Abstract
Cell specific delivery of therapeutic agents using ligand targeting is gaining interest due to the potential for increased efficacy and reduced side effects. The challenge is to develop a suitable ligand of a cell surface receptor that is selectively expressed on the desired cell. Sialoadhesin (Sn/Siglec-1/CD169) a sialic-acid-binding Ig-like lectin (Siglec) expressed on subsets of resident and inflammatory macrophages is an attractive target for development of a ligand-targeted delivery system. Here, we report the development of a high affinity and selective ligand of Sn capable of targeting liposomal nanoparticles to Sn expressing cells in vivo comprising an analog of the natural ligand. An efficient in silico screen of a library of ~8400 carboxylic acids was key to identify novel 9-N-acyl Neu5Ac substituents as potential lead compounds. From the screen a small panel of targets was selected and synthesized to evaluate their affinity and selectivity. The most potent of these Sn ligands, 9-N-(4H-thieno[3,2-c]chromene-2-carbamoyl)-Neu5Acα2-3Galβ1-4GlcNAc (TCCNeu5Ac), was conjugated to lipids for display on a liposomal nanoparticle for evaluation of targeted delivery to cells. The TCCNeu5Ac liposomes are shown to selectively target liposomes in vitro to cells expressing either murine or human Sn, and when administered to mice, exhibit in vivo targeting to Sn positive macrophages.
Ligand targeted liposomal nanoparticles offer promising applications in human medicine for selective delivery of therapeutic agents to the desired cells resulting in increased efficacy and decreased side effects.1-5 The challenge is to identify cell surface receptors that are selectively expressed on the targeted cell, and to develop ligands that target and bind that receptor with high selectivity. Siglecs, a family of sialic-acid-binding Ig-like lectins with restricted expression on one or a few immune cell types, represent attractive targets for cell-directed therapies.6,7 Among them, sialoadhesin (Sn/Siglec-1/CD169) is an endocytic surface receptor expressed on subsets of resident and inflammatory macrophages, and has a preference to bind glycan ligands with the Neu5Acα2-3Galβ1-4GlcNAc sequence.8-10 Because macrophages have both protective and pathological activities including antitumor immune response, allergy and asthma, atherosclerosis and wound healing,11,12 the restricted expression and endocytic properties of Sn make it an ideal receptor for development of a macrophage targeted delivery system for therapeutic intervention.
In general, Siglecs bind with low intrinsic affinity (0.1-1 mM) to their natural sialoside ligands.13 Several reports have demonstrated the importance of sialic acid substituents (e.g., C9 of Neu5Ac) for increasing the affinity and selectivity of ligand binding to siglecs.8,14-17 In this regard, an exemplary ligand for CD22 (Siglec-2) on B cells, 9-N-biphenylcarbamoyl-Neu5Acα2-6Galβ1-4GlcNAc (6′-BPCNeu5Ac) was found to support targeting of chemotherapeutic loaded liposomes and prolong life in a murine model of human B cell lymphoma.18 More recently we have shown that liposomes decorated with a related ligand, 9-N-biphenylcarbamoyl-Neu5Acα2-3Galβ1-4GlcNAc (1, 3′-BPCNeu5Ac), were capable of targeting Sn expressing cells in vitro, however, this ligand was not sufficiently selective for in vivo targeting.19 Here, we describe our approach aided by in silico design used for development of a high affinity ligand of Sn for in vivo targeting of macrophages.
