Abstract
The St. Jude Children's Research Hospital (St. Jude) HIV-1 vaccine program is based on the observation that multiple antigenically distinct HIV-1 envelope protein structures are capable of mediating HIV-1 infection. A cocktail vaccine comprising representatives of these diverse structures (immunotypes) is therefore considered necessary to elicit lymphocyte populations that prevent HIV-1 infection. This strategy is reminiscent of that used to design a currently licensed and successful 23-valent pneumococcus vaccine. Three recombinant vector systems are used for the delivery of envelope cocktails (DNA, vaccinia virus, and purified protein), and each of these has been tested individually in phase I safety trials. A fourth FDA-approved clinical trial, in which diverse envelopes and vectors are combined in a prime-boost vaccination regimen, has recently begun. This trial will continue to test the hypothesis that a multi-vector, multi-envelope vaccine can elicit diverse B- and T-cell populations that can prevent HIV-1 infections in humans.
Keywords: clinical trial, envelope, HIV-1 vaccine, Immunology, multi-vector
INTRODUCTION
The human immune system consists of billions of cells (lymphocytes), subdivided into B- and T-cell populations. As lymphocytes develop, each undergoes a sophisticated process of recombination/splicing at the nucleic acid level that leads to expression of a unique cell surface receptor [B-cells express antibodies while T-cells express T-cell receptors (TCR)].1 As illustrated in Figure 1A, every B-cell bears a unique antibody with the capacity to bind and destroy a specific foreign pathogen by using a lock-and-key interaction. Similarly, each T-cell (not shown) bears a unique TCR that can bind a specific foreign peptide associated with a major histocompatibility complex (MHC) molecule. B-cells attack pathogens directly, while T-cells destroy infected target cells and assist other lymphocytes in their effector functions. De novo B-cell and T-cell development continues throughout life, providing an impressive surveillance system against an array of viruses and other human pathogens. The harnessing of this enormously diverse natural defense system forms the basis of our approach to HIV-1 vaccine development.
Figure 1.
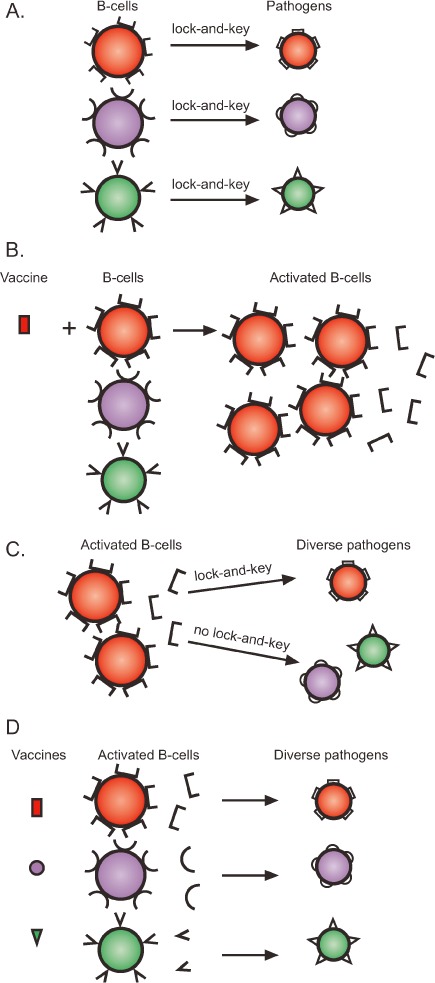
Rationale for the design of a multi-envelope HIV-1 vaccine. (A) B-cells have evolved to bear unique surface antibodies that bind and destroy pathogens with a lock-and-key interaction. (B) Vaccines can be designed to mimic pathogens and thereby induce specific B-cells to divide and secrete antibodies that target the pathogen. (C) The activation of B-cells (or T-cells) with a single-component vaccine harnesses only a fraction of the human immune potential. (D) A multi-component vaccine will activate a diverse population of immune cells that can counteract a broad array of target viruses.
