Abstract
Benzene exposure is associated with acute myeloid leukemia (AML), myelodysplastic syndromes (MDS), and probably lymphoma and childhood leukemia. Biological plausibility for a causal role of benzene in these diseases comes from its toxicity to hematopoietic stem cells (HSC) or progenitor cells, from which all leukemias and related disorders arise. The effect of this toxicity is manifest as lowered blood counts (hematotoxicity), even in individuals occupationally exposed to low levels of benzene. Benzene can induce AML/MDS via several well-characterized pathways associated with these diseases. Through its metabolites, benzene induces multiple alterations that likely contribute to the leukemogenic process, and appears to operate via multiple modes of action. To improve mechanistic understanding and for risk assessment purposes, it may be possible to measure several of the key events in these modes of action in an in vitro model of the bone marrow stem cell niche. Even though benzene is leukemogenic at relatively low occupational levels of exposure, it seems unlikely that it is a major cause of leukemia in the general population exposed to benzene in the ppb range. Other established non-genetic causes of AML, e.g. smoking, ionizing radiation and cancer chemotherapy, also only explain about 20% of AML incidence, leaving ~80% unexplained. The question arises as to how to find the causes of the majority of de novo AMLs that remain unexplained. We propose that we should attempt to characterize the ‘exposome’ of human leukemia by using unbiased laboratory-based methods to find the unknown ‘environmental’ factors that contribute to leukemia etiology.
Keywords: Benzene, leukemia, myeloid, AML, mode of action, mechanism, blood, biomarker, metabolism, hydroquinone, stem cell niche
1. Introduction
Benzene is a ubiquitous environmental chemical that causes acute myeloid leukemia (AML), myelodysplastic syndromes (MDS), and probably other hematological cancers, such as non-Hodgkin lymphoma, which includes chronic lymphocytic leukemia (CLL) [1, 2]. Epidemiological studies have also provided evidence for an association with childhood leukemia [3, 4]. The mechanism by which benzene produces leukemia has not been fully elucidated, but comprehensive research over many years has revealed that benzene acts through multiple mechanisms. Recently, Meek and Klaunig presented a relatively simple, hypothesized mode of action with proposed key events for benzene-induced leukemia [5]. Below, we describe a more comprehensive “mode of action” based on our current understanding of benzene-induced leukemia that involves multiple key events and modifying factors. We discuss the implications of this mechanism for risk assessment and describe an unbiased approach to finding the causes of leukemia other than benzene.
2. Role of Metabolism in Benzene-Induced Leukemia
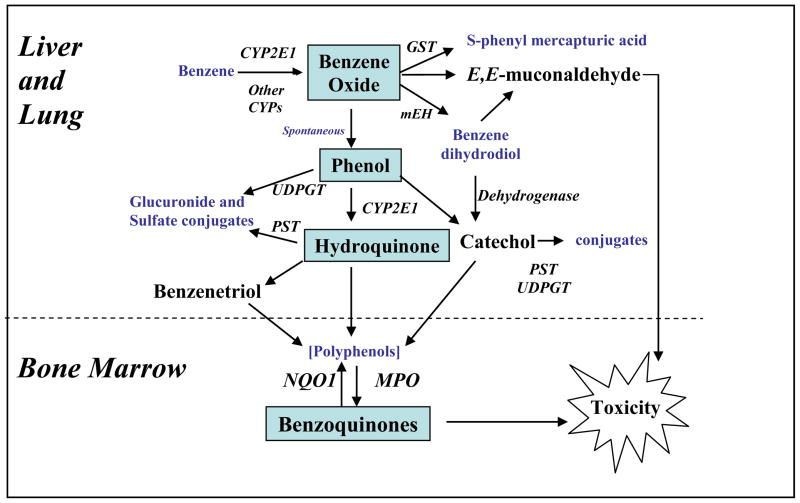
In order to become carcinogenic and cause leukemia, it is understood that benzene must be metabolized to toxic metabolites [6, 7], the general scheme of which is summarized in Figure 1. The initial metabolic step involves cytochrome P450 (CYP)-dependent oxidation of benzene to benzene oxide, which exists in equilibrium with its tautomer oxepin. Most benzene oxide spontaneously rearranges to phenol (PH), which is either excreted or further metabolized to hydroquinone (HQ), 1,4-benzoquinone (BQ) and 1,2,4-benzetriol (BT). The remaining benzene oxide is either hydrolyzed to produce catechol (CAT) and 1,2-benzoquinone or reacts with glutathione to produce S-phenylmercapturic acid (S-PMA). Metabolism of oxepin is thought to open the aromatic ring, yielding the reactive muconaldehydes and E,E-muconic acid (MA). Human exposures to benzene at air concentrations between 0.1 and 10 ppm,, result in urinary metabolite profiles with 70–85% PH, 5–10% each of HQ, MA and CAT, and less than 1% of S-PMA [8]. Benzene oxide, the benzoquinones, muconaldehydes, and benzene diol epoxides (formed from CYP oxidation of benzene dihydrodiol) are electrophiles that readily react with peptides and proteins [9–12] and can thereby interfere with cellular function [13].
