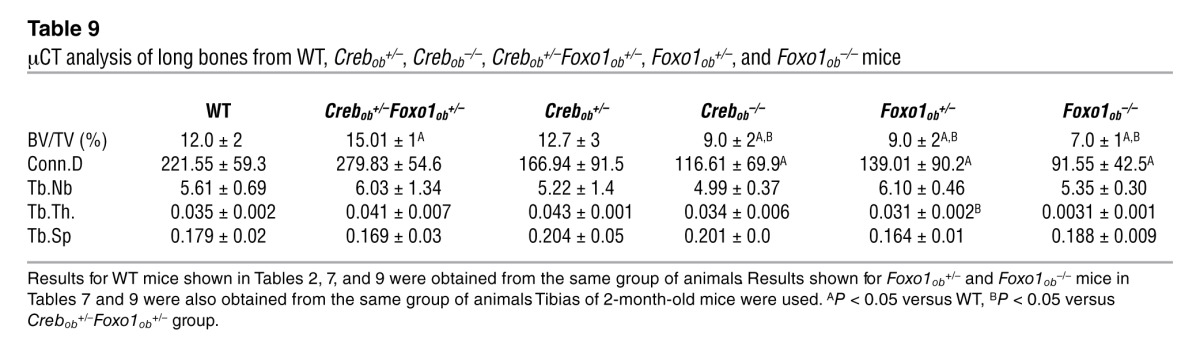
As expected, FOXO1 inactivation in osteoblasts decreased bone volume and bone formation by decreasing osteoblast numbers. Mice haploinsufficient for both Foxo1 and Atf4 (Atf4+/–Foxo1ob+/– mice) showed a worsening of the bone phenotype of Atf4+/– and Foxo1ob+/– mice and a decrease in bone volume, bone formation rate, and osteoblast numbers that was similar to that of Foxo1ob–/– animals (Supplemental Figure 2A and Supplemental Table 1). The same changes were observed in the expression of cyclin genes (Supplemental Figure 2B). With the exception of Bsp, which is known to be suppressed in ATF4 deficiency, there were no alterations in the expression of any of the osteoblast differentiation markers examined (Supplemental Figure 2C).
This genetic evidence supports the notion that FOXO1 synergizes with ATF4 to maintain osteoblast function. On the other hand, its interaction with CREB inhibits the proliferative actions of FOXO1 in osteoblasts.
Discussion
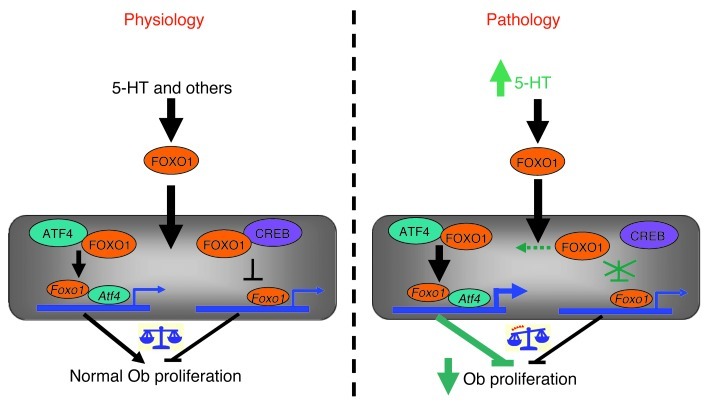
Here, we describe a mechanism by which FOXO1 transcriptional activity is regulated by serotonin that affects osteoblast proliferation and bone formation (Figure 8). In addition, to our knowledge, we have uncovered a bimodal action of FOXO1 that differentially regulates bone formation in health and disease. This mechanism relies on the ability of FOXO1 to interact with ATF4 and CREB under physiological conditions. The interaction with ATF4 promotes, whereas the one with CREB suppresses, the transcriptional activity of FOXO1 and therefore expression of FOXO1-dependent genes. The opposing actions of the 2 transcription factors balance and maintain a basal level of FOXO1 activity, which in turn maintains normal osteoblast proliferation. In pathological situations of high serotonin levels, such as in LRP5 deficiency, the association between FOXO1 and CREB is disrupted. This event relieves the suppressive actions of CREB on FOXO1 and increases its association with ATF4, thus upregulating ATF4-mediated responses. These events subsequently lead to suppression of cyclin genes required for proliferation and result in cell cycle arrest. Other proteins may be involved in transcriptional suppression of cyclin genes by activated FOXO1. Indeed, FOXO1 is known to suppress expression of cyclin genes in other cell systems. However, the mechanism of transcriptional repression is not known, but it has been shown that it is not exerted through FOXO1 binding to any of the known FOXO1-binding sites (17, 30).
