Abstract
The initial period of mammalian embryonic development is primarily devoted to cell commitment to the pluri-potent lineage, as well as to the formation of extraembryonic tissues essential for embryo survival in utero. This phase of development is also characterized by extensive morphological transitions. Cells within the preimplantation embryo exhibit extraordinary cell plasticity and adaptation in response to experimental manipulation, highlighting the use of a regulative developmental strategy rather than a predetermined one resulting from the non-uniform distribution of maternal information in the cytoplasm. Consequently, early mammalian development represents a useful model to study how the three primary cell lineages; the epiblast, primitive endoderm (also referred to as the hypoblast) and trophoblast, emerge from a totipotent single cell, the zygote. In this review, we will discuss how the isolation and genetic manipulation of murine stem cells representing each of these three lineages has contributed to our understanding of the molecular basis of early developmental events.
Keywords: Blastocyst, mouse development, preimplantation, cell fate, lineage allocation, trophectoderm, epiblast, primitive endoderm, stem cells, ES cell, TS cell, XEN cell, pluripotency
I. THE BLASTOCYST EMBRYO COMPRISES THREE CELL LINEAGES
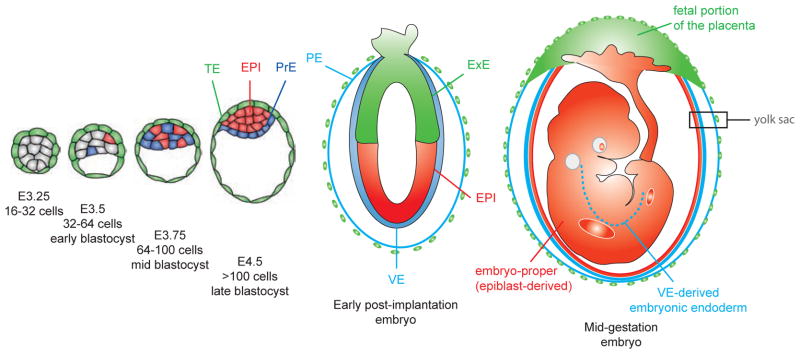
Eutherian embryonic development takes place in a protective and nutritive environment. Unlike other phyla, mammalian oocytes are alecithal eggs that lack the large nutrient reserves that are usually accumulated during oogenesis. Early mammalian development is comparatively slow, taking place within the maternal uterine fluid and is mainly devoted to the establishment of tissues specialized for interactions with the maternal uterus. As a consequence, the first cell fate decisions occur early during embryogenesis before embryo implantation into the uterus. These cell fate decisions involve the specification and spatial segregation of extraembryonic tissues from the pluripotent epiblast (EPI) that will give rise to most of the embryo-proper. The first extraembryonic tissue specified is the trophectoderm (TE), which forms the fetal portion of the placenta, thereafter the primitive endoderm (PrE) will be specified which gives rise to the visceral (VE) and parietal (PE) endoderm layers of the yolk sac (Fig. 1). Importantly, the VE, which encapsulates the epiblast after implantation, is critical for axis formation [1, 2]. At gastrulation, the distal VE (also referred to as emVE) is incorporated into the gut endoderm tissue layer of the fetus in contrast to it previously being ascribed as having an exclusively extraembryonic fate [3].
Fig. 1. Schematic representation of the formation and fate of the first cell lineages of the mouse embryo.
The pluripotent epiblast (red) will form the embryo proper. The trophectoderm (green) will give rise to the fetal portion of the placenta and contribute to the yolk sac. The primitive endoderm (blue) will differentiate into the parietal and visceral endoderm that will constitute the yolk sac. The visceral endoderm will also be incorporated into the embryonic endoderm (blue dashed line). EPI, epiblast; ExE, extraembryonic ectoderm; PE, parietal endoderm; PrE, primitive endoderm; TE, trophectoderm; VE, visceral endoderm.
By the end of the preimplantation period, the embryo is composed of three distinct cell lineages that are defined by their position within the embryo, their developmental potential and their expression of molecular markers (Fig. 1 and 4). During the last decade, several factors exhibiting lineage-specific expression have been identified, many of them required for lineage establishment, maintenance or differentiation. The trophectoderm emerges as an outer epithelial cell layer and expresses Cdx2, Eomes, Gata3 and Krt8. The PrE also forms an epithelial cell layer at the interface between EPI and the blastocoel cavity and expresses Dab2, Fgfr2, Gata4, Gata6, Pdgfra, Sox7 and Sox17. The pluripotent EPI is encapsulated by these two nascent extraembryonic tissues and expresses markers such as Fgf4, Nanog, Oct4 and Sox2.
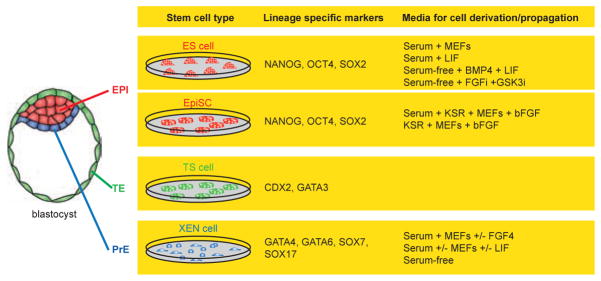
Fig. 4. Stem cells that can be derived from blastocyst stage mouse embryos.
Three stem cells can be isolated from a mouse blastocyst embryo. Embryonic stem (ES) cells and Epi stem cells (EpiSCs) from the epiblast (red), trophoblast stem (TS) cells from the trophectoderm (green) and extraembryonic endoderm (XEN) cells from the primitive endoderm (blue). These stem cells are characterized by their distinct morphologies, their developmental potential in chimeras, as well as the expression of different sets of lineage-specific transcription factors (the transcription factors listed can be recognized by commercially available antibodies) and by the conditions used to isolate and propagate them in culture.
Early mammalian development can therefore be viewed as a process by which totipotency is progressively lost, and where initial asymmetries reinforced by positional information, direct the formation of the first three cell lineages. Lineage-specific markers have been extensively used to follow cell lineage commitment and segregation. Our current view of the segregation of the first lineages, TE vs. ICM and subsequently within the ICM EPI vs. PrE, has suggested that early lineage segregation occurs in two successive phases. In the first phase, markers of derivative lineages are co-expressed in individual cells at variable levels. In the second phase, marker expression becomes mutually exclusive as cells are lineage committed. In the following section, we review our current understanding on how these lineages might be specified and segregated during mouse embryonic development.
2. TE vs. ICM SPECIFICATION
Upon fertilization, the first three cell divisions (1 -> 2 -> 4 -> 8 cell stage) give rise to 8 blastomeres that generally appear morphologically indistinguishable. At the 8-cell stage, embryos undergo compaction, a process where cell-cell contacts between blastomeres increase. E-cadherin relocalization plays a major role in this process. Zygotic E-cadherin mutant embryos compact normally [4], while maternal removal of E-cadherin delays compaction [5]. Removal of both maternal and zygotic contribution prevents compaction [6]. As compaction proceeds, blastomeres become polarized along their apical-basal axis, such that the apical surface of blastomeres which faces outside is free of cell-cell contacts, contains microvilli and is enriched in aPKC isoforms (such as PKCζ, PKCδ and PKCλ), PAR3, PAR6 and EZRIN. By contrast, the basolateral regions of compacting blastomeres are enriched in LGL1, JAM1 and PAR1. This localization of proteins is associated with the formation of gap junctions, adherens and tight junctions between neighboring blastomeres, and occurs concomitant with epithelialization.
It has been proposed that, during the next two rounds of cell division (8 -> 16 -> 32 cell stage), cleavage plane orientation underlies the emergence of two cell types that differ in their position within the morula, such that outside cells will form the TE, whereas inside cells will form the ICM (Figure 2). Symmetric cell divisions (with a cleavage plane parallel to the basolateral axis) will give rise to two identical polarized daughter cells having an outside position, whereas an asymmetric cell division (with a cleavage plane perpendicular to the basolateral axis) will generate an outer polarized cell and an inner non-polarized cell.
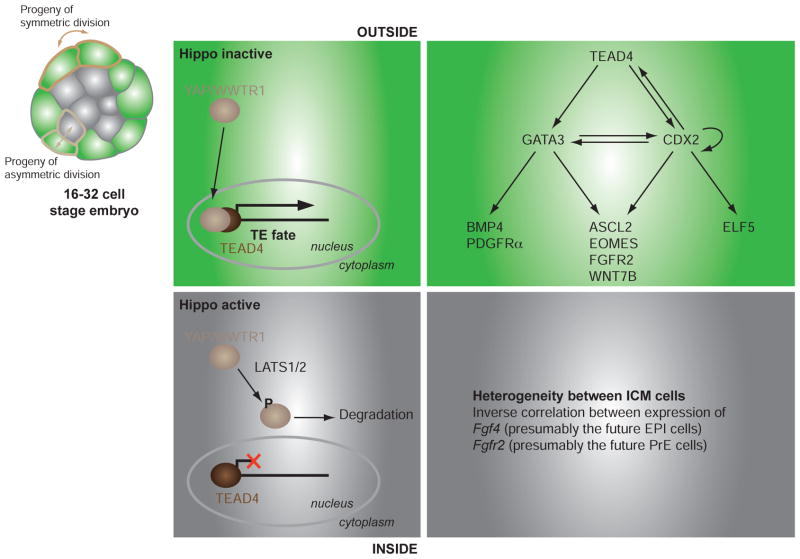
Fig. 2. The first cell lineage decision in the mouse embryo.
While symmetric divisions starting from the 8-cell stage give rise to two outside (green) polarized cells, asymmetric divisions give rise to an outside cell and an inside (grey) apolar cells. In outside cells, TEAD4 transcription factor and its cofactors YAP/WWTR1 regulate the expression of a TE-specific transcription program that ultimately commits outside cells to a trophectoderm fate. This program is not achieved in inside cells because of the activity of the Hippo pathway that leads to the degradation of YAP/WWTR1. ICM cells are not homogenous and an inverse correlation of the expression of Fgf4 and Fgfr2 may presage the divergence between PrE and EPI lineages.
