Abstract
Magnetic Interactions: The dissociation constant (KD) of various molecular interactions was determined using a novel competition assay with binding magnetic relaxation nanosensors (bMRs). In this assay, changes in the magnetic relaxation of an aqueous suspension of the nanosensors facilitate the fast determination of KD values using nanomolar protein concentrations.
Keywords: protein-protein interaction, protein-ligand interaction, membrane receptors, dissociation constant, magnetic nanoparticles
Molecular interactions, particularly those involving a ligand and a protein, play a vital role in multiple biological processess.[1–5] The study of these interactions, notably those associated with disease, is important for the development of novel therapeutics and sensing technologies. These interactions are typically evaluated by a dissociation constant (KD); an equilibrium constant that describes how strongly a ligand binds to a particular target protein by measuring the propensity of the ligand to dissociate from the protein. The smaller the KD, the higher the affinity between the ligand and protein. Techniques such as surface plasmon resonance (SPR)[1], titration calorimetry[6, 7] and radioligand binding assays[8] among others have been developed to assess these molecular interactions and measure KD values. However these techniques have some drawbacks. For example, SPR requires the binding of either the ligand or the protein on a solid support, which affect the binding kinetics and it is not representative of the binding affinity in solution. Titration calorimetry and radioligand assays, even though they are performed in solution either use expensive instrumentation, radioactive materials, or solubilized receptors instead of living cells to perform the assay. Furthermore, as most reported KD values have been determined using different methods, comparative studies are difficult as variations in KD values can be seen using different techniques.[9]
The conjugation of targeting ligands to iron oxide nanoparticles has been extensively utilized to fabricate nanosensors and targeting imaging agents for the detection of various molecular targets.[10–13] In particular, we recently reported that binding of a protein target to a ligand attached to magnetic iron oxide nanoparticle in solution resulted in an increase in the spin-spin relaxation times (T2) of the water protons in solution.[14] This observation facilitated the development of magnetic relaxation nanosensors that can quantitatively sense the presence of a target by measuring the increase in the water T2 upon target binding. To differentiate our nanosensors from previously described magnetic nanosensors that cluster upon target addition resulting in a decrease (not an increase) in T2, we herein denote our nanosensors as binding magnetic relaxation (bMR) nanosensors and explore their utility as sensors to interrogate molecular interactions. Specifically, we hypothesized that a targeting bMR nanosensor can be used in a competition assay format to determine the KD of a particular molecular interaction as it occurs on the surface of a nanoparticle.
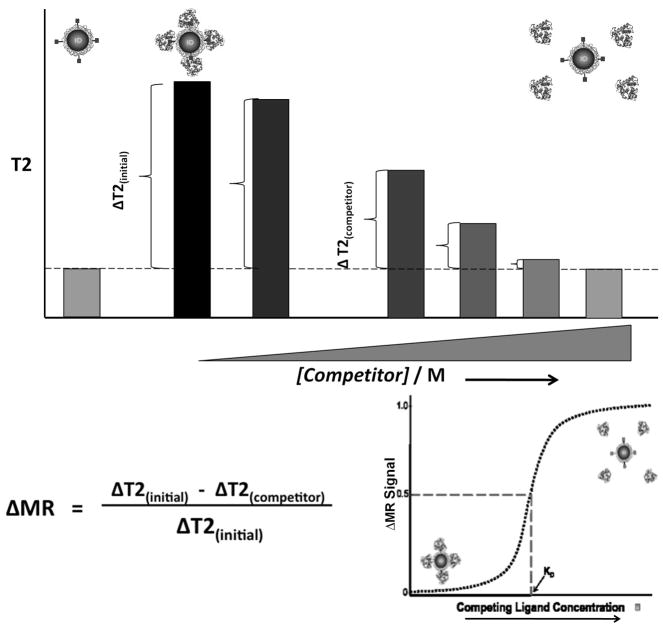
In our magnetic-relaxation-based competition assay, a constant amount of bMR nanosensors in a series of aqueous solutions containing increasing amounts of free competing ligand is mixed with the targeting protein. In the absence of free ligand, the target protein binds to ligands on the bMR nanosensors, causing an increase in the T2 of the solution. This change in T2 represents the initial state of our assay and herein we denote it as ΔT2 (initial). The addition of increasing amounts of free ligand (competitor) in the sample lowers the observed change in T2, as the added free ligands in solution compete with the bMR nanosensors for binding to the target protein. We denote this change in T2 in the presence of a competitor as ΔT2 (competitor). As the amount of free ligand increases, the magnetic relaxation signal change (ΔMR signal) defined herein as the [ΔT2 (initial) − ΔT2 (competitor)]/ΔT2 (initial) increases, reaching a plateau when ΔMR-signal approaches a value of one (Scheme 1). The initial ΔMR signal in the absence of a competitor will be zero, since there is no competing ligand present in the sample to compete for binding to the target protein. Thus, low competitor concentrations will yield low ΔMR signals while high competitor concentrations will cause a disruption in the interaction between the target protein and the bMR nanosensors causing larger ΔMR signal values.
