Abstract
The prevalent DNA modification in higher organisms is the methylation of cytosine to 5-methylcytosine (5mC), which is partially converted to 5-hydroxymethylcytosine (5hmC) by the Tet (ten eleven translocation) family of dioxygenases. Despite their importance in epigenetic regulation, it is unclear how these cytosine modifications are reversed. Here, we demonstrate that 5mC and 5hmC in DNA are oxidized to 5-carboxylcytosine (5caC) by Tet dioxygenases in vitro and in cultured cells. 5caC is specifically recognized and excised by thymine-DNA glycosylase (TDG). Depletion of TDG in mouse embyronic stem cells leads to accumulation of 5caC to a readily detectable level. These data suggest that oxidation of 5mC by Tet proteins followed by TDG-mediated base excision of 5caC constitutes a pathway for active DNA demethylation.
Cytosine methylation is directly involved in the modulation of transcriptional activity and other genome functions (1), and DNA demethylation therefore plays important roles in transcriptional activation of silenced genes (2, 3). Multiple mechanisms have been proposed to achieve DNA demethylation in mammals, which include direct removal of the exocyclic methyl group from the cytosine via C-C bond cleavage, enzymatic removal of the 5-hydroxylated methyl group as formaldehyde (4), and replacement of the methylated cytosine base and nucleotide through DNA base-excision repair (BER) and nucleotide excision repair pathways, respectively (5). In theory, all these processes can be triggered by hydroxylation of 5-methylcytosine (5mC) by the recently identified Tet (ten eleven translocation) dioxygenases (6, 7).
The discovery of Tet proteins capable of hydroxylating 5mC to afford 5-hydroxymethylcytosine (5hmC) (8) prompted us to search for previously unknown enzymatic activities that modify 5mC and/or 5hmC in mammalian nuclear extracts. Base modification may prevent digestion by restriction enzymes used in thin-layer chromatography (TLC) detection methods. To circumvent this problem, we used the EcoNI restriction enzyme, which recognizes two trinucleotides separated with a spacer of any five nucleotides (5′ CCTNN/NNNAGG 3′; N is any nucleotide). Thus further modification of 5mC placed at the third N position should not block EcoNI digestion. We tagged the 5mC-containing DNA substrate with biotin and, by following the procedure summarized in fig. S1A, were able to detect an additional spot (designated “X”) on TLC plates in the DNA sample treated with nuclear extract from human embryonic kidney (HEK) 293T cells transfected with Tet2 (fig. S1B). This spot migrated much more slowly than all other nucleotides, and its amount was proportional to the decrease in 5mC. A similar spot also appeared from the 5hmC-containing DNA but not the “C” control DNA samples upon incubation with the nuclear extract (fig. S1B).
Because Tet dioxygenases catalyze oxidation of 5mC to 5hmC (8), we surmised that protein(s) associated with Tet2 or Tet2 itself might be responsible for the generation of the unknown nucleotide in spot X. To address this, we purified Flag-tagged full-length Tet2 protein from transfected 293T cells (fig. S2) and tested its activity on 5mC-containing DNA substrates. TLC analysis revealed that spot X could be generated by incubating the 5mC or 5hmC substrate with the purified Tet2 protein (Fig. 1A).
Fig. 1. Purified Tet2 catalyzes the modification of 5mC and 5hmC.

(A) The 32P spot X on a TLC plate generated from 5mC and 5hmC DNA substrates incubated with the full-length Flag-Tet2 protein. The top spot in lanes 3 to 8 was 5′ end-labeled deoxyadenosine monophosphate (dAMP) resulting from the incomplete EcoNI digestion of the DNA substrate (fig. S1A). (B) TLC confirmation of the origin of spot X from 5mC with a 14C-labeled methyl group. (C) HPLC detection of a new nucleoside generated from 5mC and 5hmC DNA substrates upon incubation with Flag-Tet2. AU indicates absorption units.
To ascertain that spot X on TLC plates does arise from 5mC, we performed an isotope-tracing experiment by labeling DNA substrate in the methyl group of 5mC using the CpG-specific bacterial methyltransferase M.SssI and [methyl-14C] S-adenosylmethionine. A 14C spot was detected with the same migration rate as the 32P spot X (Fig. 1B), demonstrating that the unknown nucleotide in spot X originated from 5mC.