Figure 1.
Metabolism of Benzene to Toxic Metabolites
The identification of metabolic susceptibility factors has confirmed the importance of metabolism in benzene toxicity. CYP2E1, which catalyzes the first step in benzene metabolism, represents a key metabolic susceptibility factor [14]. Other cytochrome P450s, such as CYP2F1 and CYP2A13 in the lung may also be involved in benzene metabolism [15–17]. Other metabolic susceptibility factors include epoxide hydrolase, glutathione-S-transferases (GSTT1, GSTM1), myeloperoxidase (MPO) and NAD(P)H:Quinone Oxidoreductase (NQO1) [18, 19]. In cellular studies, the levels of MPO and NQO1 have been suggested to modulate the toxicity of phenolic metabolites of benzene particularly in stromal cells where multiple cell types exist with varying enzyme activities [20, 21].
It remains unclear what role these different metabolites play in benzene carcinogenicity, but BQ formation from HQ via MPO in the bone marrow has been suggested as being key in benzene carcinogenicity as shown in Figure 1 [13]. Further, the BQ-detoxifying enzyme NQO1 protects mice against benzene-induced myelodysplasia [22, 23] and humans against benzene hematotoxicity [18, 19, 24]. However, this does not rule out adverse effects from other metabolites, such as the muconaldehydes [25, 26].
Mechanisms of Benzene-Induced Leukemia
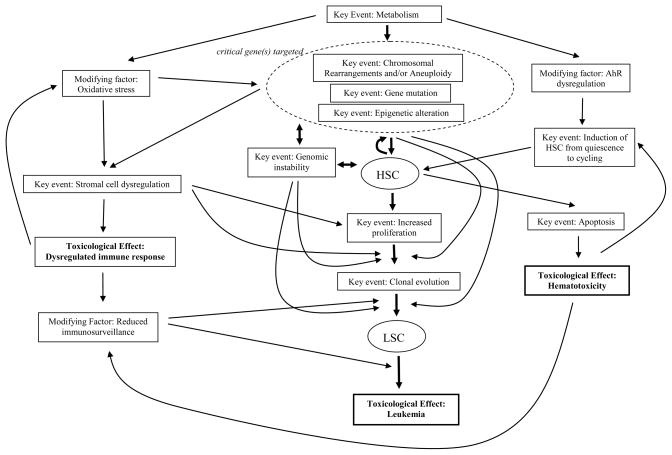
In order to produce leukemia, the reactive metabolites of benzene probably mutate a critical gene or set of genes related to proliferation and differentiation in human stem cells (HSC) by causing chromosome aberrations (aneuploidy, translocations, inversions, and deletions), aberrant mitotic recombination, gene mutations, and/or epigenetic alterations [4]. Ensuing genomic instability, or continued exposure to benzene, may result in the acquisition of additional alterations (Figure 2). Initiated HSC express these mutations as they enter the cycling state from quiescence, a process triggered by benzene exposure through the aryl hydrocarbon receptor (AhR) [27], generating leukemic stem cells (LSC in Figure 2). Concomitantly, adverse effects of benzene on the marrow stromal cells that regulate hematopoiesis can promote inappropriate survival/proliferation of the initiated HSC (Figure 2). Further, benzene metabolites and NQO1 deficiency can potentially disrupt the vascular stem cell niche by interfering with endothelial cell adhesion molecules [28, 29]. Oxidative stress resulting from benzene metabolism can cause both DNA damage and altered hematopoietic cell signaling. Reduced immunosurveillance could allow pre-leukemic clones to escape detection and elimination (Figure 2). Hence, there are multiple key events and modifying factors involved in benzene-induced leukemia suggesting that it has multiple modes of action.