2.1. Inside-Outside Model
To explain this first lineage divergence, it had been proposed that fate might be determined by cell position, this being inside vs. outside, within the morula. Inside cells adopt an ICM fate, whereas outside cells become TE [7]. Such a divergence could result from differences in the microenvironment and promote two distinct cell fate decisions. Alternatively, differences in the nature of the cell-cell contacts could impact on lineage commitment, since inner cells are encapsulated by neighboring cells, whereas outer cells have a free apical surface which is exposed to the outside environment.
This “inside-outside” model is supported by the observation that cell fate can be altered by experimentally repositioning cells within the embryo [8, 9]. Furthermore, ICMs isolated from early blastocysts are able to regenerate an outer TE epithelial layer, and go on to form a blastocyst structure [10–12]. By demonstrating that early blastomeres are not irreversibly committed to particular fates, and that topological position within the developing embryo may be important for lineage determination, these observations reveal an intrinsic cell fate plasticity present within the early mouse embryo.
2.2. Polarity Model
In an alternative, but not necessarily mutually exclusive model, Johnson and colleagues proposed that allocation of TE vs. ICM takes place between the 8 to 16-cell stage, prior to the emergence of inside and outside cells [13]. They suggested that one determinant likely results from the acquisition of cell polarity. Support for this model comes from the demonstration that experimental perturbation of cell polarity affects cell position, and presumably cell fate. Indeed, down-regulation of aPKC or PAR3 promotes cell localization to the inside of the embryo [14]. However, it is still not clear how polarity is established at the time of compaction.
If cell-cell contacts are a key determinant for cell polarization, the adhesion molecule E-cadherin is likely to be involved. Analysis of zygotic mutant embryos has revealed that maternal stores are clearly sufficient to polarization. However, it was recently reported that in maternal/zygotic E-cadherin mutants polarization still occurs, at least partially, leading to the establishment of both TE and ICM cells even if the allocation of blastomeres to the inside or outside compartments is impaired [6].
2.3. Key Lineage Determinants of TE vs. ICM Identity
Presently, the earliest identified signaling event involves components of the Hippo signaling pathway and specifically requires the TEAD transcription factor family member TEAD4 (Fig. 2). Mutants lacking Tead4 die at preimplantation; they fail to form TE and never reach the blastocyst stage [15, 16]. Even so, polarity is maintained in Tead4 mutant embryos indicating that TEAD4 acts either downstream or in parallel to the mechanisms that establish cell polarity. Interestingly, CDX2 was detected in early morula stage Tead4 mutant embryos, but its expression was not maintained (in Nishioka study [15] but not Yagi study [16]). In addition, all cells of Tead4 mutant embryos expressed the epiblast-specific OCT4 and NANOG transcription factors, but blastomeres exhibited heterogenous levels of these proteins [15]. This is intriguing, and if confirmed, would indicate that the initial period of stochastic marker expression may be initiated in Tead4 mutants, but that subsequent marker restriction concomitant with cell fate choice is impaired.
Surprisingly at these early stages, Tead4 is widely expressed and TEAD4 protein localizes to the nucleus of all blastomeres, indicating that its role in TE specification might be mediated by its co-factor(s). Indeed, the TEAD co-activators YAP and TAZ (also called WWTR1) specifically localize to the nuclei of outside blastomeres, not to inside blastomeres. The current model proposes that the Hippo pathway is regulating the subcellular localization of YAP/TAZ proteins. The pathway is active in inside cells, leading to the phosphorylation and subsequent degradation of YAP/TAZ. Conversely, the pathway is inactive in outside cells, leading to the nuclear accumulation of YAP/TAZ and regulation of TEAD4 activity. This leads to the question of how might the Hippo pathway, which plays a major role in the regulation of tissue growth [17], be involved in early lineage determination? A recent study suggests that the Hippo pathway is regulated by positional information and cell polarity, and that therefore it might function to integrate major developmental signaling pathways, including TGFβ signaling. This process may rely on a cell’s ability to sense its environment, for example cell density inputs through the Crumbs polarity complex [18]. In turn, active TEAD4 regulates the expression of two TE-specific transcription factors: CDX2 and GATA3 [15, 16, 19].
Cdx2 encodes a Caudal-type homeodomain transcription factor essential for TE maintenance. Indeed, zygotic Cdx2-deficient embryos progress to the blastocyst stage but the blastocoel, even though initially formed, is not maintained due to a failure to maintain epithelial integrity [20, 21]. A recent report described the presence of maternal Cdx2 mRNAs at the 8-cell stage [22]. However the role of this maternal pool remains unclear. Jedrusik and colleagues reported that Cdx2 knockdown affects cell polarization, leading to preferential contribution to inside cells suggesting a feedback loop between Cdx2 and cell polarity. Consequently, the phenotype of maternal/zygotic Cdx2 mutants is more severe than zygotic mutants [22]. This finding contrasts with other studies which suggest that Cdx2 knockdown does not affect polarization, and that the phenotypes of zygotic and maternal/zygotic mutants are undistinguishable, thus arguing against the idea that Cdx2 may be involved in TE specification [23, 24]. Interestingly, Jedrusik and colleagues reported the asymmetric localization of Cdx2 mRNA at the apical side of the 8-cell stage blastomeres which may be inherited by outside as opposed to inside cells [22].
Even so, it is widely accepted that CDX2 and OCT4 exert mutual repressive effects leading to the establishment of a CDX2-positive; OCT4-negative outside compartment (the future TE) and a CDX2-negative; OCT4-positive inside compartment, that will give rise to the ICM. Thus in the absence of Cdx2, outside cells maintain Oct4 expression. However, Oct4 mutant blastocysts initially exhibit a normal restriction of Cdx2 expression in the TE outside cell layer, but this pattern is not maintained by the time of implantation, and mutant ICM cells upregulate TE markers [19, 25]. Taken together, these observations suggest that additional mechanisms must be in place to restrict the localization of key lineage-specific regulators to their appropriate compartments [19, 26]. Of note, the kinetics of establishment of a mutually exclusive expression between Cdx2 and Oct4 varies among mammals, and the co-repression between OCT4 and CDX2 does not seem to be conserved in cattle, suggesting that mechanisms described in the mouse may perhaps be unique to rodents, but not exploited by other mammals [27]. Recently, live imaging of the kinetics of an OCT4 protein fused to a phosphoactivatable PA-GFP predicted division and cell allocation to the outside and the inside compartments [28] arguing in favor of early asymmetries, existing in 4-cell to 8-cell stage embryos, that might influence cell fate specification (reviewed in [29, 30]).
GATA3, a member of the GATA transcription factor family, is involved in this first cell lineage decision. GATA3 is first expressed at around the 4-cell stage and becomes progressively restricted to the TE lineage concomitant with the restriction in Cdx2, in a process which, by contrast, does not appear to require repression by OCT4 [19]. Interestingly, while Gata3-deficient embryos survive until mid-gestation [31, 32], Gata3 knockdown by RNA interference at the 2-cell stage impairs the morula-to-blastocyst transition suggesting an early role for GATA3 in TE formation [33]. Thus, one might predict that a maternal/zygotic Gata3 mutant will exhibit an earlier lethal phenotype.
3. ACQUISITION OF EPI OR PrE CELL FATE WITHIN THE ICM
3.1. Specification of PrE vs. EPI
The divergence within the ICM between EPI and PrE lineages is thought to occur at the early blastocyst stage and leads to the formation of a morphologically-distinct epithelial PrE cell layer in contact with the blastocoel. The fact that ICMs isolated from blastocyst stage embryos generate a PrE layer on their surface has led to the suggestion that cell position may also play a role in this cell fate choice [34]. However, the identification of lineage-specific markers, as well as the use of live imaging, have revealed that lineage commitment may occur earlier than previously thought, and that cell position may be involved in later refining lineage commitment.
The current model posits at least two distinct steps involving lineage specification and segregation. The first step corresponds to the specification of cell identity and occurs around the 64-cell stage. It involves a progressive restriction in the expression of key transcription factors such as GATA6 (PrE-specific) and NANOG (EPI-specific). By the 64-cell stage, EPI and PrE markers that were initially co-expressed by all ICM cells become mutually exclusive, such that the ICM comprises a mixture of EPI and PrE precursor cells organized in an apparent “salt-and-pepper” distribution (Fig. 3) [35–37]. The idea that lineage commitment likely occurs around this time has been suggested by testing the developmental potential of isolated ICM cells from early or late stage blastocysts [35]. While cells from early stage blastocysts could contribute to both EPI and PrE lineages, cells from late stage blastocysts were restricted to either EPI or PrE lineage.
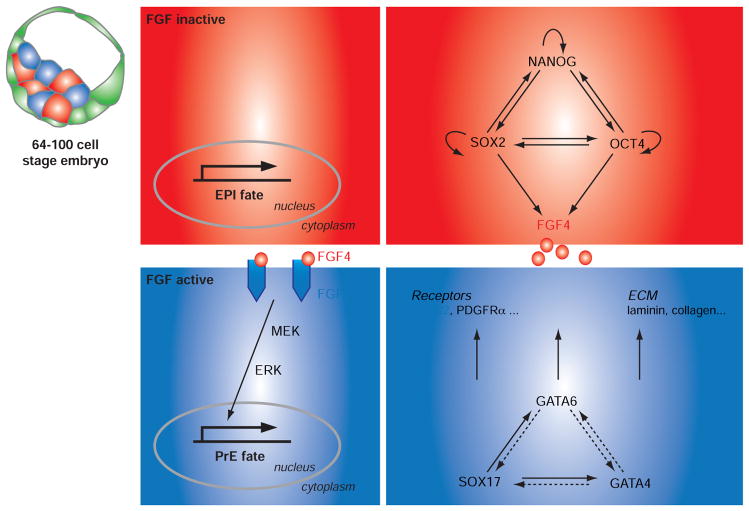
Fig. 3. Lineage specification within the inner cell mass of the preimplantation mouse embryo.
Lineage commitment of PrE and EPI cells is thought to occur around the 64-cell stage and is driven by a core-regulatory network of lineage-restricted transcription factors. FGF4 signaling pathway is a major determinant of PrE specification. FGF4 ligand is synthesized by EPI cells whereas FGFR2 is expressed by PrE cells. Activation of ERK signaling is predicted to be necessary for PrE commitment.
