Abstract
We introduce a simple, versatile and robust one-step technique that enables real-time imaging of multiple intracellular caspase activities in living cells without the need for complicated synthetic protocols. Conventional fluorogenic probes or recently reported activatable probes have been designed to target various proteases but are limited to extracellular molecules. Only a few have been applied to image intracellular proteases in living cells because most of these probes have limited cell-permeability. Our platform does not need complicated synthetic processes; instead it involves a straightforward peptide synthesis and a simple mixing step with a commercial transfection agent. The transfection agent efficiently delivered the highly quenched fluorogenic probes, comprised of distinctive pairs of dyes and quenchers, to the initiator caspase-8 and the effector caspase-3 in MDA-MB-435 cells, allowing dual-imaging of the activities of both caspases during the apoptotic process induced by TNF-related apoptosis induced ligand (TRAIL). With the combination of multiple fluorogenic probes, this simple platform can be applied to multiplexed imaging of selected intracellular proteases to study apoptotic processes in pathologies or for cell-based high throughput screening systems for drug discovery.
Keywords: caspase, activatable probe, fluorescence imaging, peptide, transfection agent
1. Introduction
Apoptosis, or programmed cell death, is a controlled sequence of events leading to discharge of cells without releasing toxic materials to the surroundings [1]. In physiological conditions, apoptosis plays pivotal roles in regulating embryonic development, growth and differentiation and causes tumor cell death once triggered by chemotherapeutic drugs [2, 3]. One of the key players in apoptosis is a family of cysteine-dependent aspartate-directed proteases, named caspases. There are twelve known caspases found in humans which can be classified into two distinct types: the initiators (e.g. caspase-8 and −9) and the effectors (e.g. caspase-3 and −7) [4-6]. Initiators are in charge of activating effectors by cleaving the pro-forms of effector caspases. Subsequently, effectors will cleave protein substrates in the death pathway at specific sites containing aspartic acid followed by triggering the apoptotic process. Since successful anticancer therapeutics such as chemo- and radiation-therapy rely on apoptosis, a robust apoptosis monitoring and screening method has the potential to accelerate the process of drug discovery and development. Various strategies based on different imaging modalities have been reported to image apoptotic processes in cells and in vivo, which are well summarized elsewhere [7-11].
The gold standard method to assess apoptosis utilizes dye-labeled annexin V, a Ca2+ dependent phospholipid-binding protein which binds to phosphatidylserine exposed on the cell membrane during the stages of apoptosis [12, 13]. Although binding of annexin V to the apoptotic cell surface has proven valuable and simple, it is an indirect method that is not completely specific to apoptosis. Another approach is to monitor specific apoptosis signaling molecules such as caspases. Caspases have attracted particular interests in apoptosis imaging because these enzymes are direct and crucial mediators of apoptosis [5, 14]. Recently, significant efforts have focused on the development of optical molecular probes targeting altered activities of caspases during apoptotic processes. Measuring the levels of different types of activated caspases from a single cell lysate can be done easily with a commercialized assay kit comprised of fluorogenic substrates specific to certain caspases. However, monitoring caspases’ activities in living cells can be difficult because caspases are intracellular enzymes and fluorogenic substrates are typically cell-impermeable [15]. Furthermore, multiplexed imaging of multiple caspases in living cells is more complicated [16]. To target key intracellular caspases, various strategies are reported to enhance the membrane-permeability of caspase-sensitive probes [17]. Among them, cell-permeable activatable optical imaging probes have become an attractive platform for real-time imaging of apoptosis [18, 19].
Activatable probes are designed to amplify fluorescence signals in response to specific biomolecular recognition in real-time. These probes are typically based on fluorogenic substrates comprised of a peptide substrate specific for a target enzyme, a dye and quencher pair and a carrier to optimize stability or enhance the cellular uptake of the probe. A variety of activatable probes have been introduced by a combination of substrate, dye/quenchers, polymers or inorganic nanoparticles [20-25]. Most reported probes have been designed to target extracellular enzymes but only a few have ability to target intracellular enzymes like caspases in living cells [17]. Recently, we and others reported a series of cell-permeable, caspase-activatable probes based on the use of cell-penetrating peptides, self-assembled polymeric nanoparticles, polymer-coated gold nanoparticles, or silica nanoparticles [16-19, 26-28]. Each design displays promising results either in vitro or in vivo; however, the reported systems are not necessarily practical for routine monitoring the activities of multiple caspases in living cells due to the need for complicated multistep synthesis and purifications. Alternatively, engineered fluorescent and luminescent proteins can be applied to multiplexed caspase imaging in living cells, but these strategies require genetically modified cells, which are time-consuming and costly [29, 30].
In this study, we report a simple, versatile and robust method using a one-step technique that is capable of boosting multi-fluorescence signals upon apoptosis in living cells, enabling real-time imaging of the caspase cascade. The formula is based on the mixture of highly quenched fluorogenic probes consisting of substrates targeting different caspases and diverse pairs of dyes and dark quenchers (Fig. 1A). Instead of modifying these probes through complicated multi-step synthesis, we simply improved its cell-permeability by blending fluorogenic probes with a commercially available transfecting agent, PULSin®. PULSin® is a cationic amphiphilic-based delivery agent. It forms a complex with negatively charged peptides or proteins by electrostatic or hydrophobic interactions. [31, 32]. As a proof-of-concept, we demonstrate the synthesis, preparation and characterization of the activatable formula and apply it to real-time imaging of two representative initiator and effector caspases, namely caspase-8 and −3, in single living cells undergoing apoptosis.
Figure 1.