Figure 8. Model illustrating the mechanism of serotonin/FOXO1 interaction in osteoblasts.
Under physiological conditions, FOXO1 interacts in the nucleus with both ATF4 and CREB to maintain normal osteoblast proliferation. The association of FOXO1 with ATF4 promotes, whereas the one with CREB suppresses, expression of FOXO1-regulated transcriptional targets. An increase in circulating serotonin levels disrupts the interaction of FOXO1 with CREB. This favors the formation of ATF4-FOXO1 heterodimers. These ATF4-dependent or -independent interactions lead to increased transcriptional activity of FOXO1 and suppression of cell-cycle progression genes. Additionally, following release from its complex with CREB, high levels of available FOXO1 may interact with other transcription factor(s).
Supporting a bimodal FOXO1 action, although a basal level of FOXO activity is required for a given cellular process, prolonged nuclear localization or increased protein stability can have the opposite effect and inhibit it. Remarkably, the best known example of such, seemingly paradoxical, effects of FOXOs has been described in FOXO1 control of cell proliferation. Indeed, although FOXO1 is required for basal levels of cell proliferation, stress increases the threshold of FOXO1 transcriptional activity and evokes a FOXO1-dependent antiproliferative action (29, 32, 33). Our findings indicate that the interaction of FOXO1 with ATF4 or CREB under the control of serotonin can alter the outcome on osteoblast proliferation by shifting to ATF4-dependent responses, although they do not provide a direct mechanistic explanation at the gene regulation level for the suppressive effect of the ATF4/FOXO1 complex in the expression of cyclins. This is because, to this day and although similar suppressive effects of FOXO1 on cyclins have been well established for other cell models, FOXO1-binding sites mediating such suppressive effects have not been identified (17, 30).
Similar to PTH, luteinizing hormone, and follicle-stimulating hormone, serotonin levels cycle and have been shown to vary depending on the time of the day that the blood sample is obtained. In healthy subjects and in patients with depression as well as in rodents, serotonin levels show a clear, distinct pattern of diurnal rhythm (34–39). In fact, in both patients and in rodents, serotonin levels peak in the morning and decline in the afternoon, with differences in serotonin levels between different groups being readily detectable in the morning but not in the afternoon. Differences in the circadian fluctuations of serotonin have been reported among patients and control healthy subjects. In addition to the variations in diurnal rhythm, serotonin levels also depend on gender and genetic background (39).
Recently, Cui et al., using a mouse model of Lrp5 inactivation in osteocytes, reported that they failed to detect an association among LRP5 deficiency, increased circulating serotonin levels, and bone loss (40). These results raised the possibility that deletion of Lrp5 in a later, more mature stage of the osteoblastic lineage may affect bone mass. However, inactivation of Lrp5 in late osteoblasts failed to induce any changes in bone mass, as assessed by several μCT parameters and by osteoblast numbers (41). In contrast, deletion of Lrp6 in the same cells decreased osteoblast numbers and bone mass. Differences in the genetic background between experimental animals may have been a factor involved in the differential effects of LRP5. Therefore, we measured serotonin levels and bone changes in Lrp5+/–, Lrp5–/–, Lrp5+/–Foxo1+/–, and Lrp5/–/–Foxo1+/– mice from 2 different genetic backgrounds, C57BL/6 and mixed C57BL/6/Sv129. We found no differences in any of these parameters between the 2 backgrounds. Even more important than all the different mouse models used by different investigators are the emerging data in humans indicating a link between serum serotonin levels and low bone mass. Directly linking LRP5 to suppression of serotonin, Fort et al. showed that serotonin levels decrease in patients with the high bone mass Lrp5 mutation (42). In addition, circulating serotonin levels are inversely associated with body and spine areal BMD, with femur neck total and trabecular volumetric BMD, and with bone volume, trabecular numbers, and trabecular thickness in women (43). We believe that, taken together our observations, the observations of other investigators using different mouse models and more importantly the clinical data suggest that Lrp5 is a potent regulator of serotonin synthesis and that gut-derived serotonin suppresses bone formation.