It has been suggested that, akin to TE vs. ICM specification, a reciprocal inhibition between different lineage-specific transcription factors might provide the mechanistic determinant for lineage commitment and divergence. The initial analysis of Nanog knockout embryos suggested that NANOG is involved in maintaining pluripotency by inhibiting PrE formation. Outgrowths from Nanog mutant ICMs could not be maintained in culture as they differentiated into PE-like cells [38]. Recently however, an in depth analysis of Nanog mutant embryos revealed that PrE formation is impaired in absence of Nanog [39, 40]. This phenotype could be rescued when wild type ES cells were injected into mutant recipient embryos. These observations therefore suggest that Nanog acts non-cell autonomously in the EPI to promote PrE formation [39].
Gata4 and Gata6 encode GATA transcription factors expressed in the PrE. GATA6 is one of the earliest markers of the cells of the developing embryo, whereas GATA4 is expressed starting at the 64-cell stage in cells likely to be committed to the PrE lineage [37]. Surprisingly however, Gata4 and Gata6 mutants affect PrE differentiation rather than formation [41–43]. However, given potential redundancy between these two GATA factors, the double mutant may reveal an earlier phenotype but it has yet to be reported. Even so, one study has reported a complete absence of a PrE layer in E4.5 Gata6 mutant embryos supporting the idea that GATA6 may play an earlier role [44].
So far, the only reported mutants that exhibit severely impaired PrE formation are in genes encoding components of the FGF signaling pathway. These include the FGF ligand FGF4 ([45, 46]; A. Piliszek, J.A., M. Kang and A.-K.H. unpublished data), the FGF receptor FGFR2 [47] and the adaptor molecule GRB2 [35, 48]. PrE commitment of ICM cells is strictly dependent on the level of FGF activity (Fig. 3). Perturbation of this activity directly affects the ratio of EPI and PrE cells formed within the ICM. Pharmacological inhibition of the FGFR (PD173074, SU5402) and/or MEK effector (PD0325901, PD184352) prevents PrE formation [49, 50]. Conversely, when cultured in high doses of recombinant FGF4, a majority of ICM cells will adopt a PrE fate [50]. Furthermore, modifying the heparan sulfation of FGF receptors leads to a similar phenotype [51].
Not only does FGF signaling play a role in cell lineage commitment, but it also appears to exert a mitogenic activity on both the PrE and the TE lineages [46, 52, 53]. A major paracrine source of FGF ligands within the blastocyst comes from EPI cells which express Fgf4 as a direct target of pluri-potency factors OCT4 and SOX2 [54], while extraembryonic lineages express FGF receptors (Fig. 3). Single-cell expression analyses have revealed an inverse correlation between Fgfr2/Fgf4 expression starting at the 32-cell stage. Intriguingly, this receptor/ligand reciprocal expression precedes the reciprocal expression of lineage-specific transcription factors such as Nanog/Gata4 or Nanog/Gata6 [55]. Thus, FGF signaling might act upstream of the lineage restriction of lineage-specific transcription factors. However, the mechanism(s) by which a mutually exclusive expression of this ligand/receptor combination might be established remains to be determined.
A relationship between the timing of formation of inner cells and a tendency towards commitment to a particular lineage has recently been suggested [56]. In this way, the expression of Sox2 in first-born inner cells would lead to an upregulation of Fgf4 and might affect neighboring cells. Thus, early inner cells formed by the first wave of asymmetric cell divisions (between 8-cell -> 16-cell stage) would preferentially adopt an EPI fate (due to high levels of SOX2, and resulting upregulation of Fgf4). By contrast, inner cells formed during the next two waves of cell divisions (16-cell -> 32-cell and 32-cell -> 64-cell stage) would preferentially adopt a PrE identity (due to lower levels of SOX2, down-regulation of Fgf4 and upregulation of Fgfr2). Even though no single study has so far revealed an absolute tendency, this “time-on-outside” model contrasts with results from Yamanaka and colleagues who failed to discern a relationship between time of origin of ICM cells and their allocation to EPI or PrE lineages [50].
In addition to FGF, other receptor tyrosine kinases (RTKs) may participate in the regulation of MEK/ERK activity required for the establishment and maintenance of extraembryonic lineages. Hence, an Egfr mutation leads to a periimplantation lethality on a CF-1 genetic background leaving open the possibility that EGF signaling may to some extent be involved in PrE formation [57]. Importantly, Platelet derived growth factor alpha (Pdgfra) is, along with Gata6, the earliest expressed PrE marker [37]. A role for PDGF signaling has been demonstrated in the expansion of the PrE lineage in late and implantation delayed blastocysts [58]. It is therefore possible that sustained MEK/ERK activity is required for the specification and the maintenance of the PrE lineage, and that its activity could be initially regulated by FGF signaling, and then reinforced by PDGF signaling.
3.2. Sorting of PrE Cells
The ICM eventually undergoes cell rearrangements that ultimately lead to the localization of PrE cells on the surface adjacent to the blastocoel where they will form a polarized epithelial layer [59]. The process that drives PrE vs. EPI spatial segregation is often referred to as cell sorting. The current model suggests that cell sorting involves multiple cell behaviors including actin-dependent cell movements and retention of position by PrE committed cells initially in contact with the blastocoel cavity [37, 60]. The few cells expressing PrE markers which do not sort to the surface and instead retain an inside position within the ICM usually have two alternative fates. They can either downregulate PrE marker expression, and presumably acquire an EPI fate, suggesting that there is still some plasticity of the ICM cells at the late blastocyst stage. Alternatively, cells that fail to sort can undergo selective apoptosis [37, 56, 60]. PrE cells that sort to their correct location on the ICM’s surface may receive positional information indicating they have reached their final destination which will eventually lead to their epithelialization. Accordingly, Sox7 expression is acquired in PrE cells once sorted, indicating that sorted and unsorted PrE cell populations are indeed distinct from one another [61].
The molecular basis for early lineage specification and segregation remains poorly understood but likely relies to some extent on differences in the adhesive properties of PrE vs. EPI cells. For example, in embryos lacking the cargo protein adaptor DAB2 [62] or the extracellular matrix protein LAMC1 [63], PrE cells are specified, but either fail to sort, or cannot be maintained at the ICM’s surface. In addition, integrins α5 and β1 have been shown to play a role in extraembryonic endoderm (ExEn) formation in embryoid bodies (EBs) [64], and Itgb1 mutant embryos die shortly after implantation with defects in the specification or differentiation of the PrE [65, 66]. Reanalyzing the phenotype of some of these mutants with the current cohort of lineage-specific markers combined with live imaging will likely help to gain further insight into the mechanisms of cell sorting.
4. ISOLATION OF THREE STEM CELL POPULATIONS REPRESENTATIVE OF THE EARLY EMBRYONIC LINEAGES
Stem cell lines can be isolated from the three first lineages of the mouse blastocyst, these being embryonic stem (ES) cells which are likely derived from and represent the EPI [67, 68], the trophoblast stem (TS) cells from the TE [69] and extraembryonic endoderm (XEN) cells from the PrE [70] (Fig. 4). These cells are often considered as stem cells based on their ability to: (i) proliferate for extensive periods in culture, (ii) remain undifferentiated when cultured under appropriate conditions, and (iii) retain their developmental potential, and so differentiate in vivo and ex vivo into cell types that normally arise from their cognate lineage.
Stem cells provide not only a basis for regenerative medicine, but are also powerful tools for investigating the molecular mechanisms involved in specification, maintenance and differentiation of these first cell lineages. Our knowledge of these early developmental events has largely been hampered by the small size of the embryo, the limited number of embryos that can be recovered, and their inaccessibility. In this way, access to cognate stem cells overcomes some of these limitations.
In this context, evaluating the role of a given gene in these early lineages can be determined by the ability to establish stem cell lines (ES, TS, XEN) from mutant embryos. Furthermore, candidate factors involved in lineage specification, for example TE or PrE, can be validated by determining if their misexpression in ES cells can drive differentiation towards TE or PrE lineages. Elucidating the factors involved in stem cell maintenance can be resolved by two complementary approaches: loss-of-function should lead to differentiation, while forced expression should impair differentiation. Lastly, if the expression of a gene is sufficient to induce differentiation, it is likely a good indication that this gene is involved in differentiation. In the following section, we highlight the specificity and common features of three stem cell lines that can be derived from mouse blastocysts, based on the extrinsic and intrinsic molecules involved in their self-renewal, as well as discussing some emergent ideas on their stem cell properties.
4.1. Signaling Molecules Required for Maintenance and Self-Renewal of Early Stem Cells
ES cells were originally isolated on mitotically inactivated fibroblast feeder cells [67, 71]. Feeders can be replaced by supplementing the serum-containing medium with LIF, a molecule that belongs to IL-6 family [72, 73]. LIF signals through the LIFR and GP130 leading to phosphorylation of the transcription factor STAT3. STAT3 eventually translocates into the nucleus where it regulates gene expression. ES cells constitutively expressing a STAT3-ER fusion protein can be grown in the absence of LIF only when the fusion is nuclear-localized by the presence of tamoxifen [74]. By contrast, expression of a STAT3 dominant-negative protein impairs ES cell self-renewal [75]. STAT3 is a main regulator of Myc expression and ES cells stably expressing a stabilized form of the MYC protein can be maintained in absence of LIF [76]. Serum can be replaced by BMP4, such that ES cells can be derived and propagated in serum-free culture conditions supplemented with LIF and BMP4 [77]. In this situation, the LIF and BMP signaling pathways cooperate to maintain pluripotency [78]. Furthermore, it has been elegantly demonstrated that self-renewal does not require activation of signaling pathways, but rather the inhibition of proteins involved in differentiation, namely FGF/ERK signaling and GSK3 [79]. This double inhibitor (also referred to as 2i) strategy permits the efficient derivation of ES cells from genetic backgrounds that are normally not permissive for ES cell establishment [49, 79, 80], as well as the establishment of ES cell from other mammalian species including rat [81, 82]. Thus, in the absence of FGF signaling, ES cells are blocked in a “naïve” state that is more refractory to differentiation. In this way, FGF signaling appears to be essential for epiblast lineage commitment [51]. The effect of GSK3 inhibition is not entirely clear, but it may facilitate cell survival during the derivation process [79].
