Abstract
A high-density phylogenetic microarray (PhyloChip) was applied to track bacterial and archaeal populations through different phases of remediation at Ft. Lewis, WA, a trichloroethene (TCE)-contaminated groundwater site. Biostimulation with whey, and bioaugmentation with a Dehalococcoides-containing enrichment culture were strategies implemented to enhance dechlorination. As a measure of species richness, over 1300 operational taxonomic units (OTUs) were detected in DNA from groundwater samples extracted during different stages of treatment and in the bioaugmentation culture. In order to determine active members within the community, 16S rRNA from samples were analyzed by microarray and ~600 OTUs identified. A cDNA clone library of the expressed 16S rRNA corroborated the observed diversity and activity of some of the phyla. Principle component analysis of the treatment plot samples revealed that the microbial populations were constantly changing during the course of the study. Dynamic analysis of the archaeal population showed significant increases in methanogens at the later stages of treatment that correlated with increases in methane concentrations of over two orders of magnitude. Overall, the PhyloChip analyses in this study have provided insights into the microbial ecology and population dynamics at the TCE-contaminated field site useful for understanding the in situ reductive dechlorination processes.
Keywords: 16S rRNA, PhyloChip, ecology, microbial community, bioremediation, chlorinated solvents
Introduction
Chlorinated ethenes such as tetrachloroethene (PCE), trichloroethene (TCE), isomers of dichloroethene (DCE), and vinyl chloride (VC) are frequently detected contaminants in groundwater aquifers in the United States.1 These compounds are toxic and carcinogenic2 and removing them from the environment is a priority. There have been decades of efforts to remediate chlorinated ethene-contaminated sites and in situ bioremediation is a promising strategy.3 Bioremediation is a viable clean-up option because bacteria in the genus Dehalococcoides can reduce chlorinated ethenes completely to the innocuous end product ethene.4-10 In the energy-generating dehalorespiration process of Dehalococcoides spp., chlorinated ethenes serve as electron acceptors, coupling hydrogen oxidation with growth (see references for comprehensive review).4,11,12 To date, the capability of dechlorinating beyond DCE to VC and ethene has been found to be restricted to the Dehalococcoides genus, even though a wide variety of other bacteria can catalyze the first two steps of PCE reduction.13
The importance of Dehalococcoides spp. to bioremediation was demonstrated in field projects where the presence of Dehalococcoides spp. was correlated to the complete reduction of chlorinated ethenes to ethene.14 In field sites where Dehalococcoides spp. with the appropriate reductive dehalogenases (RDases) are present indigenously, biostimulation via carbon sources and nutrients can lead to complete reduction to ethene,15,16 whereas in cases where Dehalococcoides spp. are absent, bioaugmentation with Dehalococcoides-containing cultures is required before the conversion to ethene occurs.17,18
Microbial interactions that occur within microbial consortia can potentially supplement Dehalococcoides spp. with essential macro and micro nutrients which are otherwise limiting. For example, Dehalococcoides spp. are obligated to utilize hydrogen as electron donor and acetate as carbon source,19,20 and these essential substrates can be generated by fermentation of exogenous carbon sources by various microbial species.4 Known Dehalococcoides strains are also unable to synthesize corrinoids such as vitamin B12, that serve as crucial prosthetic groups of RDases, the proteins that allow Dehalococcoides to respire chlorinated ethenes.19-21 Dehalococcoides spp. can also benefit other species within communities; for example, the low hydrogen threshold of Dehalococcoides spp.22 can create a thermodynamically favorable environment for some fermentative organisms that might otherwise be inhibited by high concentrations of hydrogen.
Because of the importance of microbial interactions to sustained activity within bioremediation communities, a number of studies have reported the community structure of field samples and laboratory enrichment cultures undergoing reductive dechlorination.16,23-37 Culture-independent methods such as gene sequencing, fluorescence in situ hybridization (FISH), terminal restriction-fragment length polymorphism (T-RFLP), chemical or temperature denaturing gradient gel electrophoresis (DGGE), and oligonucleotide microarray targeting the bacterial and archaeal 16S rRNA genes have commonly been used. These studies have shown that microbial communities undergoing dechlorination are diverse with membership governed by factors such as geochemistry, enrichment conditions and treatment strategies. While results from these studies have provided insights into microbial community structures, they tend to underestimate community diversity since the applied techniques target dominant members of the communities.
In this study, analyses of community DNA and RNA using a high-density oligonucleotide microarray (PhyloChip) were coupled with a 16S rRNA clone library to study the in situ microbial ecology of a TCE dense-non-aqueous-phase-liquid (DNAPL) contaminated site at Fort Lewis, WA. The dynamics and gene expression of the functionally important Dehalococcoides populations at this site have previously been reported.38 The purpose of this study was to examine the membership and dynamics of the microbial community that supports Dehalococcoides spp. during different phases of treatment in terms of relative gene abundance (DNA) and relative expression activity (RNA). Biostimulation with whey and bioaugmentation with a Dehalococcoides-containing culture were strategies implemented to promote bioremediation at this site.38 Previous studies39-42 have demonstrated that the PhyloChip can provide high resolution for elucidating microbial community structure and detecting active constituents; thus, providing a comprehensive analysis of the community dynamics.
Materials and methods
Site description and treatment procedures
Detailed description of the Ft. Lewis treatment site, treatment strategies implemented, procedures for sample collection, and the location and number of samples collected have been reported previously in Lee et al.38 Briefly, two treatment plots were set-up at a location where there were high concentrations of TCE-DNAPL and groundwater samples were collected from monitoring wells that were screened at different depths over a one year period. A total of 16 samples were collected from the two treatment plots. In addition, a single sample was taken during the February 2006 sampling from monitoring wells located 80 and 100 feet upstream and downstream of the treatment plot, respectively. The upstream well was isolated from any treatment manipulation and served as the background control against other samples that had been subjected to various levels of treatment manipulations. No deliberate treatment action was implemented near the downstream well and it only received residual carbon that was carried downstream by ambient groundwater flow from the treatment plot.
Biostimulation with whey (composed of 10 to 13% protein and 70 to 75% lactose) was performed on a monthly basis starting in June 2005 with different concentrations (10 or 100 g/L) during different treatment phases to plots 1 and 2. Bioaugmentation with 18 L of culture occurred one month after the initiation of biostimulation. The sampling schedule (Supporting Information Figure S1) within the treatment plots for molecular analyses was as follows: a baseline sample was collected two months prior to biostimulation (April 2005), and samples were collected 1 month after the initiation of biostimulation (July 2005), 1 month after bioaugmentation (August 2005), 4 months after bioaugmentation (November 2005), 7 months after bioaugmentation when whey injection was discontinued (February 2006), and finally 2 months after discontinuing whey injection (April 2006). Samples for RNA analysis were only collected during the February 2006 and April 2006 sampling periods.