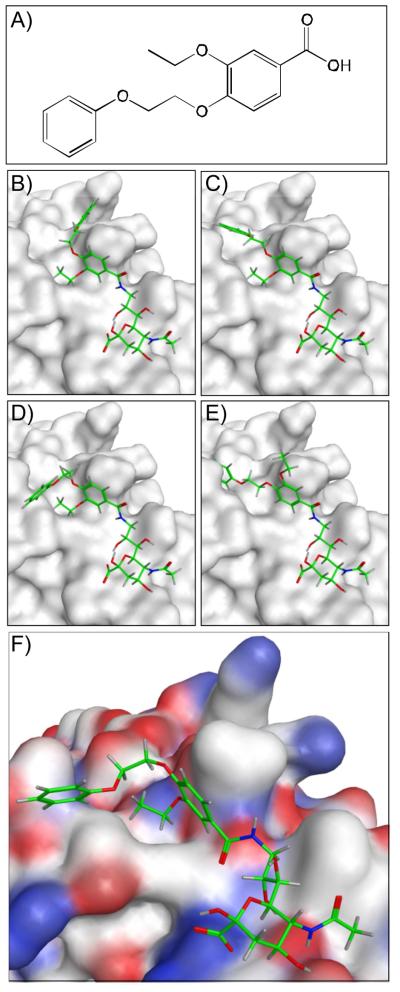
In order to develop ligands of higher selectivity and affinity for Sn we adopted a strategy that takes advantage of the existing crystal structure to identify novel 9-N-acyl Neu5Ac substituents which could favorably bind to Sn in the hydrophobic pocket occupied by the previously described biphenyl substituent. To this end we conducted a virtual screen of a library of carboxylic acids guided by the co-crystal structure of murine Sn and BPCNeu5Ac-OMe.8 As an alternative to solution based screening methods, which would require extensive synthetic effort, the in silico approach has the advantage of rapidly screening large compound libraries to identify lead structures.20,21 Figure 1 outlines the in silico screening approach for one representative carboxylic acid (Figure 1a). Initially, up to 250 conformers were calculated for each of ~8400 carboxylic acids from a commercial building block library. The resulting conformations were treated as unique acid structures then virtually coupled to the amino group of 9-NH2-Neu5Ac fixed within the binding pocket. An aromatic ring pharmacophore was implemented using the coordinates of the first benzene ring of the biphenyl substituent in BPCNeu5Ac-OMe. The tethered docking of the acid conformers was scored based on London dispersion energy using Molecular Operating Environment (MOE). Four representative solutions from this tethered docking approach are shown in Figure 1b-e. From this preliminary evaluation the top 3000 poses were selected for further inspection using AutoDock 4.2.22 Figure 1f depicts a tethered AutoDock solution for the representative carboxylic acid. The final non-tethered docking solutions resembled the canonical sialic acid binding pose and provided a ranking of the acids based on calculated binding energies. From this ranking a small panel of six target structures were selected from the top ranked 100 (2-7) for their structural diversity based on the computed 2D molecular fingerprints of the corresponding acids (Table 1). In addition, as “non-ranked’ controls, two additional sialosides (8 and 9) were selected with N-acyl substituents that were eliminated from the screen at an early stage.
Figure 1.
Representative structures from the in silico screening strategy to identify high affinity ligands of Sialoadhesin (Sn). (A) Representative carboxylic acid from a commercial building block library that were screened as potential substituents of 9-NH2-Neu5Ac. (B-E) Four representative solutions from the tethered docking approach highlighting varying docking poses of the tethered acid substituent. Conformations of the respective substituents were calculated and conjugated via in silico amide coupling to 9-NH2-Neu5Ac, which was fixed within the binding site. The top 3000 hits from this initial docking evaluation were further inspected using AutoDock. (F) Representative AutoDock solution for the selected carboxylic acid coupled to 9-NH2-Neu5Ac. Oxygen, carbon, and nitrogen atoms are highlighted in red, white, and blue, respectively.
Table 1. Inhibitory potencies of sialoside analogues (1-9) against murine Sn.a.

| Compound | R = | IC50 (μM) |
|---|---|---|
| 1 |

|
4.82 ±0.14 |
| 2 |
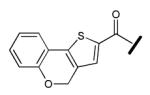
|
0.38 ±0.04 |
| 3 |
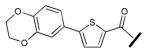
|
7.25 ±0.61 |
| 4 |
|
21.6 ±2.1 |
| 5 |

|
25.8 ±1.4 |
| 6 |

|
>200 |
| 7 |

|
>400 |
| 8 |
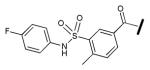
|
26.5 ±3.8 |
| 9 |
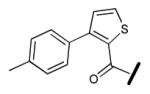
|
>500 |
All titrations were performed in triplicate, and standard deviation is given from three independent measurements.
All of the targets were synthesized chemo-enzymatically (Scheme 1). Briefly, Galβ1-4GlcNAc-ethyl azide 10 was reacted with CMP-9-NH2-Neu5Ac using Pasteurella multocida α2-3-sialyltransferase 1 (PmST1)23 to afford the trisaccharide scaffold 11. Divergent reaction of 11 with the panel of NHS activated carboxylic acids afforded the final targets (2-9). The reference ligand substituted with BPC (1) was also prepared.