TS cells can be derived and expanded on fibroblast feeder cells in serum-containing medium supplemented with FGF4 and heparin [69]. Moreover, feeders can be replaced with feeder-conditioned medium. Members of the Tgfβ superfamily are key regulators in TS cell maintenance such that Activin and TGFβ, but not Nodal, can replace feeder cell conditioned medium [83, 84].
In contrast to ES and TS cells, which require specific conditions for their derivation, mouse XEN cell lines have successfully been isolated using at least three different protocols [70]. XEN cell lines can be established by placing blastocysts or isolated ICMs in TS cell conditions (feeders + FGF4), and then routinely cultured on feeders in the absence of FGF4. XEN cell lines have also been derived using ES cell conditions (feeders + LIF). Moreover, XEN cells have reproducibly been derived not only from mouse embryos [58, 70, 85, 86] but also from voles [87] and rats [87–90]. It is not entirely clear which signaling pathways are required for XEN cell maintenance, but PDGF signaling is required for XEN isolation and/or cell expansion. Indeed, XEN cell lines lacking the receptor PDGFRα cannot be established and conditional inactivation of Pdgfra in established XEN cells impairs their propagation [58]. It is likely that several RTKs act in concert to sustain a threshold level of MEK/ERK activity necessary for XEN cell maintenance. In addition to PDGFRα, possible candidates include the FGF and KIT receptors.
4.2. Regulatory Networks of Stem Cell Self-Renewal
Unraveling the molecular mechanisms underlying “stemness” has been a longstanding challenge. The elucidated gene regulatory networks with transcription factors representing core components have revealed both divergent and common regulators between these early stem cell types.
The core regulatory network involved in ES cell maintenance has been intensively studied and is relatively well characterized. It involves a triumvirate of transcription factors: OCT4, NANOG and SOX2 each of which function to both regulate positively their own expression, as well as repressing the expression of genes promoting cell differentiation [91, 92]. OCT4 levels are critical for the maintenance of pluripotency. It was elegantly shown that OCT4 downregulation in ES cells induces TE differentiation, while OCT4 upregulation induces mesoderm and PrE differentiation [93]. NANOG was originally isolated in a screen designed to identify factors that confer, when overexpressed, an ability to promote pluripotency in the absence of LIF [38, 94].
In addition to this core circuit, other components may work in parallel. This is likely to be the case for Ronin, a gene that encodes a zinc finger protein. RONIN is essential during early embryonic development and in ES cell maintenance [95]. Forced RONIN expression in ES cells impairs differentiation [95]. RONIN and its essential co-factor HCF-1 regulate the expression of a variety of genes involved in metabolism, thus sustaining the growth of undifferentiated cells [96].
The network that governs TS self-renewal remains to be dissected more precisely. CDX2 and GATA3 are thought to be at the top of the hierarchy as each of which, when misexpressed in ES cells, is sufficient to drive differentiation towards a TE identify [19, 97]. Interestingly, comparative transcriptome analyses of Cdx2-misexpressing and Gata3-misexpressing ES cells has revealed that while they share common targets, GATA3 and CDX2 can act in parallel pathways [19, 97]. In addition, several studies have determined a role of various transcription factors in TS self-renewal including EOMES, ELF5, ETS2, TCFAP2C and SMARCA4 [98–100]. Recent analyses of promoter binding sites has established that SMARCA4, TCFAP2C and EO-MES are likely to be part of the core transcriptional machinery in TS cells [98].
For XEN cells, even less is known, but several studies have suggested a role for the transcription factors GATA4, GATA6, SOX7 and SOX17 in cell maintenance [101, 102].
An emerging concept is that these early stem cell types may require some common regulators. This for example is the case for SOX2, which is expressed by both ES and TS cells. In ES cells, a critical role of SOX2 is to regulate the level of Oct4 [103]. In vivo, removal of both maternal and zygotic Sox2 expression leads to developmental arrest at the morula stage and a failure to form TE [104]. The exact role of SOX2 in TS cell remains to be determined, but Avilion and colleagues reported a failure to establish Sox2-deficient TS cells, suggesting a critical requirement for TS cell derivation [105]. Similarly, SALL4 is expressed in ES and XEN cells where it regulates the expression of distinct lineage regulators [101]. In vivo SALL4 is essential for the development of ICM lineages [106].
Taken together, recent studies suggest some regulators are co-opted to participate in different gene regulatory networks, and that specificity is likely to be context dependent, for example, though the availability of co-factors as well as through accessibility of binding sites.
4.3. Pluripotent Stem Cells from Periimplantation and Early Postimplantation Stage Mouse Embryos
Various pluripotent cells have been isolated from early postimplantation stage mouse embryos, for example epiblast stem cells (EpiSC) have been isolated from the epiblast [107, 108]. Mouse EpiSCs differ from mouse ES cells not only by their flattened colony morphology, molecular signature and ability to contribute cellular progeny to chimeras but also in the extrinsic conditions (Activin and bFGF) necessary for their isolation and propagation. Interestingly, EpiSCs have also been isolated from mouse blastocyst embryos [80, 109] leading the question of whether EpiSC lines are established from a particular subset of epiblast cells resident within the ICM or result from the progressive maturation of ICM cells into an epiblast-like state in culture (Fig. 4).
Interestingly, Bao and colleagues reported the isolation of pluripotent reprogrammed epiblast ES cell-like cells (rESCs) from the epiblast of perigastrulation stage mouse embryos resembling to ES cells [110] indicating that the mature state of the epiblast around the time of gastrulation may still be reversible to a more immature state. Additional types of pluripotent stem cells have been isolated from mouse blastocyst embryos cultured in the presence of bFGF, Activin, BIO (a GSK3 inhibitor) and a LIF blocking antibody [111]. These cell lines, referred to as FAB-SCs according to the culture conditions in which they were isolated, express pluripotent markers, but fail to differentiate in teratoma or embryoid body assays if not stimulated with LIF and BMP4 beforehand.
Lastly, embryonic germ (EG) cells which have been isolated from primordial germ cells at later stages of postimplantation development and share common properties to other pluripotent cell types [112–117].
Notably, human ES cells which are derived from blastocyst embryos [118] share greater similarities to mouse EpiSCs than ES cells. hES cells, like mouse (m)EpiSCs, grow as flattened colonies and require FGF and Activin signaling to self-renew. Interestingly, while SMAD2/3 directly controls Nanog expression in both mEpiSC and hES cells, FGF2 only supports self-renewal in hES cells [119]. However, hES cells also share some features with mES cells but not mEpiSCs, suggesting they may represent an intermediate pluripotent state. For example, they also express some ICM markers (such as REX1 or ZFP42), but not the epiblast marker FGF5, in contrast to mEpiSCs. For more details, we invite the readers to dedicated reviews on this topic [120, 121].
4.4. Heterogenous and Interconvertible Stem Cell Sub-populations Coexist in Culture
It is becoming increasingly apparent that stem cells are heterogenous in culture. This is relatively well documented in mouse ES cells based on detection of various markers including Nanog [122–124], Pecam [125, 126], Rex1 [127] and Stella [128]. Interestingly, these sub-populations reflect cell states that are interconvertible, but share distinct patterns of gene expression and exhibit distinct differentiation properties. For example, expression profile comparisons of NANOG-high expressing cells vs. NANOG-low expressing cells revealed that NANOG-low cells expressed genes enriched in the PrE [123, 124]. Similarly, PECAM-positive ES cells preferentially contributed to the epiblast when injected into recipient embryos whereas PECAM-negative cells expressed markers of differentiation and primarily contributed to extraembryonic lineages [125, 126]. More recently, Canham and colleagues reported low levels of expression of Hex (a marker of distal/anterior visceral endoderm) in ES cells in culture [129]. Using a sensitive reporter, they showed that Hex-positive and Hex-negative cell populations are interconvertible. When allowed to differentiate, these cells have different properties: Hex-negative cells contribute to embryonic lineages, while Hex-positive cells differentiate into extraembryonic lineages, suggesting they likely represent an early PrE state. Similar observations have been made with the endoderm marker Sox17 [102] and the mesoderm marker Brachyury [130].
Of note, several subpopulations have also been described in mouse EpiSCs [131, 132] and in hES cells [133–136]. An emerging idea is that pluripotency is associated with metastable states, which are usually interconvertible, and which are defined by differences in gene expression and developmental potential. These fluctuations of (or metastable) cell states in a cultured population may extend to other early stem cell types, including TS and XEN cells. Indeed, XEN cells undergo morphological transitions [70] and heterogenously express some markers such as Sox7 [137]. These observations may reflect fluctuations in cell states and an inherent metastability. Moreover, we recently demonstrated that activation of BMP signaling in XEN cells drives their differentiation toward a stable state resembling the extraembryonic VE [138]. In this state, we observed fluctuations of a VE reporter transgene, indicating that at least two subpopulations coexist in BMP-treated XEN cell cultures, and that they are interconvertible. Additional studies will be required to address the origin and significance of the fluctuations that are observed and stably maintained in XEN cells, as well as establishing whether a similar heterogeneity exists in TS cell cultures.
Even though the molecular mechanisms and significance of such heterogeneity is not fully understood, it reveals that some cells in a cultured population may be more undifferentiated than others. This concept was first developed by Austin Smith and colleagues who defined pluripotency as a “ground state” in which, in the absence of GSK3 and FGF activity, ES cells remain undifferentiated. ES cells can be isolated and maintained in this naïve state of pluripotency in 2i conditions. FGF signaling is required to convert naïve cells to a state where they are primed to differentiate, a state that may resemble the more mature periimplantation epiblast and its ex vivo counterpart, the EpiSC. Indeed, ES cells cultured in the presence of an FGF inhibitor (or lacking Fgf4) are generally refractory to differentiation [139, 140]. Thus, both naïve ES cell subpopulation, as well as one poised for differentiation, may coexist in culture. Interestingly, ES cells can be converted into EpiSC simply by changing their culture conditions to those for EpiSCs [141]. Reciprocally, EpiSC can be reprogrammed to a more naïve ES-like state by overexpression of various factors such as Klf2, Klf4, Nanog, Nr5a2, Nr5a1 or Stat3 [40–144] or by culturing in ES cell conditions [80, 110, 119].
