Abstract
Bipolar disorder (BD) is characterized by disruptions in circadian rhythms such as sleep and daily activity that often normalize after lithium treatment in responsive patients. Since lithium is known to interact with the circadian clock, we hypothesized that variation in circadian “clock genes” would be associated with lithium response in BD. We determined genotype for 16 variants in 7 circadian clock genes, and conducted a candidate gene association study of these in 282 Caucasian BD patients who were previously treated with lithium. We found that a variant in the promoter of NR1D1 encoding Rev-Erbα (rs2071427) and a second variant in CRY1 (rs8192440) were nominally associated with good treatment response. Previous studies have shown that lithium regulates Rev-Erbα protein stability by inhibiting GSK3β. We found that GSK3β genotype was also suggestive of a lithium response association, but not statistically significant. However, when GSK3β and NR1D1 genotypes were considered together, they predicted lithium response robustly and additively in proportion to the number of response-associated alleles. Using lymphoblastoid cell lines from BD patients, we found that both the NR1D1 and GSK3β variants are associated with functional differences in gene expression. Our findings support a role for Rev-Erbα in the therapeutic mechanism of lithium and suggest that the interaction between Rev-Erbα and GSK3β may warrant further study.
Keywords: Lithium, Bipolar Disorder, Circadian Rhythm, CRY1, glycogen synthase kinase, GSK3B, Rev-Erb-alpha, NR1D1
INTRODUCTION
Bipolar disorder (BD) is a psychiatric illness characterized by severe, episodic mood disturbances that include distinct periods of mania and depression. In both phases of the illness, BD perturbs daily rhythms in sleep, energy, appetite and activity. Furthermore, mood symptoms can vary with time of day and season, and bright light is both an effective antidepressant and a potential trigger for mania. For these reasons, circadian clock abnormalities have been suspected in BD for decades (reviewed by McClung, 2007). Previous studies from our laboratory and others have supported this hypothesis, reporting that polymorphisms in a number of genes with circadian rhythm roles are associated with BD including ARNTL, CRY1, CRY2, NR1D1, and RORB among others (Kripke et al. 2009; Mansour et al. 2005; Mansour et al. 2009; McGrath et al. 2009; Nievergelt et al. 2006; Shi et al. 2008; Sjöholm et al. 2010, Soria et al 2010). Moreover, models for mania have been developed based upon experimental manipulations of the circadian clock in animals (Mukherjee et al. 2010; Le-Niculescu et al. 2008; Roybal et al. 2007).
The circadian clock is comprised of a complex, interacting network of genes (“clock genes”) that collectively maintains rhythmic expression over ~24 hr cycles (reviewed by Takahashi et al. 2008). The protein products of CLOCK and ARNTL are the primary transcriptional activators, whereas the protein products of PER1/2, CRY1/2 and NR1D1/2 (Rev-Erbα/β) are the major transcriptional inhibitors that complete negative feedback loops within the clock (Kume et al. 1999, Liu et al. 2008). Interestingly, these genes are expressed widely throughout the brain and peripheral tissues maintaining rhythms throughout the body.
Among the effective treatments for BD, lithium (Li) is commonly used. However, a good treatment response to lithium is not universal and among BD patients, ~50% fail Li by the second year of treatment (Vestergaard and Schou 1988). The clinical profiles of Li responsive (Li-R) and non-responsive (Li-NR) BD patients have been reported to differ. For example, euphoric mania and a positive family history are considered to be factors favoring Li response, while dysphoric mania, the presence of co-morbid psychiatric conditions and a negative family history are considered poor prognostic indicators (Kukopulos et al. 1980, Grof et al. 2003). Based on these and other observations, it has been proposed that Li-R and Li-NR BD may be genetically distinct forms of illness (Grof et al. 2003; 2009). However, the biological basis for these differences in Li responsiveness remains unknown.
Despite its common use for over 50 years, the precise therapeutic mechanism of Li remains unknown, but may involve the circadian clock. In support of this notion, various molecular components of the circadian clock are known to be Li sensitive. Specifically, Li has been reported to lengthen the period of circadian rhythms in several species (Kripke et al. 1979; Kripke and Wyborney 1980; Welsh and Moore-Ede et al. 1990) and to alter circadian rhythms in neuronal firing (Abe et al. 2000). Li also inhibits glycogen synthase kinase 3β (GSK3β, Klein and Melton 1996), which in turn regulates protein turnover and stability in the clock network by phosphorylating target proteins, thereby modulating circadian rhythms (Iitaka et al. 2005, Yin et al. 2006). Of particular interest to the current study, GSK3β inhibition by Li has been shown to facilitate the degradation of Rev-Erbα, an important clock signaling molecule (Yin et al. 2006). This evidence suggests that Li affects clock genes, and that within the clock, the relationship between GSK3β and Rev-Erbα (transcribed from the gene NR1D1) may be of particular interest. Supporting this notion, the variant rs2314339 within NR1D1 has recently been associated with Li response (Campos-de-Sousa et al. 2010). However, in this study the relationship of NR1D1 to GSK3β was not examined.
Presently, we sought to identify functional genetic variants in the circadian clock that may be useful in predicting lithium response in BD. We found clock gene variants that are nominally associated with clinical Li response in BD patients, including two variants from the well characterized clock signaling pathway that combine for a more robust association. We then used lymphoblastoid cell lines (LCLs) to model the effects of genetic variation within the clock on Li-induced gene expression in Li-R and Li-NR patients with BD.
METHODS
Subjects
Subjects (N=282) were recruited for genetic studies through a mood disorder clinic at the Veterans Affairs San Diego Healthcare System (VASDHS). Research was approved by the UCSD/VASDHS IRB and all participating subjects provided informed consent. BD can be subdivided into types I and II, based primarily upon illness severity and the presence of mania (in BD I) or hypomania (in BD II), whereas BD not otherwise specified (NOS) is used when one or more factors limit the ability to make a specific BD I/II diagnosis (American Psychiatric Association 2000). Since Li is an accepted therapy for all forms of BD, subjects with any BD sub-type were included. The majority (91%) had BD I, whereas a minority had BD II (7%) or BD NOS (2%). All diagnoses were established using one or more standardized instruments including the Structured Clinical Interview for DSM-III-R or DSM-IVTR (SCID), and the Diagnostic Interview for Genetic Studies (DIGS). All subjects were of self-declared Caucasian ancestry.
Clinical assessment
Li response was determined retrospectively as described previously (Bremer et al. 2007). Briefly, a detailed review of medication trials was conducted using information from the patient, medical records and other informants. A panel of clinicians blind to genotype then rated this information. Episode frequency and severity were compared between periods on/off Li. Only Li trials of three months or longer were considered. Response was rated as good if there was a 50% reduction in the frequency and/or severity of symptoms on Li. These subjects were classified as Li responders (Li-R, N=148). Those with < 50% symptom reduction were considered Li non-responders (Li-NR, N=134). The majority of subjects were also scored on the 10 point Alda scale of Li response (Grof et al. 2002). Average Alda A criterion score (± SD) for Li-R (N=115) was a 6.6 ± 1.5, corresponding to a response of ~81.5%, and for Li-NR (N= 118) was 2.5 ± 1.5, corresponding to a response of ~22.5%. In each group, average age (yr ± SD) was similar (Li-R: 45 ± 12.9, Li-NR: 44 ± 12.0). Among the Li-R group, 82/148 (55%) were men, compared to 60/134 (45%) men in the Li-NR group [χ2=3.1(1), p=0.08].