Analytical methods
Concentrations of chlorinated ethenes, organic acids, redox parameters, and geochemical composition were measured according to methods described elsewhere.43
Nucleic acids extraction
Groundwater samples intended for genomic DNA (gDNA) extraction were collected in autoclaved 1-liter bottles and shipped overnight on ice to the laboratory at the University of California, Berkeley as described previously.38 A total of 16 groundwater samples were collected for gDNA analysis from both treatment plots during each sampling period. Groundwater samples intended for RNA extraction were filtered on site from selected monitoring wells at the time of sampling as described previously and shipped on dry ice.38 The procedures to extract gDNA and total RNA have been described previously.38 For the purpose of this study, equal masses of gDNA or total RNA were pooled from all samples collected from both treatment plots into a single sample and analyzed via PhyloChips to obtain a representative picture of the treatment plot at each sampling time point.
Amplification of 16S rRNA genes for PhyloChip analysis
The 16S rRNA genes were amplified from 105 ng of gDNA in each sample using the universal primers 27F (5′- AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) for bacteria and 4Fa (5′- TCCGGTTGATCCTGCCRG-3′) and 1492R for archaea.39,42 Each 25 μL PCR reaction mix contained 1× Ex Taq buffer, 0.8 mM dNTP mix, 0.3 μM each of forward and reverse primers, 1 μg/ μL of BSA, and 0.625 U Ex Taq hot-start DNA polymerase (Takara Mirus Bio, Madison, WI). A total of eight PCR reactions were set up for each sample for the respective bacterial and archaeal primer sets and each reaction was run at a different annealing temperature between 48 to 58°C to maximize the number of sequences that could be amplified. The PCR cycling protocol included: an initial denaturation step at 95°C (3 min), followed by 25 cycles of denaturation (95°C, 30 s), annealing (48 to 58°C, 25 s), and extension (72°C, 120 s), with a final extension at 72°C for 10 min. Keeping the bacterial and archaeal sets separate, the amplified products from the eight reactions for each sample were combined and concentrated by precipitation. The re-suspended 16S rRNA PCR products were visualized and quantified on an Agilent 2100 bioanalyzer (Agilent, Palo Alto, CA) using the DNA 7500 chips according to the manufacturer’s protocol.
16S rRNA clone library construction
In order to identify active bacterial members in the microbial community, a clone library was constructed based on the expressed 16S rRNA genes. Total RNA collected from selected monitoring wells during the February 2006 sampling was pooled and two 0.24 μg-samples were reverse-transcribed in parallel using the SuperScript III First-Strand synthesis system (Invitrogen, Carlsbad, CA) as described previously.38 Following reverse-transcription (RT), the two cDNA samples were combined and 2 μL of cDNA products were amplified in eight separate reactions with annealing temperature between 48 to 58°C using the universal bacterial primers, PCR reaction mixture and thermo cycling protocol listed above. The amplified products were combined and the 16S rRNA band was quantified as described above. A parallel no reverse-transcriptase sample was prepared and no band was visible on the gel electrophoresis after RT-PCR, indicating no DNA contamination.
The clone library of the 16S rRNA gene PCR products was constructed with the TOPO TA cloning kit (with the pCR2.1-TOPO vector) (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. A total of 384 transformants were randomly picked and inserts were sequenced bidirectionally using M13-vector specific primers at the University of California, Berkeley sequencing center. Paired-end sequencing reads were vector trimmed, assembled and quality-checked via the Joint Genome Institute’s GeneLib software package (Kirton, E. unpublished data).
Phylogenetic analysis of 16S rRNA sequences
The 351 16S rRNA sequences that passed quality check were pre-aligned using Greengenes’ NAST aligner, and subsequently imported into the Greengenes database44 in ARB. The alignment was manually refined and a distance matrix was calculated. This distance matrix was used in DOTUR45 to dereplicate the dataset on 97% sequence identity. From the obtained 199 operational taxonomic units (OTUs), one representative each was taken for the subsequent analyses. The sequences were checked for chimeras using Bellerophon46 and a partial treeing approach47 and 102 OTUs were considered non-chimeric. A phylogenetic tree was calculated using the neighbor-joining algorithm and the filter lanemaskPH in ARB in order to exclude highly variable positions of the alignment.
PhyloChip analysis of DNA and RNA
The bacterial (500 ng) and archaeal (100 ng) PCR products of each DNA sample were combined and spiked with known concentrations of synthetic 16S rRNA gene fragments and non-16S rRNA gene fragments as internal standards for normalization, with quantities ranging from 5.02 × 108 to 7.29 × 1010 molecules applied to the final hybridization mix. The procedures to fragment, biotin-label and hybridize the targets on the PhyloChips as well as the staining and scanning methods were as previously reported.39 The algorithm for background subtraction, noise calculation, and detection and quantification criteria were also performed as reported.39 Analysis of active microbial OTUs was carried out by direct hybridization of the total RNA onto the PhyloChips according to the method previously described48 except the arrays were scaled to 2500.
A total of ten PhyloChips were analyzed in this study, eight for DNA and two for RNA (Supporting Information Tables S1 and S2). Technical replication on the PhyloChip has an average coefficient of variation of 10% for an OTU across arrays,40 indicating low variation and high reproducibility, and technical replicates were therefore not analyzed here. DNA-PhyloChip analyses were performed on the five samples collected within the treatment plot (July 05, August 05, November 05, February 06, and April 06), the upstream (background) and downstream samples, and the Dehalococcoides-containing enrichment culture used for bioaugmentation. An insufficient mass of PCR products was generated from the baseline sample collected within the treatment plot prior to biostimulation (April 05) to analyze. RNA-PhyloChip analyses were performed on the samples collected during the February 2006 and April 2006 sampling periods. Throughout this study, the samples were referred to by their date or location with ‘April’ referring to the 2006 sample and ‘FTB’ referring to the bioaugmentation culture.
A taxon was considered present in a sample when 90% or more of its assigned probe pairs in a given probe set were positively detected (positive-fraction (pf) ≥ 0.9). When multiple samples were considered, the data set was filtered accordingly based on the positive-fraction cutoff. Unless otherwise stated, analyses in this study were carried out at the subfamily level as a conservative estimate of array specificity and the probe set with the highest normalized array intensity (HybScore) was used as a representative output of a subfamily. Present calls were propagated upwards through the taxonomic hierarchy by considering any node (subfamily, family, order, etc) as present if at least one of its subordinate OTUs was present.41 Hierarchical clustering of samples and subfamilies was performed within the R statistical environment as previously described.39 Principal component analysis (PCA) ordination of samples was performed using the software package PC-ORD v5.0 (MjM Software, Gleneden Beach, OR).
Nucleotide sequence accession numbers
The 16S rRNA gene sequences of each representative OTU were deposited in GenBank (Accession no.: HM481300 - HM481401).