All our mouse models of genetic or chemical inhibition of circulating serotonin synthesis show that serotonin suppresses bone mass by suppressing osteoblast numbers, without affecting osteoclastogenesis. A recent study on Tph1–/– mice finds that serotonin also affects bone resorption by promoting RANKL-induced osteoclast formation (44). However, the reported decrease in bone volume in 16-week-old Tph1–/– mice in this study is in contrast with other observations showing that 16-week-old Tph1–/– mice have increased femoral bone volume (8, 40). More importantly, bone marrow transplantation experiments used to examine a cell-autonomous role of TPH1 in osteoclasts are inconclusive, since serotonin compromises hematopoietic stem cell function and numbers (45). Thus, the defect in osteoclastogenesis in mice with global deletion of Tph1 may be a consequence of reduced HSC numbers or compromised HSC lineage determination.
We show that inactivation of the Htr1b serotonin receptor in osteoblasts increased bone mass by increasing osteoblast numbers and mediated the effects of serotonin on FOXO1. This mode of osteoblast regulation by Htr1b is relevant to studies reporting an opposite, bone-protective effect of the Htr2b receptor in osteoblast function (46, 47). To exclude the possibility that the anabolic effect of Htr1b deletion could be due to upregulation of Htr2b, we measure its expression in our mouse models. We found that Htr2b expression remained unchanged in Htr1b- and Foxo1-deficient osteoblasts (data not shown). Therefore, the bone phenotype of Htr1b inactivation is not due to a bimodal response conferred by Htr2b.
FOXO1, by its actions on bone as well as through bone, has emerged as a crucial regulator of osteoblast physiology. It is a transcriptional potentiator of the ability of the skeleton to regulate glucose metabolism (28), and it regulates osteoblast formation by either responding to oxidative stress (18) or, as shown here, by receiving and translating signals from other organs. In this latter role, our data have uncovered a mechanism by which different levels of FOXO1 transcriptional activity have opposing effects on cell proliferation, by identifying CREB and ATF4 as molecular interactors of FOXO1. Given the ubiquitous pattern of expression of this transcription factor, it is possible that similar interactions of FOXO1 with CREB or ATF4, or even with additional proteins, may mediate the effects of other hormones on osteoblasts or that this mechanism of action may operate in other cell systems.
Methods
Animals.
Generation of Foxo1fl/fl, Col1a1-Cre, Foxo1+/–, Htr1b+/–, Crebfl/fl, Atf4+/–, Lrp5fl/fl, and Lrp5+/– mice has previously been reported (8, 20, 48–52). All the floxed models (Foxo1fl/fl, Crebfl/fl, and Lrp5fl/fl) have normal expression and activity of the floxed gene. Mice in which Foxo1 has been replaced with a Foxo1 locus carrying exon 2 flanked by 2 loxP sites have been fully characterized (48). Mice lacking one Htr1b allele display an increase in osteoblast numbers, bone formation rate, and bone mass at adulthood (51). Lrp5+/– mice mirror the phenotype of Htr1b+/– mice by showing a decrease in osteoblast numbers, bone formation rate, and bone mass at the same age (53). Conditional inactivation in osteoblasts was performed using the Col1a1-Cre transgenic mice expressing Cre under the control of the osteoblast-specific 2.3-kb sequence of the rat collagen type 1A1 promoter. These mice have been used to direct strong osteoblast- or osteoclast-specific expression of various genes in transgenic mice (54–57). The promoter is active in osteoblast precursors, osteoblasts, and osteocytes (58). Mice with osteoblast-specific deletion of Foxo1 (Foxo1ob–/–), Creb (Crebob–/–), and Lrp5 (Lrp5ob–/–) were generated by crossing Foxo1fl/fl, Crebfl/fl, and Lrp5fl/fl mice with Col1a1-Cre mice. Osteoblast-specific inactivation of Foxo1, Lrp5, and Creb in mice was confirmed (Supplemental Figure 3). Conditional inactivation of FOXO1 in osteoblasts resulted in 77% decrease in the expression of Foxo1. In each experiment, the mice used were littermates. All mice were in C57BL/6 background, with the exception of the mice used in the experiments shown in Figure 1, A–D, and Tables 1 and 2 as well as Htr1b+/–, Htr1b–/–, and Htr1b+/–Foxo1+/– mice, which were in C57BL/6/Sv129 background. Female mice at 2 months of age were used for the bone histomorphometric and the μCT analyses. Inhibition of duodenal-derived serotonin synthesis was carried out by oral gavage administration of the serotonin synthesis inhibitor LP533401 (Dalton Chemical Laboratories Inc.), as described in Results. Mice treated with the serotonin synthesis inhibitor had normal body weight and appeared healthy, as they exhibited normal physical activity and appetite (Supplemental Figure 4).
Histomorphometry.