Scheme 1. Chemo-enzymatic synthesis of C-9 N-acyl modified sialoside analogues.a.
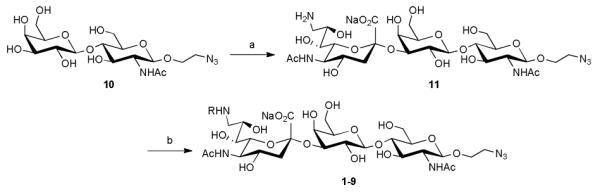
aReagents and conditions: a) CMP-9-NH2-Neu5Ac, PmST1; b) RC(O)NHS, THF, H2O (1-9, see Table 1 for R).
The inhibitory potencies (IC50) of the glycan derivatives (1-9) were evaluated in a flow cytometry assay based on the competitive binding of murine Sn-Fc chimera to beads decorated with the natural ligand Neu5Acα2-3Galβ1-4GlcNAc. The IC50 values required to displace the bound mSn-Fc were determined with serial dilutions of the competitors (Table 1 and Figure 2).
Figure 2.

Inhibitory potencies of selected sialoside analogs in a murine Sn competitive bead binding assay. Each compound was analyzed in triplicate for inhibition of the binding of mSn-Fc chimera to Neu5Acα2-3-Galβ1-4GlcNAc coated magnetic beads as described in the Supporting information. See Table 1 for glycans 1, 2 and 4.
A wide range of binding affinities was observed for the sialosides containing the top ranked substituents (2-7). Relative to the reference compound 1 with an IC50 of 4.8 μM, two were weak inhibitors (6 and 7; IC50 >200 μM), three were of comparable affinity (3-5; IC50 of 7.3-26 μM) and one was about 1-fold higher in affinity (7; IC50 0.38 QM). This result is particularly notable in view of the affinity of the natural non-substituted ligand, Neu5Acα2-3Galβ1-4GlcNAc, which gives an IC50 of 1300 μM in a comparable assay.13 Analysis of 7 in complex with Sn suggests that the bioisostere replacement of the first benzyl group in 1 provides improved shape complementarity and additional contacts to the protein (data not shown). Of the two unranked substituents, one was poorly active (9; IC50 >500 μM) and the other showed relatively high affinity 8 (IC50 of 26.5 μM). Thus, while our approach of using the in silico screen to identify target substituents requires further investigation to validate its potential and general utility, it provided a significant lead with minimal investment in synthetic resources. Accordingly, we proceeded to further evaluate the high affinity lead structure, 9-N-(4H-thieno[3,2-c]chromene-2-carbamoyl)-Neu5Acα 2-3Galβ1-4GlcNAc 2 (TCCNeu5Ac).
We next tested the selectivity of TCCNeu5Ac (2) for Sn when incorporated into targeted liposomes. To accomplish this, fluorescent targeted liposomes were formulated to include high affinity glycan ligand 2 coupled to pegylated lipid (see Supporting information), which were then tested for binding to a panel of cell lines expressing human and murine Siglecs.18,19 Cells expressing Siglecs were incubated with fluorescently labeled targeted liposomes then following a washing step were analyzed by flow cytometry. The results reveal that the TCCNeu5Ac ligand is highly selective for Sn (Figure 3). Only cells expressing murine or human Sn bound the targeted liposomes, while non-targeted “naked” liposomes did not bind any of the cells.
Figure 3.
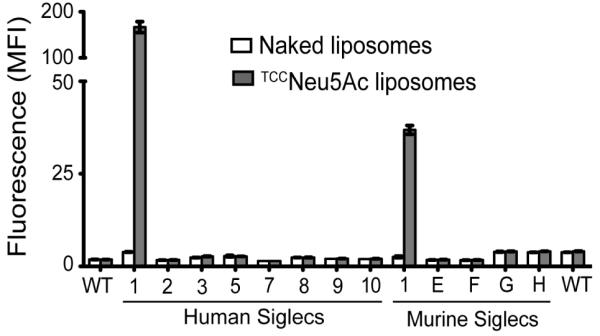
Fluorescence assisted cell sorting (FACS) analysis for in vitro binding of targeted TCCNeu5Ac or non-targeted “naked” liposomes to cells expressing murine and human Siglecs. Binding is shown as mean fluorescence intensity (MFI) ±SD (n = 3).