Figure 2.
Probable Mechanism of Benzene-Induced Leukemia
3. Implications for the risk assessment of benzene
Quantification of the key events and modifying factors described above will be challenging and the generation of a biologically-based risk model for risk assessment purposes will require additional mechanistic research. Key events in benzene-induced leukemia include the induction of genetic and epigenetic changes in HSC, altered proliferation and differentiation of HSC, and apoptosis, with reduced immunosurveillance being a key modifying factor (Figure 2). It may be possible to measure several of these key events if a suitable in vitro model of the bone marrow and stem cell niche could be generated. Such a model, in which the HSC interact with stromal and endothelial cells, could perhaps be generated from induced pluripotent stem cells or fresh CD34-positive umbilical cord blood cells. In such a model it may be possible to measure genetic and epigenetic changes in the HSC using microfluidic technologies [30],along with determination of the rates of apoptosis, differentiation and proliferation. Several groups are developing in vitro models of the bone marrow niche [31–33].
Another approach to identifying key events in benzene-induced leukemogenesis would be to model dose-response relationships of particular biomarkers at different levels of benzene exposure in both humans and experimental animals. In cancer immunosurveillance, lymphocytes act as sentinels in recognizing and eliminating transformed cells [34], which in the case of leukemia, are the leukemic stem cells (LSC) (Figure 2). Suitable biomarkers for use in the dose-response modeling of benzene-induced leukemia would, therefore, include lymphocyte counts, genetic and epigenetic damage of relevance to leukemia, proliferation rates of blood stem and progenitor cells, and levels of HSC and LSC apoptosis. Several of these biomarkers have been measured in human populations exposed to a broad range of benzene concentrations.
Studies in Chinese workers have shown that benzene affects white blood cell and lymphocyte counts at low levels of occupational exposure and that there is no evidence of a threshold [18, 35]. Indeed, Lan and co-workers have reported that a supra-linear, or at least linear, dose-dependent effect is observed on lymphocyte counts at low occupational/high environmental levels of exposure [36]. Further, linear dose-dependent effects have been observed on colony formation from myeloid stem and progenitor cells [18]. More research is needed on the dose-dependent effects of benzene on genetic damage at lower levels of exposure. However, Bollatti et al. have shown that epigenetic changes in the global DNA methylation and methylation of the p15 gene (CDKN2B) promoter are linearly related to low levels of exposure to benzene [37].
Thus, for the biomarkers of mechanistic relevance to the leukemic process measured to date, the effects of benzene are present at low levels of exposure and increase monotonically with increasing doses, suggesting that a “no threshold” model is most appropriate for risk extrapolation.
4. Causes of Leukemia in the General Population
Even though benzene is probably leukemogenic at relatively low occupational levels of exposure, it seems unlikely to be a major cause of leukemia in the general population which is usually exposed in the few ppb range. Other known causes of AML, i.e. smoking, ionizing radiation, cancer chemotherapy and formaldehyde, also only explain perhaps ~20% of the non-genetic factors that influence AML incidence, leaving ~80% unexplained (Table 1). Genetic factors and family history also contribute to AML risk, but probably account for less than 10% of overall risk. The question, therefore, arises as to how to find the ‘environmental’ causes of the large proportion of AMLs that remains unexplained.
Table 1.