Histomorphometric analyses were performed as previously described on vertebral column specimens collected from 2-month-old mice using undecalcified sections according to standard protocols using the osteomeasure analysis system (Osteometrics) (18). Images were taken using a ×10 objective on a Leica microscope outfitted with a camera. Six to 10 animals were analyzed for each group.
μCT analysis.
Trabecular bone architecture of the distal tibia was assessed by using a μCT system (VivaCT 40; SCANCO Medical AG) as described previously (59).
Cell culture and treatments.
Calvaria-derived osteoblasts were obtained and cultured as described previously (18). OB-6 osteoblastic cells, a bone marrow–derived osteoblastic cell line, have been previously described (60). Calvaria or OB-6 cells were treated with serotonin (Sigma-Aldrich) at a concentration of 50 μM or 100 μM every 6 hours for 24 and 48 hours.
In vivo and in vitro osteoblast proliferation.
Osteoblast proliferation was assessed in vivo by injecting thymidine analog bromodeoxyuridine (BrdU) intraperitoneally as previously described (18). Toluidine blue staining was performed in adjacent sections for osteoblast morphology. Only those cells lining the bone with osteoblast morphology in toluidine blue–stained sections and with dark brown DAB nuclear staining in BrdU-stained sections were considered positive for BrdU-labeled osteoblasts. Proliferating osteoblasts were analyzed using an OsteoMeasure Analysis System (OsteoMetrics). Cell proliferation in vitro was assessed using a commercially available BrdU proliferation assay kit (Calbiochem) or a Cell Titer One system (Promega).
Transient transfection and luciferase assay.
Calvaria or OB-6 cells, plated at 104 cells per well in 48-well plates, were cotransfected with FOXO1, ATF4, CREB, LRP5, HTR1B plasmids, and FOXO-Luc reporter construct using Lipofectamine 2000 (Invitrogen) as previously described (22). pRL-CMV Renilla luciferase control vector (20 ng; Promega) was cotransfected as an internal standard to normalize for transfection efficiency. Transient transfections were repeated twice on separate days, and each time they were carried out in triplicates.
siRNA knockdown.
Primary osteoblasts (calvaria cells) or OB-6 cells were transfected with siRNA (JNK, PKA) and control scrambled oligos using siRNA transfection reagents from Dharmacon. The JNK1-specific siRNA sequence is 5′-TAAATACGCTGGATATAGC-3′. siRNA-resistant JNK1 was generated by replacing 3 residues in the JNK1 siRNA target region (underlined): 5′-TAAAGACGCGGGCTATAGC-3′.
Subcellular localization of FOXO1.
Subcellular localization of FOXO1 was detected by a fluorescence-based assay in calvaria cells transfected with JNK1 siRNA oligos and control scrambled oligos. Following knockdown of JNK1, cells were cotransfected with a FOXO1 plasmid fused to green fluorescence protein (FOXO1-GFP) and a plasmid expressing red fluorescent protein fused to a nuclear localization sequence (nRFP) using Lipofectamine 2000 (Invitrogen). Cells were viewed under fluorescence microscope, and the percentage of FOXO1-GFP in the cytoplasm and nucleus was calculated. Images were taken at ×20 magnification.
Protein interaction analysis.
Full-length His-CREB and fragments encompassing aa 1–113, 80–300, and 270–341 were PCR amplified, subcloned into the pET14b vector (Novagen), and verified by sequencing. GST-FOXO1 (aa 1–300, 290–655) constructs and His-ATF4 (aa 1–151, 110–121, 186–349) constructs have been described previously (20, 23, 24). The GST and His fusion proteins were expressed in Escherichia coli strain BL21pLys by iso-propyl-thio-galactose induction. Extracts of the GST-fusion-protein–transformed cells were coupled with glutathione-Sepharose beads. Following SDS-PAGE and Coomassie Brilliant Blue staining for monitoring coupling efficiency, GST fusion protein bound beads were incubated with purified His fusion proteins overnight, washed extensively, and eluted by boiling in SDS-PAGE loading buffer. Bound proteins were visualized by Western blot using anti-His antibody.
Quantitative real-time PCR.
Total RNA was isolated from tissues or cultured osteoblastic cells using TRIzol reagent. Bone marrow cells were removed from bones by flushing the femurs or tibias with PBS. Total RNA (2 μg) was reversed transcribed at 42°C with SuperScript II (Invitrogen). Quantitative real-time PCR was performed using the SYBR Green Master Mix (Bio-Rad) as previously described (18, 28). β-Actin was used as reference gene. The primer sequences are shown in Supplemental Table 2.