Genotyping
Seven genes were selected for analysis based on their critical roles in the core clock mechanism, both in the positive and negative feedback limbs of the system (Table 1). Sixteen single nucleotide polymorphisms (SNPs) were selected based on predicted function and position within the gene. We focused on regions showing epigenetic histone marks associated with regulatory elements and/or phylogenetic conservation (UCSC Genome Browser: http://genome.ucsc.edu, March 2006 Assembly). We also used previous evidence from a case-control study performed previously by our group (Kripke et al 2009), and unpublished association data. This set of markers does not provide comprehensive haplotype coverage, but tests multiple candidate gene regions while minimizing multiple comparisons. All SNPs were transmitted independently across genes. Within ARNTL, rs1982350 and rs2279287 were correlated (r2 = 0.36), and all 6 SNPs in PER3 showed varying degrees of correlation (r2 = 0.21 to 0.84). Thus, the Bonferroni correction employed for multiple testing was considered conservative (Nyholt 2004).
Table 1.
Variants selected from circadian clock genes and Li response association results. Gene and SNP information including the minor allele frequency (MAF) for the rare allele is indicated. Additive, dominant and recessive genetic transmission models were used, the nominal p-values (P) and odds ratios (OR) are shown for each (Add, Dom, Rec). Uncorrected p<0.05 was considered nominally statistically significant with one degree of freedom for each analysis. In some cases, exclusions for quality control slightly reduced total sample size.
| Gene | SNP | Chr | Position | Location | N= | Minor allele |
Major allele |
MAF | Wald Stat |
OR Add | P Add |
OR Dom |
P Dom | OR Rec |
P Rec |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ARNTL | rs2279287 | 11 | 13298485 | Enhancer | 281 | A | G | 0.27 | 0.56 | 1.11 | 0.57 | 1.10 | 0.68 | 1.30 | 0.57 |
| ARNTL | rs1982350 | 11 | 13350131 | Enhancer | 281 | T | C | 0.35 | −0.18 | 0.97 | 0.86 | 1.04 | 0.89 | 0.80 | 0.56 |
| ARNTL | rs2278749 | 11 | 13397878 | Promoter | 282 | A | G | 0.19 | 0.72 | 1.18 | 0.47 | 1.11 | 0.69 | 2.67 | 0.24 |
| CLOCK | rs1801260 | 4 | 56301369 | Exon | 281 | C | T | 0.26 | 0.32 | 1.07 | 0.75 | 1.32 | 0.26 | 0.45 | 0.13 |
| CLOCK | rs3736544 | 4 | 56309992 | Exon | 279 | A | G | 0.36 | −1.64 | 0.75 | 0.10 | 0.68 | 0.12 | 0.68 | 0.27 |
| CLOCK | rs34897046 | 4 | 56325365 | Exon | 282 | C | G | 0.05 | 0.75 | 1.34 | 0.45 | 1.52 | 0.31 | NA | NA |
| CRY1 | rs8192440 | 12 | 107395106 | Exon | 278 | A | G | 0.41 | −2.50 | 0.64 | 0.01 | 0.54 | 0.02 | 0.59 | 0.11 |
| GSK3B | rs6438552 | 3 | 119631814 | Enhancer | 281 | C | T | 0.42 | 1.38 | 1.28 | 0.17 | 1.17 | 0.54 | 1.82 | 0.08 |
| NR1D1 | rs2071427 | 17 | 38254492 | Promoter | 282 | A | G | 0.28 | 1.95 | 1.45 | 0.05 | 1.90 | 0.01 | 0.91 | 0.83 |
| PER2 | rs2304672 | 2 | 239186589 | Exon | 281 | G | C | 0.09 | −1.65 | 0.59 | 0.10 | 0.60 | 0.12 | NA | NA |
| PER3 | rs228729 | 1 | 7845695 | Intron | 282 | A | G | 0.31 | 0.69 | 1.14 | 0.49 | 1.25 | 0.36 | 0.97 | 0.95 |
| PER3 | rs228642 | 1 | 7863293 | Intron | 282 | C | T | 0.43 | 0.56 | 1.11 | 0.58 | 1.06 | 0.83 | 1.27 | 0.46 |
| PER3 | rs228666 | 1 | 7868725 | Intron | 282 | C | T | 0.37 | −0.51 | 0.92 | 0.61 | 0.95 | 0.84 | 0.78 | 0.46 |
| PER3 | rs228697 | 1 | 7887579 | Exon | 281 | G | C | 0.11 | 0.44 | 1.12 | 0.66 | 1.10 | 0.75 | 1.58 | 0.61 |
| PER3 | rs2859388 | 1 | 7888215 | Promoter | 282 | G | A | 0.36 | 0.26 | 1.05 | 0.79 | 0.99 | 0.97 | 1.26 | 0.55 |
| PER3 | rs2640909 | 1 | 7890117 | Exon | 277 | C | T | 0.33 | −0.72 | 0.88 | 0.47 | 0.83 | 0.45 | 0.87 | 0.73 |
For genotyping, we used SNPlex with an ABI 3730 48-capillary DNA analyzer following the manufacturer’s protocols (Applied Biosystems). Three SNPs (rs2304679, rs3736544 and rs8192440) were genotyped by Taqman PCR (Applied Biosystems) using an ABI 7900 thermocycler. Among our cohort of 282 BD subjects, 184 (92 Li-R and 92 Li-NR, 65% of subjects) have been genotyped previously using a distinct and non-overlapping set of markers (Bremer et al. 2007). A different set of 56 BD subjects (26 Li-R, 30 Li-NR, 20% of subjects) was genotyped previously using some of the same markers as part of a BD case-control association study (Kripke et al. 2009). In the present case-case analysis, no healthy controls were included, and all samples were re-genotyped. Using the latter 56 samples as a quality control measure, we found 100% agreement in genotype calls between our previous study and the results presently reported. All of the 16 SNPs used for the Li association experiment also passed the following quality controls: call rate >0.98, minor allele frequency (MAF) >0.05, and genotypes in Hardy-Weinberg equilibrium.
Genetic association analysis
We used a panel of 41 ancestry-informative genetic markers to control for population stratification. Continental ancestry was determined using 944 Human Genome Diversity Panel subjects as a reference (Nievergelt, in preparation). Within subjects of European ancestry, the multidimensional scaling method (Purcell et al. 2007) was used to infer continuous axes of genetic variation to be used as covariates to control for residual stratification.
Genetic association was determined using logistic regression. The Wald test was used to calculate odds ratios (OR) for each variant with Li-R/Li-NR as the dependent variable, allelic frequencies of the SNPs as the predictors and gender with two MDS components as covariates. Since we had no a priori hypothesis regarding the specific inheritance patterns that determine lithium response, we explored three common mechanisms, analyzing our genetic association data using additive, recessive, and dominant genotypic models for each SNP. A haplotype analyses using a moving window approach was performed but did not result in any significant associations (not shown). All associations were performed in PLINK v. 1.0.7 and SPSS v. 16.
