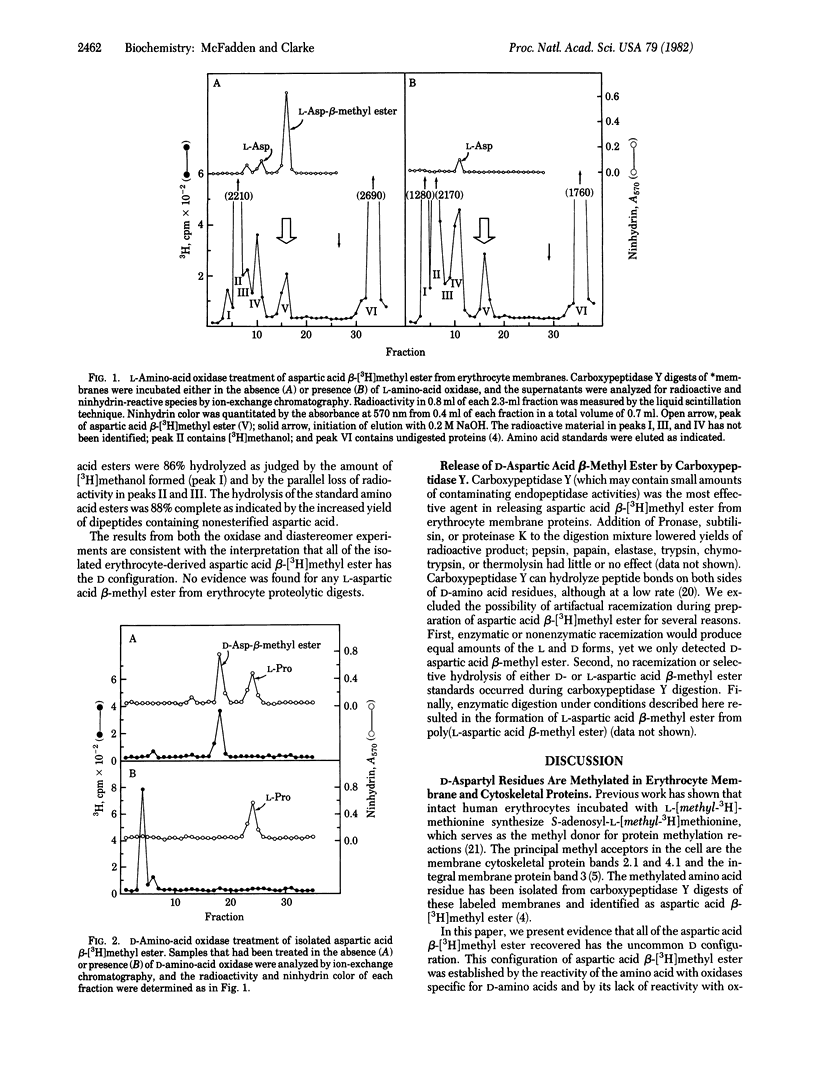
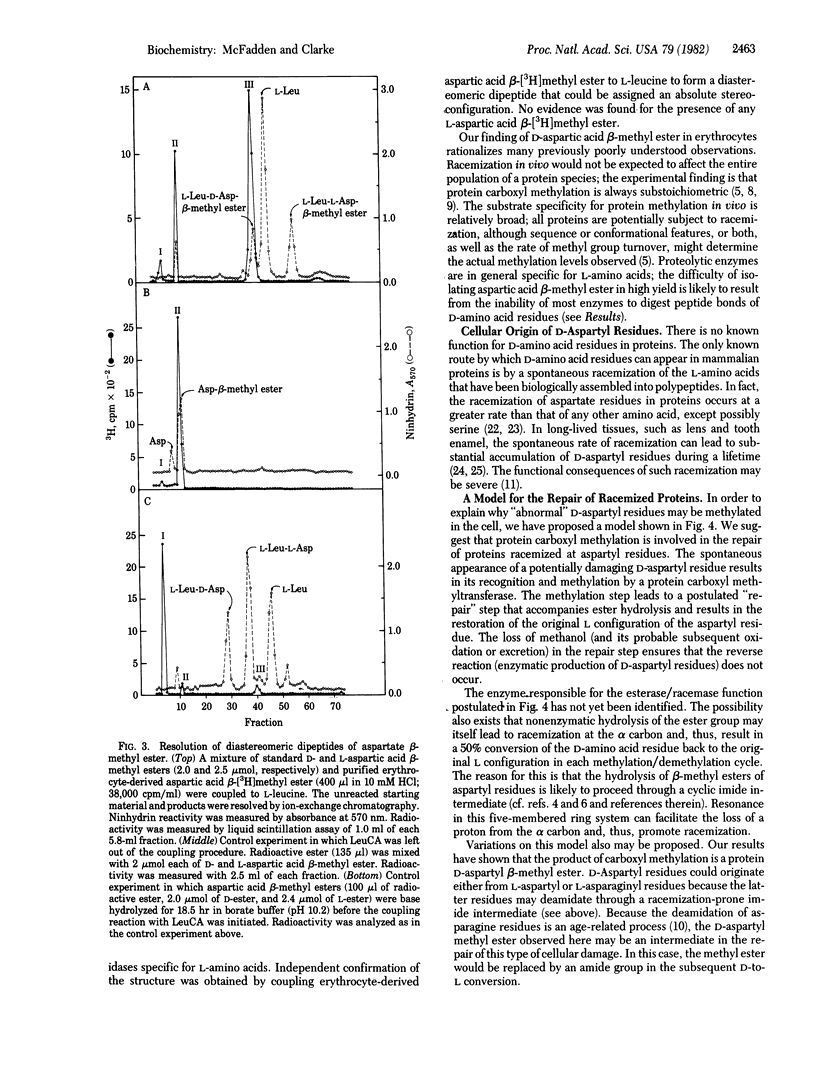
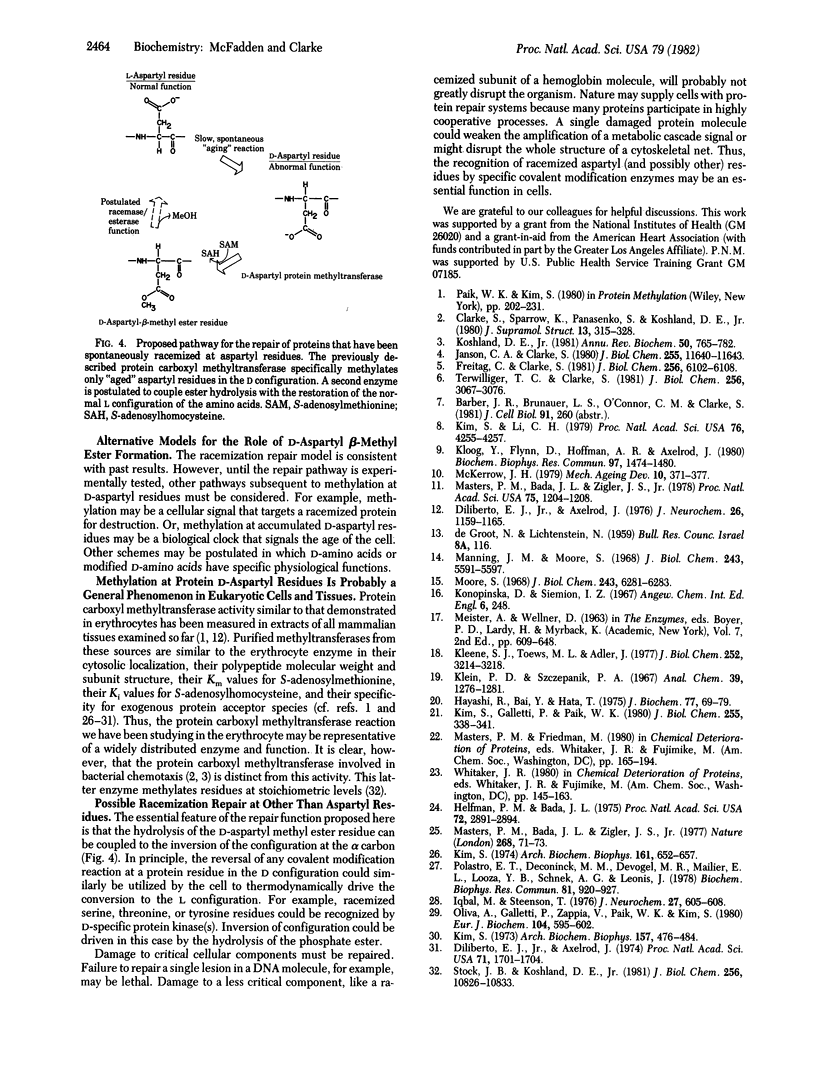
Abstract
Reversibly methylated aspartyl residues in human erythrocyte membrane proteins are shown to be in the "unnatural" D configuration. This is demonstrated by treatment of proteolytically derived aspartic acid beta-[3H]methyl ester with L- and D-amino-acid oxidases and by the resolution of diastereomeric L-leucyl dipeptides containing either L- or D-aspartic acid beta-methyl ester by ion-exchange chromatography. Based on this observation, we propose a novel role for eukaryotic protein carboxyl methyltransferases (protein O-methyltransferase; S-adenosyl-L-methionine:protein O-methyltransferase, EC 2.1.1.24). We suggest that these widely distributed enzymes function to recognize aspartyl residues that have racemized spontaneously for a subsequent repair reaction. This repair function is postulated to couple ester hydrolysis with the restoration of the original L configuration of the aspartyl residue. It is possible that similar types of racemization repair processes may occur by reversible covalent modifications at other residues. Other possible functions for D-aspartic acid beta-methyl ester residues in proteins are considered.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clarke S., Sparrow K., Panasenko S., Koshland D. E., Jr In vitro methylation of bacterial chemotaxis proteins: characterization of protein methyltransferase activity in crude extracts of Salmonella typhimurium. J Supramol Struct. 1980;13(3):315–328. doi: 10.1002/jss.400130305. [DOI] [PubMed] [Google Scholar]