A derivative of 5mC upon incubation with Tet2 could also be detected by high-performance liquid chromatography (HPLC) analysis of nucleosides. Wild-type Tet2 converted over 90% of 5mC or 5hmC into a new nucleoside (peak X′) (Fig. 1C). No X′ peak was generated by the mutant enzyme harboring substitutions of the key catalytic residues (HxD) of the conserved Fe2+-binding motif. Similarly, both Tet1 and Tet3 showed activity (fig. S3). The C-terminal regions containing the catalytic domain showed a weaker but clearly detectable activity (figs. S4 and S5).
The HPLC peak X′ appeared only when 5mC and 5hmC DNA substrates had been incubated with Tet enzymes in the presence of Fe2+ and 2-oxoglutarate (fig. S6), the two cofactors required by this superfamily of dioxygenases. This cofactor requirement, together with the retention of the 14C-labeled methyl group attached to cytosine (Fig. 1B), suggested that the new modification arose from oxidation of the 5-methyl group of 5mC (6). Stepwise oxidation of 5mC would result in the formation of 5hmC, 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC). The unknown 5mC derivative generated by Tet2 eluted in HPLC at the same time point as the chemically synthesized 5caC standard (Fig. 2A). Moreover, TLC analysis showed that the X spot detected from the Tet-mediated oxidation of 5mC in DNA comigrated with an authentic 5caC nucleotide labeled with 32P (Fig. 2B). Furthermore, we collected the HPLC peak X′ and subjected it to high-resolution mass spectrometric analysis. Mass spectra in a negative-ion mode identified an ion with a mass/charge ratio (m/z) of 270.0731, which matched the monoisotopic mass of the ion derived from 5caC (m/z of 270.0726) with a deviation of only 1.8 parts per million (ppm) (Fig. 2C, top). The overall fragmentation ion pattern obtained matched well that of the authentic 5-carboxylcytidine (Fig. 2C, bottom). Together, these observations confirmed that Tet2 catalyzes oxidation of 5mC and 5hmC to 5-carboxylcytosine in DNA. 5fC, the other potential oxidation intermediate of 5mC or 5hmC that is detectable in mouse embryonic stem (ES) cell genomic DNA (9), was not found in the DNA product under the same reaction and detection conditions.
Fig. 2. The modification product of 5mC is 5caC.

The 5mC derivative generated by Flag-Tet2 was analyzed by HPLC analysis (A), TLC (B), and mass spectrometry (C) using a 5caC standard as a reference. (A) HPLC analysis of the nucleoside derived from 5mC in the DNA substrate treated with Flag-Tet2. (B) TLC identification of the modification product of Tet2. The 5mC substrate used was the same as in Fig. 1A. 5caC DNA (lane 3), a synthetic oligonucleotide duplex with the same sequence but containing a 5-caC 3′ of the cleavage site of EcoNI, provides a reference spot for 5-carboxycytidine monophosphate (5ca-dCMP). (C) Mass spectrometry analysis of a HPLC fraction corresponding to the peak X′ in (A). Structural formulas deduced are shown for the major peaks.
To characterize the enzymatic properties of Tet2, we carried out bisulfite sequencing analysis on methylated DNA upon oxidation in vitro. 5caC behaved as an unmethylated cytosine in bisulfite conversion (fig. S7). The distribution pattern of 5caCs formed upon the reaction (fig. S8) suggested that Tet2 is quite processive with regard to the oxidation of 5mC sites.
We then examined whether Tet enzymes can catalyze the oxidation of endogenous 5mC in genomic DNA in cultured cells. HEK 293T cells were transfected with the full-length Tet2 or the C-terminal catalytic domain (fig. S9). HPLC analysis of genomic DNA isolated from the transfected cells showed a new peak with the same retention time as the 5caC standard, in addition to the 5hmC peak (Fig. 3A). This new peak was not detected when the cells were transfected with a catalytically inactive mutant of Tet2.
Fig. 3. Tet2 catalyzes formation of 5caC in genomic DNA in vivo.

