Abstract
BACKGROUND & AIMS
Wilson disease is a severe disorder of copper metabolism caused by mutations in ATP7B, which encodes a copper-transporting adenosine triphosphatase. The disease presents with a variable phenotype that complicates the diagnostic process and treatment. Little is known about the mechanisms that contribute to the different phenotypes of the disease.
METHODS
We analyzed 28 variants of ATP7B from patients with Wilson disease that affected different functional domains; the gene products were expressed using the baculovirus expression system in Sf9 cells. Protein function was analyzed by measuring catalytic activity and copper (64Cu) transport into vesicles. We studied intracellular localization of variants of ATP7B that had measurable transport activities and were tagged with green fluorescent protein in mammalian cells using confocal laser scanning microscopy.
RESULTS
Properties of ATP7B variants with pathogenic amino-acid substitution varied greatly even if substitutions were in the same functional domain. Some variants had complete loss of catalytic and transport activity, whereas others lost transport activity but retained phosphor-intermediate formation or had partial losses of activity. In mammalian cells, transport-competent variants differed in stability and subcellular localization.
CONCLUSIONS
Variants in ATP7B associated with Wilson disease disrupt the protein’s transport activity, result in its mislocalization, and reduce its stability. Single assays are insufficient to accurately predict the effects of ATP7B variants the function of its product and development of Wilson disease. These findings will contribute to our understanding of genotype–phenotype correlation and mechanisms of disease pathogenesis.
Keywords: P-Type ATPase, Genetic Analysis, Liver Disease, Golgi
Wilson disease (WD) is a hereditary disease due to mutations of the copper-transporting P-type adenosine triphosphatase (ATPase) ATP7B. WD is associated with copper accumulation resulting in liver damage and/or neurologic symptoms.1,2 The manifestations of liver disease vary from clinically asymptomatic with only biochemical abnormalities, to acute liver failure.3 Similarly, the spectrum of neurologic manifestations ranges from normal or mild disturbances to a rapid and severe progression of neurologic disability.4 The mechanisms behind this variability are likely to be complex because even monozygotic twins can have different disease manifestations.5 Genetic studies showed some association between several mutations and the age of onset/disease severity6–9; however, robust genotype–phenotype correlation in the case of missense mutations has not been found so far, although complete loss of protein expression due to non–sense mutations is expected to result in a more severe phenotype. This study focuses on one potential source of phenotypic variability—the effect of mutations in ATP7B on the function of the corresponding Cu-transporting Wilson ATPase.
To date, >500 different mutations associated with WD are known (http://www.wilsondisease.med.ualberta.ca/database.asp).10 The high prevalence of compound heterozygosity prevents a clear correlation between genotype and phenotype in affected individuals. A large fraction of mutations are missense mutations producing a single amino-acid change in Wilson ATPase ATP7B. ATP7B is a large membrane protein located in the trans-Golgi network (TGN). In normal liver, ATP7B has these 2 functions: it transports copper into the TGN for incorporation into ceruloplasmin and exports excess copper by sequestering metal in vesicles for subsequent biliary excretion. The second function requires ATP7B trafficking from TGN to endocytic vesicles in response to elevation of intracellular copper concentration.
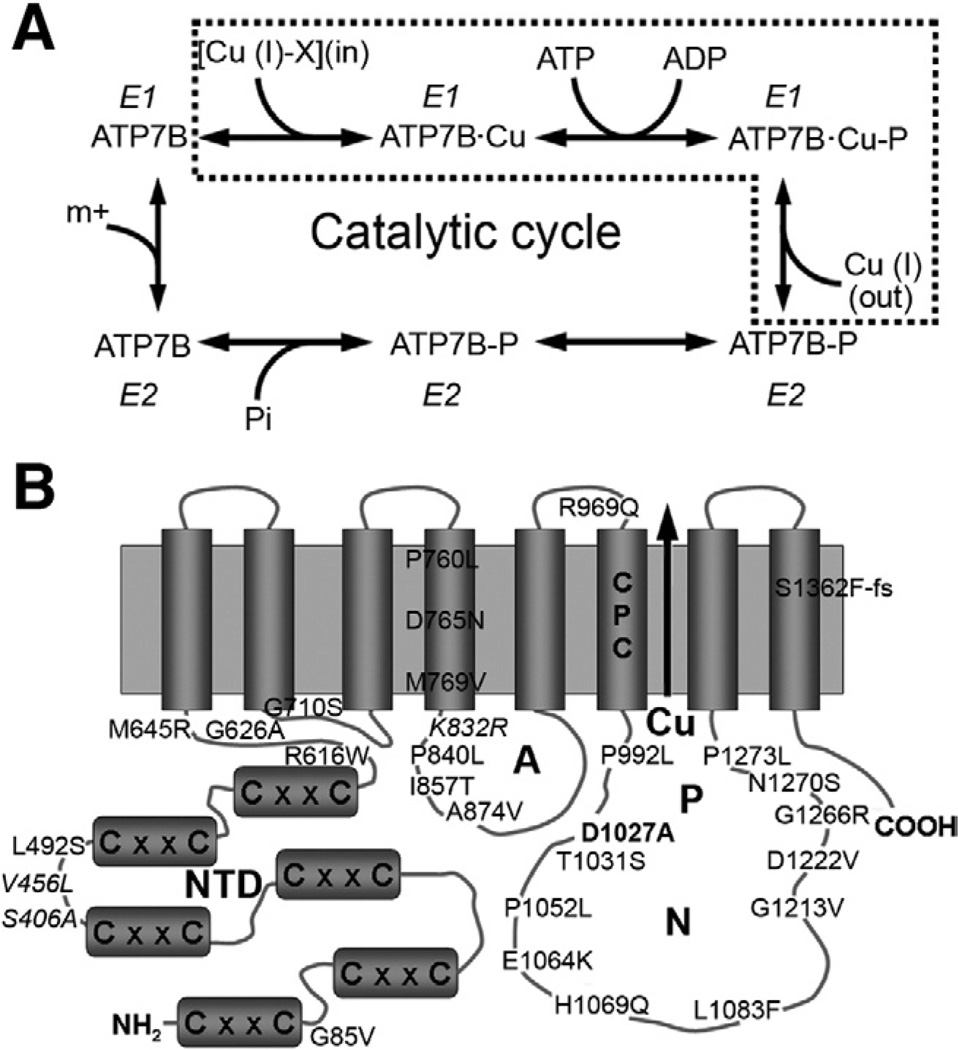
The ATP7B-mediated transport of copper involves several steps. First, ATP7B binds copper via its cytosolic N-terminal domain and ATP via the nucleotide-binding domain. ATP is then hydrolyzed, and ATP7B becomes transiently phosphorylated at the residue D1027 located in the P-domain (catalytic phosphorylation). Subsequent dephosphorylation releases energy necessary to transfer copper across membrane (transport step) (Figure 1A and B). Each of these steps can be affected by WD-causing mutations.11 The effect could be severe, resulting in complete loss of ATP7B function, if mutated residues are critical for binding of ATP or copper and/or conformational transitions during catalysis. The inactivation of ATP7B could also be partial if mutations diminish affinity for substrates, slow down conformational transitions, or interfere with precise protein targeting to TGN or vesicles. Understanding of phenotypic diversity in WD requires knowledge of how causative mutations alter protein stability, activity, and localization in the cell.
Figure 1.