Scheme 1.
Approach for the determination of the dissociation constant (KD) via changes in magnetic relaxation. In the absence of a competing ligand, the target protein interacts with ligands on the bMR nanosensors, resulting in a low ΔMR signal. As the concentration of competing ligand increases, the intercation between the protein and the nanosensor is disrupted, resulting in an increase in the ΔMR signal.
Therefore, as the concentration of the competitor increases in the system, the ΔMR-signal values are expected to increase, reflecting the competition that the free ligand is effecting on the system. The concentration of free ligand at which a 50% change in the MR value is observed is then defined as the dissociation constant. Specifically at this concentration, an equilibrium is achieved, where 50% of the target protein interacts with a free ligand and the remaining portion associates with the nanosensors, inducing quantifiable MR changes.
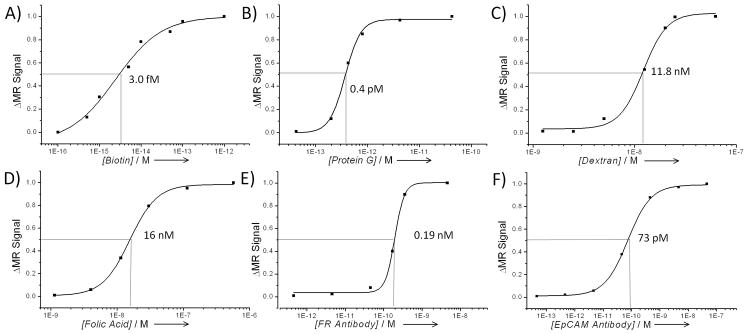
In order to test our assay, we measured the KD via magnetic relaxation of a broad range of protein-ligand interactions with different degrees of affinities. As a model system, we first used the avidin - biotin interaction, a well-studied strong interaction, with reported KD values in the femtomolar range.[15] For these studies, we designed a bMR nanosensor consisting of avidin-conjugated iron oxide nanoparticles (76 nm and an r2 relaxivity of 116 mM−1s−1 at 0.47T). Samples containing the bMR nanosensor (0.015 mg Fe/mL) in a solution containing different concentrations (10 fM – 1 pM) of free avidin (competitor) were prepared, followed by the addition of biotin (200 nM). Within 15 minutes, T2 measurements and calculation of the ΔMR signal values at various concentrations of inhibitor revealed a concentration-dependent trend (Figure 1A). A 50% change in the ΔMR signal values was observed at an avidin concentration of 3 fM, indicating that the avidin-biotin KD was equal to this value. This KD value is in close agreement with a reported KD value of 1fM, determined via titration calorimetry studies.[15] In control studies with bovine serum albumin, the bMR nanosensors yielded nominal changes, suggesting that the observed changes in magnetic relaxation were target-specific.
Figure 1. Determination of the dissociation constant for various molecular interactions via magnetic relaxation and bMR nanosensors.
A) Avidin – Biotin, B) Protein G – IgG, C) Dextran –Concanavalin A, D) Folic acid – Folate receptor (FR) expressed on HeLa cells, E) anti-folate-receptor antibody – Folate receptor on HeLa cells), F) anti-EpCAM antibody – EpCAM Receptor (MCF-7 Cells). (Errors were within 1–2%, which are too small to depict).
