Abstract
Understanding the transcriptional mechanisms that underlie pancreas formation is central to the efforts to develop novel regenerative therapies for type 1 diabetes. Recently, mutations in the transcription factor GATA6 were unexpectedly shown to be the most common cause of human pancreas agenesis. In this issue of the JCI, Carrasco et al. and Xuan et al. investigate the role of Gata6 and its paralogue Gata4 in mouse embryonic pancreas and show that GATA factors are essential regulators of the proliferation, morphogenesis, and differentiation of multipotent pancreatic progenitors.
Type 1 diabetes results from autoimmune destruction of insulin-producing β cells. The major goal of current research efforts to reverse this disease is focused on learning how to generate new β cells to replace those that have been destroyed while at the same time blocking the autoimmune process. This challenge has fueled intense research to uncover the extracellular signals and transcriptional regulators that promote the generation of β cells during embryonic development (1–3). Proof-of-concept experiments have shown that this type of knowledge can indeed be exploited to derive insulin-producing cells from multipotent cells in vitro (4) or through transdifferentiation of somatic cells (5).
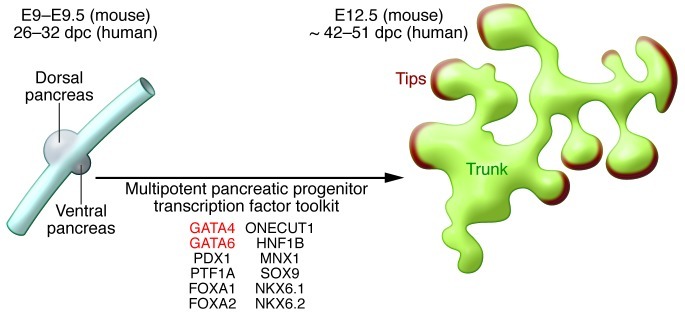
One of the critical stages of the developmental process that leads to β cell differentiation is the formation and orderly expansion of pancreatic multipotent progenitors. The latter originate from the foregut endoderm at around E8.5 in mice (6), and prior to 26 days post conception (dpc) in humans (ref. 7 and Figure 1). Nearly two decades ago, mouse genetic knockout experiments first proved that Pdx1 is essential for the expansion of the early pancreatic bud (8, 9). Since then, numerous transcription factors have been shown to be essential at this initial developmental stage, including Pdx1, Ptf1a, Mnx1, Sox9, and Hnf1b (3). In mice, only homozygous null mutations of these factors (either germline, chimeric, or conditional) have revealed a pancreatic phenotype. For some transcription factors, such as Foxa1/2, Onecut1, or Nkx6.1, critical regulatory roles have been uncovered by the complete inactivation of more than one gene, reflecting a degree of functional redundancy (10–12). Although most of these discoveries have relied on mouse genetics, mutations in PDX1, PTF1A, and HNF1B have also been identified in human patients with monogenic diabetes and pancreatic aplasia or hypoplasia, consistent with the notion that pancreatic developmental programs are fundamentally conserved in humans and mice (3).
Figure 1. Representation of early pancreatic development.
At approximately E9 in the mouse, pancreatic progenitors form dorsal and ventral buds off of the developing gut tube. Just a few days later, these multipotent progenitors give rise to a branched structure in which the tips are committed to an acinar cell fate, and the trunk contains duct-endocrine progenitors. The box lists transcription factors required for this remarkable transition.
Human genetics uncovers a key pancreatic regulator
Genome sequencing technologies have recently provided a major unexpected discovery in this field. Allen et al. examined 27 human patients who had neonatal diabetes due to pancreas agenesis and found that only one had a homozygous mutation in a known pancreatic regulator. An exome sequencing strategy revealed that 15 of the remaining patients had heterozygous loss of function mutations in the gene encoding the zinc finger transcription factor GATA6 (13). Patients also had developmental cardiac defects, in some cases mild, consistent with earlier reports of GATA6 mutations causing cardiac outflow defects. Several patients with mutations had endoderm developmental abnormalities, including thyroid, pituitary, gut, and biliary tract defects. In short, most humans with pancreatic agenesis have mutations in a gene that had so far been largely left off the radar screen in the pancreas development field. Intriguingly, another case report described a mutation in the paralogue GATA4 in a single patient with pancreatic agenesis and a cardiac malformation (14). The human genetics findings therefore establish that GATA6 (and less conclusively GATA4) is an essential regulator of pancreas development, although they do not shed light on the underlying molecular mechanism, nor do they define the precise cell types or developmental stages in which the essential role takes place.