(A) HPLC detection of 5caC in genomic DNA of HEK 293T cells expressing wild-type Flag-Tet2. Synthetic 2′-deoxy-5-carboxylcytidine (5caC) and the nucleoside hydrolysate of a synthetic DNA containing cytosine (C), 5hmC, and 5mC were used as standards. Arrows point to the 5caC peaks detected in DNA isolated from cells transfected with wild-type Tet2. (B) HPLC–tandem mass spectrometry (MS/MS) detection of genomic 5caC. Shown are MRM elution profiles of negative-ion mass transitions from a precursor to its three product ions as shown in Fig. 2C for a synthetic 2′-deoxy-5-carboxycytidine standard (blue) and a DNA hydrolysate [isolated peak indicated with a red arrow in (A)] from cells transfected with full-length Tet2 (black). Red line was from a control DNA sample isolated from cells transfected with the inactive Tet2 mutant.
The identity of the endogenous 5caC product was further confirmed by analyzing HPLC chromatograms with a triple quadrupole mass spectrometer that was set in a MRM (multiple-reaction-monitoring) mode and optimized for the detection of three expected ion transitions (270→110, 270→154, and 270→227). Co-elution of the MRM signals on a reversed-phase column (Fig. 3C) provided an unambiguous identification of the Tet2 product generated in cells as 5caC. Similarly, Tet1 was also able to generate 5caC in the genomic DNA of the transfected cells.
5-Carboxylcytosine is not detectable in mouse ES cells and neurons that express high levels of Tet enzymes (10), yet it is chemically stable and does not spontaneously decarboxylate to cytosine under physiological conditions. This raises the possibility that 5caC might be actively removed from genomic DNA immediately after its generation in cells. Because BER has been implicated in DNA demethylation (6), we tested whether nuclear extracts of mammalian cells contain base-excision activity toward 5caC (fig. S10A). A 5caC-specific glycosylase activity was detected in mouse ES cell nuclear extract (Fig. 4A). Incubation of a 20-nucleotide oligomer (20-mer) 5caC substrate with the extract resulted in a 9-mer cleavage product, because removal of the 5caC base generated an abasic site that was broken by a hot alkaline treatment. Incubation with the nuclear extracts did not lead to excision of 5hmC from the substrate DNA in the same assay.
Fig. 4. TDG glycosylase recognizes and excises 5caC from DNA.
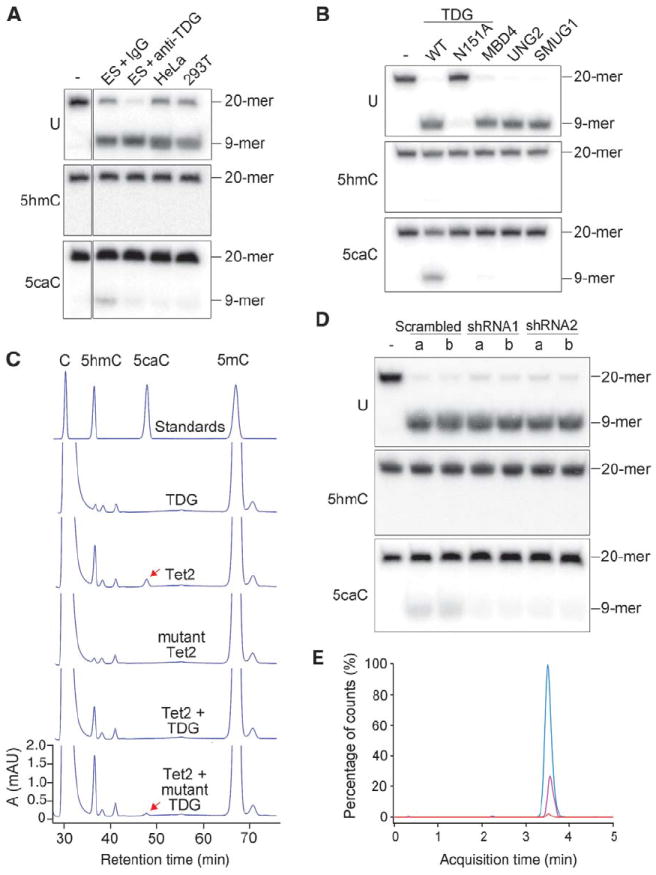
(A) ES cell nuclear extract contains 5caC-specific base-excision activity. The activity in the nuclear extract of mouse ES cells to generate alkaline-sensitive sites was assayed by using 5caC-containing oligonucleotide duplexes. Shown are the results obtained with 20-mer DNA duplexes containing either G/U (U), G/5hmC (5hmC), or G/5caC (5caC) base pairs in the middle. (B) Excision of 5caC from DNA by Flag-TDG but not by Flag-MBD4, Flag-UNG, or GST-SMUG1. Asn151→Ala151 (N151A) is a catalytically inactive mutant of TDG. Proteins used are shown in fig. S10B. 6xHis-TDG purified from bacteria was also active in 5caC excision. TDG displayed a much stronger glycosylase activity for the “hemi-carboxylated” DNA substrate containing 5caC only on one strand (fig. S11). WT, wild type. (C) Reduced 5caC formation by cotransfection of TDG in HEK 293T cells expressing ectopic Tet2. The mutant TDG was as in (B). (D) Lack of 5caC base-excision activity in Tdg knockdown ES cells. Nuclear extracts prepared from the two independent cell lines (a and b) containing shRNA knockdown construct 1, 2, or scramble control were tested as in (A). (E) HPLC-MS/MS detection of 5caC in ES cells depleted of TDG. MRM profiles of hydrolysates of genomic DNA from control (red) and TDG-depleted ES cells (pink) were analyzed. Synthetic 5caC nucleoside was used as a positive control (blue). Depletion of TDG was confirmed by Western analysis of independent stable knockdown ES cell lines (fig. S12).