Established Non-Genetic Causes of Leukemia
| Known Causes | Percentage Range (%) | Probable (%) |
|---|---|---|
| Benzene | <1 – 5 | 1 |
| Ionizing Radiation (+ cosmic) | <1 – 20 | 2 |
| Cancer Chemotherapy | <10 | 4 |
| Smoking | 5 – 25 | 10 |
| Formaldehyde | <1 – 20 | 1 |
| Subtotal | (< 8 – 80) | 18 |
|
| ||
| Unknown Causes | 82 | |
Recent studies suggest that excess body weight and dietary factors are associated with increased risk of AML [38–40]. Obesity promotes chronic low-grade inflammation, altered immune responses through production of pro-inflammatory adipokines [41] and hormonal modulation, particularly in insulin. These pro-inflammatory and hormonal mediators can activate anti-apoptotic and proliferative signaling pathways in B- and T-cells that may promote tumor development. Recent studies also suggest a role for cholesterol and high-density lipoprotein (HDL) in regulating the proliferation of bone marrow myeloid progenitors [42] which may influence AML risk. However, few studies have been conducted to determine the influence of diet on the risk of adult AML.
A large U.S. cohort study found that meat intake was positively associated with risk of AML [43]. There was no evidence of an increased risk associated with preference for well-done meat [43]. These findings were consistent with those of a U.S. case-control study, particularly for beef consumption [40]. High meat intake may influence cancer risk through its effects on hormonal and metabolic responses to cell growth and survival, through exposure to dietary carcinogens such as polycyclic aromatic hydrocarbons, or by alteration of the gut microbiome, which may cause the elevated formation of the benzene metabolites, phenol and hydroquinone [44]. In support of the latter idea is the finding of high background levels of BQ-protein adducts in the blood of human control populations that may arise from the dietary ingestion of benzene’s phenolic metabolites and their formation as a side-product of tyrosine metabolism by the gut microflora [44, 45]. Thus, further investigation of the roles of diet and natural internal processes, such as inflammation, is warranted in studying the etiology of leukemia. However, such investigations would be highly challenging using traditional epidemiological approaches in which exposures are gleaned from self-reported questionnaires.
5. A Hypothesis-Free “Exposomic” Approach to Studying the Causes of Leukemia
In exploring possible unknown environmental determinants of leukemia etiology, we favor development and use of unbiased methods to determine these exposures using quantitative laboratory-based analyses. This approach aims to characterize the ‘exposome’, representing the totality of all exposures, first conceived by Wild in 2005 [46]. Under this view, the assessment of exposures should not be restricted to chemicals entering the body from air, water, food, smoking, etc., but should also include internally-generated toxicants produced by the gut flora, inflammation, oxidative stress, lipid peroxidation, infections, and other natural biological processes. In other words, we must focus upon the ‘internal chemical environment’ arising from all exposures to bioactive chemicals inside the body [47, 48].
Although it will be challenging to fully characterize the internal chemical environment throughout life, it should be possible to generate snapshots of exposomes during important stages of life by measuring a combination of omic endpoints and legacy biomarkers in repeated blood samples [47, 48]. We refer to this strategy as ‘top-down exposomics’ and stress its unbiased approach to discovering the causes of disease. In top-down exposomics, the exposome would comprise a profile of the most prominent classes of toxicants that are known to cause disease, namely, reactive electrophiles, endocrine (hormone) disruptors, modulators of immune responses, agents that bind to cellular receptors, and metals [47]. Exposures to these agents can be monitored in the blood either by direct measurement or by looking for their effects on physiological processes (such as receptor-based signaling). Some “omics” methods also offer unbiased means of characterizing exposures to drugs [49], metals (metallomics) [50], small metabolic products (metabolomics) [51] and reactive electrophiles (adductomics) [52]. These omic technologies could help generate signatures of exposures in the blood. By comparing exposomic patterns between de novo leukemia cases and controls, preferably from longitudinal studies, it should be possible to identify key exposures associated with the leukemia and then to develop appropriate interventions for reducing those exposures.