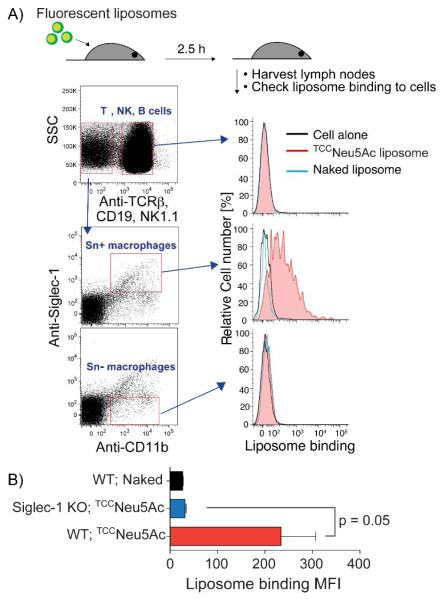
To evaluate the specificity of the TCCNeu5Ac (2) ligand in an in vivo setting, we sought to see if TCCNeu5Ac (2) decorated liposomes could target Sn expressing macrophages in peripheral lymph nodes. Accordingly, wild type or Sn knockout mice were subcutaneously injected in the flank with Alexa-647 labeled TCCNeu5Ac liposomes. After 2.5 hours, immune cells from neighboring lymph nodes on the same side were harvested and analyzed by flow cytometry. Populations of T, B, and NK lymphocytes and myeloid cells were identified using specific antibodies (Figure 4). Furthermore, Sn positive myeloid cells (macrophages) were identified using an anti-Sn antibody. The gated cell populations revealed that the TCCNeu5Ac liposomes effectively targeted Sn positive myeloid cells, observed as a right shift in the histogram (Figure 4a). There was no binding of targeted liposomes to T-, B-, NK, or Sn negative macrophages. Naked liposomes did not bind to any cells. Furthermore, no binding of targeted liposomes to macrophages from Sn knockout mice was detected indicating uptake of liposomes was Sn dependent (Figure 4b).
Figure 4.
TCCNeu5Ac liposomes target Sn+ macrophages in vivo. A) Wild type mice were injected subcutaneously with fluorescently labeled naked or TCCNeu5Ac liposomes and the neighboring lymph nodes were harvested after 2.5 h. Cells were stained with various antibodies as described in the Supporting Information and analyzed by flow cytometry. B) Wild type and Sn KO mice (n = 3 in each group) were injected with the liposomes. The binding of liposomes to macrophages (TCRb−NK1.1−CD19−CD11b+) were analyzed by flow cytometry. The geometric mean of fluorescent intensity (MFI) were plotted.
In summary, we describe the successful development of a high affinity ligand of Sn suitable for targeting liposomal nanoparticles to Sn expressing cells in vivo. An efficient virtual screen of a commercial building block library was performed to identify novel substituents of potential lead compounds. From this screen, a small panel of selected target structures was translated into sialoside derivatives with limited synthetic efforts owing to the design strategy. Consequently, a novel high affinity ligand of Sn was identified. Targeted liposomal nanoparticles displaying the ligand showed high selectivity for human and murine Sn expressing cells in vitro. Further evaluation of the targeted liposomes in mice showed effective in vivo targeting to Sn positive macrophages. We anticipate that delivery systems incorporating this novel Sn ligand will provide a powerful means for active in vivo delivery of antigens or therapeutic agents to macrophages.
Supplementary Material
ACKNOWLEDGMENT
C.R. wishes to thank EMBO for a long-term fellowship. This work was supported by the National Institute of Health (NIH) - P01HL107151 & R01AI05143.