5. EMBRYOID BODY FORMATION: AN EX VIVO MODEL OF PERIIMPLANTATION DEVELOPMENT
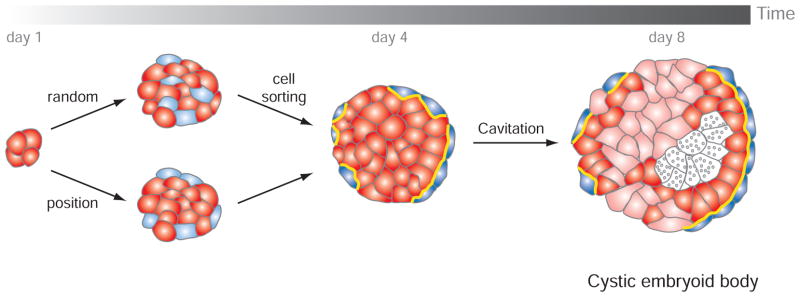
Pluripotent ES cells have the tremendous potential to generate virtually all the cell types of the body, and this feature directly impacts their use in regenerative medicine. Two key differentiation protocols have been defined and reflect the culture of cells in two or 3 dimensions (3D). In 3D dimensions, cells are cultured in non-adherent conditions promoting the formation of small aggregates called embryoid bodies (EB). EBs can also be generated from embryonal carcinoma (EC) cells derived from teratocarcinoma tumors [71, 145]. EB differentiation follows a stereotypical sequence of events that mimics the differentiation of the ICM, and apposition of tissue layers during normal periimplantation and early postimplantation stages of embryonic development (Fig. 5). As a consequence, EBs are commonly used as a first step in ES cell differentiation protocols.
Fig. 5. Schematic representation of the first differentiation events occurring in an embryoid body.
An initial morphological event is the formation of an outer layer of extraembryonic endoderm cells (blue). This could be the result from the random differentiation of ES cells (red) within the embryoid body and subsequent cell sorting, and/or from the differentiation of cells residing at the periphery and exposed to the outside environment. Extraembryonic endoderm cells secrete ECM components that will participate in the maturation of an epithelial epiblast and in the cavitation (grey dots indicate cell death). This process is usually not homogenous within an embryoid body and is accompanied by cell differentiation (pink cells).
In this section, we review the first events occurring in the process of EB formation in relation to early mammalian development in vivo. The first morphologically distinct differentiation within EBs, taking place between 3 to 4 days, involves the formation of an outer layer with extraembryonic endoderm (ExEn) characteristics. By 8 days of differentiation, a cavity has formed and internal cells are arranged into a pseudostratified columnar epithelium. These cystic embryoid bodies resemble early postimplantation embryos in which the proamniotic cavity is surrounded by embryonic ectoderm (epiblast) encapsulated by a layer of VE. Cells eventually differentiate into the three germ layers of the embryo-proper (ectoderm, mesoderm and gut endoderm).
5.1. Formation of the Outer Layer of ExEn
As previously discussed, a recent model of in vivo PrE formation and segregation involves: (i) positional inductive signals that may reinforce fate, (ii) sorting of cells already committed to PrE vs. EPI lineages, and (iii) selective apoptosis. If this model could be adapted to ExEn formation during EB differentiation, then one might predict that EBs are composed of a heterogeneous population of cells committed to either EPI- or ExEn-like fates that would eventually sort and resolve. The fact that ES cells in culture fluctuate between different states might lend support to such a model. Therefore, monitoring the dynamics of these fluctuations during the early steps of EB differentiation will likely be interest with respect modeling PrE formation.
It is difficult to find evidence suggesting that EBs can be used to model cell sorting. In EBs generated from ES cells lacking Dab2 [146] or Sox17 [102], ExEn cells differentiate but are found inside the EB. In contrast, in EBs comprised of wild-type cells, ExEn cells comprise the outer layer. These observations can be interpreted as a failure of ExEn cells to sort and/or be maintained in an outer location. However, a mixed population of undifferentiated and RA-treated ES cells segregates into two distinct compartments, suggesting differential adhesive properties [147]. Transmission electron microscopy analyses has not revealed any endodermal cells at the early stages of EB differentiation arguing in favor of the positional model, where outside cells would acquire ExEn identity [145]. By contrast, Rula and colleagues reported some DAB2-positive cells inside EBs preceding outer ExEn layer differentiation [147].
Thus, the question of a positional effect in EBs and its relevance to the embryo remains to be addressed. Interestingly, it was reported that an outer layer of ExEn cells is formed upon aggregation of GATA6-overexpressing ES cells [148]. Similarly, RA-treated EBs (from ES or EC cells) are composed by an outer layer of ExEn (see for review [149] indicating that the position of cells within the EB may be important for fate decision. Strikingly, some studies reported the transient expression of ExEn markers including Afp [150] and Gata6 [58] in almost all cells within the EB prior their specific localization of endodermal cells. Therefore, the formation of the ExEn layer during EB formation is a multistep processes which might reflect aspects of the in vivo segregation of PrE and EPI lineages within the ICM of the blastocyst.
5.2. Differentiation of the ExEn Layer
The identity of endodermal cell types that constitute the outer layer of an EB was originally described through ultra-structural studies by transmission electron microscopy [145]. Cells that form the outer layer of the EB are usually composed of a mixed population of both PE and VE cells. PE cells contain rough endoplasmic reticulum (ER) usually filled with a substance resembling the Reichart’s membrane. VE cells have a thinner ER, microvillosities and vacuoles. In addition, the outer layer exhibits non-homogenous expression of some VE markers such as Afp (and a reporter trans-gene Afp::GFP [151, 152]), suggesting that several ExEn subtypes may co-exist within an EB. Our recent analysis of EBs over a period of 3 to 7 days of differentiation revealed a lack of expression of Pdgfra or Sox7, both markers of nascent PrE, PE and proximal VE [58]. Taken together, these results suggest that the outer layer if EBs might most closely resemble the distal VE overlying the epiblast, which is also referred to as emVE.
Several signaling pathways have been proposed to be important for ExEn differentiation; they include BMP, FGF, LIF and Retinoic Acid (RA). When EBs are cultured in presence of LIF, the outer layer acquires a PrE-like identity, but cells do not differentiate into either PE or VE-like derivatives indicating that LIF pathway prevents PrE differentiation [153]. Exogenous addition of BMP4 triggers differentiation of the outer layer of EB towards a VE-like fate [154]. In 2D, RA promotes differentiation of F9 EC cells into PrE while when combined with dbAMPc cells adopt a PE-like identity. In contrast, RA-treated F9 EBs differentiate into VE (reviewed in [149]). ES cells stably expressing a dominant negative FGFR2 or mutant for Grb2 neither differentiate into VE-like, or form a columnar epithelium and cavitate [48, 155]. A similar phenotype was also observed when PI3K activity is inhibited [155] and the effect observed in ES cells expressing a dominant negative FGFR2 can be rescued when cultured in matrigel or in presence of Laminin-1 [156].
5.3. Cavitation
Cavitation in EB occurs by apopototic cell death and is thought to mimic the formation of the proamniotic cavity in the embryo soon after implantation. Cavitation occurs at the periphery of an EB and then progress inward. Cavitation is concomitant with the formation of a columnar epithelium resembling the embryonic ectoderm (Fig. 5). A comparison between two EC lines that differ in their ability to form cystic EBs has revealed some insight into the mechanisms of cavitation, and highlighted the intimate dialog between neighboring tissues [154, 157]. Notably, these studies have identified the BMP signaling pathway as important for cavitation and ExEn differentiation [154].
Formation of an ExEn epithelial cell layer on the surface of an EB leads to the deposition of ECM proteins basally. ECM appears to be required for the formation of an inner columnar ectoderm and EB cavitation, as absence of Lamc1 or Itgb1, results in a failure of cavitation and epithelialization, but does not affect ExEn layer formation [158, 159].
6. CONCLUDING REMARKS
In this review, we have discussed some of the parallels which can be drawn between pluripotent stem cell biology and early mammalian embryology. Understanding the mechanisms governing stem cell self-renewal vs. differentiation is not only key for facilitating progress in regenerative medicine, but also provides clues about the critical early cell fate decisions taking place in vivo within the embryo. Conversely, knowledge gained from dissecting the processes involved in lineage specification and diversification during embryonic development is likely to directly impact our understanding of stem cell biology. We have chosen to highlight studies in the mouse, the premier experimentally tractable mammalian model. However, one long-standing question in the field concerns the inherent similarities and notable differences taking place across mammalian species. We do not currently know how far, and at what level, events correlate, and how relevant knowledge gained from mouse model will be in understanding events taking place in higher mammals, including humans. Clearly, future studies involving cross-species comparisons will be required both in embryos and in stem cells for validating molecules and signals involved in cell lineage commitment, self-renewal and differentiation.
Acknowledgments
We thank Michel Cohen-Tannoudji, Ann Foley and Panos Xenopoulos for critical comments on the manuscript. Work in our laboratory is supported by the HFSP, National Institutes of Health (RO1-HD052115 and RO1-DK084391) and NYSTEM.