LYMPHOCYTE DIVERSITY CAN BE EXPLOITED TO COUNTERACT HIV-1 DIVERSITY
Although lymphocyte populations are well equipped to destroy invading germs, they often exist in a resting state, unable to respond promptly. A pathogen mimic or “look-alike” can therefore be used as a vaccine to activate (or prime) B- and T-cell populations (Figure 1B illustrates B-cell activation). Vaccination induces the proliferation of antigen-specific lymphocytes and, in the case of B-cells, promotes secretion of antibodies into the blood and lymph. This priming process yields effector and memory cells that can persist for the lifetime of a vaccinated subject, providing an impressive barrier against future infection and disease.2
The ability of activated immune cells to prevent immunodeficiency virus infection was demonstrated in the early 1990s.3,4 As one example, Hu et al. prepared a vaccine comprising envelope glyco-protein (the outer coat protein) of simian immunodeficiency virus (SIV). Four macaques were vaccinated with the envelope-based vaccine while four macaques served as controls. When later challenged with an infectious clone of SIV that expressed an envelope protein identical to that in the vaccine, all four vaccinated animals were protected from infection. In contrast, all control animals became infected.4
Encouraged by these early successes, scientists prepared mono- or bivalent HIV-1 envelope vaccines5,6 for clinical study. However, in the human trials, unlike the situation for the non-human primates, the challenge viruses could not be pre-selected to share envelope antigens with the vaccines. When trial participants were naturally exposed to the diversity of HIV-1 isolates, their activated lymphocytes could not respond. Specifically, as illustrated in Figure 1C, the limited set of antibodies primed by mono- or bivalent-envelope vaccines could target only a subset of viruses (using lock-and-key interactions). Viruses with divergent envelope structures escaped the primed surveillance system.7,8
HIV-1 populations display impressive sequence diversity as the result of an error-prone reverse transcriptase and a lack of polymerase-related proofreading function.9,10 This sequence heterogeneity occurs throughout the HIV-1 genome and affects both internal and external viral antigens. Envelope protein, the primary target of neutralizing antibodies, encompasses five hypervariable regions that can differ substantially among isolates in both sequence and size.11–14 Constraints on envelope structure exist, as the protein must bind highly conserved molecules on human cells (e.g., CD4 and co-receptors such as CCR5).15 Nonetheless, the number of mutually exclusive envelope antigens able to mediate infection is likely greater than one or two, explaining (at least in part) the failure of mono- or bi-component vaccines to fully protect against HIV-1 infection in human clinical trials.
The variability of HIV-1 envelope proteins is reminiscent of the well-characterized antigenic variation of Streptococcus pneumoniae. Populations of bacteria comprise multiple variants, each with distinct surface antigens recognized by distinct subsets of lymphocytes. Accordingly, pneumococcus vaccine developers designed an effective 23-valent cocktail matching the predominant antigenic diversity of the target pathogen. The success of this strategy illustrates the protective capacity of an appropriately primed immune system.16 On the basis of the effectiveness of the pneumococcus vaccine and a number of other licensed cocktail vaccines, St. Jude researchers have designed an HIV-1 envelope cocktail (Figure 1D) intended to activate a lymphocyte repertoire sufficiently diverse to counter the diversity of HIV-1.17–23
ENCOURAGING IN-VIVO FINDINGS
Although no HIV-1 vaccine has yet shown signs of protection in a clinical trial, it should be emphasized that the tested vaccines have activated only a subset of immune cells, thereby harnessing only limited immune potential. The full potential of the immune system is better illustrated by studies in which monkeys were exposed to wild-type or attenuated SIV. Even though the animals could not clear resident virus from their first infection, they each generated protective immunity against subsequent challenges with SIV. Furthermore, serum from protected monkeys could be administered to naïve animals to transfer the protective effect. Essentially, the first infection acted as a natural cocktail vaccine against the second.24 How did this occur? We suggest that the following course of events provides an explanation: When virus exposure occurs before immune activation, some of the virus particles are quickly sequestered in privileged sites (e.g., brain tissue), where they