Western blotting and immunoprecipitation.
Bone lysates were obtained by homogenizing long bones in 600 μl lysis buffer (20 mM Tris-HCl, pH 7.5, 150 Mm NaCl, 200 μm sodium orthovanadate,1 mM EDTA) containing 1% Triton-X and a protease inhibitor cocktail (Roche) as previously described (18). Protein estimation was performed by the Bradford method using the Bio-Rad Protein Assay Kit. Bone or osteoblast extracts (15–60 μg) were analyzed on a SDS-polyacrylamide gel, transferred to a PVDF membrane, and immunoblotted with Akt (sc-1618; Santa Cruz Biotechnology Inc.), β-actin (sc-47778; Santa Cruz Biotechnology Inc.), GLAST (sc-15316; Santa Cruz Biotechnology Inc.), p-FOXO1 (9461; Cell Signaling Technology), p-PKA (4781; Cell Signaling Technology), PKA (4782; Cell Signaling Technology), p-AKT (4060; Cell Signaling Technology), JNK1 (3708; Cell Signaling Technology), JNK2 (4672; Cell Signaling Technology), His-Tag (2365; Cell Signaling Technology), and PSAT (96136; Abcam) antibodies. PKA phosphorylation was evaluated using a phosphor-PKA C (Thr197) antibody that detects endogenous levels of PKA C (PKA Cα, PKA Cβ, PKA Cγ) when phosphorylated at Thr197 (4781; Cell Signaling Technology). For immunoprecipitation, 100 μg bone/cell lysate protein were incubated with 2 μg specific antibodies and 20 μl protein A/G agarose beads (Santa Cruz Biotechnology Inc.) overnight at 4°C on a rotating device. Band density was quantified by densitometry analysis (ImageJ software). The ratio of phospho-to-total proteins was calculated based on the intensity of the bands determined by densitometry.
Serum serotonin levels and SOD2 activity.
For measurements of serum serotonin levels, mice were exposed to Isoflurane (Baxter) for 30 seconds, and blood was collected by cardiac puncture. Similar volumes of blood were collected from different animals for consistency in the clotting times. Serum collection from all mice was done between 9:00 AM and 12:00 PM. Blood was allowed to clot on ice for 5 minutes before centrifugation at maximum speed and 4°C for 10 minutes. The serum supernatant was snap frozen and stored at –80°C. Serum serotonin levels were quantified by ELISA (Serotonin Kit, Fitzgerald) according to the manufacturer’s instructions. Mitochondrial superoxide dismutase (SOD2) activity was measured in osteoblast cell lysates using the Superoxide Dismutase Kit II (Calbiochem).
Statistics.
Statistical analyses were performed using a 1-way ANOVA followed by Student-Newman-Keuls test, and a P value less than 0.05 was considered significant. Data are presented in all figures and tables as mean ± SEM. The epistasis of Htr1b and Foxo1ob, as well as ATF4 and Foxo1ob, on osteoblast numbers, bone formation, rate and bone volume was assessed with 1-way ANOVA and post-hoc pair-wise comparisons. In addition to the additive model, using the raw scores for osteoblast numbers, bone formation rate, and bone volume, the multiplicative model (61), using log-transformed (10) values for osteoblast numbers, bone formation rate, and bone volume, was tested with pair-wise comparison limited to the double-heterozygous and single-heterozygous groups.
Study approval.
All the protocols and experiments performed in mice were approved by the Institute of Comparative Medicine, Columbia University, New York, New York, USA.
Supplementary Material
Acknowledgments
The authors are grateful to Larry Chan for providing the Lrp5–/– and the Lrp5fl/fl mice, Guenther Schütz for providing the Crebfl/fl mice, John P. Bilezikian for helpful discussions, Don McMahon for help with statistical analysis, and Charles Duncan and Jayesh Sha for technical assistance. We thank the histology facility of the Diabetes and Endocrinology Research Center of Columbia University Medical Center (supported by NIDDK DK063608-07) for help with histological analysis. This work was supported by the NIH (P01 AG032959 and R01 AR055931 to S. Kousteni).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(10):3490–3503. doi:10.1172/JCI64906.