Further analyses of the NR1D1 and GSK3β genotypic combinations were made post-hoc, examining each of the nine genotype possibilities individually. Calculations for ORs were made for each possible genotype combination and compared to the OR of Li-R/Li-NRs for all of the other genotypes, combined in an iterative manner for each of the nine possibilities. In all analyses, a higher OR corresponds to better chance of treatment response. A chi-square test was used to calculate ORs with significance defined as p<0.05. Gene x gene interaction power analyses were performed using QUANTO v. 1.2.
Lymphoblastoid cell cultures
For the NR1D1 expression studies, lymphoblastoid cell lines (LCLs) were selected from Li-R (N=13) and Li-NR (N=18) subjects with BD I on the basis of NR1D1 genotype. For GSK3β expression studies, additional samples were required to obtain sufficient numbers of each genotype, and not all of these cells had donor’s Li response history available. In all cases, LCLs were cultured from frozen vials in standard RPMI growth medium with 10% fetal bovine serum (FBS) for 10–14 days to a density of ~1 million cells/ml. Cells were then collected, rinsed in PBS and stored at −80°C. For time course experiments, cells were serum shocked in medium containing 50% FBS for 90 min to synchronize circadian clock gene expression (Liu et al. 2007), then re-suspended into standard medium in parallel cultures. After a 20 hr equilibration period, flasks were harvested every three hours over the next 24 hr. For the Li studies, additional LCL cultures were grown in fresh medium ± Li chloride (1mM) for 72 hr, a duration that simulates the early phase of a clinically relevant drug exposure, but short enough to eliminate the need for manipulations that could introduce artifacts between cultures. Parallel cultures from the same donor ± Li were compared, allowing each cell line to serve as its own control.
RNA preparation and cDNA synthesis
RNA extraction from LCLs was performed with the RNeasy Mini Kit (Qiagen). RNA (500 ng) was used to synthesize cDNA from each sample using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems).
Assessment of clock gene expression
Expression of individual genes was measured by RT-PCR using commercial primers (Applied Biosystems) for GSK3β (Hs00275656_m1) and GAPDH (Hs99999905_m1). NR1D1 has been previously reported to contain an alternative promoter, generating two functional transcripts of different lengths. For this reason, two distinct PCR primer pairs were used to measure NR1D1 expression. The first (Hs00253876_m1) targeted the junction of the first and second exons, thereby measuring only the full length transcript. The second (Hs00897531_m1) targeted the junction of exons six and seven, thereby measuring both the full length and truncated transcripts. RT-PCR reactions were performed in an ABI 7900 thermocycler with all samples run in duplicate. Gene expression was calculated as a ratio of the target gene to GAPDH, and analyzed using the comparative Ct method (Schmittgen and Livak 2008). Where appropriate, data were normalized as a percent of baseline.
Statistical Analysis
Time course data were analyzed using a one-way ANOVA with post-hoc t tests. The least squares method was then used to determine the best line or non-linear function to fit the data. Changes in gene expression following Li were determined by two-way ANOVA with repeated measures using commercial software (Graphpad Prism).
RESULTS
Genetic association of lithium response in BD patients
Among the 16 clock gene variants, two SNPs (rs8192440 and rs2071427) were nominally associated with Li response. The rs8192440 (CRY1) association was strongest using an additive transmission model, with the common G allele favoring good response (Table 1). The rs2071427 (NR1D1) association was strongest using a dominant transmission model, with the rare A allele favoring good response (Table 1). Previous reports have suggested that GSK3β polymorphisms are associated with Li response (Benedetti et al. 2005; Adli et al. 2007). In our sample, the GSK3β variant rs6438552 showed only a suggestive trend toward association with Li response using a recessive inheritance model (Table 1), whereby homozygosity at the rare C allele favored Li response. The results obtained using a univariate analysis also revealed nominal lithium response associations with rs8192440 from CRY1 and rs2071427 from NR1D1, but no significant association with the GSK3β variant rs6438552 (Supplementary Table 1). In neither analysis did any of the nominal associations remain significant after Bonferroni correction for 16 tests.
Association analysis of combined NR1D1 and GSK3β variants
In model systems, GSK3β has been reported to phosphorylate multiple proteins within the circadian clock including Rev-Erbα (Yin 2006), ARNTL (also called BMAL, Sahar et al. 2010), PER2 (Iitaka et al. 2005) and CRY2 (Harada et al. 2005). Unlike CRY2, evidence suggests that CRY1 is not a substrate for GSK3B phosphorylation (Kurabayashi et al. 2010).
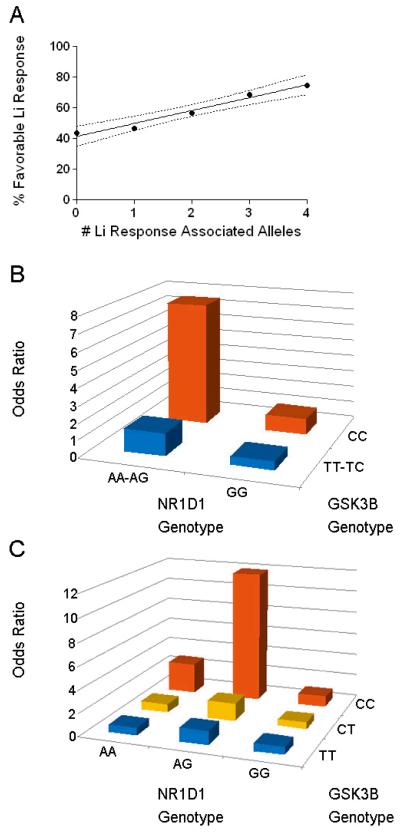
Since Rev-Erbα is a substrate for GSK3β phosphorylation, and Li has been shown to inhibit this interaction (Yin et al. 2006), we examined whether multiple genetic variants within this known signaling pathway could interact to better predict Li response outcomes. Considering the NR1D1 and GSK3β variants together, the total number of Li response-associated alleles (range 0-4) was highly correlated with clinical outcome (Figure 1A; r2=0.97, F1,3 = 98, p < 0.0025). Subjects who were doubly homozygous for the favorable A allele at rs2071427 and C allele at rs6438552 had a 75% chance of being in the Li-R group. In comparison, those who were doubly homozygous for the unfavorable G allele at rs2071427 and T allele at rs6438552 had only a 44% response rate. Those with three, two or one favorable alleles were intermediate with progressively lower chances of favorable clinical outcomes. Two additional analyses were conducted to evaluate the Li response rates of specific genotype combinations. The first, a 2×2 analysis using only the best fitting transmission models (dominant for NR1D1 and recessive for GSK3β) maximized statistical power. The second, a 3×3 analysis using each of the nine possible genotypic combinations allowed for higher resolution comparisons of the each genotypic combination (Figure 1B, 1C). In the 2×2 analysis, those with NR1D1:GSK3β genotypes of AA-AG:CC responded significantly more often than did the comparison group [χ2(1)= 9.36, OR = 7.5, 95% CI 1.68 – 33.46, p< 0.0025]. Conversely, those with GG:TT-TC genotypes responded significantly less often [χ2(1)=7.40, OR = 0.52, 95% CI 0.32 - 0.83, p< 0.0075]. The more detailed 3×3 analysis suggested that the rs6438552 CC genotype at GSK3β nominally favored Li response, regardless of NR1D1 genotype. The statistical analysis revealed that subjects with NR1D1:GSK3β genotype of AG:CC fared best [χ2(1) = 8.75, OR 11.87, 95% CI 1.52 - 92.25, p< 0.003] whereas those with GG:TC genotype fared worst [χ2(1) = 3.85, OR 0.58, 95% CI 0.33 - 1.00, p<0.05]. Only four subjects had the doubly rare AA:CC genotype. While this combination showed a trend toward favorable Li response its OR was not significantly different from unity [χ2(1) = 0.84, OR 2.77, 95% CI 0.28 - 26.98, p < 0.36]. Similarly, while the Li response rate of the AG:CC genotype was nominally higher than the AA:CC genotype, these groups had overlapping confidence intervals and could not be statistically discriminated. None of the other genotype combinations differed significantly from unity in the post-hoc OR analyses (range 0.66 – 2.77).