Results and discussion
Dechlorination activity and groundwater chemistry
The dechlorination profile of chlorinated ethenes at Ft. Lewis has been reported previously38 and is shown in Supporting Information Figure S2. Briefly, during the year-long active remediation process, dechlorination of TCE to mostly cDCE was observed during the first six months of treatment and conversion to VC and ethene was achieved during the last four months. The growth of three distinct populations of Dehalococcoides, measured via quantitative PCR (qPCR) of the unique RDaseencoding genes (tceA, vcrA, bvcA) within each population, was concomitant with the dechlorination of chlorinated ethenes.38
A number of groundwater chemistry parameters indicative of the prevailing biogeochemical conditions were analyzed (Supporting Information Figure S3). Prior to biostimulation with whey, the subsurface groundwater environment was mostly aerobic with close to 2 mg/L of dissolved oxygen. Following whey injection, significant increases in chemical oxygen demand (COD) and organic acids such as lactate, acetate and butyrate were observed along with simultaneous decreases in dissolved oxygen. Changes in indigenous terminal electron acceptor concentrations such as nitrate, sulfate and dissolved iron were also measured at the site. Whey that was applied in this study contained sulfate as a byproduct (approximately 13 and 130 mg/L in the 10 g/L and 100 g/L injected whey solutions respectively) that likely caused the sulfate concentrations to fluctuate while stimulating the sulfate-reducing populations. The pH range at the site tended to remain between 5 and 6.
Richness and taxonomic representation
PhyloChip analyses of the DNA in groundwater samples from the Ft. Lewis site were used to comprehensively evaluate the phylotypes represented there, including those present at low abundances. With 8741 bacterial and archaeal taxa on the G2 PhyloChip, over 1300 OTUs were positively detected in DNA from the groundwater, with a similar richness detected in the laboratory-grown bioaugmentation culture containing Dehalococcoides (Table 1). As a conservative estimate, the OTUs were summarized to the subfamily level and with this restriction, ~300 subfamilies were detected (Table 1). An OTU on the PhyloChip is defined as exhibiting a 97 to 100% sequence homology, while a subfamily consists of a group of OTUs with typically no less than 94% sequence homology.
Table 1.
Richness of samples as represented by the number of OTUs or subfamilies that was positively present.
| DNA |
RNAa |
|||
|---|---|---|---|---|
| Samples | OTUc | Subfamily | OTUc | Subfamily |
| FTBb | 1334 | 285 | ||
| Background | 1520 | 314 | ||
| Down-stream | 1448 | 307 | ||
| Treatment plot | ||||
| July | 1402 | 278 | ||
| Aug | 1445 | 297 | ||
| Nov | 1432 | 297 | ||
| Feb | 1458 | 297 | 583 | 174 |
| April | 1340 | 289 | 631 | 194 |
RNA samples were only collected during February and April 2006;
Bioaugmentation culture
An OTU is considered present in a sample when 90% or more of its assigned probe pairs in a given probe set are positive.
While analyzing a microbial community via DNA reveals the overall genetic diversity and relative abundance of the constituent OTUs, the metabolic activity of the community members are expected to differ. The activity of different populations can be inferred from the expressed 16S rRNA.42,49 When RNA from groundwater samples were hybridized onto the PhyloChip to analyze for the expressed 16S rRNA genes, ~600 OTUs or ~180 subfamilies were detected in the February or April samplings (Table 1). Of the 154 subfamilies that were detected in the rRNA from both the February and April samplings, 98 were also detected in the DNA from the same time point while 146 were detected in DNA from one or more of the time points, demonstrating as theoretically expected, that the subfamilies that were detected in the RNA were also present in the DNA. Table 2 presents a summary of the phylum-level composition of the groundwater analyzed during the February and April samplings, listing the 37 bacterial and 2 archaeal phyla detected in the DNA fraction for both sampling periods. There were sequences from five bacterial phyla that were detected in the RNA fraction but not in the DNA fraction, but only one subfamily was detected for each of these phyla.
Table 2.
Phyla that were detected by PhyloChips or clone library.
| Phylochipa |
cDNA Clone | ||
|---|---|---|---|
| Phylum | DNA | RNA | Library b |
| Archaea | |||
| Crenarchaeota | × (4) | × (3) | |
| Euryarchaeota | × (14) | × (5) | |
| Bacteria | |||
| Acidobacteria | × (8) | × (2) | × |
| Actinobacteria | × (14) | × (5) | |
| AD3 | × (1) | × (1) | |
| Aquificae | × (1) | ||
| Bacteroidetes | × (16) | × (16) | × |
| BRC1 | × (1) | × (2) | |
| Caldithrix | × (2) | × (1) | |
| Chlamydiae | × (2) | × (1) | |
| Chlorobi | × (4) | × (3) | × |
| Chloroflexi | × (12) | × (5) | × |
| Coprothermobacteria | × (1) | × (1) | |
| Cyanobacteria | × (13) | × (3) | |
| Deinococcus-Thermus | × (3) | × (1) | |
| Dictyoglomi | × (1) | ||
| DSS1 | × (1) | ||
| Firmicutes | × (28) | × (25) | × |
| Gemmatimonadetes | × (1) | ||
| Lentisphaerae | × (1) | × (1) | × |
| LD1PA group | × (1) | ||
| Marine group A | × (2) | × (2) | |
| NC10 | × (1) | × (2) | |
| Natronoanaerobium | × (1) | ||
| Nitrospira | × (2) | ||
| OD1 | × (1) | × | |
| OP10 | × (3) | × (3) | |
| OP11 | × (3) | × | |
| OP3 | × (2) | × (2) | |
| OP8 | × (1) | × (1) | |
| OP9/JS1 | × (1) | × (1) | |
| Planctomycetes | × (3) | × (1) | |
| Proteobacteria | |||
| α-Proteobacteria | × (28) | × (14) | |
| β-Proteobacteria | × (7) | × (9) | × |
| δ-Proteobacteria | × (13) | × (10) | × |
| ε-Proteobacteria | × (2) | × (2) | × |
| γ-Proteobacteria | × (21) | × (13) | × |
| Unclassified | × (1) | × (2) | |
| SPAM | × (1) | × (1) | |
| Spirochaetes | × (2) | × (2) | |
| SR1 | × (1) | ||
| Synergistes | × (1) | × (1) | |
| Thermodesulfobacteria | × (1) | ||
| TM6 | × (1) | ||
| TM7 | × (1) | × | |
| Unclassified | × (6) | × (5) | × |
| Verrucomicrobia | × (8) | × (3) | |
| WS3 | × (2) | ||
| WS5 | × (1) | ||
A phylum was considered present (indicated by ×) when it was present in both the February and April 2006 samples. The number of subfamilies present in each phylum is indicated in parentheses.
cDNA clone library was constructed using the February RNA sample. Only bacterial primers were used.