References
- 1.Ottaviani E, Franceschi C. The neuroimmunology of stress from invertebrates to man. Prog Neurobiol. 1996;48(4–5):421–440. doi: 10.1016/0301-0082(95)00049-6. [DOI] [PubMed] [Google Scholar]
- 2.Azmitia EC. Serotonin neurons, neuroplasticity, and homeostasis of neural tissue. Neuropsychopharmacology. 1999;21(2 suppl):33S–45S. doi: 10.1016/S0893-133X(99)00022-6. [DOI] [PubMed] [Google Scholar]
- 3.Goodrich JT, Bernd P, Sherman D, Gershon MD. Phylogeny of enteric serotonergic neurons. J Comp Neurol. 1980;190(1):15–28. doi: 10.1002/cne.901900103. [DOI] [PubMed] [Google Scholar]
- 4.De-Miguel FF, Trueta C. Synaptic and extrasynaptic secretion of serotonin. Cell Mol Neurobiol. 2005;25(2):297–312. doi: 10.1007/s10571-005-3061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoyer D, et al. International Union of Pharmacology classification of receptors for 5-hydroxytryptamine (Serotonin). Pharmacol Rev. 1994;46(2):157–203. [PubMed] [Google Scholar]
- 6.Lucki I. The spectrum of behaviors influenced by serotonin. Biol Psychiatry. 1998;44(3):151–162. doi: 10.1016/S0006-3223(98)00139-5. [DOI] [PubMed] [Google Scholar]
- 7.Yadav VK, et al. A serotonin-dependent mechanism explains the leptin regulation of bone mass, appetite, and energy expenditure. Cell. 2009;138(5):976–989. doi: 10.1016/j.cell.2009.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yadav VK, et al. Lrp5 controls bone formation by inhibiting serotonin synthesis in the duodenum. Cell. 2008;135(5):825–837. doi: 10.1016/j.cell.2008.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato M, et al. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J Cell Biol. 2002;157(2):303–314. doi: 10.1083/jcb.200201089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babij P, et al. High bone mass in mice expressing a mutant LRP5 gene. J Bone Miner Res. 2003;18(6):960–974. doi: 10.1359/jbmr.2003.18.6.960. [DOI] [PubMed] [Google Scholar]
- 11.Boyden LM, et al. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med. 2002;346(20):1513–1521. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- 12.Little RD, et al. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am J Hum Genet. 2002;70(1):11–19. doi: 10.1086/338450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yadav VK, et al. Pharmacological inhibition of gut-derived serotonin synthesis is a potential bone anabolic treatment for osteoporosis. Nat Med. 2010;16(3):308–312. doi: 10.1038/nm.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang B, Moussaif M, Kuan CJ, Gargus JJ, Sze JY. Serotonin targets the DAF-16/FOXO signaling pathway to modulate stress responses. Cell Metab. 2006;4(6):429–440. doi: 10.1016/j.cmet.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24(50):7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 16.Arden KC. Multiple roles of FOXO transcription factors in mammalian cells point to multiple roles in cancer. Exp Gerontol. 2006;41(8):709–717. doi: 10.1016/j.exger.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 17.Zhao HH, et al. Forkhead homologue in rhabdomyosarcoma functions as a bifunctional nuclear receptor-interacting protein with both coactivator and corepressor functions. J Biol Chem. 2001;276(30):27907–27912. doi: 10.1074/jbc.M104278200. [DOI] [PubMed] [Google Scholar]
- 18.Rached MT, et al. FoxO1 is a positive regulator of bone formation by favoring protein synthesis and resistance to oxidative stress in osteoblasts. Cell Metab. 2010;11(2):147–160. doi: 10.1016/j.cmet.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitamura YI, et al. FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metab. 2005;2(3):153–163. doi: 10.1016/j.cmet.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 20.Yang X, et al. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell. 2004;117(3):387–398. doi: 10.1016/S0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- 21.Harding HP, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11(3):619–633. doi: 10.1016/S1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 22.Kode A, et al. FoxO1 cooperates with ATF4 in osteoblasts to control glucose homeostasis. J Biol Chem. 2012;287(12):8757–8768. doi: 10.1074/jbc.M111.282897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitamura T, et al. A Foxo/Notch pathway controls myogenic differentiation and fiber type specification. J Clin Invest. 