Next, we investigated the affinity between Protein G and IgG (Figure 1B), as well as Concanavalin A and dextran (Figure 1C), as model interactions for protein-protein and protein-carbohydrate interactions. For the Protein G – IgG interaction, a bMR nanosensor composed of Protein G-carrying iron oxide nanoparticles was incubated with various amounts of free Protein G (0.43 fM – 0.43 pM), before addition of mouse IgG (50 nM) as target protein. Our magnetic relaxation results indicated that the KD of this interaction was in the picomolar range (0.4 pM). The reported KD value for a similar interaction, using a rabbit IgG in a fluorescence binding assay, is also in the picomolar range (50 pM), although slighthly higher.[16] This is expected as the interaction with Protein G and IgG has been reported to be different depending on the source of IgG used (rabbit vs mouse).[17] Meanwhile, for the interaction between dextran and the carbohydrate-binding protein Concanavalin A (Figure 1C), dextran-coated iron oxide nanoparticles were used as bMR sensors and introduced to a solution containing various amounts of free dextran (1.25 nM – 62.5 nM), before addition of Concanavalin A (50 nM). Our measurements indicated that a KD value for this interaction was is in the nanomolar range (18.8 nM), within the same range of the reported value in the literature (90 nM).[18] The use of different sources and batches of Concanavalin A (a lectin) and dextran (a complex carbohydrate) might affect the interaction between these two macromolecules, resulting in different KD values. Furthermore, control experiments using bovine serum albumin (BSA) does not reveal a concentration-dependent signal change confirming the specificity of our assays (Supporting Information, Figure S1A–D). Taken together, these results indicate that our bMR-based method can rapidly measure the dissociation constant of different protein–ligand interactions within a wide range of affinities.
After validating the ability of our magnetic relaxation method to measure KD values of molecular interactions in solution, we investigated whether our method can be applied to study interactions with transmembrane proteins in intact cells. Membrane proteins and cellular receptors control key biological processes within the cell and are the target for a wide variety of therapeutics.[19–21] Most of the current methods to determine Kd values use purified membrane receptors in solution or attached to a flat surface.[9, 22–24] Therefore, we reasoned whether one could use the bMR assay to determine the KD between a ligand and a cell receptor using cells in suspension. To validate our hypothesis, we used the folate receptor (FR) as a model system and investigated the affinity of this receptor to its natural targeting ligand, folic acid, using HeLa cells as the source of FR.[25] For these studies, a folic-acid-conjugated iron oxide nanoparticle was used as the bMR nanosensor. Following a 30-minute incubation of the HeLa cells with the bMR nanosensors and increasing amounts of free folic acid (1.1 nM – 0.56 μM), a similar behavior to that observed with the soluble protein targets was observed (Figure 1D). At low amount of competitor, a low ΔMRS-signal was observed that increased at higher concentrations and eventually reached a plateau, allowing us to calculate the KD as 16 nM for this particular interaction. This value compares with the reported values of 0.1 nM[26] and 30 nM[25] for folate receptor/folic acid interactions. However, these values were obtained using the solubilized receptor in a radioligand-binding assay, instead of using HeLa cells in suspension. It is worth noticing the different KD values reported in the literature for the same molecular interaction, suggesting that these values depend on the nature of the assay and experimental conditions. [9]
The interaction between the folate receptor and an anti folate receptor antibody (Santa Cruz Biotechnology, sc-28997) was also studied (Figure 1E). For this study, anti folate antibody conjugated nanoparticles were used as bMR sensors. Results revealed a KD value in the low nanomolar range (0.19 nM), while the reported value in the literature for a system using a recombinant solubilized receptor and a totally different antobody was 2.23 nM.[22] In control studies, substituting the FR antibody with the EpCAM antibody (Santa Cruz Biotechnology, sc-73491) resulted in no significant increase in the ΔMR signal value with increasing concentration of EpCAM antibody. Similarly, no response was observed when MCF-7 cells were used, as this cell line does not express the folate receptor assays (Supporting Information, Figure S1C–D. These results indicate that the observed changes in ΔMR signal are specific to a folate receptor/anti-folate antibody interaction and not to a non-specific interaction between the designed bMR nanosensors and the HeLa cells. In additional experiments, we used the anti EpCAM antibody conjugated iron oxide nanoparticles as bMR nanosensors to measure the KD between the EpCAM antibody and EpCAM receptors in MCF-7 cells (Figure 1F). Our results indicated a KD of 73 pM while the reported value in the literature is 550 pM.[23] Again, these studies were performed using the extracelluar domain of EpCAM (expresed and purified from yeast) and attached to a solid support for SPR studies. Taken together, these results indicate that our magnetic relaxation method can be used to study the interaction of small molecule ligand and proteins with cell surface receptors using intact cells with comparable accuracy to current methods, yet at a higher speed and potentially lower cost. As these experiments are performed within 30 minutes with cells in suspension, any endocytosis of the nanoparticles is minimized.