- Diliberto E. J., Jr, Axelrod J. Characterization and substrate specificity of a protein carboxymethylase in the pituitary gland. Proc Natl Acad Sci U S A. 1974 May;71(5):1701–1704. doi: 10.1073/pnas.71.5.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diliberto E. J., Jr, Axelrod J. Regional and subcellular distribution of protein carboxymethylase in brain and other tissues. J Neurochem. 1976 Jun;26(6):1159–1165. doi: 10.1111/j.1471-4159.1976.tb07001.x. [DOI] [PubMed] [Google Scholar]
- Freitag C., Clarke S. Reversible methylation of cytoskeletal and membrane proteins in intact human erythrocytes. J Biol Chem. 1981 Jun 25;256(12):6102–6108. [PubMed] [Google Scholar]
- Hayashi R., Bai Y., Hata T. Kinetic studies of carboxypeptidase Y. I. Kinetic parameters for the hydrolysis of synthetic substrates. J Biochem. 1975 Jan 1;77(1?):69–79. [PubMed] [Google Scholar]
- Helfman P. M., Bada J. L. Aspartic acid racemization in tooth enamel from living humans. Proc Natl Acad Sci U S A. 1975 Aug;72(8):2891–2894. doi: 10.1073/pnas.72.8.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal M., Steenson T. Purification of protein carboxymethylase from ox brain. J Neurochem. 1976 Aug;27(2):605–608. doi: 10.1111/j.1471-4159.1976.tb12289.x. [DOI] [PubMed] [Google Scholar]
- Janson C. A., Clarke S. Identification of aspartic acid as a site of methylation in human erythrocyte membrane proteins. J Biol Chem. 1980 Dec 25;255(24):11640–11643. [PubMed] [Google Scholar]
- Kim S., Galletti P., Paik W. K. In vivo carboxyl methylation of human eruthrocyte membrane proteins. J Biol Chem. 1980 Jan 25;255(2):338–341. [PubMed] [Google Scholar]
- Kim S., Li C. H. Enzymatic methyl esterification of specific glutamyl residue in corticotropin. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4255–4257. doi: 10.1073/pnas.76.9.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. Purification and properties of protein methylase II. Arch Biochem Biophys. 1973 Aug;157(2):476–484. doi: 10.1016/0003-9861(73)90665-6. [DOI] [PubMed] [Google Scholar]