2007;117(9):2477–2485. doi: 10.1172/JCI32054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puigserver P, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423(6939):550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto M, et al. Regulation of oxidative stress by the anti-aging hormone klotho. J Biol Chem. 2005;280(45):38029–38034. doi: 10.1074/jbc.M509039200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiong S, et al. PGC-1 alpha serine 570 phosphorylation and GCN5-mediated acetylation by angiotensin II drive catalase down-regulation and vascular hypertrophy. J Biol Chem. 2010;285(4):2474–2487. doi: 10.1074/jbc.M109.065235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furukawa-Hibi Y, Yoshida-Araki K, Ohta T, Ikeda K, Motoyama N. FOXO forkhead transcription factors induce G(2)-M checkpoint in response to oxidative stress. J Biol Chem. 2002;277(30):26729–26732. doi: 10.1074/jbc.C200256200. [DOI] [PubMed] [Google Scholar]
- 28.Rached MT, et al. FoxO1 expression in osteoblasts regulates glucose homeostasis through regulation of osteocalcin in mice. J Clin Invest. 2010;120(1):357–368. doi: 10.1172/JCI39901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgering BM, Kops GJ. Cell cycle and death control: long live Forkheads. Trends Biochem Sci. 2002;27(7):352–360. doi: 10.1016/S0968-0004(02)02113-8. [DOI] [PubMed] [Google Scholar]
- 30.Burgering BM, Medema RH. Decisions on life and death: FOXO Forkhead transcription factors are in command when PKB/Akt is off duty. J Leukoc Biol. 2003;73(6):689–701. doi: 10.1189/jlb.1202629. [DOI] [PubMed] [Google Scholar]
- 31.Noda M, Higashida H, Aoki S, Wada K. Multiple signal transduction pathways mediated by 5-HT receptors. Mol Neurobiol. 2004;29(1):31–39. doi: 10.1385/MN:29:1:31. [DOI] [PubMed] [Google Scholar]
- 32.Trinei M, et al. A p53-p66Shc signalling pathway controls intracellular redox status, levels of oxidation-damaged DNA and oxidative stress-induced apoptosis. Oncogene. 2002;21(24):3872–3878. doi: 10.1038/sj.onc.1205513. [DOI] [PubMed] [Google Scholar]
- 33.Nemoto S, Finkel T. Redox regulation of forkhead proteins through a p66shc-dependent signaling pathway. Science. 2002;295(5564):2450–2452. doi: 10.1126/science.1069004. [DOI] [PubMed] [Google Scholar]
- 34.Pietraszek MH, et al. Diurnal variations of whole blood serotonin content in patients with depression and neurosis. J Neurol Neurosurg Psychiatry. 1992;55(4):336. doi: 10.1136/jnnp.55.4.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humphries LL, Shirley P, Allen M, Codd EE, Walker RF. Daily patterns of serotonin uptake in platelets from psychiatric patients and control volunteers. Biol Psychiatry. 1985;20(10):1073–1081. doi: 10.1016/0006-3223(85)90005-8. [DOI] [PubMed] [Google Scholar]
- 36.Rausch JL, Shah NS, Burch EA, Donald AG. Platelet serotonin uptake in depressed patients: circadian effect. Biol Psychiatry. 1982;17(1):121–123. [PubMed] [Google Scholar]
- 37.Modai I, Malmgren R, Asberg M, Beving H. Circadian rhythm of serotonin transport in human platelets. Psychopharmacology (Berl). 1986;88(4):493–495. doi: 10.1007/BF00178513. [DOI] [PubMed] [Google Scholar]
- 38.Baron M, Barkai A, Kowalik S, Fieve RR, Quitkin F, Gruen R. Diurnal and circannual variation in platelet 3H-imipramine binding: comparative data on normal and affectively ill subjects. Neuropsychobiology. 1988;19(1):9–11. doi: 10.1159/000118425. [DOI] [PubMed] [Google Scholar]
- 39.Jernej B, Cicin-Sain L, Kveder S. Physiological characteristics of platelet serotonin in rats. Life Sci. 1989;45(6):485–492. doi: 10.1016/0024-3205(89)90098-2. [DOI] [PubMed] [Google Scholar]
- 40.Cui Y, et al. Lrp5 functions in bone to regulate bone mass. Nat Med. 2011;17(6):684–691. doi: 10.1038/nm.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zylstra CR, et al. Lrp6 and Lrp5 exert distinct roles in regulating bone acquisition in the mature osteoblast. J Bone Miner Res. 25(suppl 1):1078. [Google Scholar]
- 42.Frost M, et al. Levels of serotonin, sclerostin, bone turnover markers as well as bone density and microarchitecture in patients with high-bone-mass phenotype due to a mutation in Lrp5. J Bone Miner Res. 2011;26(8):1721–1728. doi: 10.1002/jbmr.376. [DOI] [PubMed] [Google Scholar]