remain permanently hidden from the immune system. The subsequent immune response clears much of the virus from the body but cannot clear the virus from privileged sites. Over the course of months, sequestered virus thrives, mutates, and sheds escape variants into the periphery, where respective immune responses are activated.25,26 Repetitive cycles of immune response, virus mutation, and virus escape continue until the peripheral immune system has been presented with a broad diversity of antigens. This process introduces the immune system to essentially all ‘shapes’ of HIV-1 compatible with infection. At this point, the diversity of primed B- and T-cell populations can successfully counter heterogeneous viruses from an exogenous source. The strength of the primed immune system explains the protection against superinfection associated with both SIV and HIV-1 infections.27,28
The association of chronic virus infection with protective immunity is not unique to SIV or HIV-1 but is characteristic of numerous pathogens, such as varicella zoster virus (VZV), Epstein-Barr virus (EBV), and cytomegalovirus (CMV). Each of these viruses can elicit immune cells capable of preventing superinfections, even though the very same cells cannot clear virus from privileged sites. Often, such viruses coexist with the host for years or decades without incident. If, however, the immune system is compromised (often as the result of an unrelated illness), virus reactivation can overwhelm the host. The fact that there is now a licensed, successful VZV vaccine demonstrates the necessity of activating immune cells as a prophylactic measure. By blocking virus entry, vaccine-induced immunity can prevent both chronic infection and virus-mediated disease.29
HETEROGENEOUS ANTIBODIES CAN WORK SYNERGISTICALLY TO NEUTRALIZE HIV-1
To demonstrate the value of combining diverse immune activities, we conducted antibody neutralization assays with single versus mixed monoclonal antibodies. For example, we tested the ability of two neutralizing monoclonal antibodies, B24 and 114-9C10, to inhibit HIV-1IIIB growth in tissue culture. When used at limiting concentrations, each antibody neutralized a small percentage of virus infectivity (Figure 2). When the two antibodies were mixed, their virus inhibition was synergistic: the absolute percent inhibition effected by the mixture was greater than the sum of the individual activities. This result might be ascribed to at least two, non-mutually exclusive mechanisms. First, the two antibodies are known to bind different portions of HIV-1 envelope and could thus simultaneously tag each virion and enhance pathogen elimination. Second, given that the HIV-1IIIB stock was a mixture of non-identical viruses,14 the antibodies could preferentially bind different viruses within the pool, increasing the size of the targeted population. Monoclonal antibody synergism has also been demonstrated by other investigators.30,31 Results emphasize the advantage of recruiting diverse effectors to inhibit HIV-1 growth.
Figure 2.
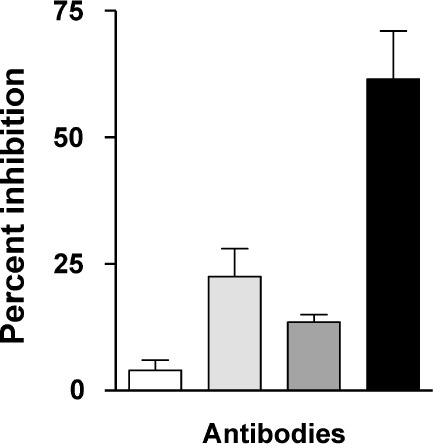
(□) Control. ( ) B24 only. (
) B24 only. ( ) 114-9C10 only. (▪) B24 + 114-9C10. A combination of antibodies mediates synergistic virus neutralization. Mice were vaccinated with a single HIV-1IIIB–derived envelope or with a combination of envelopes including envelope from HIV-1IIIB. Delivery vehicles included recombinant DNA, vaccinia virus, and protein. Splenic cells from vaccinated mice were then harvested for B-cell hybridoma production. Once stable hybridomas (B24 and 114-9C10) were derived and cloned, respective antibodies were harvested from hybridoma culture media and purified by affinity chromatography with protein G sepharose. The neutralization assay was initiated by incubating HIV-1IIIB (approximately 10 TCID-50 per well in a 96-well microtiter plate) with or without antibodies in R10 medium (RPMI 1640 plus 10% heat-treated fetal bovine serum, penicillin, streptomycin and 4mM glutamine). Antibodies were used at concentrations selected to yield fractional virus inhibition, in order to measure additive or synergistic effects (final concentrations of B24 and 114-9C10 were 0.05 μg/mL and 10 μg/mL, respectively). After overnight incubation, the contents of wells were transferred to confluent GHOST-CXCR4 cells in 96-well plates and incubated overnight. Cells were washed with R10 medium and incubated an additional 3 days, after which supernatants were analyzed for virus growth with a Coulter HIV-1 p24 antigen assay (Beckman-Coulter, Miami, FL). Percent inhibition was determined by comparing test wells with wells containing virus and no antibody. Control wells were with virus and HIV-1 negative human serum.