Modeling GATA function in mice
In this issue of the JCI, Carrasco et al. and Xuan et al. (15, 16) report two mouse genetic studies that define a key regulatory role of Gata6 and Gata4 in pancreas development. In both reports, the two Gata genes were inactivated in embryonic multipotent pancreatic progenitors using Cre/LoxP technology. Conditional mutations were required because germline homozygote null mutations cause early embryonic lethality, whereas heterozygote mice have no known pancreatic defect (17, 18). Unexpectedly, the inactivation of either Gata4 or Gata6 in pancreatic progenitors did not cause severe pancreatic defects. However, the simultaneous deletion of both factors caused a major reduction in the pancreatic size related to a severe block in the proliferation of multipotent pancreatic progenitors. Furthermore, there was abnormal branching morphogenesis and a severe defect in differentiation, including failure to separate into peripheral acinar-committed progenitors and central endocrine-committed progenitors. Xuan et al. performed an additional experiment in which they deleted Gata genes in the endoderm prior to the appearance of the pancreas and again found that the pancreatic bud was formed, yet failed to expand (16). This means that, at least with the genetic tools that were used in these studies, there is no indication that GATA factors in foregut endoderm cells have a cell-autonomous role in pancreas specification. Instead, the data show that Gata4 and Gata6 play essential functions in embryonic pancreatic progenitors.
Although the most severe phenotypes result from loss of GATA function in pancreatic embryonic multipotent progenitors, the studies also suggest that they may have important functions beyond this stage. For example, Carrasco et al. show that mice with pancreatic deletions of both Gata4 alleles and one Gata6 allele have severe loss of acinar cells (15). Another recent report has now shown that mice in which only Gata6 is completely inactivated in pancreatic progenitors are histologically normal at birth, but eventually develop a severe loss of acinar cells (19). Thus, GATA factors appear to exert distinct essential functions throughout different steps of pancreatic cell formation.
Why does GATA6 haploinsufficiency cause pancreatic agenesis in humans, whereas in mice, three or four GATA alleles need to be deleted to elicit a severe pancreatic phenotype? It is important to note that haploinsufficiency of transcription factor genes is not uncommon in human developmental disorders. In pancreatic disease, heterozygous loss-of-function mutations in HNF1B, HNF4A, and HNF1A genes cause diabetes in humans but not in mice (2). The exact reason for this mouse-specific tolerance to haploinsufficiency of transcription factor genes has not been clarified. One candidate explanation is that some developmental stages in human last weeks longer than in mice. This could increase the likelihood that there are deleterious consequences from fluctuations in transcriptional levels that are not sufficiently buffered by the presence of the other healthy allele. In the case of GATA6 and GATA4, four alleles are in play. It is possible that the gene expression patterns of these two genes exhibit greater overlap at critical steps of early pancreas development in mouse versus human embryos, thus rendering mice more resilient to genetic perturbations of a single GATA gene.
GATA factors regulate Pdx1
How do GATA factors exert their essential functions in pancreatic progenitors? GATA4 and GATA6 transcription factors have been shown to initiate developmental programming steps important for heart, gut, and liver development (17, 18). The current studies now show that GATA factors are important for the activation of Pdx1, which encodes another essential pancreatic developmental transcription factor (8, 9). Carrasco et al. created a transgenic mouse containing the upstream sequence of Pdx1 linked to a reporter and showed that deleting GATA-binding sites prevented the activation of the reporter in early embryonic multipotent pancreatic progenitors (15). This is consistent with the observation that Pdx1 expression is reduced in pancreatic progenitors of mutant mice, although it is slightly at odds with the experiments from Xuan et al., suggesting that GATA factors are not required in the endoderm to initiate the expression of Pdx1 and the inception of pancreatic buds (16). It is possible, however, that enhancers of the Pdx1 gene that were not included in the transgenic construct and do not require GATA factors are sufficient to initiate Pdx1 expression in foregut endoderm.
An organ-building tool kit
Gata6 and Gata4 are broadly expressed in endodermal and mesodermal lineages (20), in contrast with Pdx1 and Ptf1a, which are expressed in the pancreatic anlage and only few other embryonic cell types. Early studies showing that Pdx1 and Ptf1a have a selective expression pattern coupled with the fact that their knockout mice develop pancreas agenesis suggested that these two factors might work as master regulators of pancreas organogenesis (8, 21). It has now become clear, however, that pancreatic multipotent progenitors require a larger transcription factor tool kit, which the current work suggests includes GATA4 and GATA6 in addition to PDX1, PTF1A, MNX1, ONECUT1, NKX6.1/NKX6.2, FOXA1/2, SOX9, and HNF1B (Figure 1). The inactivation of each one of these factors, either alone or in combination, disrupts the progenitor program and thereby blocks pancreas development. Due to the combinatorial nature of transcription factor function, it is the unique coalescence of factors that drives the pancreatic progenitor program, and therefore it does not matter whether a particular transcription factor is more or less specific to the pancreatic lineage. Based on the genetic findings, we can surmise that all of these factors are required to bind to regulatory elements that control the transcription of genes that are important for all of the things that pancreatic multipotent progenitors need to do, namely to proliferate, undergo morphogenetic changes, and differentiate. This likely also involves conferring responsiveness to the extracellular signals that promote developmental processes. In short, an interesting combination of mouse and human genetics has provided an inventory of the key transcription factors for pancreas organogenesis. This represents an important step in untangling the mechanisms of pancreas organogenesis, which can now be pursued with novel technologies to manipulate whole animal and in vitro human models and to analyze them with genome-scale tools. Knowledge of the components of this developmental tool kit and the instructions followed are invaluable in allowing us to recapitulate the process for cell replacement therapies. The studies from Carrasco et al. and Xuan et al. provide a landmark advance in this direction (15, 16).