Among the known glycosylases that recognize and excise modified bases (11), thymine-DNA glycosylase (TDG) is the most likely candidate to process 5caC because the enzyme is essential for embryo development (12) and capable of removing cytosine analogs as well as thymines, the deamination product of 5mC (13). We therefore prepared recombinant TDG and performed a glycosylase assay with U-, 5hmC-, or 5caC-containing DNA as substrates (fig. S10). TDG was able to cleave 5caC but not 5hmC (Fig. 4B). MBD4, another DNA glycosylase that removes U or T opposite to G in the CpG sequence context (14), exhibited no activity toward 5caC. Similarly, no 5caC excision activity could be detected for the uracil-DNA glycosylase (UNG) and the single-strand-selective monofunctional uracil DNA glycosylase 1 (SMUG1), both of which remove uracils and 5-hydroxyuracils (the deamination product of 5hmC) from DNA (15, 16) (Fig. 4B). The 5caC-excision activity of TDG was further confirmed in vivo in transfected HEK 293T cells. Ectopic expression of wild-type TDG diminished the amount of 5caC generated by cotransfected Tet2 but did not significantly reduce 5hmC (Fig. 4C). Expression of a catalytically inactive TDG mutant had no effect. Consistently, nuclear extract from the Tdg knockdown ES cells had little 5caC excision activity (Fig. 4D). Moreover, immunodepletion of TDG from the ES cell nuclear extract greatly reduced the 5caC excision activity (Fig. 4A, lane 3). These results indicate that TDG is able to recognize and excise 5caC, an oxidation product of 5mC, in duplex DNA.
Stable ES cell lines expressing a Tdg-specific small interfering RNA were established, and TDG depletion was confirmed by Western analysis (fig. S12). By using triple quadrupole mass spectrometry, we could detect 5caC in genomic DNA isolated from TDG-depleted ES cells, but no reliable signal was detected in TDG-proficient control cells expressing scramble short hairpin RNA (shRNA) (Fig. 4E). Similarly, 5caC was detectable in mouse induced pluripotent stem (iPS) cells when the Tdg gene was knocked out (fig. S13). Judging from our calculation based on the measurement of a 5caC standard, the number of 5caC per genome is ~9000 in Tdg-depleted ES or iPS cells but below 1000 in wild-type cells.
TDG has been implicated in DNA demethylation for its function in excising the deamination product of 5mC, 5hmC, or 5mC itself from DNA (17-19), yet mammalian TDG lacks glycosylase activity toward 5mC (6, 12). Although TDG is able to excise 5hmU (19), the deamination product of 5hmC, our work provides evidence that the Tet dioxygenases oxidize 5mC and 5hmC to 5caC, which becomes a substrate for TDG. Therefore, Tet-mediated conversion of 5mC and 5hmC to 5caC could trigger TDG-initiated BER, as indicated here. These sequential events would lead to DNA demethylation, because unmethylated cytosines are inserted into the repaired genomic region (fig. S14).