) 114-9C10 only. (▪) B24 + 114-9C10. A combination of antibodies mediates synergistic virus neutralization. Mice were vaccinated with a single HIV-1IIIB–derived envelope or with a combination of envelopes including envelope from HIV-1IIIB. Delivery vehicles included recombinant DNA, vaccinia virus, and protein. Splenic cells from vaccinated mice were then harvested for B-cell hybridoma production. Once stable hybridomas (B24 and 114-9C10) were derived and cloned, respective antibodies were harvested from hybridoma culture media and purified by affinity chromatography with protein G sepharose. The neutralization assay was initiated by incubating HIV-1IIIB (approximately 10 TCID-50 per well in a 96-well microtiter plate) with or without antibodies in R10 medium (RPMI 1640 plus 10% heat-treated fetal bovine serum, penicillin, streptomycin and 4mM glutamine). Antibodies were used at concentrations selected to yield fractional virus inhibition, in order to measure additive or synergistic effects (final concentrations of B24 and 114-9C10 were 0.05 μg/mL and 10 μg/mL, respectively). After overnight incubation, the contents of wells were transferred to confluent GHOST-CXCR4 cells in 96-well plates and incubated overnight. Cells were washed with R10 medium and incubated an additional 3 days, after which supernatants were analyzed for virus growth with a Coulter HIV-1 p24 antigen assay (Beckman-Coulter, Miami, FL). Percent inhibition was determined by comparing test wells with wells containing virus and no antibody. Control wells were with virus and HIV-1 negative human serum.
ASSEMBLY OF A COCKTAIL VACCINE REPRESENTING ANTIGENICALLY DISTINCT VIRAL PROTEINS
History has shown that a diverse pathogen can be adequately represented with a manageable cocktail of vaccine antigens. This is in part due to the fact that non-identical amino acid sequences often fold to yield similar three-dimensional conformations. Thus, while a researcher may be overwhelmed by overall amino acid sequence differences between certain proteins, the immune system may view the same proteins as being the same. Antibody binding/neutralization studies can be used by the researcher to visualize antigenic versus sequence differences. Figure 3 provides an illustration of this concept. Here are shown six hypothetical HIV-1 envelope proteins, each with a different amino acid sequence (represented as a string of colored beads). In this hypothetical example, the first three proteins fold to juxtapose amino acids 5 and 18, a configuration required both for envelope binding to CD4 molecules and for envelope binding to an inhibitory antibody. Binding experiments with the inhibitory antibody may be used to identify the three-member protein group and to select one member as a representative. The second set of three proteins might fold differently to support a variant mechanism of infection. This group might be bound and inhibited by a different antibody. Again, antibody experiments may be used to identify the group and select a representative. Upon completion of experiments, a mixed vaccine with only two components may be formulated to represent all six proteins.
Figure 3.
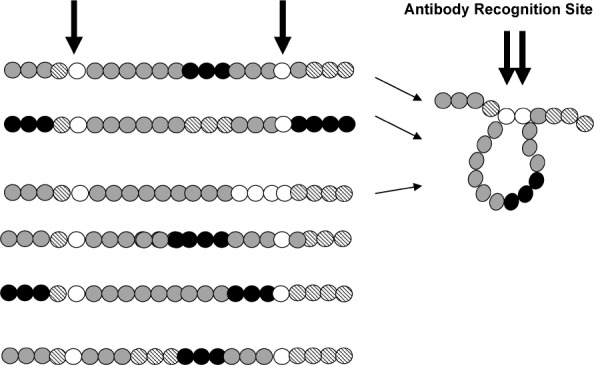
Antibodies can recognize conformationally similar antigenic determinants among proteins with diverse amino acid sequences. Each of these six hypothetical envelope proteins is represented as a string of beads, with different colors representing different amino acid residues. The successful capture of HIV-1 envelope as a crystal structure has been difficult and the prediction of three-dimensional protein structure cannot easily be predicted based solely on amino acid sequences. Antibodies may thus be used to assist in the characterization and categorization of envelopes. For example, the binding and neutralization of the first three envelopes (among six) by a specific monoclonal antibody may reveal a shared conformational structure that may be pertinent to a specific mechanism of infectivity. Envelopes with such similar conformational structures might be represented by a single component in a vaccine cocktail, and envelopes with other common structures (e.g., envelopes 4–6 in this series) might require a different single vaccine component as a representative. Information from antibody-antigen analyses has assisted the design of successful cocktail vaccines in other research fields.