Acknowledgments
Work in the authors’ laboratory is funded by the Ministerio de Economía y Competitividad, the Innovative Medicines Initiative, the European Commission Seventh Framework Programme, and the Beta Cell Biology Consortium (National Institute of Diabetes and Digestive and Kidney Diseases).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(10):3469–3471. doi:10.1172/JCI65751.
See the related articles beginning on pages 3504 and 3516.
References
- 1.Zaret KS, Grompe M. Generation and regeneration of cells of the liver and pancreas. Science. 2008;322(5907):1490–1494. doi: 10.1126/science.1161431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Servitja JM, Ferrer J. Transcriptional networks controlling pancreatic development and beta cell function. Diabetologia. 2004;47(4):597–613. doi: 10.1007/s00125-004-1368-9. [DOI] [PubMed] [Google Scholar]
- 3.Oliver-Krasinski JM, Stoffers DA. On the origin of the beta cell. Genes Dev. 2008;22(15):1998–2021. doi: 10.1101/gad.1670808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kroon E, et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26(4):443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- 5.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455(7213):627–U630. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jorgensen MC, Ahnfelt-Ronne J, Hald J, Madsen OD, Serup P, Hecksher-Sorensen J. An illustrated review of early pancreas development in the mouse. Endocr Rev. 2007;28(6):685–705. doi: 10.1210/er.2007-0016. [DOI] [PubMed] [Google Scholar]
- 7.Piper K, et al. Beta cell differentiation during early human pancreas development. J Endocrinol. 2004;181(1):11–23. doi: 10.1677/joe.0.1810011. [DOI] [PubMed] [Google Scholar]
- 8.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371(6498):606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 9.Offield MF, et al. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122(3):983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 10.Gao N, LeLay J, Vatamaniuk MZ, Rieck S, Friedman JR, Kaestner KH. Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev. 2008;22(24):3435–3448. doi: 10.1101/gad.1752608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson SB, Schaffer AE, Sander M. The transcription factors Nkx6.1 and Nkx6.2 possess equivalent activities in promoting beta-cell fate specification in Pdx1+ pancreatic progenitor cells. Development. 2007;134(13):2491–2500. doi: 10.1242/dev.002691. [DOI] [PubMed] [Google Scholar]
- 12.Jacquemin P, Yoshitomi H, Kashima Y, Rousseau GG, Lemaigre FP, Zaret KS. An endothelial-mesenchymal relay pathway regulates early phases of pancreas development. Dev Biol. 2006;290(1):189–199. doi: 10.1016/j.ydbio.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 13.Allen HL, et al. GATA6 haploinsufficiency causes pancreatic agenesis in humans. Nat Genet. 2011;44(1):20–22. doi: 10.1038/ng.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Amato E, et al. Genetic investigation in an Italian child with an unusual association of atrial septal defect, attributable to a new familial GATA4 gene mutation, and neonatal diabetes due to pancreatic agenesis. Diabet Med. 2010;27(10):1195–1200. doi: 10.1111/j.1464-5491.2010.03046.x. [DOI] [PubMed] [Google Scholar]
- 15.Carrasco M, Delgado I, Soria B, Martín F, Rojas A. GATA4 and GATA6 control mouse pancreas organogenesis. J Clin Invest. 2012;122(10):3504–3515. doi: 10.1172/JCI63240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xuan S, et al. Pancreas-specific deletion of mouse Gata4 and Gata6 causes pancreatic agenesis. . J Clin Invest. 2012;122(10):3516–3528. doi: 10.1172/JCI63352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuo CT, et al. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11(8):1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 18.Morrisey EE, et al. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 1998;12(22):3579–3590. doi: 10.1101/gad.12.22.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martinelli P, Canamero M, del Pozo N, Madriles F, Zapata A, Real FX. Gata6 is required for complete acinar differentiation and maintenance of the exocrine pancreas in adult mice. Gut. In press [DOI] [PubMed] [Google Scholar]
- 20.Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem. 2000;275(50):38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- 21.Krapp A, et al. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 1998;12(23):3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]