(A) Catalytic cycle of ATP7B (the dotted frame highlights partial reactions, relevant for this study). (B) Transmembrane organization of ATP7B and representation of mutants and variants examined in this study. The 6 metal-binding sites in the N-terminal Cu-binding unit domain (NTD) of ATP7B are indicated by letters CxxC. ATP is bound to the nucleotide binding domain (N) and phosphorylated during ATP hydrolysis. The site of ATP-hydrolysis is the invariant residue D1027 (D1027A mutant = bold). The gene variants S406A, V456L, and K832R are presented in italic type. The CPC motif located in the sixth transmembrane domain is thought to form the intramembrane copper-binding site(s). A, actuator domain; P, phosphorylation domain.
Presently, for most of the WD-causing mutations, such detailed information is not available. Yeast complementation assay was used to segregate ATP7B mutants into severe and mild categories based on their ability to restore the growth of yeast strain that lacks the endogenous copper transporter.12–14 Studies in mammalian cells revealed decreased protein levels and mislocalization for several mutants,15–17 with a surprisingly small effect on copper transport, which was evaluated indirectly.17 So far, no studies have directly assessed the transport or catalytic activity of disease-causing mutants. Such information is necessary for future mechanism-based attempts to correct the ATP7B function. Here, we directly measured catalytic and transport activity of 28 ATP7B variants representing all protein domains (Table 1, Figure 1B) and tested the intracellular localization for mutants, for which transport activity was observed. The mutants and variants were chosen based on clinical findings (see WD database10) and recent literature; gene variants found in the population with no WD symptoms were also characterized.
Table 1.
Copper Transport and Phosphorylation Activity of Wild-Type, Mutant, and Variant ATP7B
| Protein domain | Cu-uptake at 120 min, nmol/mg protein (SD) |
Transport | Phosphorylation activity, % of wt (SD) |
Activity | |
|---|---|---|---|---|---|
| wt-ATP7B | — | 1.33 (0.18) | Normal | 100.0 | Normal |
| Artificial mutant of phosphorylation site | |||||
| D1027A | P-domain | 0.04 (0.01) | No | 25.6 (8.4) | Inactive |
| Human mutants | |||||
| G85V | NTD | 0.11 (0.02) | Low | 27.3 (25.3) | Inactive |
| L492S | NTD | 0.11 (0.02) | Low | 43.6 (16.5) | Inactive |
| R616W | NTD | 0.14 (0.01) | Low | 223.1 (28.2) | Hyperphosphorylation |
| G626A | NTD | 0.28 (0.01) | Partial | 86.3 (37.9) | Normal |
| M645R | NTD | 1.18 (0.06) | Normal | 167.3 (74.5) | Hyperphosphorylation |
| G710S | Loop 2–3 | 0.08 (0.01) | Low | 69.1 (24.6) | Normal |
| P760L | TMD 4 | 0.09 (0.07) | Low | 152.1 (46.8) | Hyperphosphorylation |
| D765N | TMD 4 | 0.61 (0.02) | Partial | 148.0 (97.9) | Hyperphosphorylation |
| M769V | TMD 4 | 0.65 (0.01) | Partial | 203.3 (32.9) | Hyperphosphorylation |
| P840L | A-domain | 0.08 (0.01) | Low | 292.9 (83.7) | Hyperphosphorylation |
| I857T | A-domain | 0.65 (0.03) | Partial | 202.4 (50.0) | Hyperphosphorylation |
| A874V | A-domain | 0.71 (0.02) | Partial | 162.1 (57.3) | Hyperphosphorylation |
| R969Q | Loop 5–6 | 0.37 (0.00) | Partial | 291.4 (72.9) | Hyperphosphorylation |
| P992L | P-domain | 0.23 (0.03) | Partial | 129.0 (20.9) | Normal |
| T1031S | P-domain | 0.09 (0.03) | Low | 126.0 (57.9) | Normal |
| P1052L | P-domain | 0.02 (0.00) | No | 90.7 (70.0) | Normal |
| E1064K | N-domain | 0.03 (0.01) | No | 16.9 (11.0) | Inactive |
| H1069Q | N-domain | 0.13 (0.01) | Low | 51.4 (29.9) | Inactive |
| L1083F | N-domain | 0.22 (0.02) | Partial | 98.6 (25.1) | Normal |
| G1213V | P-domain | 0.03 (0.00) | No | 119.8 (20.2) | Normal |
| D1222V | P-domain | 0.11 (0.02) | Low | 17.9 (6.0) | Inactive |
| G1266R | P-domain | 0.08 (0.01) | Low | 38.3 (35.3) | Inactive |
| N1270S | P-domain | 0.02 (0.02) | No | 254.2 (126.9) | Hyperphosphorylation |
| P1273L | P-domain | 0.18 (0.01) | Partial | 245.6 (166.2) | Hyperphosphorylation |
| S1362F-fs | TMD 8 | 0.17 (0.01) | Partial | 53.8 (13.2) | Inactive |
| Human variants | |||||
| S406A | NTD | 1.08 (0.01) | Normal | 137.5 (45.1) | Normal |
| V456L | NTD | 0.50 (0.02) | Partial | 115.5 (58.2) | Normal |
| K832R | A-domain | 0.92 (0.00) | Partial | 130.0 (58.6) | Normal |
NOTE. Copper uptake was considered impaired (no) (<5% of wt at 120 minutes, 0–0.07 nmol/mg); low (5%–10% of wt, 0.08–0.14 nmol/mg); partial (11%–75% of wt, 0.15–0.99 nmol/mg); normal (>75% of wt, >1.0 nmol/mg). The ability to form phosphorylated intermediate (catalytic phosphorylation) was considered normal (60%–140% of wt); inactive (<60% of wt); hyperphosphorylated (>140% of wt). Mutations highlighted in bold are predicted to be most severe (markedly affect copper transport or both functionalities), whereas mutations in italics are likely to be less severe or potentially correctable. Copper transport rates are given in Supplementary Table 2.
A-domain, actuator domain; NTD, N-terminal Cu-binding domain; N-domain, nucleotide-binding domain; P-domain, phosphorylation domain; TMD, transmembrane domain (see Figure 1B).
Materials and Methods
Reagents and Cell Lines
All chemicals, unless otherwise specified, were purchased from Sigma (Deisenhofen, Germany). Sf9 cells (Invitrogen, Carlsbad, CA) were maintained at 27°C in suspension cultures in Sf-900 II (Gibco, Grand Island, NY). HEK293 T-REx cells (Invitrogen) were maintained at 37°C in adherent cultures in minimal essential medium (Gibco, Carlsbad, CA) media with 10% fetal bovine serum, 3% penicillin-streptomycin, 1% nonessential amino acids, 0.1% zeocin, and 0.1% blasticidin.
Generation of Recombinant Baculovirus ATP7B Variants
The plasmid encoding the full-length 4.4-kilobase ATP7B complementary DNA (pFastBacDual-wt-ATP7B) and the catalytically inactive D1027A mutant were described previously18 and used as a template. The ATP7B mutations were introduced with the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and appropriate primers (Supplementary Table 1). The entire coding sequence of all constructs was verified by automated sequencing.