These considerations, while oversimplified here for the purpose of illustration, have assisted researchers in other fields to develop successful vaccines. Additional considerations include: 1) the existence of multiple epitopes within a single protein, 2) the effect of context on epitope structure/function/exposure,32–35 3) differences between antigenicity and immunogenicity, and 4) the importance of capturing qualitatively diverse T-cell, as well as B-cell, epitopes in a vaccine.1
While vaccine formulation is complex in nature, there is consolation in the fact that (as stated above) HIV-1 envelope structures are constrained by function. Indeed, other pathogens characterized by high sequence diversity have been prevented by relatively simple cocktail vaccines.17,36,37 Clinical studies of HIV-1 multi-envelope vaccines may ultimately show that the number of antigenic components in a successful cocktail, while more than one or two, need not be vast.
SELECTION OF VACCINE DELIVERY VEHICLES
The selection of appropriate vectors with which to deliver multi-envelope vaccines must also be considered. St. Jude investigators have therefore examined a number of vehicles including recombinant DNA (D), recombinant vaccinia virus (V), and recombinant purified envelope proteins (P). It was found that when delivery vehicles were alternated and when vaccines were delivered successively (approximately once per month) in prime-boost regimens, the most potent and durable immune responses could be elicited. This strategy induced both B- and T-cell responses that could be identified at systemic and mucosal sites and that remained detectable for the lifetime of the vaccinated animals.38–40
TESTING OF A MULTI-VECTOR, MULTI-ENVELOPE VACCINE IN A NON-HUMAN PRIMATE SYSTEM
Following the preparation of envelope cocktails and the testing of D, V, and P delivery vehicles, St. Jude investigators initiated a combination vaccine study at the Tulane Regional Primate Center. Six macaques received sequential inoculations of recombinant D (intramuscularly), recombinant V (subcutaneously), and recombinant P (intramuscularly). The immunogenicity of this regimen was immediately evident: all six animals scored positively for HIV-1-specific antibodies in a standard clinical ELISA (Abbott Laboratories). Antibody-dependent cell-mediated cytotoxicity responses, antibody-mediated neutralizing responses, and T-cell responses (measured by interferon-gamma production) were also identified in every animal. In addition, upon challenge with a SHIV 89.6P retrovirus (a chimeric virus created by combining HIV-1 and SIV components), vaccinated animals showed statistically significant control of disease compared to unvaccinated controls, both in terms of CD4+ T-cell maintenance and reduced viral load. The result provided proof-of-concept that at least one envelope in the cocktail, although heterologous to the 89.6P envelope, was of sufficient similarity to elicit protective B- and T-cell responses. This result must be viewed with some caution, as 89.6P SHIV is a manmade virus grown in an unnatural host.41 Nonetheless, this experiment was the first to reveal control of 89.6P SHIV-mediated disease with a vaccine that contained no SIV components or 89.6P envelope sequences.23 The success provided further incentive to test the multi-envelope vaccine concept in human trials.
CLINICAL EVALUATION OF THE SJCRH MULTI-ENVELOPE HIV-1 VACCINE
Thus far, three FDA-approved clinical trials have demonstrated (individually) the safety of the D, V, and P vaccine components. In the P vaccine trial, every participant responded with HIV-1-specific immune activity. An FDA-approved, multi-envelope D-V-P prime-boost clinical trial has now begun. Researchers predict that the D-V-P multi-envelope vaccine will elicit durable B-cell and T-cell responses in virtually every volunteer.
CONCLUSION
The administration of cocktail vaccines is a logical approach for protection against HIV-1, because it exploits the inherent capacity of the immune system to recognize an enormous number of variant epitopes. St. Jude researchers have shown that HIV-1 envelope protein cocktails delivered with three different vehicles yield durable B- and T-cell responses. The multi-vector, multi-envelope prime-boost protocol was also shown to protect monkeys from SHIV-induced disease. Ongoing clinical trials will continue to test the hypothesis that envelope cocktails can ultimately confer complete protection against HIV-1 infections in humans.