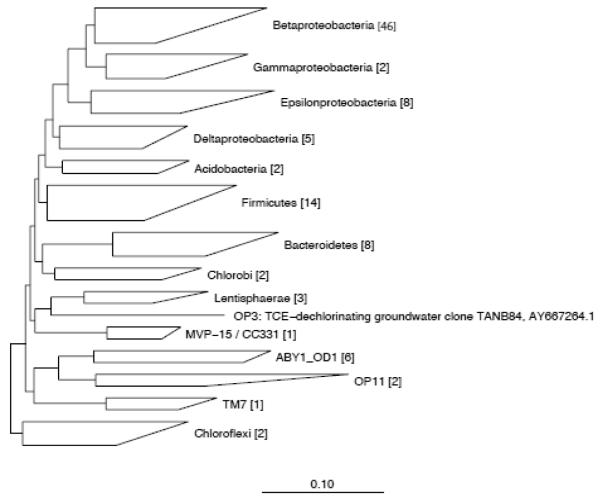
In addition to the PhyloChip analysis, a cDNA clone library of the expressed 16S rRNA genes was constructed to further analyze the active community members. The 237 non-chimeric sequences were grouped into 102 OTUs with 97% sequence homology across 14 phyla (Figure 1), indicating a broad diversity and supporting the richness detected via PhyloChip (Table 1). In the clone library, a majority of the phylotypes (60%) belonged to the four classes of Proteobacteria with β-Proteobacteria being the disproportionately dominant group (Figure 1). A relatively large number of phylotypes were also identified from the phyla Firmicutes and Bacteroidetes. Two phylotypes from the phylum Chloroflexi were identified, but neither were identified as Dehalococcoides sequences (closest database matches were to uncultured environmental sequences). The lack of Dehalococcoides detection in clone libraries constructed via universal primers of environmental dechlorinating communities has been previously observed.33 Given that the more sensitive qPCR method detected only 108 Dehalococcoides cells per liter of groundwater at this site,38 it is clear that even though Dehalococcoides spp. were functionally important at this site, they represent only a small proportion of the community and therefore are difficult to detect with clone libraries. However, Dehalococcoides was detected in the Ft. Lewis samples via the more sensitive PhyloChip in both DNA and RNA (Supporting Information Tables S1 and S2). Detailed phylogenetic analysis also showed that a number of the phylotypes were not closely related to any previously identified sequences (Supporting Information Figure S4) and the clone library significantly under-sampled species diversity, identifying only a small subset of the bacterial phyla that were detected by PhyloChip (Table 2 and Figure 1).
Figure 1.
Phylogenetic tree of the expressed 16S rRNA sequences. Neighbor-joining algorithm with the filter lanemaskPH was used for phylogenetic analysis in order to exclude highly variable positions of the alignment. A total of 102 phylotypes were found (number in brackets) based on 97% sequence homology. Scale bar indicates 10% estimated sequence divergence.
A number of studies have previously reported the microbial ecology of reductively dechlorinating laboratory enrichment cultures and environmental samples using molecular techniques that target the dominant members of the communities. Bowman et al.23 summarized the bacterial phylotypes from 16S rRNA clone libraries of four different chlorinated-ethene dechlorinating microbial communities, reporting around ten bacterial phyla per community. The diversity captured by the PhyloChip was far more comprehensive than previously reported, and was similar to a PhyloChip-analyzed soil sample (~1250 OTUs detected) that was collected from a uranium contaminated site at the NABIR Field Research Center at Oak Ridge, TN.41 Interestingly, the observed diversity was not restricted to the environmental samples, but was also a characteristic of the laboratory enrichment culture used for bioaugmentation at Ft. Lewis. Examination of both the DNA and RNA from samples indicated that a relatively large number of subfamilies were from the different classes of Proteobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, and Firmicutes (Table 2), representing five of the nine phyla that have been typically identified to account for greater than 90% of soil library sequences.50 The other dominant phyla in soils that were not found to be well represented at Ft. Lewis include Acidobacteria, Verrucomicrobia, Planctomycetes, and Gemmatimonadetes.
Dynamics of the bacterial populations within the treatment plot
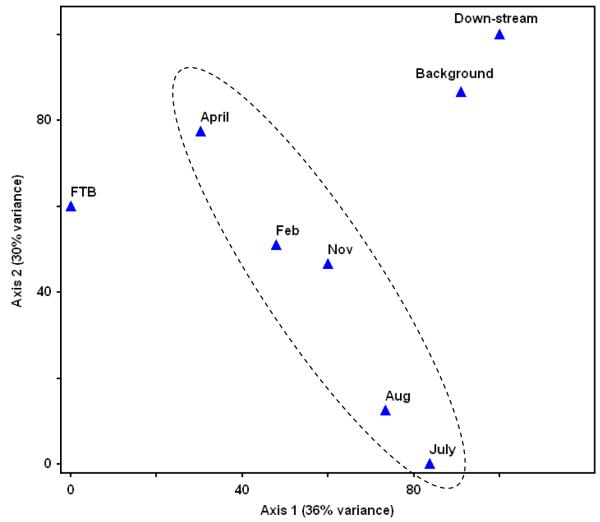
Principle component analysis (PCA) of the DNA hybridization intensity (HybScore) that included any subfamilies that were positively detected in at least one of the samples showed that the eight analyzed samples were unique and that 66% of the dataset variance could be explained by two axes (Figure 2). The two communities that were closely associated were the background sample and the sample collected downstream of the treatment plot, outside the zone of direct whey influence, highlighting the significant influence of exogenous carbon amendment on the microbial community. The time series samples that were collected within the treatment plot showed that over the course of treatment, the community structure was changing continuously, with separation along both axes on the PCA plot relative to the July sample (Figure 2).
Figure 2.
Principle component analysis (PCA) of the PhyloChip-analyzed samples. The dotted circle highlights the samples that were collected within the treatment plot at different time points.
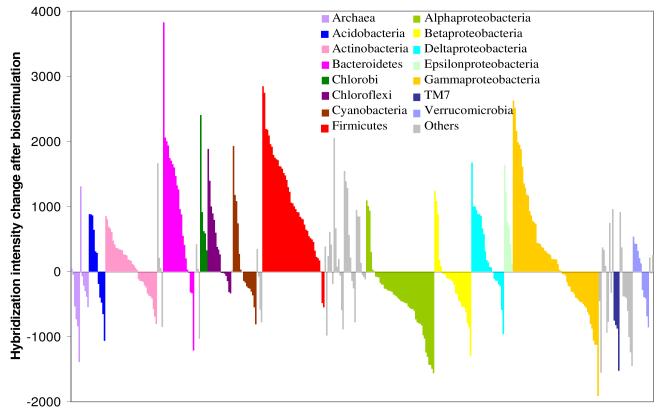
The immediate responses of the microbial community to biostimulation can be seen in changes in HybScores between the background sample and the July treatment plot sample, the first sampling period after whey injection (Figure 3). Changes in HybScores are positively correlated to relative abundance, with estimates that a 1000-unit change in HybScore is roughly proportional to a 10-fold change in abundance.39,51 Of the 393 subfamilies that responded to whey, 193 showed changes in HybScore of 500 units or more with 121 and 72 subfamilies showing increases and decreases, respectively (Supporting Information Table S3). Subfamilies from Acidobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, Cyanobacteria, Firmicutes, and all classes of Proteobacteria accounted for 76% of this dynamic subset. Specifically, bacteria in the Bacteroidetes and Firmicutes and those with diverse metabolic capabilities to utilize electron acceptors in the δ-Proteobacteria and ε-Proteobacteria were enriched. Functionally, the presence and activity of the Bacteroidetes and Firmicutes (particularly the Clostridia) likely reflect their roles as fermenters, and this is commonly found in chlorinated-ethene communities.23,26,33,34 The significant increases in organic acid concentrations (Supporting Information Figure S3) also reflected that these fermentative members were active functional members of the community. Dehalococcoides sequences, which are within the Chloroflexi phylum,13 increased significantly (μ HybScores = 1400 units). Three OTUs (31, 816, 10439) in the Firmicutes and ε-Proteobacteria that contain known TCE-dechlorinators13 also showed significant increases in intensity. A relatively large fraction of members that exhibited decreases in abundance following whey injection were from the α-Proteobacteria and β-Proteobacteria, while γ-Proteobacteria was a group that exhibited both increases and decreases in populations (Figure 3).