Expression in Sf9 Insect Cells and Preparation of Membrane Fractions
Maintenance, infection with recombinant viruses, harvesting of Sf9 insect cells, and isolation of membrane fractions were carried out as described earlier.18,19 To obtain a total microsomal fraction, cells from a 50-mL culture were pelleted by centrifugation at 500g (10 minutes, 4°C) and then resuspended in 5 mL homogenizing buffer: 25 mM imidazole (pH 7.4), 0.25 M sucrose, 1 mM dithiothreitol, 1 mM 4-(2-Aminoethyl)benzenesulfonyl fluoride hydrochloride. One tablet of complete protease inhibitor mixture without EDTA (Roche, Basel, Switzerland) was added per 50 mL buffer solution. Cells were homogenized by 20 strokes in a Dounce glass homogenizer, and then centrifuged (10 minutes, 500g). The supernatant was subjected to an additional centrifugation (30 minutes, 20,000g) to pellet microsomal membranes that were then resuspended in homogenizing buffer and stored at −80°C. Protein concentration in membrane fraction was determined by the method of Lowry.20 The expression of wild-type (wt) and mutant ATP7B was analyzed by separating 50 µg total membrane protein on a 7.5% Laemmli gel,21 followed by Coomassie staining and Western blotting with polyclonal antibody a-ABD (1:20,000).18,19
Preparation of Vesicles for Copper Transport
Sf9 cells were infected with virus encoding wt-ATP7B, empty vector (mock), or ATP7B variants and harvested after 3 days. Cells from a 50-mL culture were pelleted, resuspended in 6 mL ice-cold buffer containing 50 mM Tris (pH 7.0), 50 mM Mannitol, 2 mM ethylene glycol-bis-(2-aminoethyl)-N,N,N′, N′-tetraacetic acid, antipain, and leupeptin (each diluted 1:1000), phenylmethylsulfonyl fluoride (diluted 1:200), homogenized 20× in a semi-automatic homogenizer (Schütt Homgen Plus, Göttingen, Germany) at 3000 rpm, and then centrifuged (10 min, 500g, 4°C). Subsequently, the supernatant was centrifuged (1 hour, 100,000g) to sediment the membranes. The membranes were resuspended in a sterile filtered buffer (SMS): 50 mM sucrose, 100 mM potassium nitrate, 10 mM HEPES/Tris (pH 7.4). Vesicle formation was facilitated by vortexing and passing several times through a 25-gauge 5/8-inch needle. The vesicles were stored in liquid nitrogen. Protein concentration was determined by the method of Bradford.22 To confirm protein expression, all samples were analyzed by Western analysis using polyclonal antibody a-ABD.18,19
64Cu-Uptake in Vesicles
The transport assay was performed according to Gmaj et al.23 The vesicle solution was thawed, diluted to 5 µg protein/µL with sterile filtered SMS buffer and kept on ice until further use. For the transport assay, 20 µL vesicle solution was preincubated at 37°C for 1 minute. The reaction was started at 37°C by addition of 80 µL sterile filtered radioactive incubation solution: 50 mM sucrose, 100 mM KNO3, 10 mM HEPES/Tris (pH 7.4), 12.5 mM Mg (NO3)2, 12.5 mM dithiothreitol, 6.25 mM ATP, 2.5 µM CuCl2 (final copper concentration 2 µM), 6 µCi/ml 64Cu Rotop Pharmaka, Radeberg, Germany). For inhibition experiments, orthovanadate was added to the incubation solution to a final concentration of 200 µM. The copper transport reaction was stopped after 2, 5, 20, 45, 120, or 240 minutes by addition of 3 mL ice-cold sterile filtered stopping solution (50 mM sucrose, 100 mM KCl, 10 mM HEPES/Tris [pH 7.4], 0.5 mM EDTA) and filtered immediately through a 0.45-µm nitrocellulose-vacuum-filter-system. The filter was washed with 3 mL stopping solution before and after filtration, and its radioactivity was measured by the γ-Counter Cobra Quantum (PerkinElmer, Rodgau-Jügesheim, Germany). To measure the background radioactivity, 3 mL stopping solution was added to the vesicle solution on ice before the addition of 80 µL incubation solution. Vesicles with an empty vector (mock) and inactive variant D1027A were used as negative controls. The data were analyzed by a nonlinear regression using SigmaPlot software.
[γ-32P]ATP Phosphorylation
Membrane preparations (50 µg) containing wt-ATP7B or mutants were resuspended in 200 µL phosphorylation buffer: 20 mM bis-Tris propane (pH 7), 200 mM KCl, 5 mM MgCl2. Radioactive [γ-32P]ATP (5 µCi, specific activity 20 mCi/µmol; PerkinElmer, Waltham, MA) was added to a final concentration of 1 µM, and the mixture was incubated on ice for 4 minutes. The reaction was stopped by addition of 50 µL ice-cold 1 mM NaH2PO4 in 50% trichloroacetic acid and then centrifuged (10 minutes, 20,000g). The protein pellet was washed once with 1 mL ice-cold water and centrifuged a second time for 5 minutes. The pellet was dissolved in 40 µL sample buffer (5 mM Tris-PO4 [pH 5.8], 6.7 M urea, 0.4 M dithiothreitol, 5% sodium dodecyl sulfate, and Bromphenol blue) and 30 µL were loaded on a 7% acidic gel (stacking gel: 5.5% acrylamide, 41.3 mM Tris, pH 5.8, 1% sodium dodecyl sulfate, 5% ammonium persulfate, 5% tetramethylethylenediamine; separating gel: 7% acrylamide, 64.5 mM Tris, pH 6.8, 1% sodium dodecyl sulfate, 6,25% ammonium persulfate, 6.25% tetramethylethylenediamine). After electrophoresis, gels were fixed in 10% acetic acid for 10 minutes and dried on blotting paper. Dried gels were exposed overnight to either a screen for the Fuji BAS reader (Fuji, Düsseldorf, Germany) or Kodak BioMax MR film (Kodak, Rochester, NY). The photon-stimulated luminescence intensity of bands was quantified using Aida Image Analyzer (Raytest, Straubenhardt, Germany). To test ATP-dependent dephosphorylation, ATP was added after [γ-32P]ATP phosphorylation to a final concentration of 1 mM at room temperature and incubated for 10 minutes before the reaction was stopped and samples were processed in the same manner as described here.
Expression and Localization of ATP7B Variants in HEK 293 T-REx Cells
Selected mutations (L1083F, R969Q, and A874V) were introduced into a green fluorescent protein–ATP7B construct (kindly provided by Dr Ann Hubbard, Johns Hopkins University) using the QuikChange Site Directed Mutagenesis kit and appropriate primers (Supplementary Table 1). The presence of the desired mutation and the absence of additional mutations were confirmed by DNA sequencing. For transfection into mammalian cells, plasmids were isolated using a Zyppy Endo-free Plasmid Miniprep Kit (Zymo Research, Irvine, CA). HEK 293 TREx cells were seeded on flame-sterilized coverslips in 12-well plates and allowed to grow until approximately 70% to 80% confluency. Transfections of 2500 ng plasmid DNA were performed in a serum-free minimum essential medium for 6 hours using 4 µL TurboFect reagent (Fermentas, Glen Burnie, MD) per well. The medium was then replaced with Gibco minimum essential medium (with 10% fetal bovine serum, 3% penicillin-streptomycin, 1% nonessential amino acids) and left overnight at 37°C or 28°C. Cells were fixed with chilled 1:1 aceton:methanol mixture for 30 seconds and blocked overnight in blocking buffer (1% gelatin, 1% bovine serum albumin in phosphate-buffered saline [PBS]).