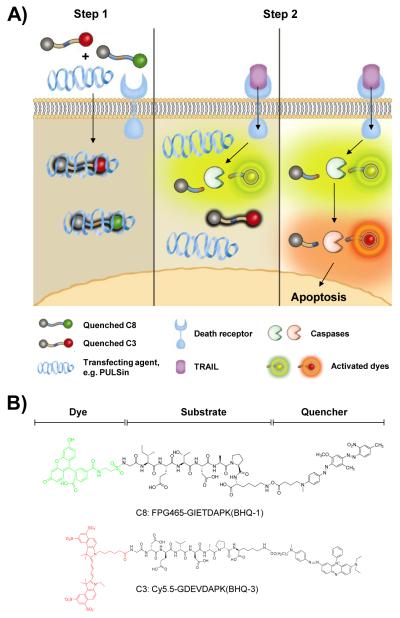
A) Schematic diagram of imaging the caspase cascade. Conventional dark-quenched, cell-impermeable fluorogenic probes for caspases can be efficiently delivered into the cells followed by simple incubation with a commercially available transfection agent such as PULSin® (Step 1). Once delivered into the cells, highly quenched probes are sequentially activated by expressed target caspases triggered by different initiator pathways, e.g. TRAIL-induced caspases activation (Step 2). B) Chemical structures of fluorogenic probes targeting caspase-8 and caspase-3, C8 and C3, respectively. Each probe consists of a substrate and a pair of dye/dark quencher with no significant spectral overlap of emission between the two probes.
2. Materials and Methods
2.1. Chemicals
Cy5.5 N-hydroxy succinimide (NHS) ester was purchased from GE Healthcare (Piscataway, NJ) and FPG456 was a gift from BioActs (Incheon, Korea). TNF-related apoptosis induced ligand (TRAIL) was obtained from Sigma-Aldrich (St. Louis, MO). Caspase-3 and caspsase-8 inhibitors and their polyclonal antibodies were obtained from R&D Systems Inc. (Minneapolis, MN). MDA-MB-435 cells were purchased from the American Type Culture Collection (Manassas, VA). Peptide/protein transfection reagent, PULSin®, was purchased from VWR International (Radnor, PA). 35-mm glass bottom dishes and 8-well chambers were purchased from MatTek Corporation (Ashland, MA, USA). The amino acids and peptide synthesis related reagents were obtained from CS Bio. Co. (Menlo Park, CA). All other solvents and chemicals were purchased from Sigma-Aldrich.
2.2. Synthesis of fluorogenic probes
A synthetic scheme is described in the Supplementary Data (Scheme S1 and S2, Fig. S1 and S2). Briefly, caspase-3 activatable probe (denoted as C3) was prepared from the side chain protected caspase-3 substrate 3, NH2-Gly-Asp(tBu)-Glu(tBu)-Val-Asp(tBu)-Ala-Pro-Lys(Boc)-OH. 3 was synthesized using an automatic peptide synthesizer by Fmoc chemistry at a 0.1 mmol scale using a Gly-2-ClTrt resin 1 (0.43 meq/g). O-Benzotriazole-N,N,N’,N’-tetramethyl-uronium-hexafluoro-phosphate (HBTU) and hydroxybenotriazole (HOBt) were used as the activating reagents. Crude peptide on resin 2 was cleaved by 1% trifluoroacetic acid (TFA) in dichloromethane, followed by precipitation in cold ethyl ether. Peptide 3 was purified by preparative reversed-phase high performance liquid chromatography (RP-HPLC, Dionex, CA), using 20% to 90% acetonitrile containing 0.1% TFA versus distilled water containing 0.1% TFA over 30 minutes at a flow rate of 10 mL/min (C18 column, 5 μm, 250 × 20 mm). The proper fraction was collected and lyophilized. Then the fluorescence donor, Cy5.5 NHS ester (Cy5.5-NHS, 10 μM), was reacted with 3 in anhydrous N, N-dimethylformamide (DMF, 200 μL) containing 2% diisopropylethyalamine (DIPEA) at room temperature in the dark. The reaction was monitored by analytical RP-HPLC, using 5% to 65% acetonitrile containing 0.1% TFA versus distilled water containing 0.1% TFA over 30 minutes at a flow rate of 1 mL/min (C18 column, 5 μm, 120A°, 250 × 4.6 mm). Crude product of 4 was precipitated by addition of cold ethyl ether and then lyophilized. The side chain protecting groups were removed by a TFA/water/triisopropylsilane/1,2-ethanedithiol (92.5/2.5/2.5/2.5, v/v/v/v) cleavage cocktail. Cy5.5 labeled caspase-3 substrate 5 was purified by preparative RP-HPLC, 10% to 55% acetonitrile/water (0.1% TFA), for 30 min at a 10 mL/min flow rate, and lyophilized as described above. The dried compound gave a blue color and was kept in dark for the further experiments. Finally, the NHS ester of Black Hole Quencher® 3 (BHQ-3-NHS) was coupled to the ε-amino group of lysine and product was purified and lyophilized. The purity and mass of C3, 6, was confirmed by analytical RP-HPLC and LC/MS (Fig. S1). Similarly, caspase-8 activatable probes (denoted as C8) with and without quencher were prepared as described above using the peptide sequence of Gly-Ile-Glu(OtBu)-Thr(tBu)-Asp(OtBu)-Ala-Pro-Lys(Boc) and FPG456 (ex/em, 475/522 nm) and BHQ-1-NHS as a quenching pair (Fig. S2).