Figure 3.
Changes in hybridization intensity relative to the background sample of the 393 subfamilies immediately after biostimulation with whey (July 2005) in the treatment plot. Phyla are color-coded and ordered alphabetically from left to right starting with the archaeal domain followed by the bacterial domain. Each bar represents a subfamily with positive bars indicating subfamilies that increased in abundance relative to the background after receiving whey and negative bars represent subfamilies that decreased in abundance. A 1000-unit change in hybridization intensity is equivalent to a 10-fold change in relative abundance.
Injection of the laboratory-grown bioaugmentation culture did not result in significant immediate changes as the July and August samples were relatively similar to each other according to the PCA plot. Further, the PCA plot shows that the bioaugmentation culture was different from any of the samples collected at Ft. Lewis throughout the study. The bioaugmentation culture was originally seeded with groundwater from the TCE-contaminated Bachman Road aquifer located in Oscoda, MI and the culture was enriched on TCE and lactate for over two years. A pairwise comparison between DNA from the bioaugmentation culture and the sample collected after biostimulation (July) indicated that 202 subfamilies were detected in both communities, while 83 and 76 subfamilies were unique to the bioaugmentation culture and the July sample, respectively.
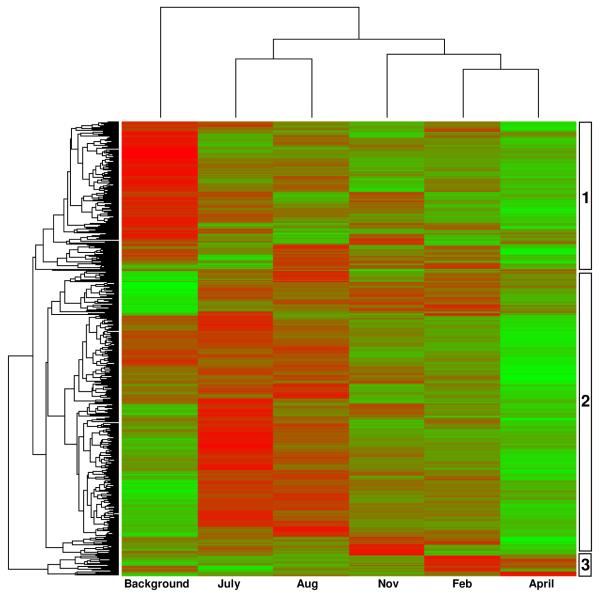
Clustering analysis of the HybScores of each of the samples indicates that the bacterial populations within the treatment plot after biostimulation with whey were significantly different from the background sample, with the latter forming a separate cluster (Figure 4, top-axis). As is consistent with the PCA analysis, the July and August samples form a cluster that is separate from the three later samples. Overall, clustering analysis of all the 478 bacterial subfamilies found during the course of treatment identified three distinct groups (Figure 4, y-axis and Supporting Information Table S4).
Figure 4.
Hierarchical clustering analysis of samples and subfamilies in the bacterial domain over the course of treatment. The color gradient from green to red of the heatmap represents increasing array hybridization intensity. Each row represents a subfamily and each column represents a sample with labeling at the bottom. Three main dynamic groups were identified and labeled on the right.
Cluster group 1 consisted of sequences from 157 subfamilies from 26 phyla whose amplicons exhibited a high relative abundance in the background sample followed by a sharp decline upon whey injection that continued over the course of treatment (Figure 4). The dominant members in cluster group 1 were from different classes of Proteobacteria (88 subfamilies), especially the α-Proteobacteria (36 subfamilies). Members from the orders of Bradyrhizobiales, Consistiales and Rhizobiales within the α-Proteobacteria, Burkholderiales within the β-Proteobacteria, and Legionellales and Thiotrichales within the γ-Proteobacteria were within this cluster.
Cluster group 2 is the largest group consisting of 300 subfamilies distributed over 35 phyla. This cluster had a low relative abundance initially, followed in the first month by an increase then faded towards the end, indicating that these bacteria were initially stimulated by whey but were not sustained at higher relative abundances over the course of treatment (Figure 4). The dominant members in cluster group 2 were from Actinobacteria (24 subfamilies), Bacteroidetes (25 subfamilies), Firmicutes (66 subfamilies), γ-Proteobacteria (40 subfamilies), and δ-Proteobacteria (25 subfamilies). Fermenters within the orders of Bacteroidales, Flavobacteriales and Sphingobacteriales in the Bacteroidetes along with Bacillales, Clostridiales and Lactobacillales in the Firmicutes were within this cluster. Bacteria in the families of Enterobacteriaceae and Pseudomonadaceae within γ-Proteobacteria and Desulfoarculaceae, Geobacteraceae and Nitrospinaceae in the δ-Proteobacteria were also in cluster group 2.
Cluster group 3 is relatively small with only 21 subfamilies and they remained at a low relative abundance until the February sampling when significant increases were observed (Figure 4). Members in the Campylobacteraceae family within the ε-Proteobacteria and the Desulfobulbaceae family within the δ-Proteobacteria were representatives of this group. Interestingly, three subfamilies within the candidate phyla OP11 were also in cluster group 3. Cluster group 3 was the only set of bacterial subfamilies that were present at a high density relative to the background towards the end of treatment.
As observed, members within the five classes of Proteobacteria (α, β, δ, ε, γ) were a dynamic component of the microbial community. Members of Proteobacteria tended to show mixed responses after the injection of whey (Figure 3) in contrast with the Bacteroidetes and Firmicutes which tended to show significant increases in all subfamilies. In general, a great variety of physiology and metabolism are found within the Proteobacteria phylum, and many are typical heterotrophic soil microbes that can respire different terminal electron acceptors for growth (oxygen, nitrate, sulfate, iron).52 Significant enrichment in some of the Proteobacteria subfamilies was concomitant with the decreases in dissolved oxygen, nitrate and sulfate concentrations as well as the generation of ferrous iron after whey injection (Supporting Information Figure S3), suggesting that the diverse microbes were respiring the available terminal electron acceptors. Perhaps because the injected whey contained a background level of sulfate, sulfate-reducers from families such as Desulfobacteraceae, Desulfohalobiaceae, Desulfomicrobiaceae, and Desulfuromonaceae in the δ-Proteobacteria became more active towards the end of treatment.