Figure 1.
Additive effects of GSK3β and NR1D1 alleles on lithium response. A) All possible genotype combinations at rs2071427 and rs6438552 were determined for all Li-R and Li-NR subjects and scored (0-4) for the number of alleles associated with favorable lithium outcomes. Dashed lines indicate 95% confidence intervals for the regression line. Rates of response for each allelic combination are the following: 4 alleles: 75% (N=4), 3 alleles: 69% (N=26), 2 alleles: 57% (N=95), 1 allele: 44% (N=108), 0 alleles: 47% (N=48). B) Odds ratios for lithium response calculated for each of 2×2 GSK3β and NR1D1 genotype combinations using dominant and recessive models. Sample sizes for each subgroup are as follows: AA-AG:CC N=17, AA-AG:TT-TC N=116, GG:TT-TC N=119, GG:CC N= 29 C) Odds ratios for lithium response calculated for each of 3×3 GSK3β and NR1D1 genotype combinations. Sample sizes for each subgroup are as follows: AA:CC N=4, AA:TC N=13, AA:TT N=7, AG:CC N=13, AG:TC N=59, AG:TT N=37, GG:CC N=29, GG:TC N=71, GG:TT N=48.
Examination for epistatic interaction
To more precisely define the combinatorial nature of the relationship between NR1D1 and GSK3β, a logistic regression analysis was performed to predict Li response including both rs2071427 and rs6438552. For each gene this analysis used the most significant genetic model based on the results from the single SNP association analyses (dominant model for rs2071427, recessive model for rs6438552, see Table 1) and included covariates for gender and population stratification. This analysis showed a significant additive effect [χ2(5) = 15.92, p<0.007], but fell short of providing evidence for a synergistic gene x gene interaction [χ2(1) = 2.228, p=0.136]. We conducted a power analysis to assess our ability to detect epistasis using the model parameters that corresponded to the strongest single SNP associations (rs2071427: dominant model, MAF 0.28, OR 1.90; rs6438552: recessive model, MAF 0.42, OR 1.82) for 148 Li-R and 134 Li-NR at a baseline Li response rate of 0.47. Under these assumptions, we had 80% power to detect only very large interaction effects of OR >25. To detect interaction effects in the range of 7 - 12, as seen in Figure 1B and 1C, we would need at least 246 Li-R and 224 Li-NR, or 187 Li-R and 171 Li-NR, respectively.
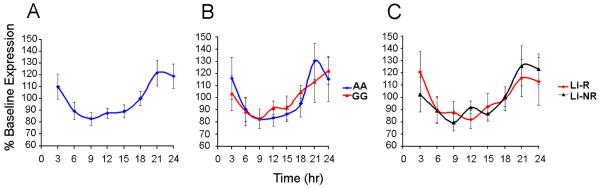
Variant rs2071427 does not affect rhythmic NR1D1 expression
We next examined whether rs2071427 was associated with functional effects on NR1D1 expression. As a clock gene, expression of NR1D1 (encoding Rev-Erbα) was expected to oscillate rhythmically (Liu et al. 2008). Indeed, we were able to show that NR1D1 expression did vary significantly over time (F7,88 = 3.55, p<0.005) after synchronization of the cells, and that a non-linear curve fit the data better than did the best fit line, suggesting a ~24 hr periodic expression rhythm (Figure 2A). When NR1D1 expression was analyzed by rs2071427 genotype and by Li treatment outcome (Figure 2B and 2C), the expression profiles of NR1D1 did not differ significantly between groups. Both NR1D1 PCR primer sets (directed towards exon junctions 1/2 or 6/7) revealed similar results, and the ratios of NR1D1 alternate transcripts did not differ by time (not shown).
Figure 2.
Time course of NR1D1 expression. A) Expression in LCLs (N=12) over 24 hrs after serum shock varies in a non-linear manner suggestive of a ~24 hr periodic rhythm. Expression is similar when analyzed by B) Genotype (N= 6/ group) or C) Li response history (N= 5 Li-R and 7 Li-NR). In all cases data are presented as % baseline expression ± SEM.
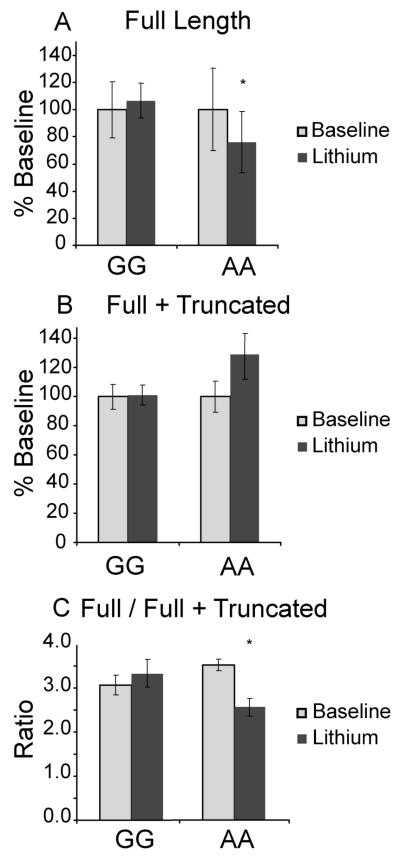
Variant rs2071427 and NR1D1 expression after lithium
Genetic variation in NR1D1 might also affect expression in response to Li. In order to assess this possibility, we measured NR1D1 transcripts in LCLs after 72 hr of Li exposure. Under these circumstances, rs2071427 genotype was associated with several differences in NR1D1 expression. First, in samples homozygous for the A allele (AA), expression of the full length NR1D1 transcript was decreased by ~20% following Li, whereas expression in samples homozygous for the G (GG) allele did not change. Statistical analysis revealed a significant interaction of genotype x Li treatment, favoring decreased expression selectively in the AA group (F1,18 = 5.4, p<0.05, Figure 3A). When expression of both the full and truncated NR1D1 transcripts was measured, the AA samples had a non-significant ~15% trend towards increased NR1D1 expression after Li treatment, whereas the GG samples again did not show any change in expression (Figure 3B).
Figure 3.