Acknowledgments
This work was supported in part by the U.S. Environmental Protection Agency under order number EP09H000461 (Disclaimer: The views expressed in this article are those of the authors and do not necessarily reflect the views or policies of the U.S. Environmental Protection Agency). Additional support was provided by NIH grants P42ES004705 (MTS), P42ES005948 (SMR), and U54ES016115 (SMR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Khalade A, Jaakkola MS, Pukkala E, Jaakkola JJ. Exposure to benzene at work and the risk of leukemia: a systematic review and meta-analysis. Environ Health. 2010;9:31. doi: 10.1186/1476-069X-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steinmaus C, Smith AH, Jones RM, Smith MT. Meta-analysis of benzene exposure and non-Hodgkin lymphoma: biases could mask an important association. Occup Environ Med. 2008;65(6):371–378. doi: 10.1136/oem.2007.036913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eden T. Aetiology of childhood leukaemia. Cancer treatment reviews. 2010;36(4):286–297. doi: 10.1016/j.ctrv.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 4.Smith MT. Advances in understanding benzene health effects and susceptibility. Annual review of public health. 2010;31:133–148. doi: 10.1146/annurev.publhealth.012809.103646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meek ME, Klaunig JE. Proposed mode of action of benzene-induced leukemia: Interpreting available data and identifying critical data gaps for risk assessment. Chem Biol Interact. 2010;184(1–2):279–285. doi: 10.1016/j.cbi.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Snyder R. Xenobiotic metabolism and the mechanism(s) of benzene toxicity. Drug metabolism reviews. 2004;36(3–4):531–547. doi: 10.1081/dmr-200033445. [DOI] [PubMed] [Google Scholar]
- 7.Ross D. The role of metabolism and specific metabolites in benzene-induced toxicity: evidence and issues. Journal of toxicology and environmental health Part A. 2000;61(5–6):357–372. doi: 10.1080/00984100050166361. [DOI] [PubMed] [Google Scholar]
- 8.Kim S, Vermeulen R, Waidyanatha S, Johnson BA, Lan Q, Smith MT, Zhang L, Li G, Shen M, Yin S, Rothman N, Rappaport SM. Modeling human metabolism of benzene following occupational and environmental exposures. Cancer Epidemiol Biomarkers Prev. 2006;15(11):2246–2252. doi: 10.1158/1055-9965.EPI-06-0262. [DOI] [PubMed] [Google Scholar]
- 9.Bechtold WE, Willis JK, Sun JD, Griffith WC, Reddy TV. Biological markers of exposure to benzene: S-phenylcysteine in albumin. Carcinogenesis. 1992;13(7):1217–1220. doi: 10.1093/carcin/13.7.1217. [DOI] [PubMed] [Google Scholar]
- 10.Henderson AP, Bleasdale C, Delaney K, Lindstrom AB, Rappaport SM, Waidyanatha S, Watson WP, Golding BT. Evidence for the formation of Michael adducts from reactions of (E,E)-muconaldehyde with glutathione and other thiols. Bioorganic Chemistry. 2005;33(5):363–373. doi: 10.1016/j.bioorg.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 11.McDonald TA, Waidyanatha S, Rappaport SM. Production of benzoquinone adducts with hemoglobin and bone-marrow proteins following administration of [13C6]benzene to rats. Carcinogenesis. 1993;14(9):1921–1925. doi: 10.1093/carcin/14.9.1921. [DOI] [PubMed] [Google Scholar]