References
- 1.Arnold SJ, Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol. 2009;10(2):91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- 2.Rossant J, Tam PP. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136(5):701–13. doi: 10.1242/dev.017178. [DOI] [PubMed] [Google Scholar]
- 3.Kwon GS, Viotti M, Hadjantonakis AK. The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Dev Cell. 2008;15(4):509–20. doi: 10.1016/j.devcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larue L, Ohsugi M, Hirchenhain J, Kemler R. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc Natl Acad Sci USA. 1994;91(17):8263–7. doi: 10.1073/pnas.91.17.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Vries WN, Evsikov AV, Haac BE, et al. Maternal beta-catenin and E-cadherin in mouse development. Development. 2004;131(18):4435–45. doi: 10.1242/dev.01316. [DOI] [PubMed] [Google Scholar]
- 6.Stephenson RO, Yamanaka Y, Rossant J. Disorganized epithelial polarity and excess trophectoderm cell fate in preimplantation embryos lacking E-cadherin. Development. 2010;137(20):3383–91. doi: 10.1242/dev.050195. [DOI] [PubMed] [Google Scholar]
- 7.Tarkowski AK, Wroblewska J. Development of blastomeres of mouse eggs isolated at the 4- and 8-cell stage. J Embryol Exp Morphol. 1967;18(1):155–80. [PubMed] [Google Scholar]
- 8.Hillman N, Sherman MI, Graham C. The effect of spatial arrangement on cell determination during mouse development. J Embryol Exp Morphol. 1972;28(2):263–78. [PubMed] [Google Scholar]
- 9.Mintz B. In: Experimental genetic mosaicism in the mouse. Wolstenholme GEWOCM, editor. London: J. A. Churchill Ltd; 1965. [Google Scholar]
- 10.Handyside AH. Time of commitment of inside cells isolated from preimplantation mouse embryos. J Embryol Exp Morphol. 1978;45:37–53. [PubMed] [Google Scholar]
- 11.Hogan B, Tilly R. In vitro development of inner cell masses isolated immunosurgically from mouse blastocysts. I. Inner cell masses from 3.5-day p.c. blastocysts incubated for 24 h before immunosurgery. J Embryol Exp Morphol. 1978;45:93–105. [PubMed] [Google Scholar]
- 12.Spindle AI. Trophoblast regeneration by inner cell masses isolated from cultured mouse embryos. J Exp Zool. 1978;203(3):483–9. doi: 10.1002/jez.1402030315. [DOI] [PubMed] [Google Scholar]
- 13.Johnson MH, Ziomek CA. The foundation of two distinct cell lineages within the mouse morula. Cell. 1981;24(1):71–80. doi: 10.1016/0092-8674(81)90502-x. [DOI] [PubMed] [Google Scholar]
- 14.Plusa B, Frankenberg S, Chalmers A, et al. Downregulation of Par3 and aPKC function directs cells towards the ICM in the preimplantation mouse embryo. J Cell Sci. 2005;118(Pt 3):505–15. doi: 10.1242/jcs.01666. [DOI] [PubMed] [Google Scholar]
- 15.Nishioka N, Yamamoto S, Kiyonari H, et al. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech Dev. 2008;125(3–4):270–83. doi: 10.1016/j.mod.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML, et al. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. 2007;134(21):3827–36. doi: 10.1242/dev.010223. [DOI] [PubMed] [Google Scholar]
- 17.Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138(1):9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varelas X, Samavarchi-Tehrani P, Narimatsu M, et al. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-beta-SMAD pathway. Dev Cell. 2010;19(6):831–44. doi: 10.1016/j.devcel.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 19.Ralston A, Cox BJ, Nishioka N, et al. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development. 2010;137(3):395–403. doi: 10.1242/dev.038828. [DOI] [PubMed] [Google Scholar]
- 20.Ralston A, Rossant J. Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev Biol. 2008;313(2):614–29. doi: 10.1016/j.ydbio.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 21.Strumpf D, Mao CA, Yamanaka Y, et al. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132(9):2093–102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- 22.Jedrusik A, Parfitt DE, Guo G, et al. Role of Cdx2 and cell polarity in cell allocation and specification of trophectoderm and inner cell mass in the mouse embryo. Genes Dev. 2008;22(19):2692–706. doi: 10.1101/gad.486108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meissner A, Jaenisch R. Generation of nuclear transfer-derived pluripotent ES cells from cloned Cdx2-deficient blastocysts. Nature. 2006;439(7073):212–5. doi: 10.1038/nature04257. [DOI] [PubMed] [Google Scholar]
- 24.Wu G, Gentile L, Fuchikami T, et al. Initiation of trophectoderm lineage specification in mouse embryos is independent of Cdx2. Development. 2010;137(24):4159–69. doi: 10.1242/dev.056630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nichols J, Zevnik B, Anastassiadis K, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95(3):379–91. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 26.Dietrich JE, Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development. 2007;134(23):4219–31. doi: 10.1242/dev.003798. [DOI] [PubMed] [Google Scholar]
- 27.Berg DK, Smith CS, Pearton DJ, et al. Trophectoderm lineage determination in cattle. Dev Cell. 2011;20(2):244–55. doi: 10.1016/j.devcel.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 28.Plachta N, Bollenbach T, Pease S, Fraser SE, Pantazis P. Oct4 kinetics predict cell lineage patterning in the early mammalian embryo. Nat Cell Biol. 2011;13(2):117–23. doi: 10.1038/ncb2154. [DOI] [PubMed] [Google Scholar]
- 29.Zernicka-Goetz M. Cleavage pattern and emerging asymmetry of the mouse embryo. Nat Rev Mol Cell Biol. 2005;6(12):919–28. doi: 10.1038/nrm1782. [DOI] [PubMed] [Google Scholar]
- 30.Zernicka-Goetz M, Morris SA, Bruce AW. Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nat Rev Genet. 2009;10(7):467–77. doi: 10.1038/nrg2564. [DOI] [PubMed] [Google Scholar]
- 31.Ma GT, Roth ME, Groskopf JC, et al. GATA-2 and GATA-3 regulate trophoblast-specific gene expression in vivo. Development. 1997;124(4):907–14. doi: 10.1242/dev.124.4.907. [DOI] [PubMed] [Google Scholar]
- 32.Pandolfi PP, Roth ME, Karis A, et al. Targeted disruption of the GATA3 gene causes severe abnormalities in the nervous system and in fetal liver haematopoiesis. Nat Genet. 1995;11(1):40–4. doi: 10.1038/ng0995-40. [DOI] [PubMed] [Google Scholar]
- 33.Home P, Ray S, Dutta D, Bronshteyn I, Larson M, Paul S. GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J Biol Chem. 2009;284(42):28729–37. doi: 10.1074/jbc.M109.016840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossant J. Investigation of the determinative state of the mouse inner cell mass. II. The fate of isolated inner cell masses transferred to the oviduct. J Embryol Exp Morphol. 1975;33(4):991–1001. [PubMed] [Google Scholar]
- 35.Chazaud C, Yamanaka Y, Pawson T, Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev Cell. 2006;10(5):615–24. doi: 10.1016/j.devcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 36.Kurimoto K, Yabuta Y, Ohinata Y, et al. An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis. Nucleic Acids Res. 2006;34(5):e42. doi: 10.1093/nar/gkl050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plusa B, Piliszek A, Frankenberg S, Artus J, Hadjantonakis AK. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development. 2008;135(18):3081–91. doi: 10.1242/dev.021519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitsui K, Tokuzawa Y, Itoh H, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113(5):631–42. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 39.Messerschmidt DM, Kemler R. Nanog is required for primitive endoderm formation through a non-cell autonomous mechanism. Dev Biol. 2010;344(1):129–37. doi: 10.1016/j.ydbio.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 40.Silva J, Nichols J, Theunissen TW, et al. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138(4):722–37. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. The transcription factor GATA6 is essential for early extraembryonic development. Development. 1999;126(4):723–32. [PubMed] [Google Scholar]
- 42.Morrisey EE, Tang Z, Sigrist K, et al. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 1998;12(22):3579–90. doi: 10.1101/gad.12.22.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soudais C, Bielinska M, Heikinheimo M, et al. Targeted mutagenesis of the transcription factor GATA-4 gene in mouse embryonic stem cells disrupts visceral endoderm differentiation in vitro. Development. 1995;121(11):3877–88. doi: 10.1242/dev.121.11.3877. [DOI] [PubMed] [Google Scholar]
- 44.Cai KQ, Capo-Chichi CD, Rula ME, Yang DH, Xu XX. Dynamic GATA6 expression in primitive endoderm formation and maturation in early mouse embryogenesis. Dev Dyn. 2008;237(10):2820–9. doi: 10.1002/dvdy.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feldman B, Poueymirou W, Papaioannou VE, DeChiara TM, Goldfarb M. Requirement of FGF-4 for postimplantation mouse development. Science. 1995;267(5195):246–9. doi: 10.1126/science.7809630. [DOI] [PubMed] [Google Scholar]
- 46.Goldin SN, Papaioannou VE. Paracrine action of FGF4 during periimplantation development maintains trophectoderm and primitive endoderm. Genesis. 2003;36(1):40–7. doi: 10.1002/gene.10192. [DOI] [PubMed] [Google Scholar]
- 47.Arman E, Haffner-Krausz R, Chen Y, Heath JK, Lonai P. Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc Natl Acad Sci USA. 1998;95(9):5082–7. doi: 10.1073/pnas.95.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng AM, Saxton TM, Sakai R, et al. Mammalian Grb2 regulates multiple steps in embryonic development and malignant transformation. Cell. 1998;95(6):793–803. doi: 10.1016/s0092-8674(00)81702-x. [DOI] [PubMed] [Google Scholar]
- 49.Nichols J, Silva J, Roode M, Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136(19):3215–22. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamanaka Y, Lanner F, Rossant J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development. 2010;137(5):715–24. doi: 10.1242/dev.043471. [DOI] [PubMed] [Google Scholar]
- 51.Lanner F, Lee KL, Sohl M, et al. Heparan sulfation-dependent fibroblast growth factor signaling maintains embryonic stem cells primed for differentiation in a heterogeneous state. Stem Cells. 2010;28(2):191–200. doi: 10.1002/stem.265. [DOI] [PubMed] [Google Scholar]
- 52.Chai N, Patel Y, Jacobson K, McMahon J, McMahon A, Rappolee DA. FGF is an essential regulator of the fifth cell division in preimplantation mouse embryos. Dev Biol. 1998;198(1):105–15. doi: 10.1006/dbio.1997.8858. [DOI] [PubMed] [Google Scholar]
- 53.Rappolee DA, Basilico C, Patel Y, Werb Z. Expression and function of FGF-4 in peri-implantation development in mouse embryos. Development. 1994;120(8):2259–69. doi: 10.1242/dev.120.8.2259. [DOI] [PubMed] [Google Scholar]