ACKNOWLEDGMENTS
We thank Harold Stamey from the Tennessee Blood Services for providing samples for this study, Dr. R. Srinivas and the NIH AIDS Reagent Program (Rockville, MD) for providing HIV-1IIIB stock viruses, and Drs. KewalRamani and Littman and the NIH AIDS Reagent Program for providing the Ghost CXCR4 cells. We thank Julie Groff(Biomedical Communications, St. Jude) and Sharon Naron (Scientific Editing, St Jude) for artwork and critical editorial review, respectively. This work was supported in part by NIH NIAID: P01-AI45142 and R21-AI56974, NIH NCRR base grant P51-RR00164 to the Tulane National Primate Research Center, the Federated Department Stores, the Mitchell Fund, the Anderson Foundation, the Pendleton Fund, the Pioneer Fund and the American Lebanese Syrian-Associated Charities (ALSAC).
ABBREVIATIONS
- MHC
major histocompatibility complex
- SIV
simian immunodeficiency virus
- TCR
T-cell receptors
- VZV
varicella zoster virus
Footnotes
DISCLOSURE The authors declare no conflicts or financial interest in any product or service mentioned in the manuscript, including grants, equipment, medications, employment, gifts, and honoraria.
REFERENCES
- 1.Janeway CA, Jr., Travers P, Walport M. Immunobiology, the immune system in health and disease. New York, NY: Garland Publishing; 2005. et al. [Google Scholar]
- 2.Hammarlund E, Lewis MW, Hansen SG. Duration of antiviral immunity after smallpox vaccination. Nat Med. 2003;9:1131–1137. doi: 10.1038/nm917. et al. [DOI] [PubMed] [Google Scholar]
- 3.Berman PW, Gregory TJ, Riddle L. Protection of chimpanzees from infection by HIV-1 after vaccination with recombinant glycoprotein gp120 but not gp160. Nature. 1990;345:622–625. doi: 10.1038/345622a0. et al. [DOI] [PubMed] [Google Scholar]
- 4.Hu SL, Abrams K, Barber GN. Protection of macaques against SIV infection by subunit vaccines of SIV envelope glycoprotein gp160. Science. 1992;255:456–459. doi: 10.1126/science.1531159. et al. [DOI] [PubMed] [Google Scholar]
- 5.Belshe RB, Graham BS, Keefer MC. Neutralizing antibodies to HIV-1 in seronegative volunteers immunized with recombinant gp120 from the MN strain of HIV-1. JAMA. 1994;272:475–480. doi: 10.1001/jama.272.6.475. et al. [DOI] [PubMed] [Google Scholar]
- 6.Graham BS, Matthews TJ, Belshe RB. Augmentation of human immunodeficiency virus type 1 neutralizing antibody by priming with gp160 recombinant vaccinia and boosting with rgp160 in vaccinia-naive adults. J Infect Dis. 1993;167:533–537. doi: 10.1093/infdis/167.3.533. the NIAID AIDS vaccine clinical trials network. [DOI] [PubMed] [Google Scholar]
- 7.Belshe RB, Bolognesi DP, Clements ML. HIV infection in vaccinated volunteers. JAMA. 1994;272:431. doi: 10.1001/jama.272.6.431b. et al. [DOI] [PubMed] [Google Scholar]
- 8.Berman PW, Gray AM, Wrin T. Genetic and immunologic characterization of viruses infecting MN-rgp120-vaccinated volunteers. J Infect Dis. 1997;176:384–397. doi: 10.1086/514055. et al. [DOI] [PubMed] [Google Scholar]
- 9.Preston BD, Poiesz BJ, Loeb LA. Fidelity of HIV-1 reverse transcriptase. Science. 1988;242:1168–1171. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- 10.Roberts JD, Bebenek K, Kunkel TA. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242:1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 11.Rencher SD, Hurwitz JL. Effect of natural HIV-1 envelope V1–V2 sequence diversity on the binding of V3 and non-V3-specific antibodies. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;16:69–73. doi: 10.1097/00042560-199710010-00001. [DOI] [PubMed] [Google Scholar]