2.3. Characterizations of probes
The absorbance of each dye, quencher and probe were measured in PBS (10 mM, pH 7.4) buffer by Genesys 10S UV-Vis spectrophotometer. The quenching efficiency (QE) was quantified by F-7000 spectrofluorometer (Hitachi, Japan). Briefly, known concentrations of dyes (FPG456 and Cy5.5) and probes (C8 and C3) were measured by spectrofluorometer. The excitation wavelength was set at 450 nm for FPG456 and 675 nm for Cy5.5 and the emission spectra were recorded from 500 to 650 nm and 680 to 750 nm, respectively. The QE was calculated as C8 versus FPG456 signals at 520 nm and C3 versus Cy5.5 signals at 695 nm. To visualize the activated fluorescence signals, C8 and C3 were dissolved in the caspase reaction buffer (20 mM HEPES, 10% Sucrose, 100 nM NaCl, 10 mM DTT, 1 mM EDTA, 0.1% CHAPS, pH 7.2) and were imaged before and after the addition of 12 nM of caspase-8 and −3 by Maestro 2.0 imging system (Caliper Lifer Science, Hopkinton, MA) using “orange” and “blue” filters, respectively. The images were compared at the same scale.
2.4. In vitro caspases activation of probes
The fluorescence activation of C3 by caspase-3 was investigated by incubating 500 nM of C3 in a reaction buffer containing a sequentially diluted active caspase-3 (0.5, 1.5, 3, and 6 nM) for 90 min at 37 °C. The fluorescence intensity was monitored using a spectrofluorometer. Similarly, C8 was incubated with different concentrations of caspase-8 (1.5, 3, 6, and 12 nM) and fluorescence changes were monitored for 90 min. For the specificity test, 500 nM of C3 and C8 were mixed at a final concentration of 500 nM in the reaction buffer. The background of the probe mixture was monitored at respective emission wavelengths (522 nm and 695 nm for FPG456 and Cy5.5) at 10 min intervals during the first 50 min. At 50 min, 12 nM of caspase-8 was added into the mixture and fluorescence intensity from each dye was monitored up to 60 min. At 110 min, 6 nM of caspase-3 was further added into the reaction system and fluorescence intensity changes were recorded for another 1 hr at selected time points. Finally, the signals from each time point were analyzed and normalized with the signals from the starting time-point.
2.5. Cellular uptake of probes by PULSin
Human melanoma cell line, MDA-MB-435 cells (ATCC, Manassas, VA), were cultured in L-15 medium (Gibco, Grand Island, NY) containing 10% fetal bovine serum (FBS) with 1% penicillin-streptomycin at 37°C in a humidified 5% CO2 atmosphere. MDA-MB-435 cells were plated 12 h before the start of the experiment in 8-well chamber slides at a density of 5×103 cell/cm2 overnight. To prepare the probe/PULSin formula, 1 μg of Cy5.5-labeled, non-quenched C3 (Cy5.5-C3) was dissolved in 100 μL of HEPES (20 mM) then incubated with different amounts of PULSin (0, 1, 2 and 4 μL from a commercially provided stock solution) for 15 min. After that, L-15 medium without FBS was added to make a final volume of 1 mL for each mixture. The next day, the cells were washed twice with PBS and incubated with the above Cy5.5-C3/PULSin mixtures for 3 hr followed by treatment with Z-fix formalin and DAPI staining at room temperature. The cellular uptake of Cy5.5-C3 was examined with a confocal microscope configured for Cy5.5 filter.
2.6. Annexin V staining
The annexin V plasmid was kindly provided by Professor S.J. Martin from Trinity College in Ireland and expressed and purified as reported previously [33]. FITC was further conjugated to purified annexin V for apoptosis verification. Briefly, MDA-MB-435 cells were seeded into 8-wells chamber and subjected to TRAIL treatment (100 ng/mL in L-15 medium without FBS). Next, the cells were incubated with FITC-annexin V (1 μg/mL) in a binding buffer (10 mM of HEPES-NaOH, 150 mM of NaCl, 5 mM of KCl, 1 mM of MgCl2, 1.8 mM of CaCl2, pH 7.4) for 15 min in the dark at room temperature. After removing free FITC-annexin V by PBS washing, apoptotic cells were recognized by FITC-annexin V and nucleus was stained by DAPI. The labeled cells were observed under a fluorescence microscope (IX81, Olympus, Japan).
2.7. Cellular imaging of caspase cascade
For the visualization of caspase-8 and -3 cascade activations, 3×106 MDA-MB-435 cells were seeded onto 35 mm glass bottom dishes and allowed to grow until 80% confluent. Cells were washed twice with PBS before the imaging studies. To deliver C8 and C3 into the cells, 2 μg of C8 or C3 were incubated with 8 μL of PULSin in 200 μL of HEPES (20 mM) for 15 minutes at room temperature. Then, L-15 medium without FBS was added to make 1 mL solution. The C8/PULSin and C3/PULSin were mixed and the mixture, C8/C3/PULSin, was added into MDA-MB-435 cells for 4 hours at 37 °C. Afterwards, cells were washed by PBS to remove free probe/PULSin complexes. To induce apoptosis, TRAIL (100 ng/mL in L-15 medium without FBS) was added into the dish. After 4 hr of incubation, the cells were washed carefully by PBS and fixed with Z-fix formalin for 15 min at room temperature followed by adding mounting medium with DAPI. For the inhibition test, MDA-MB-435 cells were prepared in a 35-mm dish and treated with C8/C3/PULSin as described above. After 4 hr of incubation, the cells were further treated with either 50 μM of caspase-3 inhibitor (Z-D(OMe)-E(OMe)-V-D(OMe)-FMK) and both caspase-8 (Z-I-E(OMe)-T-D(OMe)-FMK) and -3 inhibitors before adding 100 ng/mL of TRAIL. The activation and inhibition of caspase expressions was visualized using confocal microscopy (FV10i-LIV, Olympus), in which the emission wavelengths were collected at 520 nm and 692 nm for FPG456 and Cy5.5, respectively. For real-time cellular imaging, cells were visualized by confocal microscopy once TRAIL (100 ng/mL in L-15 medium without FBS) was added into the dish to induce apoptosis. Images were collected at 30 min intervals over 4 hr with a cooled charged-coupled device video camera. Cells were imaged through different filters to acquire differential interference contrast (DIC) images, FPG456 and Cy5.5 fluorescence images.