Dynamics of the archaeal populations within the treatment plot
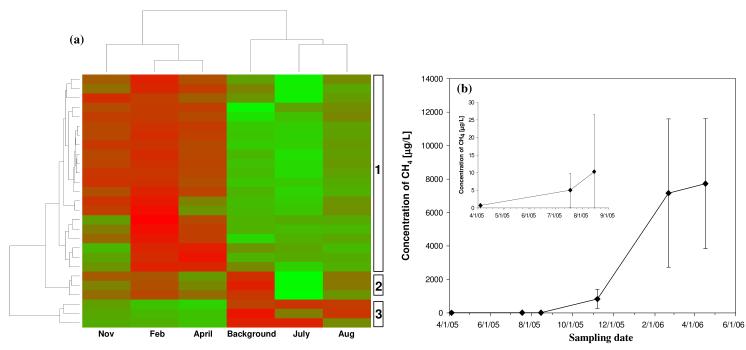
After the initial injection of whey, only a small fraction of Archaea showed responses (Figure 5a and Supporting Information Table S5). A hierarchical clustering analysis of all the 27 archaeal subfamilies detected in the DNA fraction showed that most of the Archaea remained at a relatively low abundance during the first four months of operation (up to August), but increased significantly from November to April (Figure 5a). Cluster group 1 contained many methanogens from the Euryarchaeota phylum within the Methanosaetaceae, Methanomicrobiaceae, Methanosarcinaceae, Methanocorpusculaceae, and Methanobacteriaceae families. This cluster exhibited significant increases towards the later part of treatment (Figure 5a), corresponding with over two orders of magnitude increases in methane concentrations detected in the later samples (Figure 5b) from less than 10 μg/L prior to November to over 7500 μg/L by the end of treatment, and demonstrating the strong correlation between PhyloChip results and corresponding community metabolism. Other members in cluster group 1 included representatives in the families of Thermococcaceae and Halobacteriaceae. Cluster groups 2 and 3 represent a relatively small fraction of the archaeal populations from the C1 and Thermoprotei classes of Crenarchaeota that decreased in relative abundance over time (Figure 5a).
Figure 5.
(a) Hierarchical clustering analysis of samples and subfamilies in the archaeal domain over the course of treatment. The color gradient from green to red of the heatmap represents increasing array hybridization intensity. Each row represents a subfamily and each column represents a sample with labeling at the bottom. Three main dynamic groups were identified and labeled on the right. (b) Methane concentration over the course of treatment at Ft. Lewis. The inserted graph highlights the differences in scale during the early part of treatment. The concentration at each time point is the average measurement of the 16 samples taken from monitoring wells that were spatially separated and screened to different depths on both treatment plots. The error bars represent standard deviation of the 16 concentration measurements.
Methanogens have commonly been observed in dechlorinating microbial communities33,34 and can be physically co-located with Dehalococcoides cells as biofloc.35 Although previous research has shown that hydrogen-consuming methanogens are potential competitors of Dehalococcoides spp. for hydrogen,22 some methanogens (e.g. Methanosarcina spp.) are known to synthesize corrinoids53,54 that might benefit Dehalococcoides spp. as important co-factors for RDases. Interestingly, at this contaminated site, the apparent delay in methanogensis might have been caused by the lower pH (5.2) during the first few months of operation (Supporting Information Figure S3).
Implications for contaminated sites remediation
Overall, the combination of PhyloChip analyses of DNA and RNA, together with clone library construction performed in this study have provided insights into the in situ microbial ecology and population dynamics at the TCE-contaminated field site undergoing biostimulation and bioaugmentation. Whey was injected into the treatment plots in this study to stimulate the activity of the subsurface microbial community. Of concern initially was the fact that the site was originally aerobic, but based on the PhyloChip results, diverse groups of microorganisms responded and their metabolic activities resulted in a favorable reducing condition for reductive dechlorination. Furthermore, the generated fermentation products supported growth of Dehalococcoides. These results are consistent with previously reported data38 which showed that Dehalococcoides concentrations increased following biostimulation while TCE was converted to mainly cDCE. Subsequently, during the later part of the treatment, VC and ethene were formed, and further increases in Dehalococcoides were observed.38 Throughout the study, there was no obvious indication from the molecular data that would suggest an undesirable microbial community structure for support of reductive dechlorination.
During the one-year remediation study at Ft. Lewis, the microbial community remained highly dynamic and steady-state was not reached. By using data from advanced molecular tools such as PhyloChips, practitioners will be able to obtain a comprehensive time-series view of the subsurface microbial community. Such data would complement qPCR results targeting specific key functional dechlorinators (such as Dehalococcoides). Ultimately, by tracking the overall microbial community together with key functional players, informed decisions can then be made regarding how to best manipulate the field conditions to achieve effective bioremediation of chlorinated ethenes.
Supplementary Material
Acknowledgments
We thank R. Ryan Dupont at Utah State University for generously providing the bioaugmentation culture, the staff of North Wind Inc. for conducting the field work and Ed Kirton of the Joint Genome Institute for running the GeneLib software package. This research was supported by NIEHS Superfund Basic Research Project ES04705-19, Strategic Environmental Research and Development Program (SERDP) ER-1587 and Environmental Security Technology Certification Program (ESTCP) ER-0218 and ER-0318. Part of this work was performed at Lawrence Berkeley National Laboratory, managed by the University of California under contract number DE-AC02-05CH11231 with the U.S. Department of Energy.
Footnotes
Supporting Information Available Tables S1-S5 and Figures S1-S4. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- (1).Moran MJ, Zogorski JS, Squillace PJ. Chlorinated solvents in groundwater of the United States. Environ. Sci. Technol. 2007;41(1):74–81. doi: 10.1021/es061553y. [DOI] [PubMed] [Google Scholar]
- (2).U.S. Department of Health and Human Services Report on carcinogens. 11th edition 2005.