Next, we utilized the bMR nanosensor based competition assay to study the interaction between toxins and small molecules. Recent reports describe the interaction of doxorubicin with the tetanus toxin C fragment (TTC)[27] and galactose or dextran with cholera toxin B subunit (CTB).[28] The study of these interactions is important for the development of small-molecule-based therapeutics. For the TTC-doxorubicin interaction, a doxorubicin-carrying iron oxide nanoparticle was designed. Within 15 minutes, we were able to observe a sigmoidal response with increasing ΔMR-signal value upon incubation of the bMR nanosensors with TTC (4 nM) in the presence of increasing amounts (0.9 μM– 12 μM) of doxorubicin (Supporting Information, Figure S2A). Using this data a KD of 4.1 μM was calculated, which is close to the reported value of 9.4 μM determined using a similar competition assay to ours with a fluorescence readout instead of magnetic relaxation.[29] Next, we tested if rhein, an anthracycline antibiotic structurally similar to doxorubicin, binds to TTC. Results show that indeed rhein interacts with TTC with a Kd value of 33.6 μM, which is slightly higher (weaker affinity) to the interaction with doxorubicin (Figure S2B). To our knowledge this is the first time that rhein has been reported to bind TTC. This interaction can be atributed to the fact that both molecules posess an anthraquinone group, which in the case of doxorubicin has been reported to play a key role in TTC binding.[25] This observation points toward the use of our bMR nanosensor-based competition assay in structure activity relationship (SAR) studies, where the competition of structurally similar compounds toward binding to a particular protein or cellular receptor is studied by magnetic relaxation.
Meanwhile, the interaction of the cholera toxin B subunit (CTB) with dextran was studied using dextran-coated iron oxide nanoparticles as bMR nanosensors (Supporting Information, Figure S3A). Using our competition method, we found that the KD of the dextran – CTB interaction to be 4.9 μM. This value is substantially lower that the value we have previously reported using SPR (14 mM).[28] In that report, CTB was immobolized to the SPR gold plate, using a ganglioside as a linker. Likely, this approach may block sites where dextran binds to, therefore affecting the interaction between CTB and dextran. Hence as the spatial orientation of these entities is constrained by the adhering mechanism, this may affect the KD values. Our magnetic relaxation method using bMR sensors in solution is a homogeneous assay and therefore more sensitive than those involving the attachment of the target protein to a solid support.[30, 31]
Finally, we investigated whether the dextran-coated iron oxide nanoparticles can be used as bMR nanosensors to study the interaction of CTB with other carbohydates. We reasoned that this bMR nanosensor may be able to identify the KD of similar molecules, due to their structural resemblance and chemical composition (i.e. functional groups) between the nanoparticles’ coating and the screened molecules. These studies revealed that increasing concentrations of glucose, galactose, lactose and β-cyclodextrin disrupted the association between dextran-coated iron oxide nanoparticles and CTB (Supporting Information, Figure S3B–E). Hence, we determined that glucose had a KD of 36 μM, whereas the KD of galactose, lactose, and β-cyclodextrin were 3.5 μM, 88 μM, and 5.6 μM respectively. Summarizing, we observed that dextran has a lower affinity towards CTB than galactose and β-cyclodextrin, while lactose has the least affinity followed by glucose (KDGal < KDCyclo < KDDex < KDGlu < KDLac). To our knowledge, the interaction of these carbohydates with CTB has not been previously reported. Overall, this study’s findings are summarized s in Table 1.