- 54.Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9(21):2635–45. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- 55.Guo G, Huss M, Tong GQ, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18(4):675–85. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 56.Morris SA, Teo RT, Li H, Robson P, Glover DM, Zernicka-Goetz M. Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proc Natl Acad Sci USA. 2010;107(14):6364–9. doi: 10.1073/pnas.0915063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Threadgill DW, Dlugosz AA, Hansen LA, et al. Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science. 1995;269(5221):230–4. doi: 10.1126/science.7618084. [DOI] [PubMed] [Google Scholar]
- 58.Artus J, Panthier JJ, Hadjantonakis AK. A role for PDGF signaling in expansion of the extra-embryonic endoderm lineage of the mouse blastocyst. Development. 2010;137(20):3361–72. doi: 10.1242/dev.050864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gerbe F, Cox B, Rossant J, Chazaud C. Dynamic expression of Lrp2 pathway members reveals progressive epithelial differentiation of primitive endoderm in mouse blastocyst. Dev Biol. 2008;313(2):594–602. doi: 10.1016/j.ydbio.2007.10.048. [DOI] [PubMed] [Google Scholar]
- 60.Meilhac SM, Adams RJ, Morris SA, Danckaert A, Le Garrec JF, Zernicka-Goetz M. Active cell movements coupled to positional induction are involved in lineage segregation in the mouse blastocyst. Dev Biol. 2009;331(2):210–21. doi: 10.1016/j.ydbio.2009.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Artus J, Piliszek A, Hadjantonakis AK. The primitive endoderm lineage of the mouse blastocyst: Sequential transcription factor activation and regulation of differentiation by Sox17. Dev Biol. 2011;350(2):393–404. doi: 10.1016/j.ydbio.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang DH, Smith ER, Roland IH, et al. Disabled-2 is essential for endodermal cell positioning and structure formation during mouse embryogenesis. Dev Biol. 2002;251(1):27–44. doi: 10.1006/dbio.2002.0810. [DOI] [PubMed] [Google Scholar]
- 63.Smyth N, Vatansever HS, Meyer M, Frie C, Paulsson M, Edgar D. The targeted deletion of the LAMC1 gene. Ann N Y Acad Sci. 1998;857:283–6. doi: 10.1111/j.1749-6632.1998.tb10133.x. [DOI] [PubMed] [Google Scholar]
- 64.Liu J, He X, Corbett SA, et al. Integrins are required for the differentiation of visceral endoderm. J Cell Sci. 2009;122(Pt 2):233–42. doi: 10.1242/jcs.037663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fassler R, Meyer M. Consequences of lack of beta 1 integrin gene expression in mice. Genes Dev. 1995;9(15):1896–908. doi: 10.1101/gad.9.15.1896. [DOI] [PubMed] [Google Scholar]
- 66.Stephens LE, Sutherland AE, Klimanskaya IV, et al. Deletion of beta 1 integrins in mice results in inner cell mass failure and peri-implantation lethality. Genes Dev. 1995;9(15):1883–95. doi: 10.1101/gad.9.15.1883. [DOI] [PubMed] [Google Scholar]
- 67.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292(5819):154–6. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 68.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA. 1981;78(12):7634–8. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282(5396):2072–5. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 70.Kunath T, Arnaud D, Uy GD, et al. Imprinted X-inactivation in extra-embryonic endoderm cell lines from mouse blastocysts. Development. 2005;132(7):1649–61. doi: 10.1242/dev.01715. [DOI] [PubMed] [Google Scholar]
- 71.Martin GR, Wiley LM, Damjanov I. The development of cystic embryoid bodies in vitro from clonal teratocarcinoma stem cells. Dev Biol. 1977;61(2):230–44. doi: 10.1016/0012-1606(77)90294-9. [DOI] [PubMed] [Google Scholar]
- 72.Smith AG, Heath JK, Donaldson DD, et al. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336(6200):688–90. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- 73.Williams RL, Hilton DJ, Pease S, et al. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336(6200):684–7. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 74.Matsuda T, Nakamura T, Nakao K, et al. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18(15):4261–9. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12(13):2048–60. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cartwright P, McLean C, Sheppard A, Rivett D, Jones K, Dalton S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132(5):885–96. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- 77.Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115(3):281–92. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 78.Okita K, Yamanaka S. Intracellular signaling pathways regulating pluripotency of embryonic stem cells. Curr Stem Cell Res Ther. 2006;1(1):103–11. doi: 10.2174/157488806775269061. [DOI] [PubMed] [Google Scholar]
- 79.Ying QL, Wray J, Nichols J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453(7194):519–23. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hanna J, Markoulaki S, Mitalipova M, et al. Metastable pluripotent states in NOD-mouse-derived ESCs. Cell Stem Cell. 2009;4(6):513–24. doi: 10.1016/j.stem.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Buehr M, Meek S, Blair K, et al. Capture of authentic embryonic stem cells from rat blastocysts. Cell. 2008;135(7):1287–98. doi: 10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 82.Li P, Tong C, Mehrian-Shai R, et al. Germline competent embryonic stem cells derived from rat blastocysts. Cell. 2008;135(7):1299–310. doi: 10.1016/j.cell.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Erlebacher A, Price KA, Glimcher LH. Maintenance of mouse trophoblast stem cell proliferation by TGF-beta/activin. Dev Biol. 2004;275(1):158–69. doi: 10.1016/j.ydbio.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 84.Natale DR, Hemberger M, Hughes M, Cross JC. Activin promotes differentiation of cultured mouse trophoblast stem cells towards a labyrinth cell fate. Dev Biol. 2009;335(1):120–31. doi: 10.1016/j.ydbio.2009.08.022. [DOI] [PubMed] [Google Scholar]
- 85.Golding MC, Zhang L, Mann MR. Multiple epigenetic modifiers induce aggressive viral extinction in extraembryonic endoderm stem cells. Cell Stem Cel. 2010;6(5):457–67. doi: 10.1016/j.stem.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 86.Spruce T, Pernaute B, Di-Gregorio A, et al. An early developmental role for miRNAs in the maintenance of extraembryonic stem cells in the mouse embryo. Dev Cell. 2010;19(2):207–19. doi: 10.1016/j.devcel.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 87.Shevchenko AI, Demina VV, Mazurok NA, et al. Extraembryonic endoderm stem cell lines from common voles of the genus Microtus. Genetika. 2008;44(11):1477–85. [PubMed] [Google Scholar]
- 88.Chuykin I, Lapidus I, Popova E, et al. Characterization of trophoblast and extraembryonic endoderm cell lineages derived from rat preimplantation embryos. PLoS One. 2010;5(3):e9794. doi: 10.1371/journal.pone.0009794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Debeb BG, Galat V, Epple-Farmer J, et al. Isolation of Oct4-expressing extraembryonic endoderm precursor cell lines. PLoS One. 2009;4(9):e7216. doi: 10.1371/journal.pone.0007216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Galat V, Binas B, Iannaccone S, Postovit LM, Debeb BG, Iannaccone P. Developmental potential of rat extraembryonic stem cells. Stem Cells Dev. 2009;18(9):1309–18. doi: 10.1089/scd.2009.0115. [DOI] [PubMed] [Google Scholar]
- 91.Boyer LA, Lee TI, Cole MF, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–56. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Loh YH, Wu Q, Chew JL, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38(4):431–40. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 93.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24(4):372–6. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 94.Chambers I, Colby D, Robertson M, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113(5):643–55. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 95.Dejosez M, Krumenacker JS, Zitur LJ, et al. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell. 2008;133(7):1162–74. doi: 10.1016/j.cell.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dejosez M, Levine SS, Frampton GM, et al. Ronin/Hcf-1 binds to a hyperconserved enhancer element and regulates genes involved in the growth of embryonic stem cells. Genes Dev. 2010;24(14):1479–84. doi: 10.1101/gad.1935210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Niwa H, Toyooka Y, Shimosato D, et al. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123(5):917–29. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 98.Kidder BL, Palmer S. Examination of transcriptional networks reveals an important role for TCFAP2C, SMARCA4, and EOMES in trophoblast stem cell maintenance. Genome Res. 2010;20(4):458–72. doi: 10.1101/gr.101469.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ng RK, Dean W, Dawson C, et al. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat Cell Biol. 2008;10(11):1280–90. doi: 10.1038/ncb1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wen F, Tynan JA, Cecena G, et al. Ets2 is required for trophoblast stem cell self-renewal. Dev Biol. 2007;312(1):284–99. doi: 10.1016/j.ydbio.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lim CY, Tam WL, Zhang J, et al. Sall4 regulates distinct transcription circuitries in different blastocyst-derived stem cell lineages. Cell Stem Cell. 2008;3(5):543–54. doi: 10.1016/j.stem.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 102.Niakan KK, Ji H, Maehr R, et al. Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes Dev. 2010;24(3):312–26. doi: 10.1101/gad.1833510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Masui S, Nakatake Y, Toyooka Y, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9(6):625–35. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 104.Keramari M, Razavi J, Ingman KA, et al. Sox2 is essential for formation of trophectoderm in the preimplantation embryo. PLoS One. 2010;5(11):e13952. doi: 10.1371/journal.pone.0013952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17(1):126–40. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Elling U, Klasen C, Eisenberger T, Anlag K, Treier M. Murine inner cell mass-derived lineages depend on Sall4 function. Proc Natl Acad Sci USA. 2006;103(44):16319–24. doi: 10.1073/pnas.0607884103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Brons IG, Smithers LE, Trotter MW, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448(7150):191–5. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 108.Tesar PJ, Chenoweth JG, Brook FA, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448(7150):196–9. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 109.Najm FJ, Chenoweth JG, Anderson PD, et al. Isolation of epiblast stem cells from preimplantation mouse embryos. Cell Stem Cell. 2011;8(3):318–25. doi: 10.1016/j.stem.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bao S, Tang F, Li X, et al. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature. 2009;461(7268):1292–5. doi: 10.1038/nature08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chou YF, Chen HH, Eijpe M, et al. The growth factor environment defines distinct pluripotent ground states in novel blastocyst-derived stem cells. Cell. 2008;135(3):449–61. doi: 10.1016/j.cell.2008.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.De Felici M, Farini D, Dolci S. In or out stemness: comparing growth factor signalling in mouse embryonic stem cells and primordial germ cells. Curr Stem Cell Res Ther. 2009;4(2):87–97. doi: 10.2174/157488809788167391. [DOI] [PubMed] [Google Scholar]