- Kim S. S-adenosylmethionine: protein-carboxyl methyltransferase from erythrocyte. Arch Biochem Biophys. 1974 Apr 2;161(2):652–657. doi: 10.1016/0003-9861(74)90350-6. [DOI] [PubMed] [Google Scholar]
- Kleene S. J., Toews M. L., Adler J. Isolation of glutamic acid methyl ester from an Escherichia coli membrane protein involved in chemotaxis. J Biol Chem. 1977 May 25;252(10):3214–3218. [PubMed] [Google Scholar]
- Klein P. D., Szczepanik P. A. Fine structure of isotopically labeled amino acids determined by ion exchange chromatography. Anal Chem. 1967 Sep;39(11):1276–1281. doi: 10.1021/ac60255a005. [DOI] [PubMed] [Google Scholar]
- Kloog Y., Flynn D., Hoffman A. R., Axelrod J. Enzymatic carboxymethylation of the nicotinic acetylcholine receptor. Biochem Biophys Res Commun. 1980 Dec 31;97(4):1474–1480. doi: 10.1016/s0006-291x(80)80031-3. [DOI] [PubMed] [Google Scholar]
- Konopińska D., Siemion I. Z. Synthesis of N-carboxy-alpha-amino acid anhydrides. Angew Chem Int Ed Engl. 1967 Mar;6(3):248–248. doi: 10.1002/anie.196702481. [DOI] [PubMed] [Google Scholar]
- Koshland D. E., Jr Biochemistry of sensing and adaptation in a simple bacterial system. Annu Rev Biochem. 1981;50:765–782. doi: 10.1146/annurev.bi.50.070181.004001. [DOI] [PubMed] [Google Scholar]
- Manning J. M., Moore S. Determination of D- and L-amino acids by ion exchange chromatography as L-D and L-L dipeptides. J Biol Chem. 1968 Nov 10;243(21):5591–5597. [PubMed] [Google Scholar]
- Masters P. M., Bada J. L., Zigler J. S., Jr Aspartic acid racemisation in the human lens during ageing and in cataract formation. Nature. 1977 Jul 7;268(5615):71–73. doi: 10.1038/268071a0. [DOI] [PubMed] [Google Scholar]