Primary antibody solutions were prepared by diluting into blocking buffer with 0.1% Tween. Dilutions were as follows: TGN38 (H-300) Rabbit Polyclonal Antibody (Santa Cruz Biotech, Santa Cruz, CA), 1:150; Sheep Antibody to Human TGN46 (GeneTex, Irvine, CA), 1:150; Rabbit anti-Calnexin C-terminus (Enzo Life Sciences, Plymouth Meeting, PA), 1:500, 1:250, 1:150. Sixty microliters of each antibody solution was aliquoted on Parafilm within a humidifying chamber for each coverslip. Coverslips were placed onto these primary antibody solutions cell side down and kept at room temperature for 2 hours. After two 5-minute washes with PBS and two 5-minute washes with 0.5% Tween in PBS, coverslips were treated with a secondary antibody for 1 hour and protected from light. The following dilutions were used: Alexa Fluor 555 goat anti-rabbit IgG (Invitrogen), 1:500 and Alexa Fluor 555 donkey anti-sheep IgG (Invitrogen), 1:250. Two PBS washes, three 0.5% Tween in PBS washes, and a final PBS wash were done to remove unbound antibody. Slides were rinsed in Milli-Q purified water to remove residual salts, then mounted with Vectashield Mounting Medium (Vector Laboratories, Burlingame, CA) with 4′,6-diamidino-2-phenylindole and visualized using a Zeiss LSM 5 Pascal confocal microscope with a 100× objective lens (Carl Zeiss AG, Oberkochen, Germany).
Protein Modeling
The location of the mutations in different functional domains was visualized using available nuclear magnetic resonance structures for the N-domain,24, N-terminal Cu-binding unit domain,25 and the A-domain.26 The structures of the P-domain were modeled using the ESyPred3D Web Server 1.0 and the crystallographic structures of CopA ATP-binding domain as template (RCSB accession number 2b8e).27,28
Results
In Vitro Copper Transport Assay
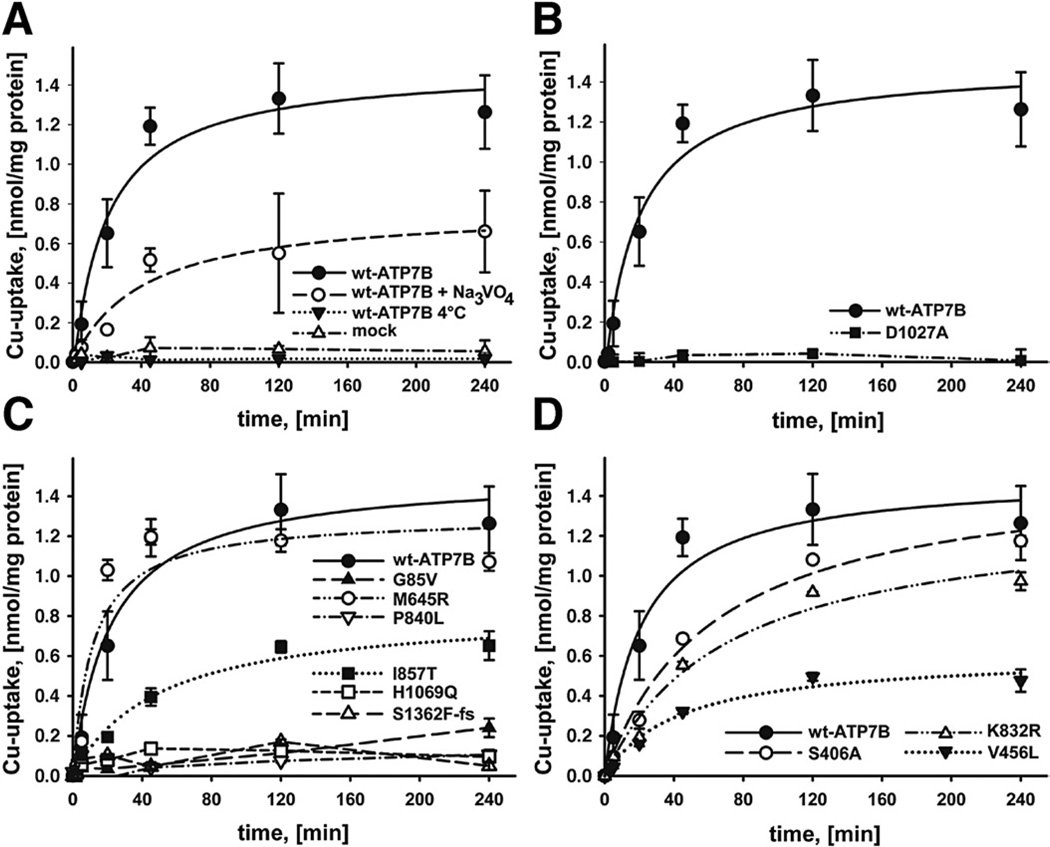
We first determined whether an ATP7B-dependent copper transport can be detected in ATP7B-containing microsomal vesicles from Sf9 cells used as a model for TGN vesicles. In the presence of ATP, the addition of 64Cu resulted in an accumulation of copper in ATP7B-containing vesicles (Figure 2); whereas no transport was observed in vesicles prepared from cells infected with empty virus (mock) (Figure 2A). To verify that copper accumulation was due to an enzymatically driven process, we repeated experiments at 4°C. No copper accumulation was observed at 4°C (Figure 2A). Orthovanadate (200 µM Na3 VO4), a general inhibitor of P-type ATPases, significantly inhibited but did not fully eliminate copper uptake (Figure 2A). Consequently, to further verify that copper accumulation was due to transport and not simply binding of copper to ATP7B, we analyzed the D1027A mutant of ATP7B, in which catalysis is disrupted but copper binding is preserved. This mutant yielded no copper accumulation in vesicles (Figure 2B).
Figure 2.
(A) Copper transport by wt-ATP7B. Means of 10 independent experiments are shown. Copper uptake into vesicles was measured in nmol/mg protein during a time period of 240 minutes. Na3VO4 was added to the incubation solution to a final concentration of 200 µM. (B) Mutation of the phosphorylation site of ATP7B (D1027A) abolishes Cu transport. (C) Copper transport by ATP7B mutants or (D) variants; means of 3 independent experiments are shown (mutants were selected to represent different functional domains; data for more mutants are shown in Table 1 and Supplementary Figure 2).
Effect of Mutations on Copper Transport
To determine the effect of mutations on ATP7B transport function, we generated 25 WD-associated variants and 3 predicted polymorphisms. Substitutions were made within different functional domains to better understand the structure–function relationships (Figure 1B). The wt-ATP7B and mutants were expressed in the Sf9 cells under identical conditions. Western blot analysis illustrated that protein expression was easily detectable for all mutants (Supplementary Figure 1). In contrast, there was a marked variation in copper transport (Table 1, Supplementary Table 2, Figure 2C and D, Supplementary Figure 2). The majority of ATP7B mutants (16 out of a total 25) had essentially no activity (−5% of wt) or a very low activity (5%–10%); the H1069Q mutant (frequently found in the white population) belonged to this latter category (Figure 2C). Unexpectedly, a sizeable fraction (8 of 25) of mutants showed partial transport activity (0.22–0.71 nmol/mg protein at 120 minutes). These mutants included A874V (frequently detected in Korean population29) and I857T. Most of the partially active mutants had slower transport rates and accumulated less copper than the wt-ATP7B, except the S406A variant, which had a slower rate compared with the wt, but with time yielded the same level of copper accumulation in vesicles (Figure 2C and D). Only one of the ATP7B mutants, M645R, displayed Cu-uptake (1.18 nmol/mg protein at 120 minutes) indistinguishable from wt-ATP7B (Figure 2C). The predicted nondisease variants (V456L and K832R) also showed reduced transport rates (Figure 2D). To further verify that these marked variations in activity were not simply due to differences in ATP7B expression levels, we selected several mutants that had similar transport activity, expressed them in parallel under identical conditions, and quantified ATP7B amounts by densitometry. Supplementary Figure 3 illustrates that a drastically different activity of these mutants is not due to difference in their expression levels.