2.8. Statistical analysis
Quantitative data were expressed as mean ± SD. A Student’s t test was used to test differences between groups. P values of less than 0.05 were considered statistically significant.
3. Results and discussion
3.1. Synthesis of caspase-specific fluorogenic probes
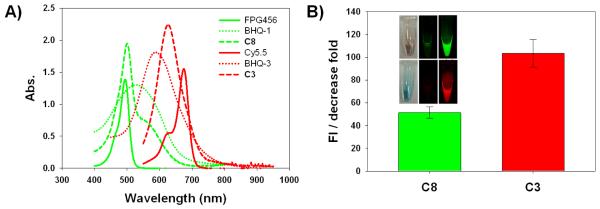
The fluorogenic probes targeting caspase-8 and −3, C8 and C3, were synthesized with a specific core substrate, Ile-Glu-Thr-Asp and Asp-Glu-Val-Asp, respectively. Gly and Ala-Pro-Lys residues were incorporated into the C-terminal and N-terminal part of each substrate to allow specific conjugation of a dye/quencher pair, FPG456/BHQ-1 and Cy5.5/BHQ-3 (Fig. 1B). FPG456 and Cy5.5 were chosen because of no significant spectral overlap of emission between two dyes and both dyes have been used frequently for cellular imaging applications. Synthesized C8 and C3 showed purity greater than 95% by RP-HPLC and mass was confirmed by LC/MS (m/z calculated/found for C8 and C3; 905.31/905.80 [M + H]2+ and 1129.38/1129.05 [M + H]2+, respectively) (Supplementary Data Fig. S1 and S2). As shown in Fig. 2A, C8 and C3 demonstrated a broad UV-Vis absorption peak over the 400 – 600 and 500 – 700 nm range covering the absorption spectra of FPG456/BHQ-1 and Cy5.5/BHQ-3 pair, respectively. The quenching efficiency of the probes was calculated based on the fluorescence emission produced by the same molar concentration of free dyes and expressed as the fold reduction from the initial value (Fig. 2B). BHQ-1 and −3 decreased fluorescence emission of FPG456 (λmax Em 522 nm) and Cy5.5 (λmax Em 695 nm) by 51.3 ± 5.1 and 103.4 ± 12.4-fold. When the quenched probes were incubated with 6 nM of caspases-8 and −3 in the reaction buffer for an hour, the fluorescence signals were significantly recovered and increased signals were imaged by an optical imaging instrument (Maestro) (Fig. 2B). Different quenching folds between C8 and C3 could be due to several factors, for example, the quenching efficiency of the quencher against a specific dye, water-solubility of dye/quencher pair or the physical structure of the fluorogenic substrate in buffer.
Figure 2.
A) Absorbance spectra of each dye, dark quencher and probe, C8 and C3, in PBS buffer. B) The quenching effect of C8 and C3 on the fluorescence produced by FPG456 (λmax ex/em 456/522) and Cy5.5 (λmax ex/em 675/695), respectively expressed as the fold decrease in emission signals from the initial values. Inset: DIC (left) and fluorescence images of vials containing C8 (upper) and C3 (lower) without (middle) or with (right) addition of caspase-8 and caspase-3, respectively.
3.2. Enzyme specificities of probes
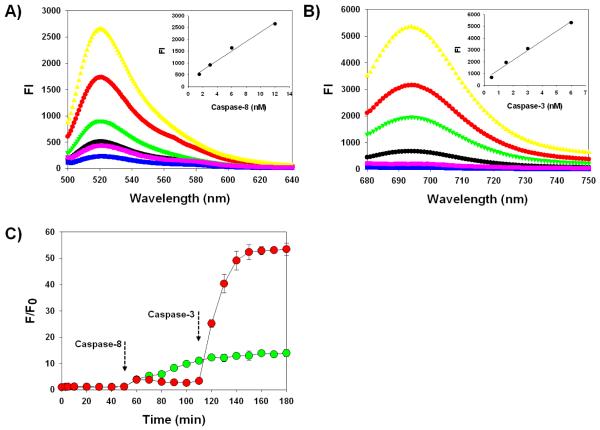
The caspase selectivity of C8 and C3 was investigated in vitro by incubating probes in the reaction buffer containing different concentrations of active caspase-8 or −3 with or without a specific caspase inhibitor. As shown in Fig. 3A and 3B, probes without caspases were in a strongly quenched state and stable in the reaction buffer. C3 with a Cy5.5/BHQ-3 pair demonstrated lower background compared to that of C8 with a FPG456/BHQ-1 pair. When the respective caspases were added, strongly quenched probes were able to produce robust fluorescence signals against the target caspase. C8 and C3 demonstrated 14.0 ± 1.5 and 53.0 ± 2.3-fold enhanced fluorescence emission signals at 522 nm and 695 nm when incubated with 12 nM of caspase-8 and −3 at 37 °C for 60 min, respectively. In contrast, the fluorescence activation was fully inhibited when the probes were incubated with caspases and their inhibitors. All probes demonstrated a linear relationship between the intensity of fluorescence signals and concentrations of active caspases (r2 > 0.95). However, due to the subtle differences in quenching properties, background signals and cleavage rates between C8 and C3, their linear range of an assay was different (for C8 and C3, 1 to 12 nM vs. 0.5 to 6 nM, respectively). Cleavage of caspase substrates is largely dependent on specific activity of caspases to their substrate and the reaction condition [34]. Commonly used caspase substrates, including the core sequences adopted in this study, are reasonably specific to their target caspase, however, the cleavage rates can vary. For example, caspsae-3 is known to cleave its substrates more rapidly than other caspases. In addition, labeling with dye/quencher pairs and inserting amino acids can also affect the substrate cleavage because they can change the overall physicochemical structures of the native substrate. Although C8 showed relatively low fluorescence activation compared to that of C3, both probes provided enough signal changes that can be visibly differentiated in the cells.