- Masters P. M., Bada J. L., Zigler J. S., Jr Aspartic acid racemization in heavy molecular weight crystallins and water insoluble protein from normal human lenses and cataracts. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1204–1208. doi: 10.1073/pnas.75.3.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKerrow J. H. Non-enzymatic, post-translational, amino acid modifications in ageing. A brief review. Mech Ageing Dev. 1979 Jul;10(6):371–377. doi: 10.1016/0047-6374(79)90019-8. [DOI] [PubMed] [Google Scholar]
- Moore S. Amino acid analysis: aqueous dimethyl sulfoxide as solvent for the ninhydrin reaction. J Biol Chem. 1968 Dec 10;243(23):6281–6283. [PubMed] [Google Scholar]
- Oliva A., Galletti P., Zappia V., Paik W. K., Kim S. Studies on substrate specificity of S-adenosylmethionine: protein-carboxyl methyltransferase from calf brain. Eur J Biochem. 1980 Mar;104(2):595–602. doi: 10.1111/j.1432-1033.1980.tb04463.x. [DOI] [PubMed] [Google Scholar]
- Polastro E. T., Deconinck M. M., Devogel M. R., Mailier E. L., Looza Y. B., Schnek A. G., Léonis J. Purification and some molecular properties of protein methylase II from equine erythrocytes. Biochem Biophys Res Commun. 1978 Apr 14;81(3):920–927. doi: 10.1016/0006-291x(78)91439-0. [DOI] [PubMed] [Google Scholar]
- Stock J. B., Koshland D. E., Jr Changing reactivity of receptor carboxyl groups during bacterial sensing. J Biol Chem. 1981 Nov 10;256(21):10826–10833. [PubMed] [Google Scholar]
- Terwilliger T. C., Clarke S. Methylation of membrane proteins in human erythrocytes. Identification and characterization of polypeptides methylated in lysed cells. J Biol Chem. 1981 Mar 25;256(6):3067–3076. [PubMed] [Google Scholar]