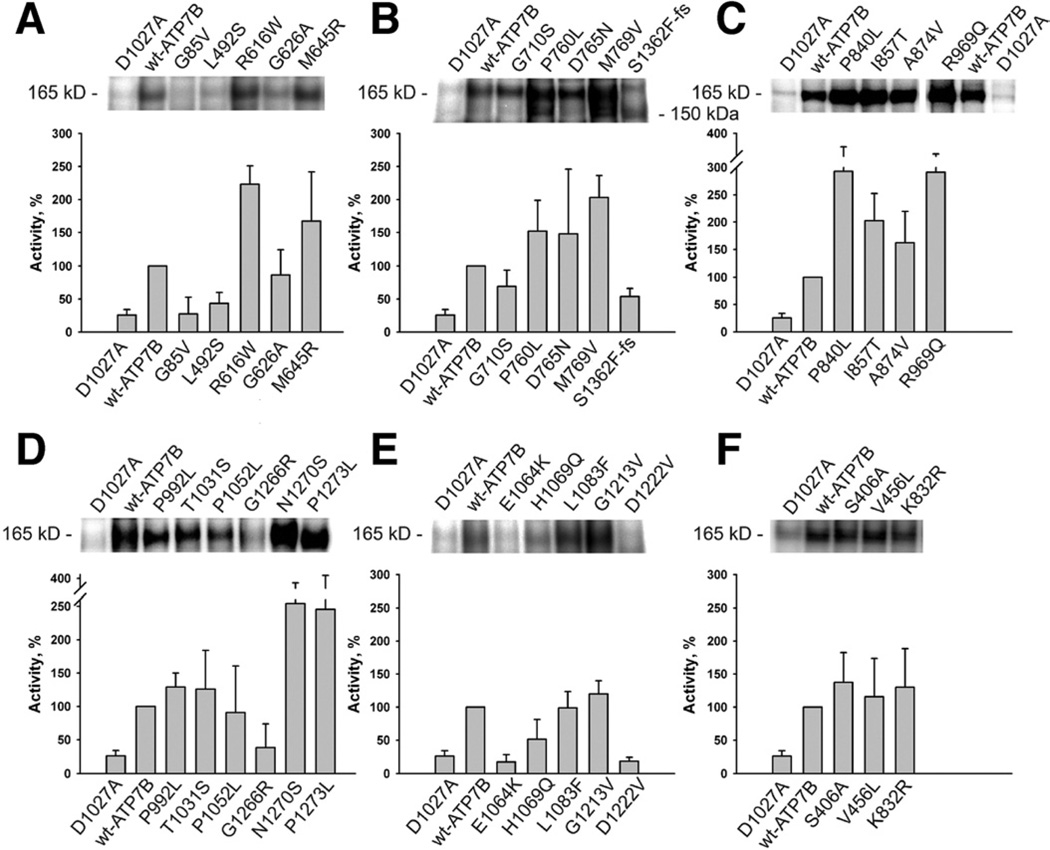
Effect of Mutations on Catalytic Activity
The loss of copper transport could be either due to disruption of copper transfer across membranes or due to the loss of ATP binding/hydrolysis. To better understand which of the steps is affected, we examined the ability of ATP7B to form a phosphorylated intermediate upon ATP binding (Figure 3, Table 1). In all A-domain mutants, phosphorylation was markedly increased (Figure 3C). This result indicated that ATP-binding is unaltered, but the well-known function of the A-domain to facilitate dephosphorylation11 was inhibited. In contrast, ATP7B variants with mutations in the N-terminal domain Figure 3A), transmembrane domain (Figure 3B), the P-, or N-domains (Figure 3D and E, respectively) showed either normal or reduced phosphorylation. All 3 ATP7B gene variants not associated with disease showed normal phosphorylation activity (Figure 3F). In our subset of mutants, disruption of ATP binding is not a major defect.
Figure 3.
Phosphorylation activity of ATP7B mutants (A–E) and variants (F): The protein gels show representative 32P autoradiograms of phosphorylated wt-ATP7B, variants, and mutants. The phosphorylation activity (bar graphs) of variant and mutant ATP7B was calculated with respect to wt-ATP7B activity (100%). Averages of 4 to 6 independent experiments are shown.
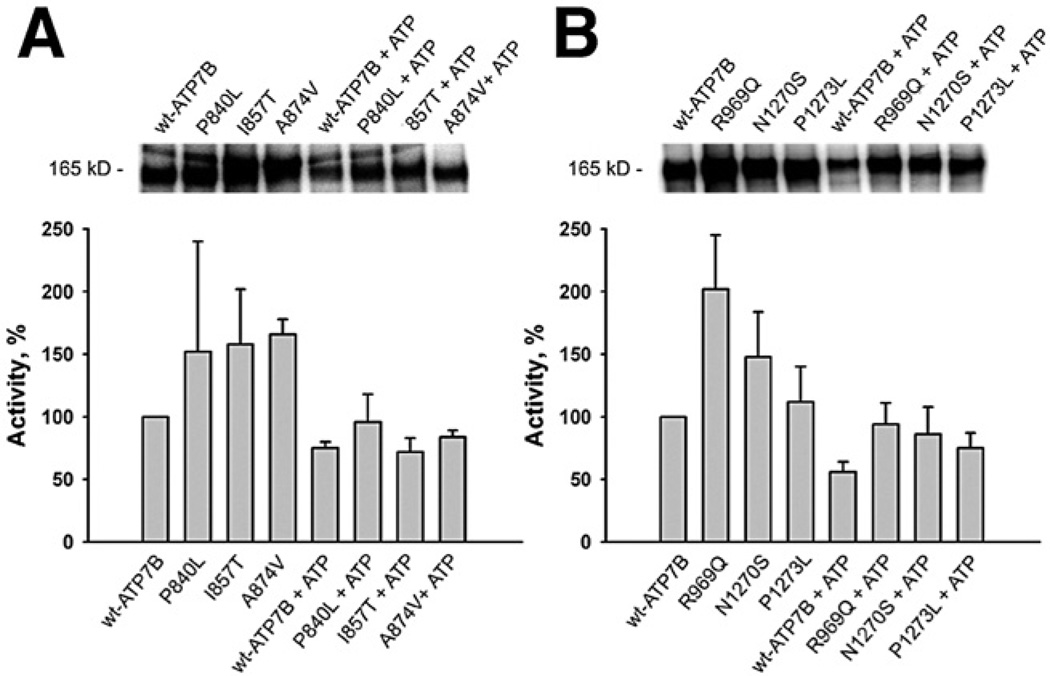
Partial transport activity along with hyperphosphorylation for several mutants suggested that these mutants have altered conformations (or defects in the communication between functional protein domains necessary for optimal protein activity). This conclusion was supported by experiments on dephosphorylation. When hyperphosphorylated mutants were incubated with cold ATP, dephosphorylation was observed for all examined mutants (Figure 4A and B). Thus, the hyperphosphorylated mutants are able to perform all the steps of the cycle, and the transport defect in these mutants is associated with structural changes affecting rates of the individual step(s).
Figure 4.
ATP-dependent dephosphorylation of the A-domain mutants (A) and P-domain mutants (B) of ATP7B at a copper concentration of 1 µM. ATP was added after [γ-32P] ATP phosphorylation to a final concentration of 1 mM and incubated for 10 minutes at room temperature before the reaction was stopped. The typical gels and the phosphorylation activities of mutant ATP7B calculated in respect to wt-ATP7B activity (100%) for 3 independent experiments are shown.