- (3).National Research Council Committee on Ground Water Cleanup Alternatives . Alternatives for groundwater cleanup. National Academy Press; Washington, D.C.: 1994. [Google Scholar]
- (4).Löffler FE, Edwards EA. Harnessing microbial activities for environmental cleanup. Curr. Opin. Biotechnol. 2006;17(3):274–284. doi: 10.1016/j.copbio.2006.05.001. [DOI] [PubMed] [Google Scholar]
- (5).Cheng D, He J. Isolation and characterization of “Dehalococcoides” sp. strain MB, which dechlorinates tetrachloroethene to trans-1,2-dichloroethene. Appl. Environ. Microbiol. 2009;75(18):5910–5918. doi: 10.1128/AEM.00767-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).He J, Ritalahti KM, Yang KL, Koenigsberg SS, Löffler FE. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature. 2003;424(6944):62–65. doi: 10.1038/nature01717. [DOI] [PubMed] [Google Scholar]
- (7).He J, Sung Y, Krajmalnik-Brown R, Ritalahti KM, Löffler FE. Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene (TCE)- and 1,2-dichloroethene-respiring anaerobe. Environ. Microbiol. 2005;7(9):1442–1450. doi: 10.1111/j.1462-2920.2005.00830.x. [DOI] [PubMed] [Google Scholar]
- (8).Maymó-Gatell X, Chien YT, Gossett JM, Zinder SH. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science. 1997;276(5318):1568–1571. doi: 10.1126/science.276.5318.1568. [DOI] [PubMed] [Google Scholar]
- (9).Müller JA, Rosner BM, von Abendroth G, Meshulam-Simon G, McCarty PL, Spormann AM. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl. Environ. Microbiol. 2004;70(8):4880–4888. doi: 10.1128/AEM.70.8.4880-4888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Sung Y, Ritalahti KM, Apkarian RP, Löffler FE. Quantitative PCR confirms purity of strain GT, a novel trichloroethene-to-ethene-respiring Dehalococcoides isolate. Appl. Environ. Microbiol. 2006;72(3):1980–1987. doi: 10.1128/AEM.72.3.1980-1987.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).McCarty PL. Microbiology: breathing with chlorinated solvents. Science. 1997;276(5318):1521–1522. doi: 10.1126/science.276.5318.1521. [DOI] [PubMed] [Google Scholar]
- (12).Maphosa F, de Vos WM, Smidt H. Exploiting the ecogenomics toolbox for environmental diagnostics of organohalide-respiring bacteria. Trends Biotechnol. 2010;28(6):308–316. doi: 10.1016/j.tibtech.2010.03.005. [DOI] [PubMed] [Google Scholar]
- (13).Smidt H, de Vos WM. Anaerobic microbial dehalogenation. Annu. Rev. Microbiol. 2004;58:43–73. doi: 10.1146/annurev.micro.58.030603.123600. [DOI] [PubMed] [Google Scholar]
- (14).Hendrickson ER, Payne JA, Young RM, Starr MG, Perry MP, Fahnestock S, Ellis DE, Ebersole RC. Molecular analysis of Dehalococcoides 16S ribosomal DNA from chloroethene-contaminated sites throughout North America and Europe. Appl. Environ. Microbiol. 2002;68(2):485–495. doi: 10.1128/AEM.68.2.485-495.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Lendvay JM, Löffler FE, Dollhopf M, Aiello MR, Daniels G, Fathepure BZ, Gebhard M, Heine R, Helton R, Shi J, Krajmalnik-Brown R, Major CL, J., Barcelona MJ, Petrovskis E, Hickey R, Tiedje JM, Adriaens P. Bioreactive barriers: a comparison of bioaugmentation and biostimulation for chlorinated solvent remediation. Environ. Sci. Technol. 2003;37(7):1422–1431. [Google Scholar]
- (16).Rahm BG, Chauhan S, Holmes VF, Macbeth TW, Sorenson KS, Jr., Alvarez-Cohen L. Molecular characterization of microbial populations at two sites with differing reductive dechlorination abilities. Biodegradation. 2006;17(6):523–534. doi: 10.1007/s10532-005-9023-9. [DOI] [PubMed] [Google Scholar]
- (17).Ellis DE, Lutz EJ, Odom JM, Buchanan RJ, Jr., Bartlett CL, Lee MD, Harkness MR, DeWeerd KA. Bioaugmentation for accelerated in situ anaerobic bioremediation. Environ. Sci. Technol. 2000;34(11):2254–2260. [Google Scholar]
- (18).Major DW, McMaster ML, Cox EE, Edwards EA, Dworatzek SM, Hendrickson ER, Starr MG, Payne JA, Buonamici LW. Field demonstration of successful bioaugmentation to achieve dechlorination of tetrachloroethene to ethene. Environ. Sci. Technol. 2002;36(23):5106–5116. doi: 10.1021/es0255711. [DOI] [PubMed] [Google Scholar]
- (19).McMurdie PJ, Behrens SF, Müller JA, Göke J, Ritalahti KM, Wagner R, Goltsman E, Lapidus A, Holmes S, Löffler FE, Spormann AM. Localized plasticity in the streamlined genomes of vinyl chloride respiring Dehalococcoides. PLoS Genet. 2009;5(11):e1000714. doi: 10.1371/journal.pgen.1000714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Seshadri R, Adrian L, Fouts DE, Eisen JA, Phillippy AM, Methe BA, Ward NL, Nelson WC, Deboy RT, Khouri HM, Kolonay JF, Dodson RJ, Daugherty SC, Brinkac LM, Sullivan SA, Madupu R, Nelson KE, Kang KH, Impraim M, Tran K, Robinson JM, Forberger HA, Fraser CM, Zinder SH, Heidelberg JF. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science. 2005;307(5706):105–108. doi: 10.1126/science.1102226. [DOI] [PubMed] [Google Scholar]
- (21).Kube M, Beck A, Zinder SH, Kuhl H, Reinhardt R, Adrian L. Genome sequence of the chlorinated compound-respiring bacterium Dehalococcoides species strain CBDB1. Nat. Biotechnol. 2005;23(10):1269–1273. doi: 10.1038/nbt1131. [DOI] [PubMed] [Google Scholar]
- (22).Yang Y, McCarty PL. Competition for hydrogen within a chlorinated solvent dehalogenating anaerobic mixed culture. Environ. Sci. Technol. 1998;32(22):3591–3597. [Google Scholar]
- (23).Bowman KS, Moe WM, Rash BA, Bae HS, Rainey FA. Bacterial diversity of an acidic Louisiana groundwater contaminated by dense nonaqueous-phase liquid containing chloroethanes and other solvents. FEMS Microbiol. Ecol. 2006;58(1):120–133. doi: 10.1111/j.1574-6941.2006.00146.x. [DOI] [PubMed] [Google Scholar]
- (24).Daprato RC, Löffler FE, Hughes JB. Comparative analysis of three tetrachloroethene to ethene halorespiring consortia suggests functional redundancy. Environ. Sci. Technol. 2007;41(7):2261–2269. doi: 10.1021/es061544p. [DOI] [PubMed] [Google Scholar]
- (25).Dennis PC, Sleep BE, Fulthorpe RR, Liss SN. Phylogenetic analysis of bacterial populations in an anaerobic microbial consortium capable of degrading saturation concentrations of tetrachloroethylene. Can. J. Microbiol. 2003;49(1):15–27. doi: 10.1139/w03-008. [DOI] [PubMed] [Google Scholar]
- (26).Duhamel M, Edwards EA. Microbial composition of chlorinated ethene-degrading cultures dominated by Dehalococcoides. FEMS Microbiol. Ecol. 2006;58(3):538–549. doi: 10.1111/j.1574-6941.2006.00191.x. [DOI] [PubMed] [Google Scholar]
- (27).Flynn SJ, Löffler FE, Tiedje JM. Microbial community changes associated with a shift from reductive dechlorination of PCE to reductive dechlorination of cis-DCE and VC. Environ. Sci. Technol. 2000;34(6):1056–1061. [Google Scholar]
- (28).Freeborn RA, West KA, Bhupathiraju VK, Chauhan S, Rahm BG, Richardson RE, Alvarez-Cohen L. Phylogenetic analysis of TCE-dechlorinating consortia enriched on a variety of electron donors. Environ. Sci. Technol. 2005;39(21):8358–8368. doi: 10.1021/es048003p. [DOI] [PubMed] [Google Scholar]