Figure 3.
A) Fluorescence emission spectra of C8 in the presence of various concentrations of caspase-8 (in nM, 0 blue, 1.5 black, 3 green, 6 red, 12 yellow) and caspase-8 with inhibitor (purple). Inset: Caspase-8 standard curve. The excitation was set at 456 nm. B) Fluorescence emission spectra of C3 in the presence of various concentrations of caspase-3 (in nM, 0 blue, 0.5 black, 1.5 green, 3 red, 6 yellow) and caspase-3 with inhibitor (purple). Inset: Caspase-3 standard curve. The excitation was set at 675 nm. C) The relative increase in fluorescence emission produced by the mixture of C8/C3 in the presence of caspases-8 and caspase-3, which were added at different time intervals. The excitation and emission was set at 456/522 nm (green) and 675/695 nm (red) and the respective emissions were recorded continuously for 180 min. Data represent means of triplicate experiments with standard deviations.
Our proposed formulation aims to simultaneously target multiple caspases, therefore, the specificity of each probe should be considered along with the different substrate activation kinetics. Many peptide-based substrates are reported to be specific; however, there are no perfect substrate for each caspase because many caspase substrate activities overlap. In other words, caspases are typically specific for their substrate, but its intended substrate can be cleaved by other caspases at different cleavage rates. For example, caspase-8 has specificity for substrates containing IETD compared to that of DEVD. The IETD substrate can also be cleaved by caspase-3 but caspase-3 has higher specificity for DEVD substrates. To determine if the probes are able to distinguish between different types of caspases, C8 and C3 were sequentially incubated with caspase-8 and caspase-3 at a 60 min time interval. As shown in Fig. 3C, both C8 and C3 in the reaction buffer without any caspases were well-quenched.
When caspase-8 was added after 50 min, the FPG456 signals from C8 gradually increased up to 11.1 ± 0.2-fold over the next 60 min whereas Cy5.5 signals from C3 showed moderate changes of 3.3 ± 0.2-fold. As expected, when caspase-3 was added at 110 min, C3 exhibited a 16.1 ± 0.7-fold increase in Cy5.5 signals at 180 min while C8 demonstrated only a 1.4 ± 0.1-fold increase (vs. fluorescence signals at 110 min). The addition of caspases-8 and −3 slightly affected the activation of both probes; however, the relative fluorescence changes induced by the respective caspases were sufficient to distinguish the specific caspases in the same buffer. Substrate specific overlap is an intrinsic limitation of reported peptide-based caspase substrates. Since our platform allows versatile modification of any substrate, the current core sequences can be further replaced by newly reported, highly specific substrates to improve its overall efficiency.
3.3 Cellular delivery of probes
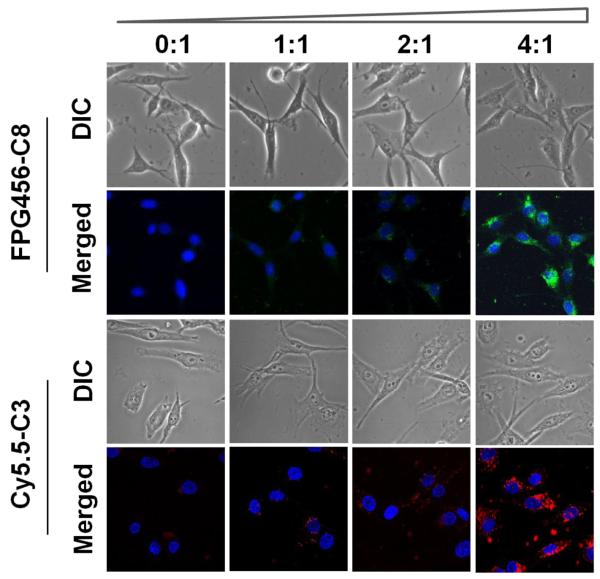
To target intracellular caspases, probes should be delivered into the cell first. In the case of C8 and C3, they are not highly cell-permeable thus cannot be directly used to image intracellular caspase activities. Instead of chemically modifying our probes to enhance the cellular internalization, we applied a commercialized transfection agent, PULSin, which is expected to enhance cellular delivery of peptides by simple mixing. To optimize the formulation, the uptake of probes was monitored by using non-quenched C8 (FPG456-C8) and C3 (Cy5.5-C3), in combination with different concentrations of PULSin in MDA-MB-435 cells. A non-quenched form of the probe was used in this assay for monitoring quick cellular uptake of a highly negatively charged peptide analog. The probe concentration was fixed between 1 - 2 μg/mL according to our optimization process and manufacturer’s protocol. As shown in Fig. 4, FPG456-C8 and Cy5.5-C3 alone (denoted as 0:1) demonstrated low cellular uptake. In contrast, when both FPG456-C8 and Cy5.5-C3 were co-incubated with PULSin, the peptides were clearly internalized into cells in a PULSin concentration-dependent manner and the uptake was maximized at the ratio of 4:1 with no sign of cytotoxicity. This indicates that the current easy-to-use platform can be exploited for live-cell imaging and quantitative analysis of apoptosis. However, it is to be remembered that PULSin is an aqueous formulation of positively charged lipids arranged into cationic liposomes; thus it works well with negatively charged peptides like C8 and C3 but may not work efficiently against positively charged peptide analogs based on electrostatic interactions. To deliver fluorogenic probes with a positive net charge, other transfecting agents should be considered.
Figure 4.