Expression and Localization of Functional ATP7B Mutants in Mammalian Cells
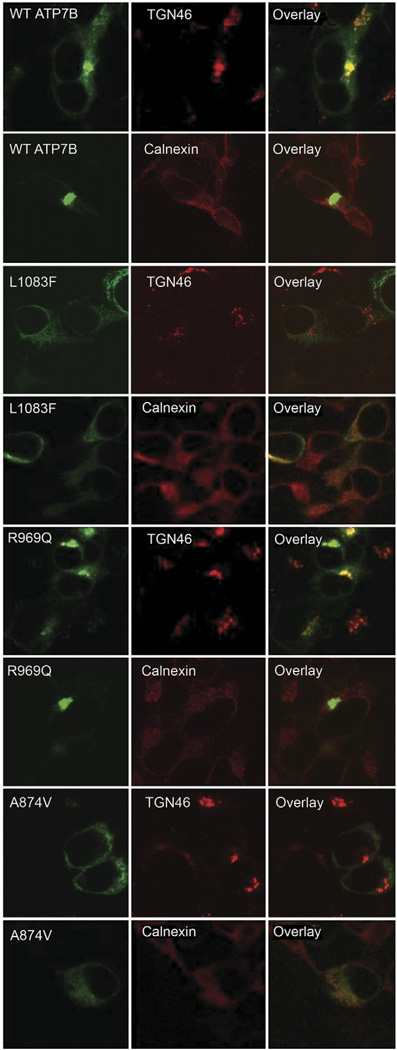
The ability of several mutants to hydrolyze ATP and transport copper in vitro suggested that in cells their properties can be altered further, exacerbating the phenotype. To test this hypothesis, 3 transport competent mutants (found in different domains: A-domain [A874V], N-domain [L1083F], and in the transmembrane domain [R969Q]) were produced as green fluorescent protein–tagged versions and their expression and localization were examined in mammalian cells. The R969Q variant showed fluorescent intensity similar to that of the wt-ATP7B and normal intracellular targeting, as illustrated by colocalization with the TGN marker TGN46 (Figure 5). In contrast, L1083F and A874V showed significant abnormalities in their subcellular localization and protein levels. When expressed at 37°C, L1083F and A874V displayed characteristic endoplasmic reticulum (ER) localization, which was confirmed by co-staining with the ER marker Calnexin (Figure 5). Both mutants also showed weaker signal intensity compared to wt-ATP7B. Thus, despite the lack of gross misfolding (indicated by the ability to perform key functional reactions in vitro), the structural changes in A874V and L1083F mutants were sufficient to produce apparent destabilization and ER retention. Thus, mislocalization is the major reason for the loss of copper delivery to the TGN by these mutants.
Figure 5.
Expression and localization of ATP7B mutants in mammalian cells. Green fluorescent protein–tagged ATP7B variants were expressed in HEK 293 T REx cells. The expression pattern of wt- and mutant ATP7B is shown in the left column (green), organelle markers in the middle (red), and the superimposition of these images on the right (yellow). TGN46 is a TGN marker; Calnexin is marker for ER.
Previous studies reported that the stability and localization of some ATP7B mutants (eg, R778L and H1069Q) can be corrected by growing cells at 30°C.17 Consequently, we tested whether the targeting of A874V and L1083F (which show copper transport activity after being expressed at 27°C in Sf9 cells (Table 1, Supplementary Figure 2B and D) can be corrected by lowering temperature during expression. For A874V, no improvement in either protein levels or targeting was seen in mammalian cells maintained at 28°C (attempts to stabilize this mutant by growing cells with increasing copper concentrations were also unsuccessful, data not shown). For the L1083F mutant, some TGN localization could be detected, but cold temperature alone was insufficient to induce TGN targeting (Supplementary Figure 4).
Discussion
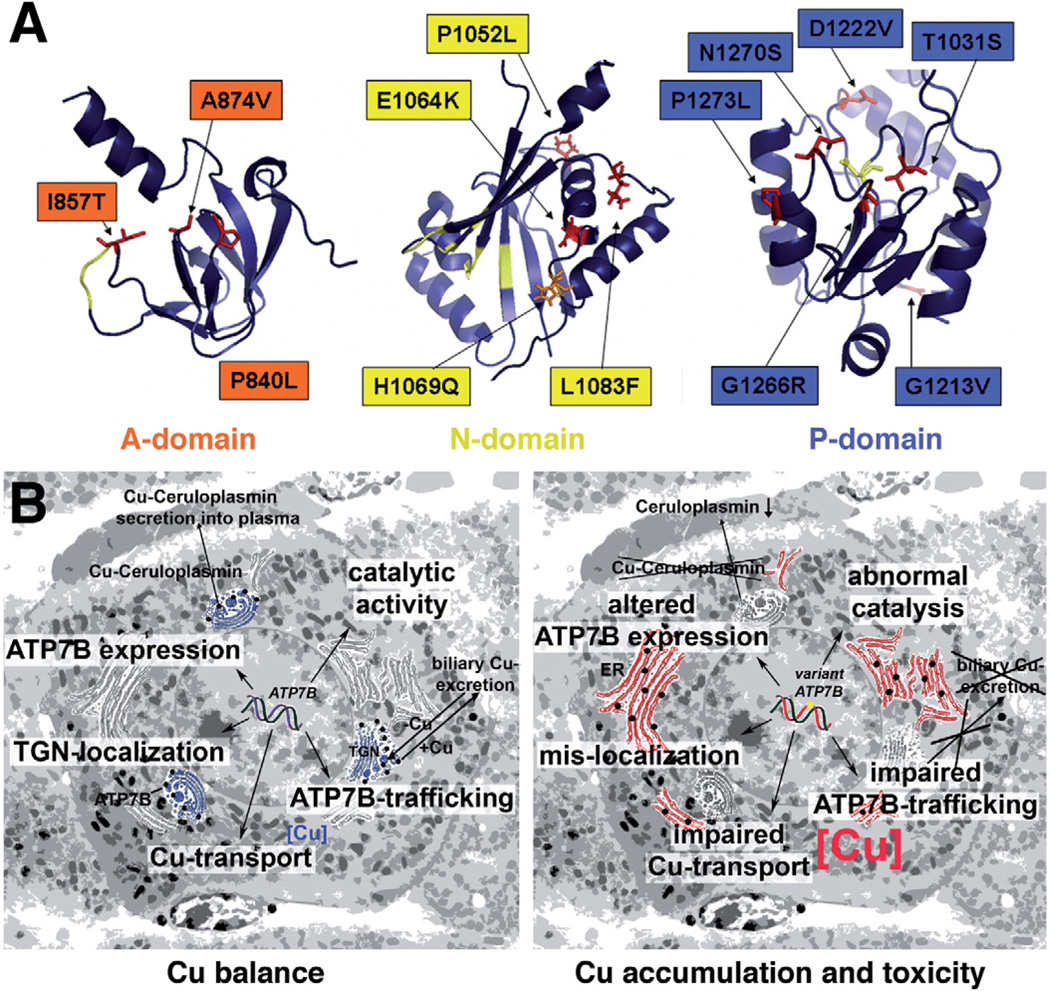
ATP7B variants have a wide spectrum of properties. In this study, we have investigated the effect of WD-causing mutations on the functional activity and intracellular functionality of ATP7B. Utilization of a heterologous expression system and membrane vesicles allowed us for the first time to directly evaluate the transport activity of a large set of clinically relevant mutants. Our results indicate that a significant diversity already exists at the molecular level, as the mutants display a markedly different stability, localization, catalytic, and transport activity. Therefore, detailed characterization of each WD-causing mutant may be necessary for better understanding of the genotype–phenotype correlations in WD. Although mutations located in the same domain can have similar properties (eg, we detected increased levels of phospho-intermediate in the A-domain mutants), the localization in a certain domain alone is not predictive of the properties of a mutant. This conclusion can be illustrated by different transport characteristics of the P840L and I857T, both situated in the A-domain (Figure 6A). The more severe effect of P840L on protein function is likely due to a more disruptive nature of Pro>Leu substitution (replacing a small and kink-inducing Pro residue with a bulky and hydrophobic Ile) compared to replacement of Ile with a smaller Thr.