Footnotes
ASSOCIATED CONTENT
Supplementary synthetic scheme, experimental protocols, synthetic methods, and compound characterization. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- (1).Torchilin VP. Nat Rev Drug Discov. 2005;4:145. doi: 10.1038/nrd1632. [DOI] [PubMed] [Google Scholar]
- (2).Trapani G, Denora N, Trapani A, Laquintana V. J Drug Target. 2012;20:1. doi: 10.3109/1061186X.2011.611518. [DOI] [PubMed] [Google Scholar]
- (3).Lepenies B, Yin JA, Seeberger PH. Curr Opin Chem Biol. 2010;14:404. doi: 10.1016/j.cbpa.2010.02.016. [DOI] [PubMed] [Google Scholar]
- (4).Jain K, Kesharwani P, Gupta U, Jain NK. Biomaterials. 2012;33:4166. doi: 10.1016/j.biomaterials.2012.02.033. [DOI] [PubMed] [Google Scholar]
- (5).Xie R, Hong SL, Feng LS, Rong J, Chen X. J Am Chem Soc. 2012;134:9914. doi: 10.1021/ja303853y. [DOI] [PubMed] [Google Scholar]
- (6).Crocker PR, Paulson JC, Varki A. Nat Rev Immunol. 2007;7:255. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- (7).O’Reilly MK, Paulson JC. Trends Pharmacol Sci. 2009;30:240. doi: 10.1016/j.tips.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Zaccai NR, Maenaka K, Maenaka T, Crocker PR, Brossmer R, Kelm S, Jones EY. Structure. 2003;11:557. doi: 10.1016/s0969-2126(03)00073-x. [DOI] [PubMed] [Google Scholar]
- (9).Delputte PL, Van Gorp H, Favoreel HW, Hoebeke I, Delrue I, Dewerchin H, Verdonck F, Verhasselt B, Cox E, Nauwynck HJ. Plos One. 2011;6 doi: 10.1371/journal.pone.0016827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Hartnell A, Steel J, Turley H, Jones M, Jackson DG, Crocker PR. Blood. 2001;97:288. doi: 10.1182/blood.v97.1.288. [DOI] [PubMed] [Google Scholar]
- (11).Murray PJ, Wynn TA. Nat Rev Immunol. 2011;11:723. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Kelly C, Jefferies C, Cryan SA. J Drug Deliv. 2011;2011:727241. doi: 10.1155/2011/727241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Blixt O, Collins BE, van den Nieuwenhof IM, Crocker PR, Paulson JC. J Biol Chem. 2003;278:31007. doi: 10.1074/jbc.M304331200. [DOI] [PubMed] [Google Scholar]
- (14).Blixt O, Han SF, Liao L, Zeng Y, Hoffmann J, Futakawa S, Paulson JC. J Am Chem Soc. 2008;130:6680. doi: 10.1021/ja801052g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Mesch S, Moser D, Strasser DS, Kelm A, Cutting B, Rossato G, Vedani A, Koliwer-Brandl H, Wittwer M, Rabbani S, Schwardt O, Kelm S, Ernst B. J Med Chem. 2010;53:1597. doi: 10.1021/jm901517k. [DOI] [PubMed] [Google Scholar]
- (16).Mesch S, Lemme K, Wittwer M, Koliwer-Brandl H, Schwardt O, Kelm S, Ernst B. ChemMedChem. 2012;7:134. doi: 10.1002/cmdc.201100407. [DOI] [PubMed] [Google Scholar]
- (17).Magesh S, Ando H, Tsubata T, Ishida H, Kiso M. Curr Med Chem. 2011;18:3537. doi: 10.2174/092986711796642580. [DOI] [PubMed] [Google Scholar]
- (18).Chen WC, Completo GC, Sigal DS, Crocker PR, Saven A, Paulson JC. Blood. 2010;115:4778. doi: 10.1182/blood-2009-12-257386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Chen WC, Kawasaki N, Nycholat CM, Han S, Pilotte J, Crocker PR, Paulson JC. Plos One. 2012;7:e39039. doi: 10.1371/journal.pone.0039039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Fadda E, Woods RJ. Drug Discov Today. 2010;15:596. doi: 10.1016/j.drudis.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Ernst B, Magnani JL. Nat Rev Drug Discov. 2009;8:661. doi: 10.1038/nrd2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. J Comput Chem. 2009;30:2785. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Yu H, Chokhawala H, Karpel R, Yu H, Wu BY, Zhang JB, Zhang YX, Jia Q, Chen X. J Am Chem Soc. 2005;127:17618. doi: 10.1021/ja0561690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.