NR1D1 expression differs by genotype after Li exposure. A) Full length transcript expression decreases after Li preferentially in AA homozygotes at the rs2071427 locus. B) Expression of full length + truncated NR1D1 transcript trends toward an increase after Li preferentially in AA homozygotes. C) Ratio of full length/(full+truncated) NR1D1 transcript is significantly and specifically decreased among AA homozygotes at rs2071427. NR1D1 expression among GG homozygotes does not change significantly after Li. N=10 LCLs/ group. Data reflect normalized averages ± SEM. (*) indicates significance of p<0.05
The ratio of truncated to full-length Rev-Erbα protein has been shown previously to affect the stability of the protein (Rambaud et al. 2009). When NR1D1 expression after Li was analyzed as the ratio of full length transcript to full + truncated transcript, the effects of drug treatment on NR1D1 transcript populations were more pronounced (Figure 3C), with a significant effect of Li (F1,18 = 5.15, p<0.05), and a highly significant interaction between Li and genotype (F1,18 =16.4, p<0.001). A post-hoc t-test revealed that Li-treated samples with the AA genotype expressed a significantly lower ratio of full length/truncated NR1D1 transcript compared to untreated samples (t = 3.58, df = 9, p<0.01), in contrast to samples with GG genotype showed no difference after Li treatment.
We examined NR1D1 expression in a smaller number of AG heterozygotes (N=5) with TT genotype at GSK3β rs6438552 and found expression levels that were on average intermediate between the homozygous groups in all conditions but did not statistically differ from either the AA or GG groups (data not shown).
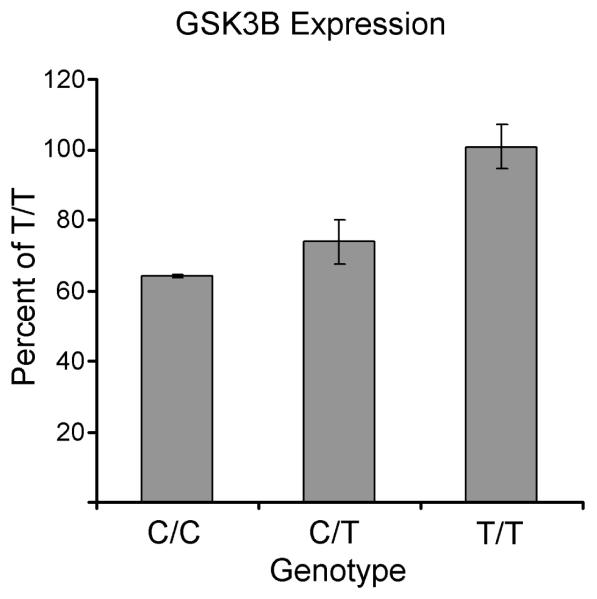
GSK3β expression: Effect of genotype, lithium and clinical history
The SNP rs334558 within GSK3β, commonly called -50 T/C (Russ et al. 2001) has previously been shown to affect gene expression and transcription factor binding (Lau et al. 1999, Kwok et al. 2005) but has not been formally studied in samples from BD patients. The rs6438552 variant that we studied has been linked to rs334558, and we confirmed this linkage in our sample, finding nearly perfect linkage disequilibrium (r2=0.95) between genotypes at rs6438552 and rs334558. We examined the association of rs6438552 with gene expression in a sample of 29 LCLs from BD patients. GSK3β expression differed significantly by rs6438552 genotype, with TT samples having the greatest expression, CC having the least, and TC intermediate (F2,26 = 5.25, p<0.05; Figure 4). In 11 LCL samples (all TT genotype) GSK3β expression after Li treatment was analyzed by clinical response history. Exposure of LCLs to Li did not have any significant effect on GSK3β expression in either Li-R (13.59 vs. 15.10) or Li-NR (13.85 vs. 15.59, data reflect normalized GSK3β expression: baseline vs. Li treated, all p-values > 0.05).
Figure 4.
GSK3β expression varies by genotype in LCLs from BD patients. Expression was measured in LCLs who were identified as CC (N=2), TC (N= 9) or TT (N= 18) genotypes at rs6438552. Data were normalized to TT genotype. Post-hoc tests showed that the differences between CC vs. TT and TT vs. TC were significant (p<0.05), but that the differences between CC and TC were not.
No interaction between GSK3β and NR1D1 genotypes on gene expression
Finally, we examined whether GSK3β genotype altered Li’s effects on NR1D1 expression. In the samples described above, NR1D1 expression for four different GSK3β:NR1D1 genotype combinations (TT:GG, TT:AA, TC:GG, TC:AA, N=5/group) was re-examined at baseline and after Li using both NR1D1 probes. We were unable to examine the rs6438552 CC genotype. No significant differences were observed (data not shown).
DISCUSSION
Overview
The circadian clock has been implicated in BD and Li response by behavioral, genetic and biochemical studies. However, few data directly address the molecular mechanisms underlying clock dysfunction in human BD patients. We performed a candidate gene association study of Li responsiveness in BD and made detailed measurements of clock gene function at the molecular level. In so doing, we have established a preliminary link between molecular clock function and clinical Li response in BD.
Study Limitations
Our work has two major limitations. First, our retrospective genetic association study was underpowered and the nominally significant findings did not survive corrections for multiple comparisons. While replication will be required, the shortcomings of our work are mitigated by several important factors. Namely, the individual variants are associated with functional differences in gene expression, have high odds ratios, are linked in a known biochemical pathway and are robust in their additive effects, together offering sufficient evidence to warrant further investigation.
The second limitation is the use of LCLs to model BD. As a model of psychiatric illness, LCLs are constrained by the absence of specialized brain circuits, clear differences in gene expression between brain and lymphocytes, and potential artifacts caused by viral transformation of the cells. Nevertheless, we believe LCLs to be a reasonable model for the following reasons. First, the circadian clock is cell-autonomous, and does not depend on brain circuits for its function (Welsh et al. 2010). Second, despite differences, there is substantial overlap among the gene expression profiles of LCLs, whole blood, and brain (Rollins et al. 2010) and while the master clock in mammals is located in the suprachiasmatic nucleus, peripheral cells including blood monocytes also express functional clock genes (Archer et al. 2008; Brown et al. 2005; Liu et al. 2007; Liu et al. 2008; Teboul et al. 2005; Yagita et al. 2001). Moreover, fibroblasts and blood cells have been used to demonstrate differences in circadian gene expression between healthy humans and patients with sleep disorders and BD, suggesting that genetic factors influencing clock function are faithfully reflected in peripheral tissues (Archer et al. 2008; Brown et al. 2005; Yang et al. 2008). We found that LCLs express >40 known clock genes (Supplementary Table 2), and our data suggest that NR1D1 expression is rhythmic. Collectively, this evidence indicates that LCLs are an appropriate model system for the study of clock genes in BD.
Genetic association of clock genes with lithium response
Using the LCL model, we found two genetic variants associated with gene expression differences. These differences plausibly link genetic variation in the circadian clock to lithium response. In addition, we also found two nominal associations of clock gene variants with Li response in BD patients: the NR1D1 variant rs2071427 and the CRY1 variant rs8192440. We found a trend for association of the GSK3β variant rs6438552 with Li response, partially supporting the findings of Benedetti et al. (2005) who previously implicated a variant within the same haplotype. While none of these findings remained significant after correcting for multiple comparisons, there was an additive effect and a trend toward epistasis between variants of NR1D1 and GSK3β, both of which were associated with relevant functional differences in gene expression. Therefore, our analysis suggests that these effects should be re-examined in larger studies of uniformly treated BD patients that have improved power to detect gene-gene interactions. Indeed, larger replication studies are planned in the near future.