- 113.Durcova-Hills G, Ainscough J, McLaren A. Pluripotential stem cells derived from migrating primordial germ cells. Differentiation. 2001;68(4–5):220–6. doi: 10.1046/j.1432-0436.2001.680409.x. [DOI] [PubMed] [Google Scholar]
- 114.Leitch HG, Blair K, Mansfield W, et al. Embryonic germ cells from mice and rats exhibit properties consistent with a generic pluripotent ground state. Development. 2010;137(14):2279–87. doi: 10.1242/dev.050427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Matsui Y, Zsebo K, Hogan BL. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell. 1992;70(5):841–7. doi: 10.1016/0092-8674(92)90317-6. [DOI] [PubMed] [Google Scholar]
- 116.Resnick JL, Bixler LS, Cheng L, Donovan PJ. Long-term proliferation of mouse primordial germ cells in culture. Nature. 1992;359(6395):550–1. doi: 10.1038/359550a0. [DOI] [PubMed] [Google Scholar]
- 117.Tada T, Tada M, Hilton K, et al. Epigenotype switching of imprintable loci in embryonic germ cells. Dev Genes Evol. 1998;207(8):551–61. doi: 10.1007/s004270050146. [DOI] [PubMed] [Google Scholar]
- 118.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 119.Greber B, Wu G, Bernemann C, et al. Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell. 2010;6(3):215–26. doi: 10.1016/j.stem.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 120.Ng HH, Surani MA. The transcriptional and signalling networks of pluripotency. Nat Cell Biol. 2011;13(5):490–6. doi: 10.1038/ncb0511-490. [DOI] [PubMed] [Google Scholar]
- 121.Pera MF, Tam PP. Extrinsic regulation of pluripotent stem cells. Nature. 2010;465(7299):713–20. doi: 10.1038/nature09228. [DOI] [PubMed] [Google Scholar]
- 122.Chambers I, Silva J, Colby D, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450(7173):1230–4. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 123.Kalmar T, Lim C, Hayward P, et al. Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 2009;7(7):e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Singh AM, Hamazaki T, Hankowski KE, Terada N. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells. 2007;25(10):2534–42. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- 125.Furusawa T, Ikeda M, Inoue F, Ohkoshi K, Hamano T, Tokunaga T. Gene expression profiling of mouse embryonic stem cell subpopulations. Biol Reprod. 2006;75(4):555–61. doi: 10.1095/biolreprod.105.049502. [DOI] [PubMed] [Google Scholar]
- 126.Furusawa T, Ohkoshi K, Honda C, Takahashi S, Tokunaga T. Embryonic stem cells expressing both platelet endothelial cell adhesion molecule-1 and stage-specific embryonic antigen-1 differentiate predominantly into epiblast cells in a chimeric embryo. Biol Reprod. 2004;70(5):1452–7. doi: 10.1095/biolreprod.103.024190. [DOI] [PubMed] [Google Scholar]
- 127.Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135(5):909–18. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- 128.Hayashi K, Lopes SM, Tang F, Surani MA. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3(4):391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Canham MA, Sharov AA, Ko MS, Brickman JM. Functional heterogeneity of embryonic stem cells revealed through translational amplification of an early endodermal transcript. PLoS Biol. 2010;8(5):e1000379. doi: 10.1371/journal.pbio.1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Suzuki A, Raya A, Kawakami Y, et al. Maintenance of embryonic stem cell pluripotency by Nanog-mediated reversal of mesoderm specification. Nat Clin Pract Cardiovasc Med. 2006;3 (Suppl 1):S114–22. doi: 10.1038/ncpcardio0442. [DOI] [PubMed] [Google Scholar]
- 131.Han DW, Tapia N, Joo JYC, et al. Epiblast stem cell subpopulations represent mouse embryos of distinct pregastrulation stages. Cell. 201;143(4):617–27. doi: 10.1016/j.cell.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 132.Hayashi K, Surani MA. Self-renewing epiblast stem cells exhibit continual delineation of germ cells with epigenetic reprogramming in vitro. Development. 2009;136(21):3549–56. doi: 10.1242/dev.037747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Enver T, Soneji S, Joshi C, et al. Cellular differentiation hierarchies in normal and culture-adapted human embryonic stem cells. Hum Mol Genet. 2005;14(21):3129–40. doi: 10.1093/hmg/ddi345. [DOI] [PubMed] [Google Scholar]
- 134.Fischer Y, Ganic E, Ameri J, Xian X, Johannesson M, Semb H. NANOG reporter cell lines generated by gene targeting in human embryonic stem cells. PLoS One. 2010;5(9) doi: 10.1371/journal.pone.0012533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hough SR, Laslett AL, Grimmond SB, Kolle G, Pera MF. A continuum of cell states spans pluripotency and lineage commitment in human embryonic stem cells. PLoS One. 2009;4(11):e7708. doi: 10.1371/journal.pone.0007708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Stewart MH, Bosse M, Chadwick K, Menendez P, Bendall SC, Bhatia M. Clonal isolation of hESCs reveals heterogeneity within the pluripotent stem cell compartment. Nat Methods. 2006;3(10):807–15. doi: 10.1038/nmeth939. [DOI] [PubMed] [Google Scholar]
- 137.Brown K, Legros S, Artus J, et al. A comparative analysis of extra-embryonic endoderm cell lines. PLoS One. 2010;5(8):e12016. doi: 10.1371/journal.pone.0012016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Artus J, Douvaras P, Piliszek A, Isern J, Baron MH, Hadjantonakis AK. BMP4 signalling directs primitive endoderm-derived XEN cells to an extraembryonic visceral endoderm identity. Dev Biol. doi: 10.1016/j.ydbio.2011.10.015. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kunath T, Saba-El-Leil MK, Almousailleakh M, Wray J, Meloche S, Smith A. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development. 2007;134(16):2895–902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- 140.Stavridis MP, Lunn JS, Collins BJ, Storey KG. A discrete period of FGF-induced Erk1/2 signalling is required for vertebrate neural specification. Development. 2007;134(16):2889–94. doi: 10.1242/dev.02858. [DOI] [PubMed] [Google Scholar]
- 141.Guo G, Yang J, Nichols J, et al. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136(7):1063–9. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Guo G, Smith A. A genome-wide screen in EpiSCs identifies Nr5a nuclear receptors as potent inducers of ground state pluripotency. Development. 2010;137(19):3185–92. doi: 10.1242/dev.052753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hall J, Guo G, Wray J, et al. Oct4 and LIF/Stat3 additively induce Kruppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell. 2009;5(6):597–609. doi: 10.1016/j.stem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 144.Yang J, van Oosten AL, Theunissen TW, Guo G, Silva JC, Smith A. Stat3 activation is limiting for reprogramming to ground state pluripotency. Cell Stem Cell. 2010;7(3):319–28. doi: 10.1016/j.stem.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Martin GR, Evans MJ. Differentiation of clonal lines of teratocarcinoma cells: formation of embryoid bodies in vitro. Proc Natl Acad Sci USA. 1975;72(4):1441–5. doi: 10.1073/pnas.72.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yang DH, Cai KQ, Roland IH, Smith ER, Xu XX. Disabled-2 is an epithelial surface positioning gene. J Biol Chem. 2007;282(17):13114–22. doi: 10.1074/jbc.M611356200. [DOI] [PubMed] [Google Scholar]
- 147.Rula ME, Cai KQ, Moore R, et al. Cell autonomous sorting and surface positioning in the formation of primitive endoderm in embryoid bodies. Genesis. 2007;45(6):327–38. doi: 10.1002/dvg.20298. [DOI] [PubMed] [Google Scholar]
- 148.Fujikura J, Yamato E, Yonemura S, et al. Differentiation of embryonic stem cells is induced by GATA factors. Genes Dev. 2002;16(7):784–9. doi: 10.1101/gad.968802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Soprano DR, Teets BW, Soprano KJ. Role of retinoic acid in the differentiation of embryonal carcinoma and embryonic stem cells. Vitam Horm. 2007;75:69–95. doi: 10.1016/S0083-6729(06)75003-8. [DOI] [PubMed] [Google Scholar]
- 150.Becker S, Casanova J, Grabel L. Localization of endoderm-specific mRNAs in differentiating F9 embryoid bodies. Mech Dev. 1992;37(1–2):3–12. doi: 10.1016/0925-4773(92)90010-h. [DOI] [PubMed] [Google Scholar]
- 151.Brown K, Doss MX, Legros S, Artus J, Hadjantonakis AK, Foley AC. eXtraembryonic ENdoderm (XEN) stem cells produce factors that activate heart formation. PLoS One. 2010;5(10):e13446. doi: 10.1371/journal.pone.0013446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hamazaki T, Terada N. In vitro differentiation of embryonic stem cells into hepatocytes. Methods Enzymol. 2003;365:277–87. doi: 10.1016/s0076-6879(03)65020-2. [DOI] [PubMed] [Google Scholar]
- 153.Murray P, Edgar D. The regulation of embryonic stem cell differentiation by leukaemia inhibitory factor (LIF) Differentiation. 2001;68(4–5):227–34. doi: 10.1046/j.1432-0436.2001.680410.x. [DOI] [PubMed] [Google Scholar]
- 154.Coucouvanis E, Martin GR. BMP signaling plays a role in visceral endoderm differentiation and cavitation in the early mouse embryo. Development. 1999;126(3):535–46. doi: 10.1242/dev.126.3.535. [DOI] [PubMed] [Google Scholar]
- 155.Chen Y, Li X, Eswarakumar VP, Seger R, Lonai P. Fibroblast growth factor (FGF) signaling through PI 3-kinase and Akt/PKB is required for embryoid body differentiation. Oncogene. 2000;19(33):3750–6. doi: 10.1038/sj.onc.1203726. [DOI] [PubMed] [Google Scholar]
- 156.Li X, Chen Y, Scheele S, et al. Fibroblast growth factor signaling and basement membrane assembly are connected during epithelial morphogenesis of the embryoid body. J Cell Biol. 2001;153(4):811–22. doi: 10.1083/jcb.153.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Coucouvanis E, Martin GR. Signals for death and survival: a two-step mechanism for cavitation in the vertebrate embryo. Cell. 1995;83(2):279–87. doi: 10.1016/0092-8674(95)90169-8. [DOI] [PubMed] [Google Scholar]
- 158.Li S, Harrison D, Carbonetto S, et al. Matrix assembly, regulation, and survival functions of laminin and its receptors in embryonic stem cell differentiation. J Cell Biol. 2002;157(7):1279–90. doi: 10.1083/jcb.200203073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Murray P, Edgar D. Regulation of programmed cell death by basement membranes in embryonic development. J Cell Biol. 2000;150(5):1215–21. doi: 10.1083/jcb.150.5.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]