Figure 6.
(A) Location of the characterized mutations in the functional domains of ATP7B. The structure of the P-domain was software-modeled using crystallographic structure of CopA (2B8E) as a template. For mutations within the N-domain, available nuclear magnetic resonance data (2ARF) were used. The nuclear magnetic resonance- based model for the A-domain was kindly provided by Dr Lucia Banci. (B) Summary of key properties of ATP7B (left panel) that are affected by various mutations examined in this study (right panel).
The comparison of A874V with the recently characterized G875R variant further illustrates the extremely delicate and precise architecture of the ATP7B molecule.30 The A874V mutation is located within the flexible loop of the A-domain (Figure 6A), and is not expected to markedly alter protein structure. This prediction is consistent with the actual functional properties of the mutant, that is, its ability to transport copper using ATP hydrolysis. And yet, in cells, the lack of precise folding prevents the A874V exit from the ER, thus disrupting copper delivery to the secretory pathway and likely causing WD phenotype. Similar ER retention of G875R (the mutation of a neighboring residue) can be corrected by increasing copper concentration in cells,30 which facilitates ER exit and proper TGN targeting (and activity). In contrast, treatment with copper does not improve the A874V stability or localization. Altogether, our studies demonstrate the need for utilizing an arsenal of methods for understanding the phenotype of disease-causing mutation, because individual assays yield only partial and nonoverlapping information about ATP7B properties.
It is useful to compare the advantages and disadvantages of various assays and information that can be generated. Yeast complementation assay is an effective way to identify ATP7B mutants, which completely lost their transport activity from those that have some activity left.12 However, it is difficult to draw more specific conclusions about properties of the WD mutants because it is unknown how much copper transport activity is necessary to allow yeast to grow. Overexpression of mutants with very low activity might be sufficient to allow copper delivery to the TGN and growth. In addition, yeast cells grow at lower temperature (30°C), which might be sufficient to stabilize proteins that unfold and degrade at 37°C. For example, H1069Q, which has low copper transport activity and is unstable in mammalian cells13 complements yeast phenotype.31
Expression in insect cells can underestimate the effects of mutation on protein stability, due to the lower temperature necessary for growth of insect cells (27°C). However, it provides a marked advantage when detailed mechanistic understanding of functional consequences of mutations is desirable. We found that measurements of catalytic phosphorylation and dephosphorylation of ATP7B mutants expressed in Sf9 cells were the least predictive of the overall functional outcomes of mutations, as many mutants that showed no transport activity were still able to undergo catalytic phosphorylation. Because the phosphorylation assay is least sensitive, we suggest that the mutants that show no activity in this assay are most severely affected. Indeed, these mutants (G85V, L492S, G1266R, E1046K, and D1222V) show neither catalytic nor transport activity. The marked effect of the last 3 mutations on ATP hydrolysis is not surprising, as they all affect conserved residues in the immediate vicinity to ATP (Figure 6A). In contrast, dramatic negative effects of G85V and L492S mutants were unexpected as both residues belong to the regulatory N-terminal domain, which is thought not to be critical for ATP binding and hydrolysis. We hypothesize that these mutants can mediate their effect indirectly through interaction with other functional domains. Because the N-domain interacts with the ATP-binding domain and these interactions affect protein activity,32 abnormal inter-domain interaction due to mutation can decrease affinity for ATP and/or trap protein in conformation not suitable for ATP hydrolysis.
The transport assay is the most direct way to evaluate ATP7B function. This assay identified several mutants that were transport competent, that is, could be functional in a cell. Identification of such mutants is particularly important because they are promising candidates for possible corrective therapy. Such variants include M769V, I857T, A874V, all located in the A-domain (Figure 6A, left). Unlike mutations in the P- and N-domains (Figure 6A, middle, right), their residues are not involved in the formation of the substrate binding sites and their substitution is more likely to alter protein stability or targeting. As described here, characterization of one of this mutant A874V revealed limitations of the in vitro assays (and possibly all heterologous systems) in predicting the mutant phenotype in mammalian cells. In mammalian cells at either 28°C or 37°C, A874V has the ER localization and low expression pointing to significant instability of this mutant. Such instability was not detected in insect cells, suggesting that mammalian cells have much more sensitive protein quality control.
Combination of in vitro and in vivo assays also discriminates between the effects of mutations on protein folding and function. Protein stability in cells is not indicative of function because catalytically inactive mutants could be structurally stable, whereas partially active mutants can degrade rapidly, producing a complete loss of function phenotype. Having information on both parameters could be particularly important for future attempts to correct the mutant phenotype by improving protein stability. For example, increasing stability of mutants such as the A874V mutant could be beneficial because A874V has a significant transport activity. Recent studies using pharmacological folding chaperones 4-phenylbutyrate and curcumin illustrate the possibility of correcting the stability/folding defects of several ATP7B mutants in cells.17 In contrast, improving stability of other mutants (such as G85V or mutants located in the vicinity of the key functional sites; Figure 6A) can be without effect because these mutants lack catalytic and transport activity. Lastly, our studies revealed that the non–disease-causing changes (V456L and K832R) can decrease ATP7B activity. Thus, the presence of such variants, although alone not causative, can have a compound effect when found in combination with other mutations, and put carriers at disadvantage.
In conclusion, mutations in ATP7B have various effects altering protein expression levels, catalytic and transport activity, as well as intracellular localization (Figure 6B). Mutants with a partial preserved transport function can result in later onset of disease or have variable manifestation if their stability and localization is modulated by the metabolic state of cells. Detailed characterization of ATP7B mutants is likely to contribute to the analysis of genotype-phenotype correlations in WD.
Supplementary Material
Acknowledgments
The authors thank L. Thiel and Dr R. Schliebs (University of Leipzig, Germany) and Dr N. Taudte (Martin-Luther-University of Halle, Germany) for advice with experimental design and technical assistance.
Funding
This work was funded by the Deutsche Forschungsgemeinschaft (HU 932/3-1), and National Institutes of Health grant R01DK071865-06 to S.L
Abbreviations used in this paper
- ATPase
adenosine triphosphatase
- ER
endoplasmic reticulum
- PBS
phosphate-buffered saline
- TGN
trans-Golgi network
- WD
Wilson disease
- wt
wild-type
Footnotes
Supplementary Materials
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at doi: 10.1053/j.gastro.2011.12.048.
Conflicts of interest
The authors disclose no conflicts.