- 12.Waidyanatha S, Rappaport SM. Investigation of cysteinyl protein adducts of benzene diolepoxide. Chemico-biological interactions. 2005;153–154:261–266. doi: 10.1016/j.cbi.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 13.Smith MT. The mechanism of benzene-induced leukemia: a hypothesis and speculations on the causes of leukemia. Environ Health Perspect. 1996;104(Suppl 6):1219–1225. doi: 10.1289/ehp.961041219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valentine JL, Lee SS, Seaton MJ, Asgharian B, Farris G, Corton JC, Gonzalez FJ, Medinsky MA. Reduction of benzene metabolism and toxicity in mice that lack CYP2E1 expression. Toxicology and applied pharmacology. 1996;141(1):205–213. doi: 10.1006/taap.1996.0277. [DOI] [PubMed] [Google Scholar]
- 15.Powley MW, Carlson GP. Cytochromes P450 involved with benzene metabolism in hepatic and pulmonary microsomes. Journal of biochemical and molecular toxicology. 2000;14(6):303–309. doi: 10.1002/1099-0461(2000)14:6<303::AID-JBT2>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 16.Rappaport SM, Kim S, Lan Q, Vermeulen R, Waidyanatha S, Zhang L, Li G, Yin S, Hayes RB, Rothman N, Smith MT. Evidence that Humans Metabolize Benzene via Two Pathways. Environmental Health Perspectives. 2009;117(6):946–952. doi: 10.1289/ehp.0800510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheets PL, Yost GS, Carlson GP. Benzene metabolism in human lung cell lines BEAS-2B and A549 and cells overexpressing CYP2F1. J Biochem Mol Toxicol. 2004;18(2):92–99. doi: 10.1002/jbt.20010. [DOI] [PubMed] [Google Scholar]
- 18.Lan Q, Zhang L, Li G, Vermeulen R, Weinberg RS, Dosemeci M, Rappaport SM, Shen M, Alter BP, Wu Y, Kopp W, Waidyanatha S, Rabkin C, Guo W, Chanock S, Hayes RB, Linet M, Kim S, Yin S, Rothman N, Smith MT. Hematotoxicity in workers exposed to low levels of benzene. Science. 2004;306(5702):1774–1776. doi: 10.1126/science.1102443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross D, Zhou H. Relationships between metabolic and non-metabolic susceptibility factors in benzene toxicity. Chem Biol Interact. 2010;184(1–2):222–228. doi: 10.1016/j.cbi.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smart RC, Zannoni VG. DT-diaphorase and peroxidase influence the covalent binding of the metabolites of phenol, the major metabolite of benzene. Molecular pharmacology. 1984;26(1):105–111. [PubMed] [Google Scholar]
- 21.Thomas DJ, Sadler A, Subrahmanyam VV, Siegel D, Reasor MJ, Wierda D, Ross D. Bone marrow stromal cell bioactivation and detoxification of the benzene metabolite hydroquinone: Comparison of macrophages and fibroblastoid cells. Molecular Pharmacology. 1990;37(2):255–262. [PubMed] [Google Scholar]
- 22.Iskander K, Jaiswal AK. Quinone oxidoreductases in protection against myelogenous hyperplasia and benzene toxicity. Chem Biol Interact. 2005;153–154:147–157. doi: 10.1016/j.cbi.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 23.Long DJ, 2nd, Gaikwad A, Multani A, Pathak S, Montgomery CA, Gonzalez FJ, Jaiswal AK. Disruption of the NAD(P)H:quinone oxidoreductase 1 (NQO1) gene in mice causes myelogenous hyperplasia. Cancer Res. 2002;62(11):3030–3036. [PubMed] [Google Scholar]
- 24.Rothman N, Smith MT, Hayes RB, Traver RD, Hoener B, Campleman S, Li GL, Dosemeci M, Linet M, Zhang L, Xi L, Wacholder S, Lu W, Meyer KB, Titenko-Holland N, Stewart JT, Yin S, Ross D. Benzene poisoning, a risk factor for hematological malignancy, is associated with the NQO1 609C-->T mutation and rapid fractional excretion of chlorzoxazone. Cancer Res. 1997;57(14):2839–2842. [PubMed] [Google Scholar]