Table 1.
Comparison of dissociation constant (KD) values determined in this study with those reported in the literature
| Interaction | KD[a] | KD [b] Reported |
|---|---|---|
| Avidin – Biotin | 3 fM | 1 fM [15] |
| Protein G – IgG | 0.4 pM | 50 pM [16] |
| Concanavalin A – Dextran | 11.8 nM | 90 nM [18] |
| Folate receptor – Folic Acid | 16 nM | 0.1 nM [26] |
| Folate receptor – anti-FR Ab | 0.19 nm | 2.2 nM [22] |
| EpCAM receptor – anti-EpCAM Ab | 73 pM | 550 pM [23] |
| TTC – Doxorubicin | 4.1 μM | 9.4 μM [29] |
| TTC – Rhein | 33.6 μM | N/A |
| CTB – Dextran | 4.9 μM | 14 mM [28] |
| CTB – Glucose | 36 μM | N/A |
| CTB – Galactose | 3.5 μM | N/A |
| CTB – Lactose | 88 μM | N/A |
| CTB – β-Cyclodextrin | 3.6 μM | N/A |
Values determined using bMR nanosensor competition assay
Values reported in the literature, corresponding refence in bracket
In summary, we have developed a novel homogeneous assay to measure KD values of molecular interactions on a nanoparticle surface, over a wide range of affinities using a simple NMR table-top relaxometer. Our method is fast, uses small amounts of targets and is performed using a magnetic nanoparticle as a ligand‘s labels. This is a unique method as it measures the interaction between ligands on the surface of a nanoparticle with either a solubilized proteins or a cell surface receptor on intact cells. When molecular interactions are studied using ligand conjugated nanoparticles and intact cells, avidity play a key role as multiple receptors can interact with multiple ligands on the nanoparticle, strenghtening the interaction and yielding a lower KD. As avidity plays a critical role in molecular recognition in nature, and multivalent nanoparticle are increasingly being used in sensing and therapeutic applications, a method, like ours, that can evaluate the contribution of these parameters in terms of KD values would provide insights in the molecular dynamics under these conditions. The contribution of avidity and nanoparticle multivalency, as well as differences in proteins used (rabbit vs goat IgG) contribute to the differences between KD values reported in the literature and those obtained by our nanoparticle based method. Furthermore, as our assay is performed using low concentrations of the bMR nanosensors with subfetomolar amounts of ligands, it could be utilized in competitive binding assays for quantification of IC50 as under these conditions Kd values are similar to IC50 values (Kd = IC50, where [labeled ligand] ≪ Kd).
Supplementary Material
Acknowledgments
The authors acknowledge funding by the National Institute of Health (GM084331) to JMP
References
- 1.Wilson WD. Science. 2002;295:2103. doi: 10.1126/science.295.5562.2103. [DOI] [PubMed] [Google Scholar]
- 2.Bornhop DJ, Latham JC, Kussrow A, Markov DA, Jones RD, Sorensen HS. Science. 2007;317:1732. doi: 10.1126/science.1146559. [DOI] [PubMed] [Google Scholar]
- 3.Gaster RS, Xu L, Han SJ, Wilson RJ, Hall DA, Osterfeld SJ, Yu H, Wang SX. Nat Nanotechnol. 2011;6:314. doi: 10.1038/nnano.2011.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mammen M, Choi SK, Whitesides GM. Angew Chem Int Ed. 1998;37:2755. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 5.MacBeath G, Schreiber SL. Science. 2000;289:1760. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 6.Ladbury JE. Biochem Soc Trans. 2010;38:888. doi: 10.1042/BST0380888. [DOI] [PubMed] [Google Scholar]
- 7.Perozzo R, Folkers G, Scapozza L. J Recept Signal Transduct Res. 2004;24:1. doi: 10.1081/rrs-120037896. [DOI] [PubMed] [Google Scholar]