Previous pharmacogenetic studies: NR1D1 and lithium
A recent candidate gene study of Li response also reported positive findings for NR1D1, associating rs2314339 with Li-NR (OR = 0.28), but reported no association with rs2071427 (Campos-de-Sousa et al. 2010). This study population (N=170) had only 7 subjects with the minor rs2071427 allele and lacked statistical power to detect a contribution from the variant. Previously implicated in BD, rs2314339 is in partial linkage with rs2071427 (Kripke et al. 2009), and thus it remains possible that the functional variant in NR1D1 lies not at rs2071427 but elsewhere in a risk-associated haplotype.
Another report failed to associate NR1D1 with Li response at two other variants previously associated with BD: rs12941497 and rs939347 (Manchia et al. 2009). These SNPS are not strongly linked with rs2071427 (Campos-de-Sousa et al. 2010; Kishi et al. 2008). The only genome-wide association study of Li response to date failed to identify any Li-associated genetic variants in NR1D1 (Perlis et al. 2009).
Structure and function of NR1D1
Rev-Erbα, transcribed from NR1D1, is of particular interest to Li’s pharmacological mechanism due to its role as a substrate for phosphorylation by GSK3β. Within NR1D1, rs2071427 is located in the first intron, a conserved region enriched in epigenetically modified chromatin (Ripperger et al 2006). This region has been shown to serve as an alternative promoter for NR1D1, generating a truncated, functional form of Rev-Erbα protein (Triqueneaux et al. 2004, Rambaud et al. 2009). Interestingly, truncated Rev-Erbα lacks the amino acids targeted for phosphorylation by GSK3β, and additional sequences that facilitate protein turnover (Rambaud et al. 2009). As a result, truncated Rev-Erbα is a poor substrate for GSK3β, but has enhanced protein stability compared to the full length protein. Li enhances proteasomal degradation of Rev-Erbα by inhibiting its phosphorylation by GSK3β (Yin et al. 2006). For this reason, Rev-Erbα truncation on the one hand opposes the action of Li by increasing protein stability, but on the other reinforces Li’s effects by preventing Rev-Erbα phosphorylation, which could affect clock function by mechanisms other than regulating protein stability. We did not assess Rev-Erbα protein content in our study, but this may be an interesting topic for future research.
In light of these past findings, we propose that differential sensitivity of NR1D1 gene expression to Li and altered ratios of NR1D1 transcripts may improve Li response among BD patients carrying the rs2071427 A allele. The direction of the NR1D1 expression changes we report in response to Li suggest that in sensitive subjects, Li increases the ratio of truncated NR1D1 transcript relative to the full-length transcript, which could alter Rev-Erbα stability, or perhaps other processes like subcellular localization (which depends on phosphorylation in other clock proteins, e.g. PER2, Vanselow et al. 2006).
Interactive genetic effects of GSK3β and NR1D1
Our genetic association results revealed that GSK3β and NR1D1 are associated with Li response additively. Our study lacked sufficient power to demonstrate epistatic gene x gene interactions, but we did find a trend in this direction. GSK3β and NR1D1 also interact biochemically in a well-characterized pathway within the circadian clock, in which Li blocks phosphorylation of Rev-Erbα by inhibiting GSK3β (Figure 5). Therefore, any process that reduces the phosphorylation of Rev-Erbα could in principle amplify the effects of Li. As discussed previously, truncated Rev-Erbα cannot be phosphorylated by GSK3β. The mood stabilizing properties of Li have been attributed to GSK3β inhibition, and lower GSK3β expression (as seen with the CC variant of rs6438552) may facilitate this inhibition by reducing the number of active GSK3β molecules available. Since both mechanisms ultimately converge upon a reduction in phosphorylated Rev-Erbα, they could be combinatorial in their effects. Our genetic association results, while preliminary, suggest that this may be the case.
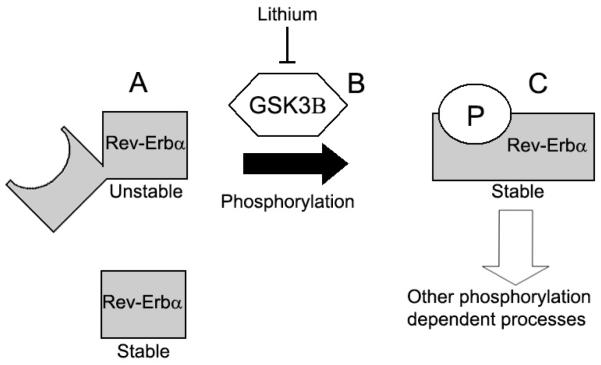
Figure 5.
Proposed model for the putative additive NR1D1-GSK3β genetic contribution to Li response. A) Variant rs2071427 is associated with altered NR1D1 transcript ratios in the presence of Li. This may lead to an increase in the truncated Rev-Erbα protein isoform which is more stable, but unable to serve as a substrate for GSK3β phosphorylation. B) Variant rs6438552 is associated with lower GSK3β gene expression, which may subsequently reduce GSK3β protein levels. C) These mechanistically independent processes could additively oppose Rev-Erbα phosphorylation, altering protein stability and/or limiting Rev-Erbα’s role in processes requiring this modification.
Conclusions
Our work provides an example of how using targeted functional studies of known biological interactions can enrich the study of genetic associations. Since many complex processes are polygenic, composed of multiple genetic effects of small size, this convergent approach may continue to be useful in the future. Because the clock system is well-characterized and accessible for direct study in samples from clinical populations, further work to clarify its role in BD may be fruitful.
Supplementary Material
Acknowledgments
This research was funded by grants to JRK from NIH (MH92758 and MH078151), and the VA (Merit Award CX-09-008); to DKW from NARSAD (Young Investigators Award); and to CN from NIH (MH092758, AG030474), VA (HSR&D) and DOD (BUMED, CDMRP). MJM was supported by NIH T32 funding (MH018399).
Footnotes
Disclosure/Conflict of Interest
None of the authors have conflicting interests to report.