- 25.Witz G, Zhang Z, Goldstein BD. Reactive ring-opened aldehyde metabolites in benzene hematotoxicity. Environ Health Perspect. 1996;104(Suppl 6):1195–1199. doi: 10.1289/ehp.961041195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golding BT, Barnes ML, Bleasdale C, Henderson AP, Jiang D, Li X, Mutlu E, Petty HJ, Sadeghi MM. Modeling the formation and reactions of benzene metabolites. Chem Biol Interact. 2010;184(1–2):196–200. doi: 10.1016/j.cbi.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Hirabayashi Y, Inoue T. Benzene-induced bone-marrow toxicity: a hematopoietic stem-cell-specific, aryl hydrocarbon receptor-mediated adverse effect. Chem Biol Interact. 2010;184(1–2):252–258. doi: 10.1016/j.cbi.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 28.Zhou H, Dehn DL, Kepa JK, Siegel D, Scott D, Tan W, Ross D. NAD(P)H:quinone oxidoreductase 1-compromised human bone marrow endothelial cells exhibit decreased adhesion molecule expression and CD34+ hematopoietic cell adhesion. J Pharmacol Exp Ther. 2010;334(1):260–268. doi: 10.1124/jpet.110.167841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou H, Kepa JK, Siegel D, Miura S, Hiraki Y, Ross D. Benzene metabolite hydroquinone up-regulates chondromodulin-I and inhibits tube formation in human bone marrow endothelial cells. Mol Pharmacol. 2009;76(3):579–587. doi: 10.1124/mol.109.057323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shuga J, Zeng Y, Novak R, Mathies RA, Hainaut P, Smith MT. Selected technologies for measuring acquired genetic damage in humans. Environ Mol Mutagen. 2010;51(8–9):851–870. doi: 10.1002/em.20630. [DOI] [PubMed] [Google Scholar]
- 31.Di Maggio N, Piccinini E, Jaworski M, Trumpp A, Wendt DJ, Martin I. Toward modeling the bone marrow niche using scaffold-based 3D culture systems. Biomaterials. 2011;32(2):321–329. doi: 10.1016/j.biomaterials.2010.09.041. [DOI] [PubMed] [Google Scholar]
- 32.Tan J, Liu T, Hou L, Meng W, Wang Y, Zhi W, Deng L. Maintenance and expansion of hematopoietic stem/progenitor cells in biomimetic osteoblast niche. Cytotechnology. 2010;62(5):439–448. doi: 10.1007/s10616-010-9297-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Butler JM, Nolan DJ, Vertes EL, Varnum-Finney B, Kobayashi H, Hooper AT, Seandel M, Shido K, White IA, Kobayashi M, Witte L, May C, Shawber C, Kimura Y, Kitajewski J, Rosenwaks Z, Bernstein ID, Rafii S. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell stem cell. 2010;6(3):251–264. doi: 10.1016/j.stem.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langenkamp U, Siegler U, Jorger S, Diermayr S, Gratwohl A, Kalberer CP, Wodnar-Filipowicz A. Human acute myeloid leukemia CD34+CD38− stem cells are susceptible to allorecognition and lysis by single KIR-expressing natural killer cells. Haematologica. 2009;94(11):1590–1594. doi: 10.3324/haematol.2009.005967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qu Q, Shore R, Li G, Jin X, Chen LC, Cohen B, Melikian AA, Eastmond D, Rappaport SM, Yin S, Li H, Waidyanatha S, Li Y, Mu R, Zhang X, Li K. Hematological changes among Chinese workers with a broad range of benzene exposures. Am J Ind Med. 2002;42(4):275–285. doi: 10.1002/ajim.10121. [DOI] [PubMed] [Google Scholar]
- 36.Lan Q, Vermeulen R, Zhang L, Li G, Rosenberg PS, Alter BP, Shen M, Rappaport SM, Weinberg RS, Chanock S, Waidyanatha S, Rabkin C, Hayes RB, Linet M, Kim S, Yin S, Rothman N, Smith MT. Benzene Exposure and Hematotoxicity: Response. Science. 2006;312(5776):998–998. doi: 10.1126/science.312.5776.998b. [DOI] [PubMed] [Google Scholar]
- 37.Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, Byun HM, Jiang J, Marinelli B, Pesatori AC, Bertazzi PA, Yang AS. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67(3):876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 38.Larsson SC, Wolk A. Overweight and obesity and incidence of leukemia: a meta-analysis of cohort studies. Int J Cancer. 2008;122(6):1418–1421. doi: 10.1002/ijc.23176. [DOI] [PubMed] [Google Scholar]