Fluorescence images of MDA-MB-435 cells incubated with non-quenched FPG456-C8 or Cy5.5-C3 formulated with various amounts of PULSin (1 μg/100 μL of peptides with 0, 1, 2 and 4 μL of PULSin from a commercially provided stock, denoted as 0:1 to 4:1). Each formula was incubated with the cells in 1 mL of L-15 medium without FBS for 3 hr at 37 °C. The cellular uptake of FPG456-C8 and Cy5.5-C3 were examined with a fluorescence microscope configured for FITC and Cy5.5 filters followed by DAPI staining at room temperature. Blue (DAPI), nucleus; green (FPG456); red (Cy5.5-C3).
3.4. Real-time visualization of caspase cascade
Finally, we investigated the potential application of C8/C3/PULSin for real-time, simultaneous imaging of two different caspases in single cells with confocal microscopy. Apoptosis was induced by treating cells with TRAIL. TRAIL is reported to stimulate caspase expressions through either the extrinsic or intrinsic pathway depending on the cell type. In MDA-MB-435 cells, TRAIL is known to activate caspase-8, −9 and −3 [35]. Before cellular imaging, the expression of caspase-8 and −3 induced by TRAIL was verified by Western blot assay (Supplementary Data, Fig. S3). TRAIL clearly activated pro-caspase-8 to form caspase-8 and contributed to the following pro-caspse-3 activation. Apoptosis was also verified by staining the cells with FITC-labeled annexin V (Supplementary Data, Fig. S4). TRAIL-treated MDA-MB-435 cells demonstrated positive annexin V staining compared to cells not treated with TRAIL and with excess amount of free annexin V for blocking test.
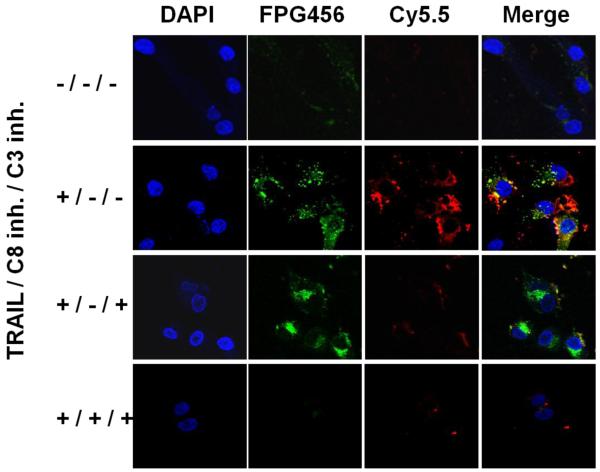
To monitor the expression profiles of caspase-8 and −3 in MDA-MB-435 cells, C8/C3/PULSin-pretreated cells were incubated with different combinations of TRAIL and caspase-8 and −3 inhibitors. After treatment, the cells were fixed at predetermined time points and were imaged through three different filters followed by staining the nucleus with DAPI. As shown in Fig. 5, the cells treated with C8/C3/PULSin alone provided low background signals. In contrast, when the cells were treated with TRAIL, each probe exhibited strong florescence signals and enabled clear visualization of apoptotic process induced by TRAIL. To confirm if recovered fluorescence signals were activated by specific caspases, the cells were pretreated with caspase-3 inhibitor or both caspase-8 and −3 inhibitors before the TRAIL treatment. When the cells were treated with both TRAIL and caspase-3 inhibitor, Cy5.5 signal from C3 was significantly reduced whereas FPG456 signal from C8 was strongly maintained. In addition, both FPG456 and Cy5.5 signals were diminished when the cells were pretreated with both inhibitors. In accordance with in vitro results, the fluorescence signals from the probes were significantly lower when the cells were treated with respective inhibitors, demonstrating that the recovered fluorescence signals were specifically activated by overexpressed target caspases.
Figure 5.
Multiplexed imaging of the caspase cascade in MDA-MB-435 cells. The cells were incubated with C8/C3/PULSin for 4 h and treated by different combinations of TRAIL (100 ng/mL) and 50 mM of caspase-3 and/or caspase-8 inhibitors. The cells were imaged using confocal microscopy followed by DAPI stating. Blue (DAPI), nucleus; green (FPG456), caspase-8; red (Cy5.5), caspase-3.
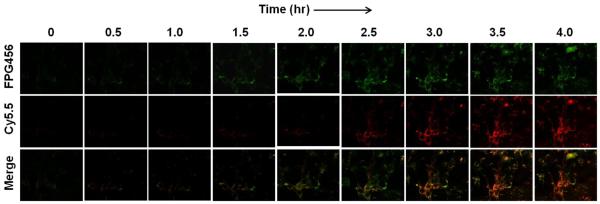
To demonstrate the utility of C8/C3/PULSin for real-time imaging of caspase cascade, MDA-MB-435 cells were treated with C8/C3/PULSin and TRAIL and continuously monitored every 30 min for 4 h using live cell imaging confocal microscopy configured for imaging of FPG456 and Cy5.5. Fig. 6 shows typical serial images. Similar to the fixed cell samples, the cells treated with C8/C3/PULSin provided low background signals, allowing real-time imaging of the caspase cascade in single cells. Upon TRAIL treatment, the cells gradually produced strong fluorescence signals depending on the incubation time. Merged images indicate the tendency of the activation of initiator caspase-8 to occur early before leading to the activation of executioner caspase-3. Overall, our results demonstrate that the simple mixture of fluorogenic probes formulated with transfecting agent can be used for real-time imaging of multiple caspase expressions at the single cell level.
Figure 6.
Activities of caspase-8 and caspase-3 in MDA-MB-435 cells serially imaged using C8/C3/PULSin. The cells were incubated with C8/C3/PULSin for 4 hr and treated with TRAIL (100 ng/mL). Images were collected at 30 min intervals over 4 hr. Green (FPG456), caspase-8; red (Cy5.5), caspase-3.