References
- 1.Pfeiffer RF. Wilson’s disease. Semin Neurol. 2007;27:123–132. doi: 10.1055/s-2007-971173. [DOI] [PubMed] [Google Scholar]
- 2.Huster D. Wilson disease. Best Pract Res Clin Gastroenterol. 2010;24:531–539. doi: 10.1016/j.bpg.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 3.Riordan SM, Williams R. The Wilson’s disease gene and phenotypic diversity. J Hepatol. 2001;34:165–171. doi: 10.1016/s0168-8278(00)00028-3. [DOI] [PubMed] [Google Scholar]
- 4.Lorincz MT. Neurologic Wilson’s disease. Ann N Y Acad Sci. 2010;1184:173–187. doi: 10.1111/j.1749-6632.2009.05109.x. [DOI] [PubMed] [Google Scholar]
- 5.Kegley KM, Sellers MA, Ferber MJ, et al. Fulminant Wilson’s disease requiring liver transplantation in one monozygotic twin despite identical genetic mutation. Am J Transplant. 2010;10:1325–1329. doi: 10.1111/j.1600-6143.2010.03071.x. [DOI] [PubMed] [Google Scholar]
- 6.Czlonkowska A, Gromadzka G, Chabik G. Monozygotic female twins discordant for phenotype of Wilson’s disease. Mov Disord. 2009;24:1066–1069. doi: 10.1002/mds.22474. [DOI] [PubMed] [Google Scholar]
- 7.Caca K, Ferenci P, Kuhn HJ, et al. High prevalence of the H1069Q mutation in East German patients with Wilson disease: rapid detection of mutations by limited sequencing and phenotype-genotype analysis. J Hepatol. 2001;35:575–581. doi: 10.1016/s0168-8278(01)00219-7. [DOI] [PubMed] [Google Scholar]
- 8.Stapelbroek JM, Bollen CW, van Amstel JK, et al. The H1069Q mutation in ATP7B is associated with late and neurologic presentation in Wilson disease: results of a meta-analysis. J Hepatol. 2004;41:758–763. doi: 10.1016/j.jhep.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 9.Ferenci P, Czlonkowska A, Merle U, et al. Late-onset Wilson’s disease. Gastroenterology. 2007;132:1294–1298. doi: 10.1053/j.gastro.2007.02.057. [DOI] [PubMed] [Google Scholar]
- 10.Kenney SM, Cox DW. Sequence variation database for the Wilson disease copper transporter, ATP7B. Hum Mutat. 2007;28:1171–1177. doi: 10.1002/humu.20586. [DOI] [PubMed] [Google Scholar]
- 11.Tsivkovskii R, Purnat T, Lutsenko S. Copper-transporting ATPases: key regulators of intracellular copper concentration. In: Futai M, Wada Y, Kaplan J, editors. Handbook of ATPases. Weinheim: Wiley-VCH Verlag GmbH & Co; 2004. pp. 99–158. [Google Scholar]
- 12.Forbes JR, Cox DW. Functional characterization of missense mutations in ATP7B: Wilson disease mutation or normal variant? Am J Hum Genet. 1998;63:1663–1674. doi: 10.1086/302163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsi G, Cullen LM, Macintyre G, et al. Sequence variation in the ATP-binding domain of the Wilson disease transporter, ATP7B, affects copper transport in a yeast model system. Hum Mutat. 2008;29:491–501. doi: 10.1002/humu.20674. [DOI] [PubMed] [Google Scholar]
- 14.Luoma LM, Deeb TM, Macintyre G, et al. Functional analysis of mutations in the ATP loop of the Wilson disease copper transporter, ATP7B. Hum Mutat. 2010;31:569–577. doi: 10.1002/humu.21228. [DOI] [PubMed] [Google Scholar]
- 15.Huster D, Hoppert M, Lutsenko S, et al. Defective cellular localization of mutant ATP7B in Wilson’s disease patients and hepatoma cell lines. Gastroenterology. 2003;124:335–345. doi: 10.1053/gast.2003.50066. [DOI] [PubMed] [Google Scholar]
- 16.Payne AS, Kelly EJ, Gitlin JD. Functional expression of the Wilson disease protein reveals mislocalization and impaired copper-dependent trafficking of the common H1069Q mutation. Proc Natl Acad Sci U S A. 1998;95:10854–10859. doi: 10.1073/pnas.95.18.10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Berghe PV, Stapelbroek JM, Krieger E, et al. Reduced expression of ATP7B affected by Wilson disease-causing mutations is rescued by pharmacological folding chaperones 4-phenylbutyrate and curcumin. Hepatology. 2009;50:1783–1795. doi: 10.1002/hep.23209. [DOI] [PubMed] [Google Scholar]
- 18.Tsivkovskii R, Eisses JF, Kaplan JH, et al. Functional properties of the copper-transporting ATPase ATP7B (the Wilson’s disease protein) expressed in insect cells. J Biol Chem. 2002;277:976–983. doi: 10.1074/jbc.M109368200. [DOI] [PubMed] [Google Scholar]
- 19.Huster D, Lutsenko S. The distinct roles of the N-terminal copper-binding sites in regulation of catalytic activity of the Wilson’s disease protein. J Biol Chem. 2003;278:32212–32218. doi: 10.1074/jbc.M305408200. [DOI] [PubMed] [Google Scholar]
- 20.Lowry OJ, Rosebrough NJ, Farr AL, et al. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 21.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Kruger NJ. The Bradford method for protein quantitation. Methods Mol Biol. 1994;32:9–15. doi: 10.1385/0-89603-268-X:9. [DOI] [PubMed] [Google Scholar]
- 23.Gmaj P, Zurini M, Murer H, et al. A high-affinity, calmodulin-dependent Ca2+ pump in the basal-lateral plasma membranes of kidney cortex. Eur J Biochem. 1983;136:71–76. doi: 10.1111/j.1432-1033.1983.tb07706.x. [DOI] [PubMed] [Google Scholar]
- 24.Dmitriev O, Tsivkovskii R, Abildgaard F, et al. Solution structure of the N-domain of Wilson disease protein: distinct nucleotide-binding environment and effects of disease mutations. Proc Natl Acad Sci USA. 2006;103:5302–5307. doi: 10.1073/pnas.0507416103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banci L, Bertini I, Cantini F, et al. Metal binding domains 3 and 4 of the Wilson disease protein: solution structure and interaction with the copper(I) chaperone HAH1. Biochemistry. 2008;47:7423–7429. doi: 10.1021/bi8004736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banci L, Bertini I, Cantini F, et al. Solution structures of the actuator domain of ATP7A and ATP7B, the Menkes and Wilson disease proteins. Biochemistry. 2009;48:7849–7855. doi: 10.1021/bi901003k. [DOI] [PubMed] [Google Scholar]
- 27.Sazinsky MH, Agarwal S, Arguello JM, et al. Structure of the actuator domain from the Archaeoglobus fulgidus Cu(+)-ATPase. Biochemistry. 2006;45:9949–9955. doi: 10.1021/bi0610045. [DOI] [PubMed] [Google Scholar]
- 28.Sazinsky MH, Mandal AK, Arguello JM, et al. Structure of the ATP binding domain from the Archaeoglobus fulgidus Cu+-ATPase. J Biol Chem. 2006;281:11161–11166. doi: 10.1074/jbc.M510708200. [DOI] [PubMed] [Google Scholar]
- 29.Yoo HW. Identification of novel mutations and the three most common mutations in the human ATP7B gene of Korean patients with Wilson disease. Genet Med. 2002;4(Suppl):43S–48S. doi: 10.1097/00125817-200211001-00009. [DOI] [PubMed] [Google Scholar]
- 30.Gupta A, Bhattacharjee A, Dmitriev O, et al. Cellular copper levels determine the phenotype of the Arg875 variant of ATP7B/Wilson disease protein. Proc Natl Acad Sci U S A. 2011;108:5390–5395. doi: 10.1073/pnas.1014959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iida M, Terada K, Sambongi Y, et al. Analysis of functional domains of Wilson disease protein (ATP7B) in Saccharomyces cerevisiae. FEBS Lett. 1998;428:281–285. doi: 10.1016/s0014-5793(98)00546-8. [DOI] [PubMed] [Google Scholar]
- 32.Tsivkovskii R, MacArthur BC, Lutsenko S. The Lys1010-Lys1325 fragment of the Wilson’s disease protein binds nucleotides and interacts with the N-terminal domain of this protein in a copper-dependent manner. J Biol Chem. 2001;276:2234–2242. doi: 10.1074/jbc.M003238200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.