- 39.Kasim K, Levallois P, Abdous B, Auger P, Johnson KC. Lifestyle factors and the risk of adult leukemia in Canada. Cancer Causes Control. 2005;16(5):489–500. doi: 10.1007/s10552-004-7115-1. [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Moysich KB, Baer MR, Weiss JR, Brasure J, Graham S, McCann SE. Intakes of selected food groups and beverages and adult acute myeloid leukemia. Leuk Res. 2006;30(12):1507–1515. doi: 10.1016/j.leukres.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 41.Marti A, Marcos A, Martinez JA. Obesity and immune function relationships. Obes Rev. 2001;2(2):131–140. doi: 10.1046/j.1467-789x.2001.00025.x. [DOI] [PubMed] [Google Scholar]
- 42.Yvan-Charvet L, Pagler T, Gautier EL, Avagyan S, Siry RL, Han S, Welch CL, Wang N, Randolph GJ, Snoeck HW, Tall AR. ATP-binding cassette transporters and HDL suppress hematopoietic stem cell proliferation. Science. 2010;328(5986):1689–1693. doi: 10.1126/science.1189731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma X, Park Y, Mayne ST, Wang R, Sinha R, Hollenbeck AR, Schatzkin A, Cross AJ. Diet, lifestyle, and acute myeloid leukemia in the NIH-AARP cohort. Am J Epidemiol. 2010;171(3):312–322. doi: 10.1093/aje/kwp371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDonald T, Holland N, Skibola C, Duramad P, Smith M. Hypothesis: phenol and hydroquinone derived mainly from diet and gastrointestinal flora activity are causal factors in leukemia. Leukemia. 2001;15(1):10–20. doi: 10.1038/sj.leu.2401981. [DOI] [PubMed] [Google Scholar]
- 45.Yeowell-O’Connell K, Rothman N, Waidyanatha S, Smith MT, Hayes RB, Li G, Bechtold WE, Dosemeci M, Zhang L, Yin S, Rappaport SM. Protein adducts of 1,4-benzoquinone and benzene oxide among smokers and nonsmokers exposed to benzene in China. Cancer Epidemiol Biomarkers Prev. 2001;10(8):831–838. [PubMed] [Google Scholar]
- 46.Wild CP. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev. 2005;14(8):1847–1850. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- 47.Rappaport SM, Smith MT. Epidemiology. Environment and disease risks. Science. 2010;330(6003):460–461. doi: 10.1126/science.1192603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rappaport SM. Implications of the exposome for exposure science. J Expo Sci Environ Epidemiol. 2010 doi: 10.1038/jes.2010.50. epub. [DOI] [PubMed] [Google Scholar]
- 49.Bushel PR, Heinloth AN, Li J, Huang L, Chou JW, Boorman GA, Malarkey DE, Houle CD, Ward SM, Wilson RE, Fannin RD, Russo MW, Watkins PB, Tennant RW, Paules RS. Blood gene expression signatures predict exposure levels. Proc Natl Acad Sci U S A. 2007;104(46):18211–18216. doi: 10.1073/pnas.0706987104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mounicou S, Szpunar J, Lobinski R. Metallomics: the concept and methodology. Chemical Society reviews. 2009;38(4):1119–1138. doi: 10.1039/b713633c. [DOI] [PubMed] [Google Scholar]
- 51.Bictash M, Ebbels TM, Chan Q, Loo RL, Yap IK, Brown IJ, de Iorio M, Daviglus ML, Holmes E, Stamler J, Nicholson JK, Elliott P. Opening up the “Black Box”: metabolic phenotyping and metabolome-wide association studies in epidemiology. Journal of clinical epidemiology. 2010;63(9):970–979. doi: 10.1016/j.jclinepi.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H, Grigoryan H, Funk WE, Lu SS, Rose S, Williams ER, Rappaport SM. Adductomics of Human Serum Albumin by Fixed-Step Selected Reaction Monitoring. Mol Cell Proteomics. 2010 doi: 10.1074/mcp.M110.004606. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]