References
- Abe M, Herzog ED, Block GD. Lithium lengthens the circadian period of individual suprachiasmatic nucleus neurons. Neuroreport. 2000;11:3261–3264. doi: 10.1097/00001756-200009280-00042. [DOI] [PubMed] [Google Scholar]
- Adli M, Hollinde DL, Stamm T, Wiethoff K, Tsahuridu M, Kirchheiner J, Heinz A, Bauer M. Response to lithium augmentation in depression is associated with the glycogen synthase kinase 3-beta -50T/C single nucleotide polymorphism. Biol Psychiatry. 2007;62:1295–1302. doi: 10.1016/j.biopsych.2007.03.023. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association . Diagnostic and statistical manual of mental disorders. Revised 4th edition Washington, DC: 2000. [Google Scholar]
- Archer SN, Viola AU, Kyriakopoulou V, von Schantz M, Dijk DJ. Inter-individual differences in habitual sleep timing and entrained phase of endogenous circadian rhythms of BMAL1, PER2 and PER3 mRNA in human leukocytes. Sleep. 2008;31:608–617. doi: 10.1093/sleep/31.5.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti F, Serretti A, Pontiggia A, Bernasconi A, Lorenzi C, Colombo C, Smeraldi E. Long-term response to lithium salts in bipolar illness is influenced by the glycogen synthase kinase 3-beta -50 T/C SNP. Neurosci Lett. 2005;376:51–55. doi: 10.1016/j.neulet.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Bremer T, Diamond C, McKinney R, Shehktman T, Barrett TB, Herold C, Kelsoe JR. The pharmacogenetics of lithium response depends upon clinical co-morbidity. Mol Diagn Ther. 2007;11:161–170. doi: 10.1007/BF03256238. [DOI] [PubMed] [Google Scholar]
- Brown SA, Fleury-Olela F, Nagoshi E, Hauser C, Juge C, Meier CA, Chicheportiche R, Dayer JM, Albrecht U, Schibler U. The period length of fibroblast circadian gene expression varies widely among human individuals. PLoS Biol. 2005;3:e338. doi: 10.1371/journal.pbio.0030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-de-Sousa S, Guindalini C, Tondo L, Munro J, Osborne S, Floris G, Pedrazzoli M, Tufik S, Breen G, Collier D. Nuclear receptor rev-erb-{alpha} circadian gene variants and lithium carbonate prophylaxis in bipolar affective disorder. J Biol Rhythms. 2010;25:132–137. doi: 10.1177/0748730410362713. [DOI] [PubMed] [Google Scholar]
- Grof P, Duffy A, Cavazzoni P, Grof E, Garnham J, MacDougall M, O’Donovan C, Alda M. Is response to prophylactic lithium a familial trait? J Clin Psychiatry. 2002;63:942–7. doi: 10.4088/jcp.v63n1013. [DOI] [PubMed] [Google Scholar]
- Grof P. Selecting effective long-term treatment for bipolar patients: monotherapy and combinations. J Clin Psychiatry. 2003;64(Suppl 5):53–61. [PubMed] [Google Scholar]
- Grof P, Duffy A, Alda M, Hajek T. Lithium response across generations. Acta Psychiatr Scand. 2009;120:378–385. doi: 10.1111/j.1600-0447.2009.01454.x. [DOI] [PubMed] [Google Scholar]
- Harada Y, Sakai M, Kurabayashi N, Hirota T, Fukada Y. Ser-557-phosphorylated mCRY2 is degraded upon synergistic phosphorylation by glycogen synthase kinase-3 beta. J Biol Chem. 2005;280:31714–31721. doi: 10.1074/jbc.M506225200. [DOI] [PubMed] [Google Scholar]
- Iitaka C, Miyazaki K, Akaike T, Ishida N. A role for glycogen synthase kinase-3beta in the mammalian circadian clock. J Biol Chem. 2005;280:29397–29402. doi: 10.1074/jbc.M503526200. [DOI] [PubMed] [Google Scholar]
- Kishi T, Kitajima T, Ikeda M, Yamanouchi Y, Kinoshita Y, Kawashima K, Okochi T, Ozaki N, Iwata N. Association analysis of nuclear receptor Rev-erb alpha gene (NR1D1) with mood disorders in the Japanese population. Neurosci Res. 2008;62:211–215. doi: 10.1016/j.neures.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Klein PS, Melton DA. A molecular mechanism for the effect of lithium on development. Proc Natl Acad Sci USA. 1996;93:8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukopulos A, Reginaldi D, Laddomada G, Serra, Tondo L. Course of the manic-depressive cycle and changes caused by treatment. Pharmakopsychiatr Neuropsychopharmakol. 1980;13:156–167. doi: 10.1055/s-2007-1019628. [DOI] [PubMed] [Google Scholar]
- Kripke DF, Judd LL, Hubbard B, Janowsky DS, Huey LY. The effect of lithium carbonate on the circadian rhythm of sleep in normal human subjects. Biol Psychiatry. 1979;14:545–548. [PubMed] [Google Scholar]
- Kripke DF, Wyborney VG. Lithium slows rat circadian activity rhythms. Life Sci. 1980;26:1319–1321. doi: 10.1016/0024-3205(80)90091-0. [DOI] [PubMed] [Google Scholar]
- Kripke DF, Nievergelt CM, Joo E, Shekhtman T, Kelsoe JR. Circadian polymorphisms associated with affective disorders. J Circadian Rhythms. 2009;7:2. doi: 10.1186/1740-3391-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Kurabayashi N, Hirota T, Sakai M, Sanada K, Fukada Y. DYRK1A and glycogen synthase kinase 3beta, a dual-kinase mechanism directing proteasomal degradation of CRY2 for circadian timekeeping. Mol Cell Biol. 2010;30:1757–1768. doi: 10.1128/MCB.01047-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok JB, Hallupp M, Loy CT, Chan DK, Woo J, Mellick GD, Buchanan DD, Silburn PA, Halliday GM, Schofield PR. GSK3B polymorphisms alter transcription and splicing in Parkinson’s disease. Ann Neurol. 2005;58:829–839. doi: 10.1002/ana.20691. [DOI] [PubMed] [Google Scholar]
- Lau KF, Miller CC, Anderton BH, Shaw PC. Molecular cloning and characterization of the human glycogen synthase kinase-3beta promoter. Genomics. 1999;60:121–128. doi: 10.1006/geno.1999.5875. [DOI] [PubMed] [Google Scholar]
- Le Niculescu H, McFarland MJ, Ogden CA, Balaraman Y, Patel S, Tan J, Rodd ZA, Paulus M, Geyer MA, Edenberg HJ, Glatt SJ, Faraone SV, Nurnberger JI, Kuczenski R, Tsuang MT, Niculescu AB. Phenomic, convergent functional genomic, and biomarker studies in a stress-reactive genetic animal model of bipolar disorder and co-morbid alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:134–166. doi: 10.1002/ajmg.b.30707. [DOI] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, Doyle FJ, 3rd, Takahashi JS, Kay SA. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchia M, Squassina A, Congiu D, Chillotti C, Ardau R, Severino G, Del Zompo M. Interacting genes in lithium prophylaxis: preliminary results of an exploratory analysis on the role of DGKH and NR1D1 gene polymorphisms in 199 Sardinian bipolar patients. Neurosci Lett. 2009;467:67–71. doi: 10.1016/j.neulet.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Mansour HA, Monk TH, Nimgaonkar VL. Circadian genes and bipolar disorder. Ann Med. 2005;37:196–205. doi: 10.1080/07853890510007377. [DOI] [PubMed] [Google Scholar]