- 12.Rencher SD, Lockey TD, Slobod KS. Drift from the GPGRAF HIV-1 envelope V3 crown sequence in a North American inner city. AIDS Res Hum Retroviruses. 1997;13:527–528. doi: 10.1089/aid.1997.13.527. et al. [DOI] [PubMed] [Google Scholar]
- 13.Slobod KS, Rencher SD, Farmer A. HIV type 1 envelope sequence diversity in an inner city community. AIDS Res Hum Retroviruses. 1994;10:873–875. doi: 10.1089/aid.1994.10.873. et al. [DOI] [PubMed] [Google Scholar]
- 14.Lockey TD, Slobod KS, Rencher SD. Fluctuating diversity in the HTLV-IIIB virus stock: Implications for neutralization and challenge experiments. AIDS Res Hum Retroviruses. 1996;12:1297–1299. doi: 10.1089/aid.1996.12.1297. et al. [DOI] [PubMed] [Google Scholar]
- 15.Siciliano SJ, Kuhmann SE, Weng Y. A critical site in the core of the CCR5 chemokine receptor required for binding and infectivity of human immunodeficiency virus type 1. J Biol Chem. 1999;274:1905–1913. doi: 10.1074/jbc.274.4.1905. et al. [DOI] [PubMed] [Google Scholar]
- 16.Biagini RE, Schlottmann SA, Sammons DL. Method for simultaneous measurement of antibodies to 23 pneumococcal capsular polysaccharides. Clin Diagn Lab Immunol. 2003;10:744–750. doi: 10.1128/CDLI.10.5.744-750.2003. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurwitz JL, Slobod KS, Lockey TD. Application of the polyvalent approach to HIV-1 vaccine development. Curr Drug Targets Infect Disord. 2005;5:143–156. doi: 10.2174/1568005054201517. et al. [DOI] [PubMed] [Google Scholar]
- 18.Slobod KS, Coleclough C, Bonsignori M. HIV vaccine rationale, design and testing. Curr HIV Res. 2005;3:107–112. doi: 10.2174/1570162053506928. et al. [DOI] [PubMed] [Google Scholar]
- 19.Zhan X, Slobod KS, Surman S. Minor components of a multi-envelope HIV vaccine are recognized by type-specific T-helper cells. Vaccine. 20;22:1206–1213. doi: 10.1016/j.vaccine.2003.09.028. et al. [DOI] [PubMed] [Google Scholar]
- 20.Slobod KS, Brown SA, Surman S. Overcoming diversity with a multi-envelope HIV vaccine. Curr Top Virol. 2004;4:159–168. et al. [Google Scholar]
- 21.Lockey TD, Slobod KS, Caver TE. Multi-envelope HIV vaccine safety and immunogenicity in small animals and chimpanzees. Immunol Res. 2000;21:7–21. doi: 10.1385/IR:21:1:7. et al. [DOI] [PubMed] [Google Scholar]
- 22.Rencher SD, Slobod KS, Dawson D. Does the key to a successful HIV vaccine lie among the envelope sequences of infected individuals? AIDS Res Hum Retroviruses. 1995;11:1131–1133. doi: 10.1089/aid.1995.11.1131. et al. [DOI] [PubMed] [Google Scholar]
- 23.Zhan X, Martin LN, Slobod KS. Multi-envelope HIV-1 vaccine devoid of SIV components controls disease in macaques challenged with heterologous pathogenic SHIV. Vaccine. 2005;23:5306–5320. doi: 10.1016/j.vaccine.2005.07.008. et al. [DOI] [PubMed] [Google Scholar]
- 24.Daniel MD, Kirchhoff F, Czajak SC. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science. 1992;258:1938–1941. doi: 10.1126/science.1470917. et al. [DOI] [PubMed] [Google Scholar]
- 25.Wrin T, Crawford L, Sawyer L. Neutralizing antibody responses to autologous and heterologous isolates of human immunodeficiency virus. J Acquir Immune Defic Syndr. 1994;7:211–219. et al. [PubMed] [Google Scholar]