4. Conclusions
Real-time monitoring of the caspase cascade in living cells will not only help to understand the complex apoptosis signaling mechanisms but also play a potentially valuable role for both preclinical and clinical applications, especially in the drug development field such as identifying and validating novel drug candidates in a high-throughput screening manner. In this study, we have introduced a simple but versatile formulation that is capable of imaging multiple caspase activities in living cells. As a proof-of-concept, a series of expression levels of initiator caspase-8 and effector caspase-3 altered by TRAIL-induced apoptosis was directly imaged in living cells. Compared to previously reported intracellular enzyme targeting imaging agents, our system does not need complicated synthetic processes but only involves a straightforward and reproducible fluorogenic peptide synthesis along with a simple mixing step with a commercially available transfecting agent. Although only two caspases were monitored in this study, more complicated multiplexed imaging, targeting three or more caspases, can be performed by designing a series of different fluorogenic probes possessing different dyes. Furthermore, this platform can be applied to cell-based high-throughput screening systems for practical applications. In addition, any intracellular enzymes in live cells can be targeted and imaged in real-time using this platform by substituting substrate sequences.
Supplementary Material
Acknowledgement
This work was supported in part by the Intramural Research Program (IRP) of the National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH), the International Cooperative Program of the National Science Foundation of China (NSFC) (81028009), Chinese Academy of Sciences professorship for Senior International Scientists (2011T2J06), and NIH Pathway to Independence (K99/R00) Award. We thank Myung Sun Lee for illustrations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
L.Z. and X. H. contributed equally.
Appendix A. Supplementary data
Supplementary data to this article can be found online.
References
- [1].Puente XS, Sanchez LM, Overall CM, Lopez-Otin C. Human and mouse proteases: a comparative genomic approach. Nat. Rev. Genet. 2003;4:544–558. doi: 10.1038/nrg1111. [DOI] [PubMed] [Google Scholar]
- [2].Fairbairn LJ, Cowling GJ, Reipert BM, Dexter TM. Suppression of apoptosis allows differentiation and development of a multipotent hemopoietic cell line in the absence of added growth factors. Cell. 1993;74:823–832. doi: 10.1016/0092-8674(93)90462-y. [DOI] [PubMed] [Google Scholar]
- [3].Brown JM, Attardi LD. The role of apoptosis in cancer development and treatment response. Nat. Rev. Cancer. 2005;5:231–237. doi: 10.1038/nrc1560. [DOI] [PubMed] [Google Scholar]
- [4].Lamkanfi M, Festjens N, Declercq W, Vanden Berghe T, Vandenabeele P. Caspases in cell survival, proliferation and differentiation. Cell Death Differ. 2007;14:44–55. doi: 10.1038/sj.cdd.4402047. [DOI] [PubMed] [Google Scholar]
- [5].Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat. Rev. Mol. Cell Biol. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- [6].Elmore S. Apoptosis: a review of programmed cell death. Toxicol. Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Vangestel C, Peeters M, Mees G, Oltenfreiter R, Boersma HH, Elsinga PH, Reutelingsperger C, Van Damme N, De Spiegeleer B, Van de Wiele C. In vivo imaging of apoptosis in oncology: an update. Mol. Imaging. 2011;10:340–358. doi: 10.2310/7290.2010.00058. [DOI] [PubMed] [Google Scholar]
- [8].Niu G, Chen X. Apoptosis imaging: beyond annexin V. J. Nucl. Med. 2010;51:1659–1662. doi: 10.2967/jnumed.110.078584. [DOI] [PubMed] [Google Scholar]
- [9].Schoenberger J, Bauer J, Moosbauer J, Eilles C, Grimm D. Innovative strategies in in vivo apoptosis imaging. Curr. Med. Chem. 2008;15:187–194. doi: 10.2174/092986708783330647. [DOI] [PubMed] [Google Scholar]
- [10].Lee S, Xie J, Chen X. Peptides and peptide hormones for molecular imaging and disease diagnosis. Chem. Rev. 2010;110:3087–3111. doi: 10.1021/cr900361p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wang K, Na MH, Hoffman AS, Shim G, Han SE, Oh YK, Kwon IC, Kim IS, Lee BH. In situ dose amplification by apoptosis-targeted drug delivery. J. Control. Release. 2011;154:214–217. doi: 10.1016/j.jconrel.2011.06.043. [DOI] [PubMed] [Google Scholar]
- [12].Narula J, Acio ER, Narula N, Samuels LE, Fyfe B, Wood D, Fitzpatrick JM, Raghunath PN, Tomaszewski JE, Kelly C, Steinmetz N, Green A, Tait JF, Leppo J, Blankenberg FG, Jain D, Strauss HW. Annexin-V imaging for noninvasive detection of cardiac allograft rejection. Nat. Med. 2001;7:1347–1352. doi: 10.1038/nm1201-1347. [DOI] [PubMed] [Google Scholar]
- [13].Boersma HH, Kietselaer BL, Stolk LM, Bennaghmouch A, Hofstra L, Narula J, Heidendal GA, Reutelingsperger CP. Past, present, and future of annexin A5: from protein discovery to clinical applications. J. Nucl. Med. 2005;46:2035–2050. [PubMed] [Google Scholar]