- 43.Modder UI, Achenbach SJ, Amin S, Riggs BL, Melton LJ, III, Khosla S. Relation of serum serotonin levels to bone density and structural parameters in women. J Bone Miner Res. 2010;25(2):415–422. doi: 10.1359/jbmr.090721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chabbi-Achengli Y, et al. Decreased osteoclastogenesis in serotonin-deficient mice. Proc Natl Acad Sci U S A. 2012;109(7):2567–2572. doi: 10.1073/pnas.1117792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang M, et al. Promoting effects of serotonin on hematopoiesis: ex vivo expansion of cord blood CD34+ stem/progenitor cells, proliferation of bone marrow stromal cells, and antiapoptosis. Stem Cells. 2007;25(7):1800–1806. doi: 10.1634/stemcells.2007-0048. [DOI] [PubMed] [Google Scholar]
- 46.Collet C, Schiltz C, Geoffroy V, Maroteaux L, Launay JM, De Vernejoul MC. The serotonin 5-HT2B receptor controls bone mass via osteoblast recruitment and proliferation. FASEB J. 2008;22(2):418–427. doi: 10.1096/fj.07-9209com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baudry A, et al. Serotonergic 5-HT(2B) receptor controls tissue-nonspecific alkaline phosphatase activity in osteoblasts via eicosanoids and phosphatidylinositol-specific phospholipase C. J Biol Chem. 2010;285(34):26066–26073. doi: 10.1074/jbc.M109.073791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paik JH, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128(2):309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dacquin R, Starbuck M, Schinke T, Karsenty G. Mouse alpha1(I)-collagen promoter is the best known promoter to drive efficient Cre recombinase expression in osteoblast. Dev Dyn. 2002;224(2):245–251. doi: 10.1002/dvdy.10100. [DOI] [PubMed] [Google Scholar]
- 50.Nakae J, et al. Regulation of insulin action and pancreatic beta-cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nat Genet. 2002;32(2):245–253. doi: 10.1038/ng890. [DOI] [PubMed] [Google Scholar]
- 51.Heath MJ, Hen R. Serotonin receptors. Genetic insights into serotonin function. Curr Biol. 1995;5:997–999. doi: 10.1016/S0960-9822(95)00199-0. [DOI] [PubMed] [Google Scholar]
- 52.Mantamadiotis T, et al. Disruption of CREB function in brain leads to neurodegeneration. Nat Genet. 2002;31(1):47–54. doi: 10.1038/ng882. [DOI] [PubMed] [Google Scholar]
- 53.Ducy P, et al. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev. 1999;13(8):1025–1036. doi: 10.1101/gad.13.8.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bogdanovic Z, et al. Upstream regulatory elements necessary for expression of the rat COL1A1 promoter in transgenic mice. J Bone Miner Res. 1994;9(2):285–292. doi: 10.1002/jbmr.5650090218. [DOI] [PubMed] [Google Scholar]
- 55.Schwartzberg PL, et al. Rescue of osteoclast function by transgenic expression of kinase- deficient Src in src–/– mutant mice. . Genes Dev. 1997;11(21):2835–2844. doi: 10.1101/gad.11.21.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hentunen TA, et al. Immortalization of osteoclast precursors by targeting Bcl -XL and Simian virus 40 large T antigen to the osteoclast lineage in transgenic mice. J Clin Invest. 1998;102(1):88–97. doi: 10.1172/JCI2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chiu WS, McManus JF, Notini AJ, Cassady AI, Zajac JD, Davey RA. Transgenic mice that express Cre recombinase in osteoclasts. Genesis. 2004;39(3):178–185. doi: 10.1002/gene.20041. [DOI] [PubMed] [Google Scholar]
- 58.Kalajzic I, et al. Dentin matrix protein 1 expression during osteoblastic differentiation, generation of an osteocyte GFP-transgene. Bone. 2004;35(1):74–82. doi: 10.1016/j.bone.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 59.Liu XS, et al. High-resolution peripheral quantitative computed tomography can assess microstructural and mechanical properties of human distal tibial bone. J Bone Miner Res. 2010;25(4):746–756. doi: 10.1359/jbmr.090822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lecka-Czernik B, et al. Inhibition of Osf2/Cbfa1 expression and terminal osteoblast differentiation by PPARgamma2. J Cell Biochem. 1999;74(3):357–371. doi: 10.1002/(SICI)1097-4644(19990901)74:3<357::AID-JCB5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 61.Rapp JP, Garrett MR, Deng AY. Construction of a double congenic strain to prove an epistatic interaction on blood pressure between rat chromosomes 2 and 10. J Clin Invest. 1998;101(8):1591–1595. doi: 10.1172/JCI2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.