Genome-wide mapping revealed that Tet1 is relatively enriched in CpG-rich active promoters that are unmethylated (20-23), but 5hmC is underrepresented in the majority of Tet1 binding sites in ES cells (24-26). These apparent paradoxes might be accounted for if active promoters with Tet1 binding sites were prevented from erroneous hypermethylation because of Tet1 oxidizing 5mC into 5caC, which could then be removed by TDG-mediated BER repair. In this case, 5mC is most likely undetectable in the active promoters because of their transient existence in a small proportion of cells. Likewise, in many of the Tet1 binding sites, 5hmC could be underrepresented because of conversion to 5caC, which is rapidly removed in cells.
Note added in proof: During the revision of this manuscript, Ito et al.’s report (www.sciencemag.org/content/early/2011/07/20/science.1210597.abstract) appeared online describing the enzymatic activity of Tet proteins in the conversion of 5mC to 5fC and 5caC, as well as the detection of these derivatives in mouse genomic DNA.
Supplementary Material
Acknowledgments
We thank C. Walsh for critical reading of the manuscript, G. Shi and S. Klimasauskas for discussions, J. Ju for providing Tet cDNA clones, T. Carell for 2′-deoxy-5-carboxylcytidine and Z. Hua for the TDG antibody. This study was supported by grants from the Ministry of Science and Technology China (2007CB947503 and 2009CB941101 to G.-L.X., 2010CB912100 to L.L.), National Science Foundation of China (30730059 to G.-L.X., 30930052 and 30821065 to L.L.), and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA01010301 to G.-L.X.) and by the NIH (GM071440 to C.H.) and (1S10RR027643-01 to K.Z.).
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/science.1210944/DC1
Materials and Methods
Figs. S1 to S14
References (27–32)
References and Notes
- 1.Jaenisch R, Bird A. Nat Genet. 2003;33(suppl):245. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 2.Simonsson S, Gurdon J. Nat Cell Biol. 2004;6:984. doi: 10.1038/ncb1176. [DOI] [PubMed] [Google Scholar]
- 3.He XJ, Chen T, Zhu JK. Cell Res. 2011;21:442. doi: 10.1038/cr.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liutkeviciute Z, Lukinavicius G, Masevicius V, Daujotyte D, Klimasauskas S. Nat Chem Biol. 2009;5:400. doi: 10.1038/nchembio.172. [DOI] [PubMed] [Google Scholar]
- 5.Walsh CP, Xu GL. Curr Top Microbiol Immunol. 2006;301:283. doi: 10.1007/3-540-31390-7_11. [DOI] [PubMed] [Google Scholar]
- 6.Wu SC, Zhang Y. Nat Rev Mol Cell Biol. 2010;11:607. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahl C, Grønbæk K, Guldberg P. Clin Chim Acta. 2011;412:831. doi: 10.1016/j.cca.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Tahiliani M, et al. Science. 2009;324:930. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfaffeneder T, et al. Angew Chem Int Ed Engl. 2011;50:7008. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- 10.Globisch D, et al. PLoS ONE. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindahl T, Wood RD. Science. 1999;286:1897. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 12.Cortázar D, et al. Nature. 2011;470:419. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 13.Bennett MT, et al. J Am Chem Soc. 2006;128:12510. doi: 10.1021/ja0634829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. Nature. 1999;401:301. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 15.Krokan HE, Standal R, Slupphaug G. Biochem J. 1997;325:1. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boorstein RJ, et al. J Biol Chem. 2001;276:41991. doi: 10.1074/jbc.M106953200. [DOI] [PubMed] [Google Scholar]
- 17.Métivier R, et al. Nature. 2008;452:45. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 18.Zhu B, et al. Proc Natl Acad Sci U S A. 2000;97:5135. doi: 10.1073/pnas.100107597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cortellino S, et al. Cell. 2011;146:67. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pastor WA, et al. Nature. 2011;473:394. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ficz G, et al. Nature. 2011;473:398. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 22.Song CX, et al. Nat Biotechnol. 2011;29:68. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu H, et al. Genes Dev. 2011;25:679. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams K, et al. Nature. 2011;473:343. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Y, et al. Mol Cell. 2011;42:451. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu H, et al. Nature. 2011;473:389. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