- 8.Frey KA, Albin RL. Curr Protoc Neurosci. 2001;Chapter 1(Unit1):4. doi: 10.1002/0471142301.ns0104s00. [DOI] [PubMed] [Google Scholar]
- 9.Hulme EC, Trevethick MA. Br J Pharmacol. 2010;161:1219. doi: 10.1111/j.1476-5381.2009.00604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaittanis C, Santra S, Perez JM. Adv Drug Deliv Rev. 2010;62:408. doi: 10.1016/j.addr.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santra S, Kaittanis C, Grimm J, Perez JM. Small. 2009;5:1862. doi: 10.1002/smll.200900389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crayton SH, Tsourkas A. ACS Nano. 2011;5:9592. doi: 10.1021/nn202863x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thorek DL, Weisshaar CL, Czupryna JC, Winkelstein BA, Tsourkas A. Mol Imaging. 2011;10:206. [PubMed] [Google Scholar]
- 14.Kaittanis C, Santra S, Santiesteban OJ, Henderson TJ, Perez JM. J Am Chem Soc. 2011;133:3668. doi: 10.1021/ja1109584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livnah O, Bayer EA, Wilchek M, Sussman JL. Proc Natl Acad Sci U S A. 1993;90:5076. doi: 10.1073/pnas.90.11.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Zhou D, Browne H, Balasubramanian S, Klenerman D. Anal Chem. 2004;76:4446. doi: 10.1021/ac049512c. [DOI] [PubMed] [Google Scholar]
- 17.Akerstrom B, Bjorck L. J Biol Chem. 1986;261:10240. [PubMed] [Google Scholar]
- 18.Liang F, Pan T, Sevick-Muraca EM. Photochem Photobiol. 2005;81:1386. doi: 10.1562/2005-02-14-RA-440. [DOI] [PubMed] [Google Scholar]
- 19.Russell RB, Eggleston DS. Nat Struct Biol. 2000;7(Suppl):928. doi: 10.1038/80691. [DOI] [PubMed] [Google Scholar]
- 20.Grisshammer R, Buchanan SK. Struc Biol of Mem Prot, RSC Publishing. 2006 [Google Scholar]
- 21.White SH. Nature. 2009;459:344. doi: 10.1038/nature08142. [DOI] [PubMed] [Google Scholar]
- 22.Ebel W, Routhier EL, Foley B, Jacob S, McDonough JM, Patel RK, Turchin HA, Chao Q, Kline JB, Old LJ, Phillips MD, Nicolaides NC, Sass PM, Grasso L. Cancer Immun. 2007;7:6. [PMC free article] [PubMed] [Google Scholar]
- 23.Ruf P, Gires O, Jager M, Fellinger K, Atz J, Lindhofer H. Br J Cancer. 2007;97:315. doi: 10.1038/sj.bjc.6603881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Shen F, Freisheim JH, Gentry LE, Ratnam M. Biochem Pharmacol. 1992;44:1898. doi: 10.1016/0006-2952(92)90089-2. [DOI] [PubMed] [Google Scholar]
- 25.Parker N, Turk MJ, Westrick E, Lewis JD, Low PS, Leamon CP. Anal Biochem. 2005;338:284. doi: 10.1016/j.ab.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 26.Sudimack J, Lee RJ. Adv Drug Deliv Rev. 2000;41:147. doi: 10.1016/s0169-409x(99)00062-9. [DOI] [PubMed] [Google Scholar]
- 27.Cosman M, Lightstone FC, Krishnan VV, Zeller L, Prieto MC, Roe DC, Balhorn R. Chem Res Toxicol. 2002;15:1218. doi: 10.1021/tx025570m. [DOI] [PubMed] [Google Scholar]
- 28.Kaittanis C, Banerjee T, Santra S, Santiesteban OJ, Teter K, Perez JM. Bioconjug Chem. 2011;22:307. doi: 10.1021/bc100442q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lightstone FC, Prieto MC, Singh AK, Piqueras MC, Whittal RM, Knapp MS, Balhorn R, Roe DC. Chem Res Toxicol. 2000;13:356. doi: 10.1021/tx000009e. [DOI] [PubMed] [Google Scholar]
- 30.Tsai CS, Yu TB, Chen CT. Chem Commun (Camb) 2005:4273. doi: 10.1039/b507237a. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe S, Yoshida K, Shinkawa K, Kumagawa D, Seguchi H. Colloids Surf B Biointerfaces. 2010;81:570. doi: 10.1016/j.colsurfb.2010.07.061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.