- [14].Vaux DL, Korsmeyer SJ. Cell death in development. Cell. 1999;96:245–254. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- [15].Pop C, Salvesen GS. Human caspases: activation, specificity, and regulation. J. Biol. Chem. 2009;284:21777–21781. doi: 10.1074/jbc.R800084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Huang X, Swierczewska M, Choi KY, Zhu L, Bhirde A, Park J, Kim K, Xie J, Niu G, Lee KC, Lee S, Chen X. Multiplex Imaging of an Intracellular Proteolytic Cascade by using a Broad-Spectrum Nanoquencher. Angew. Chem. Int. Ed. Engl. 2012;51:1625–1630. doi: 10.1002/anie.201107795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Huang X, Lee S, Chen X. Design of “smart” probes for optical imaging of apoptosis. Am. J. Nucl. Med. Mol. Imaging. 2011;1:3–17. [PMC free article] [PubMed] [Google Scholar]
- [18].Kim K, Lee M, Park H, Kim JH, Kim S, Chung H, Choi K, Kim IS, Seong BL, Kwon IC. Cell-permeable and biocompatible polymeric nanoparticles for apoptosis imaging. J. Am. Chem. Soc. 2006;128:3490–3491. doi: 10.1021/ja057712f. [DOI] [PubMed] [Google Scholar]
- [19].Maxwell D, Chang Q, Zhang X, Barnett EM, Piwnica-Worms D. An improved cell-penetrating, caspase-activatable, near-infrared fluorescent peptide for apoptosis imaging. Bioconjug. Chem. 2009;20:702–709. doi: 10.1021/bc800516n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cha EJ, Jang ES, Sun IC, Lee IJ, Ko JH, Kim YI, Kwon IC, Kim K, Ahn CH. Development of MRI/NIRF ‘activatable’ multimodal imaging probe based on iron oxide nanoparticles. J. Control. Release. 2011;155:152–158. doi: 10.1016/j.jconrel.2011.07.019. [DOI] [PubMed] [Google Scholar]
- [21].Lee S, Park K, Kim K, Choi K, Kwon IC. Activatable imaging probes with amplified fluorescent signals. Chem. Commun. (Camb.) 2008:4250–4260. doi: 10.1039/b806854m. [DOI] [PubMed] [Google Scholar]
- [22].Lee S, Xie J, Chen X. Activatable molecular probes for cancer imaging. Curr. Top. Med. Chem. 2010;10:1135–1144. doi: 10.2174/156802610791384270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Swierczewska M, Lee S, Chen X. The design and application of fluorophore-gold nanoparticle activatable probes. Phys. Chem. Chem. Phys. 2011;13:9929–9941. doi: 10.1039/c0cp02967j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Choi KY, Magdalena S, Lee S, Chen X. Protease-activated drug development. Theranostics. 2012;2:156–179. doi: 10.7150/thno.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhu L, Xie J, Swierczewska M, Zhang F, Quan Q, Ma Y, Fang X, Kim K, Lee S, Chen X. Real-time video imaging of protease expression in vivo. Theranostics. 2011;1:18–27. doi: 10.7150/thno/v01p0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lee S, Choi KY, Chung H, Ryu JH, Lee A, Koo H, Youn IC, Park JH, Kim IS, Kim SY, Chen X, Jeong SY, Kwon IC, Kim K, Choi K. Real time, high resolution video imaging of apoptosis in single cells with a polymeric nanoprobe. Bioconjug. Chem. 2011;22:125–131. doi: 10.1021/bc1004119. [DOI] [PubMed] [Google Scholar]
- [27].Sun IC, Lee S, Koo H, Kwon IC, Choi K, Ahn CH, Kim K. Caspase sensitive gold nanoparticle for apoptosis imaging in live cells. Bioconjug. Chem. 2010;21:1939–1942. doi: 10.1021/bc1003026. [DOI] [PubMed] [Google Scholar]
- [28].Edgington LE, Berger AB, Blum G, Albrow VE, Paulick MG, Lineberry N, Bogyo M. Noninvasive optical imaging of apoptosis by caspase-targeted activity-based probes. Nat. Med. 2009;15:967–973. doi: 10.1038/nm.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kanno A, Yamanaka Y, Hirano H, Umezawa Y, Ozawa T. Cyclic luciferase for real-time sensing of caspase-3 activities in living mammals. Angew. Chem. Int. Ed. Engl. 2007;46:7595–7599. doi: 10.1002/anie.200700538. [DOI] [PubMed] [Google Scholar]
- [30].Bardet PL, Kolahgar G, Mynett A, Miguel-Aliaga I, Briscoe J, Meier P, Vincent JP. A fluorescent reporter of caspase activity for live imaging. Proc. Natl. Acad. Sci. U S A. 2008;105:13901–13905. doi: 10.1073/pnas.0806983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Weill CO, Biri S, Erbacher P. Cationic lipid-mediated intracellular delivery of antibodies into live cells. Biotechniques. 2008;44:Pvii–Pxi. doi: 10.2144/000112832. [DOI] [PubMed] [Google Scholar]
- [32].Weiss A, Neuberg P, Philippot S, Erbacher P, Weill CO. Intracellular peptide delivery using amphiphilic lipid-based formulations. Biotechnol. Bioeng. 2011;108:2477–2487. doi: 10.1002/bit.23182. [DOI] [PubMed] [Google Scholar]
- [33].Logue SE, Elgendy M, Martin SJ. Expression, purification and use of recombinant annexin V for the detection of apoptotic cells. Nat. Protoc. 2009;4:1383–1395. doi: 10.1038/nprot.2009.143. [DOI] [PubMed] [Google Scholar]
- [34].McStay GP, Salvesen GS, Green DR. Overlapping cleavage motif selectivity of caspases: implications for analysis of apoptotic pathways. Cell Death Differ. 2008;15:322–331. doi: 10.1038/sj.cdd.4402260. [DOI] [PubMed] [Google Scholar]
- [35].Lu M, Strohecker A, Chen F, Kwan T, Bosman J, Jordan VC, Cryns VL. Aspirin sensitizes cancer cells to TRAIL-induced apoptosis by reducing survivin levels. Clin. Cancer. Res. 2008;14:3168–3176. doi: 10.1158/1078-0432.CCR-07-4362. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.