- 26.Richman DD, Wrin T, Little SJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsui R, Herring BL, Barbour JD. Human immunodeficiency virus type 1 superinfection was not detected following 215 years of injection drug user exposure. J Virol. 2004;78:94–103. doi: 10.1128/JVI.78.1.94-103.2004. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzales MJ, Delwart E, Rhee SY. Lack of detectable human immunodeficiency virus type 1 superinfection during 1072 person-years of observation. J Infect Dis. 2003;188:397–405. doi: 10.1086/376534. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldman GS. Incidence of herpes zoster among children and adolescents in a community with moderate varicella vaccination coverage. Vaccine. 2003;21:4243–4249. doi: 10.1016/s0264-410x(03)00459-6. [DOI] [PubMed] [Google Scholar]
- 30.Hofmann-Lehmann R, Vlasak J, Rasmussen RA. Postnatal passive immunization of neonatal macaques with a triple combination of human monoclonal antibodies against oral simian-human immunodeficiency virus challenge. J Virol. 2001;75:7470–7480. doi: 10.1128/JVI.75.16.7470-7480.2001. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mascola JR, Louder MK, VanCott TC. Potent and synergistic neutralization of human immunodeficiency virus (HIV) type 1 primary isolates by hyperimmune anti-HIV immunoglobulin combined with monoclonal antibodies 2F5 and 2G12. J Virol. 1997;71:7198–7206. doi: 10.1128/jvi.71.10.7198-7206.1997. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown SA, Lockey TD, Slaughter C. T cell epitope “hotspots” on the HIV Type 1 gp120 envelope protein overlap with tryptic fragments displayed by mass spectrometry. AIDS Res Hum Retroviruses. 2005;21:165–170. doi: 10.1089/aid.2005.21.165. et al. [DOI] [PubMed] [Google Scholar]
- 33.Brown SA, Stambas J, Zhan X. Clustering of Th cell epitopes on exposed regions of HIV envelope despite defects in antibody activity. J Immunol. 2003;171:4140–4148. doi: 10.4049/jimmunol.171.8.4140. et al. [DOI] [PubMed] [Google Scholar]
- 34.Surman S, Lockey TD, Slobod KS. Localization of CD4+ T cell epitope hotspots to exposed strands of HIV envelope glycoprotein suggests structural influences on antigen processing. Proc Natl Acad Sci USA. 2001;98:4587–4592. doi: 10.1073/pnas.071063898. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown SA, Slobod KS, Surman S. Individual HIV type 1 envelope-specific T cell responses and epitopes do not segregate by virus subtype. AIDS Res Hum Retroviruses. 2006;22:188–194. doi: 10.1089/aid.2006.22.188. et al. [DOI] [PubMed] [Google Scholar]
- 36.Clark HF, Offit PA, Plotkin SA. The new pentavalent rotavirus vaccine composed of bovine (strain WC3)-human rotavirus reassortants. Pediatr Infect Dis J. 2006;25:577–583. doi: 10.1097/01.inf.0000220283.58039.b6. et al. [DOI] [PubMed] [Google Scholar]
- 37.Offit PA. Rotavirus vaccines: round two. Hum Vaccin. 2006;2:84–85. doi: 10.4161/hv.2.2.2612. [DOI] [PubMed] [Google Scholar]
- 38.Richmond JF, Mustafa F, Lu S. Screening of HIV-1 Env glycoproteins for the ability to raise neutralizing antibody using DNA immunization and recombinant vaccinia virus boosting. Virology. 1997;230:265–274. doi: 10.1006/viro.1997.8478. et al. [DOI] [PubMed] [Google Scholar]
- 39.Caver TE, Lockey TD, Srinivas RV. A novel vaccine regimen utilizing DNA, vaccinia virus and protein immunizations for HIV-1 envelope presentation. Vaccine. 1999;17:1567–1572. doi: 10.1016/s0264-410x(98)00355-7. et al. [DOI] [PubMed] [Google Scholar]
- 40.Stambas J, Brown SA, Gutierrez A. Long lived multi-isotype anti-HIV antibody responses following a prime-double boost immunization strategy. Vaccine. 2005;23:2454–2464. doi: 10.1016/j.vaccine.2004.10.030. et al. [DOI] [PubMed] [Google Scholar]
- 41.Reimann KA, Li JT, Veazey R. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J Virol. 1996;70:6922–6928. doi: 10.1128/jvi.70.10.6922-6928.1996. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]