- (29).Gu AZ, Hedlund BP, Staley JT, Strand SE, Stensel HD. Analysis and comparison of the microbial community structures of two enrichment cultures capable of reductively dechlorinating TCE and cis-DCE. Environ. Microbiol. 2004;6(1):45–54. doi: 10.1046/j.1462-2920.2003.00525.x. [DOI] [PubMed] [Google Scholar]
- (30).Hohnstock-Ashe AM, Plummer SM, Yager RM, Baveye P, Madsen EL. Further biogeochemical characterization of a trichloroethene-contaminated fractured dolomite aquifer: electron source and microbial communities involved in reductive dechlorination. Environ. Sci. Technol. 2001;35(22):4449–4456. doi: 10.1021/es0110067. [DOI] [PubMed] [Google Scholar]
- (31).Lee PKH, Johnson DR, Holmes VF, He J, Alvarez-Cohen L. Reductive dehalogenase gene expression as a biomarker for physiological activity of Dehalococcoides spp. Appl. Environ. Microbiol. 2006;72(9):6161–6168. doi: 10.1128/AEM.01070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Lowe M, Madsen EL, Schindler K, Smith C, Emrich S, Robb F, Halden RU. Geochemistry and microbial diversity of a trichloroethene-contaminated Superfund site undergoing intrinsic in situ reductive dechlorination. FEMS Microbiol. Ecol. 2002;40(2):123–134. doi: 10.1111/j.1574-6941.2002.tb00944.x. [DOI] [PubMed] [Google Scholar]
- (33).Macbeth TW, Cummings DE, Spring S, Petzke LM, Sorenson KS., Jr. Molecular characterization of a dechlorinating community resulting from in situ biostimulation in a trichloroethene-contaminated deep, fractured basalt aquifer and comparison to a derivative laboratory culture. Appl. Environ. Microbiol. 2004;70(12):7329–7341. doi: 10.1128/AEM.70.12.7329-7341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Richardson RE, Bhupathiraju VK, Song DL, Goulet TA, Alvarez-Cohen L. Phylogenetic characterization of microbial communities that reductively dechlorinate TCE based upon a combination of molecular techniques. Environ. Sci. Technol. 2002;36(12):2652–2662. doi: 10.1021/es0157797. [DOI] [PubMed] [Google Scholar]
- (35).Rowe AR, Lazar BJ, Morris RM, Richardson RE. Characterization of the community structure of a dechlorinating mixed culture and comparisons of gene expression in planktonic and biofloc-associated “Dehalococcoides” and Methanospirillum species. Appl. Environ. Microbiol. 2008;74(21):6709–6719. doi: 10.1128/AEM.00445-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Yang Y, Pesaro M, Sigler W, Zeyer J. Identification of microorganisms involved in reductive dehalogenation of chlorinated ethenes in an anaerobic microbial community. Water Res. 2005;39(16):3954–3966. doi: 10.1016/j.watres.2005.07.010. [DOI] [PubMed] [Google Scholar]
- (37).Nemir A, David MM, Perrussel R, Sapkota A, Simonet P, Monier JM, Vogel TM. Comparative phylogenetic microarray analysis of microbial communities in TCE-contaminated soils. Chemosphere. 2010;80(5):600–607. doi: 10.1016/j.chemosphere.2010.03.036. [DOI] [PubMed] [Google Scholar]
- (38).Lee PKH, Macbeth TW, Sorenson KS, Jr., Deeb RA, Alvarez-Cohen L. Quantifying genes and transcripts to assess the in situ physiology of “Dehalococcoides” spp. in a trichloroethene-contaminated groundwater site. Appl. Environ. Microbiol. 2008;74(9):2728–2739. doi: 10.1128/AEM.02199-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Brodie EL, DeSantis TZ, Joyner DC, Baek SM, Larsen JT, Andersen GL, Hazen TC, Richardson PM, Herman DJ, Tokunaga TK, Wan JMM, Firestone MK. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl. Environ. Microbiol. 2006;72(9):6288–6298. doi: 10.1128/AEM.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Brodie EL, DeSantis TZ, Parker JPM, Zubietta IX, Piceno YM, Andersen GL. Urban aerosols harbor diverse and dynamic bacterial populations. Proc. Natl. Acad. Sci. U. S. A. 2007;104(1):299–304. doi: 10.1073/pnas.0608255104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).DeSantis TZ, Brodie EL, Moberg JP, Zubieta IX, Piceno YM, Andersen GL. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb. Ecol. 2007;53(3):371–383. doi: 10.1007/s00248-006-9134-9. [DOI] [PubMed] [Google Scholar]
- (42).Wrighton KC, Agbo P, Warnecke F, Weber KA, Brodie EL, DeSantis TZ, Hugenholtz P, Andersen GL, Coates JD. A novel ecological role of the Firmicutes identified in thermophilic microbial fuel cells. ISME J. 2008;2(11):1146–1156. doi: 10.1038/ismej.2008.48. [DOI] [PubMed] [Google Scholar]
- (43).Macbeth TW, Sorenson KS., Jr. Final report, “In situ bioremediation of chlorinated solvent source areas with enhanced mass transfer ESTCP project ER-0218”. Environmental Security Technology Certification Program. 2008 Appendix B. [Google Scholar]
- (44).DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 2005;71(3):1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Huber T, Faulkner G, Hugenholtz P. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics. 2004;20(14):2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- (47).Ludwig W, Klenk HP. Overview: a phylogenetic backbone and taxonomic framework for prokaryotic systematics. In: Boone DR, Castenholz RW, editors. The Archaea and the deeply branching and phototrophic Bacteria. vol. 1. Springer-Verlag; New York: 2001. pp. 49–65. [Google Scholar]
- (48).DeAngelis KM, Wu CH, Beller HR, Brodie EL, Chakraborty R, DeSantis TZ, Fortney JL, Hazen TC, Osman SR, Singer ME, Tom LM, Andersen GL. PCR amplification-independent methods for detection of microbial communities by the high-density microarray PhyloChip. Appl. Environ. Microbiol. 2011;77(18):6313–6322. doi: 10.1128/AEM.05262-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Kerkhof L, Kemp P. Small ribosomal RNA content in marine Proteobacteria during non-steady-state growth. FEMS Microbiol. Ecol. 1999;30(3):253–260. doi: 10.1111/j.1574-6941.1999.tb00653.x. [DOI] [PubMed] [Google Scholar]
- (50).Janssen PH. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 2006;72(3):1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).DeAngelis KM, Brodie EL, DeSantis TZ, Andersen GL, Lindow SE, Firestone MK. Selective progressive response of soil microbial community to wild oat roots. ISME J. 2009;3(2):168–178. doi: 10.1038/ismej.2008.103. [DOI] [PubMed] [Google Scholar]
- (52).Garrity GM, Brenner DJ, Krieg NR, Staley JR. Bergey’s manual of systematic bacteriology. vol 2: the Proteobacteria, parts A - C. Springer-Verlag; New York: 2005. [Google Scholar]
- (53).Mazumder TK, Nishio N, Fukuzaki S, Nagai S. Production of extracellular vitamin B-12 compounds from methanol by Methanosarcina barkeri. Appl. Microbiol. Biotechnol. 1987;26(6):511–516. [Google Scholar]
- (54).Ward DM, Mah RA, Kaplan IR. Methanogenesis from acetate: a nonmethanogenic bacterium from an anaerobic acetate enrichment. Appl. Environ. Microbiol. 1978;35(6):1185–1192. doi: 10.1128/aem.35.6.1185-1192.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.