- Mansour HA, Talkowski ME, Wood J, Chowdari KV, McClain L, Prasad K, Montrose D, Fagiolini A, Friedman ES, Allen MH, Bowden CL, Calabrese J, El Mallakh RS, Escamilla M, Faraone SV, Fossey MD, Gyulai L, Loftis JM, Hauser P, Ketter TA, Marangell LB, Miklowitz DJ, Nierenberg AA, Patel J, Sachs GS, Sklar P, Smoller JW, Laird N, Keshavan M, Thase ME, Axelson D, Birmaher B, Lewis D, Monk T, Frank E, Kupfer DJ, Devlin B, Nimgaonka,r VL. Association study of 21 circadian genes with bipolar I disorder, schizoaffective disorder, and schizophrenia. Bipolar Disord. 2009;11:701–710. doi: 10.1111/j.1399-5618.2009.00756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CA. Circadian genes, rhythms and the biology of mood disorders. Pharmacol Ther. 2007;114:222–232. doi: 10.1016/j.pharmthera.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath CL, Glatt SJ, Sklar P, Le, Niculescu H, Kuczenski R, Doyle AE, Biederman J, Mick E, Faraone SV, Niculescu AB, Tsuang MT. Evidence for genetic association of RORB with bipolar disorder. BMC Psychiatry. 2009;9:70. doi: 10.1186/1471-244X-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Coque L, Cao JL, Kumar J, Chakravarty S, Asaithamby A, Graham A, Gordon E, Enwright JF, 3rd, DiLeone RJ, Birnbaum SG, Cooper DC, McClung CA. Knockdown of Clock in the Ventral Tegmental Area Through RNA Interference Results in a Mixed State of Mania and Depression-Like Behavior. Biol Psychiatry. 2010;68:503–511. doi: 10.1016/j.biopsych.2010.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nievergelt CM, Kripke DF, Barrett TB, Burg E, Remick RA, Sadovnick AD, McElroy SL, Keck PE, Jr, Schork NJ, Kelsoe JR. Suggestive evidence for association of the circadian genes PERIOD3 and ARNTL with bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:234–241. doi: 10.1002/ajmg.b.30252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am. J. Hum. Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlis RH, Smoller JW, Ferreira MA, McQuillin A, Bass N, Lawrence J, Sachs GS, Nimgaonkar V, Scolnick EM, Gurling H, Sklar P, Purcell S. A genomewide association study of response to lithium for prevention of recurrence in bipolar disorder. Am J Psychiatry. 2009;166:718–725. doi: 10.1176/appi.ajp.2009.08111633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaud J, Triqueneaux G, Masse I, Staels B, Laudet V, Benoit G. Rev-erbalpha2 mRNA encodes a stable protein with a potential role in circadian clock regulation. Mol Endocrinol. 2009;23:630–639. doi: 10.1210/me.2008-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger JA. Mapping of binding regions for the circadian regulators BMAL1 and CLOCK within the mouse Rev-erbalpha gene. Chronobiol Int. 2006;23:135–142. doi: 10.1080/07420520500464411. [DOI] [PubMed] [Google Scholar]
- Rollins B, Martin MV, Morgan L, Vawter MP. Analysis of whole genome biomarker expression in blood and brain. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:919–936. doi: 10.1002/ajmg.b.31062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roybal K, Theobold D, Graham A, DiNieri JA, Russo SJ, Krishnan V, Chakravarty S, Peevey J, Oehrlein N, Birnbaum S, Vitaterna MH, Orsulak P, Takahashi JS, Nestler EJ, Carlezon WA, Jr., McClung CA. Mania-like behavior induced by disruption of CLOCK. Proc Natl Acad Sci U S A. 2007;104:6406–6411. doi: 10.1073/pnas.0609625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ C, Lovestone S, Powell JF. Identification of sequence variants and analysis of the role of the glycogen synthase kinase 3 beta gene and promoter in late onset Alzheimer’s disease. Mol Psychiatry. 2001;6:320–324. doi: 10.1038/sj.mp.4000852. [DOI] [PubMed] [Google Scholar]
- Sahar S, Zocchi L, Kinoshita C, Borrelli E, Sassone-Corsi P. Regulation of BMAL1 protein stability and circadian function by GSK3beta-mediated phosphorylation. PLoS One. 2010;5:e8561. doi: 10.1371/journal.pone.0008561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Shi J, Wittke-Thompson JK, Badner JA, Hattori E, Potash JB, Willour VL, McMahon FJ, Gershon ES, Liu C. Clock genes may influence bipolar disorder susceptibility and dysfunctional circadian rhythm. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1047–1055. doi: 10.1002/ajmg.b.30714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöholm LK, Backlund L, Cheteh EH, Ek IR, Frisén L, Schalling M, Osby U, Lavebratt C, Nikamo P. CRY2 is associated with rapid cycling in bipolar disorder patients. PLoS One. 2010;5:e12632. doi: 10.1371/journal.pone.0012632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria V, Martinez-Amoros E, Escaramis G, Valero J, Perez-Egea R, Garcia C, Gutierrez-Zotes A, Puigdemont D, Bayes M, Crespo JM, Martorell L, Vilella E, Labad A, Vallejo J, Perez V, Menchon JM, Estivill X, Gratacos M, Urretavizcaya M. Differential association of circadian genes with mood disorders: CRY1 and NPAS2 are associated with unipolar major depression and CLOCK and VIP with bipolar disorder. Neuropsychopharmacology. 2010;35:1279–1289. doi: 10.1038/npp.2009.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teboul M, Barrat-Petit MA, Li XM, Claustrat B, Formento JL, Delaunay F, Levi F, Milano G. Atypical patterns of circadian clock gene expression in human peripheral blood mononuclear cells. J Mol Med. 2005;83:693–699. doi: 10.1007/s00109-005-0697-6. [DOI] [PubMed] [Google Scholar]
- Triqueneaux G, Thenot S, Kakizawa T, Antoch MP, Safi R, Takahashi JS, Delaunay F, Laudet V. The orphan receptor Rev-erbalpha gene is a target of the circadian clock pacemaker. J Mol Endocrinol. 2004;33:585–608. doi: 10.1677/jme.1.01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanselow K, Vanselow JT, Westermark PO, Reischl S, Maier B, Korte T, Herrmann A, Herzel H, Schlosser A, Kramer A. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS) Genes Dev. 2006;20:2660–72. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestergaard P, Schou M. Prospective studies on a lithium cohort. 1. General features. Acta Psychiatr Scand. 1988;78:421–6. doi: 10.1111/j.1600-0447.1988.tb06361.x. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Moore-Ede MC. Lithium lengthens circadian period in a diurnal primate, Saimiri sciureus. Biol Psychiatry. 1990;28:117–126. doi: 10.1016/0006-3223(90)90629-g. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol. 2010;17:551–77. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita K, Tamanini F, Der Horst GT, Okamura H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science. 2001;292:278–281. doi: 10.1126/science.1059542. [DOI] [PubMed] [Google Scholar]
- Yan J, Wang H, Liu Y, Shao C. Analysis of gene regulatory networks in the mammalian circadian rhythm. PLoS Comput Biol. 2008;4:e1000193. doi: 10.1371/journal.pcbi.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Van Dongen HP, Wang K, Berrettini W, Bucan M. Assessment of circadian function in fibroblasts of patients with bipolar disorder. Mol Psychiatry. 2009;14:143–155. doi: 10.1038/mp.2008.10. [DOI] [PubMed] [Google Scholar]
- Yin L, Wang J, Klein PS, Lazar MA. Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science. 2006;311:1002–1005. doi: 10.1126/science.1121613. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.