Abstract
Objective
To determine the impact of methylation alteration in inflamed muscles from children with Juvenile Dermatomyositis (JDM) and other Idiopathic Inflammatory Myopathies (IIM).
Methods
MRI-directed diagnostic muscle biopsies (MBx) from 20 JDM children and 4 healthy controls were used for genome-wide DNA methylation profiling (IRB# 200813457). Bisulfite pyrosequencing confirmed methylation status in JDM and other IIM. Immunohistochemistry defined localization and expression levels of WT1.
Results
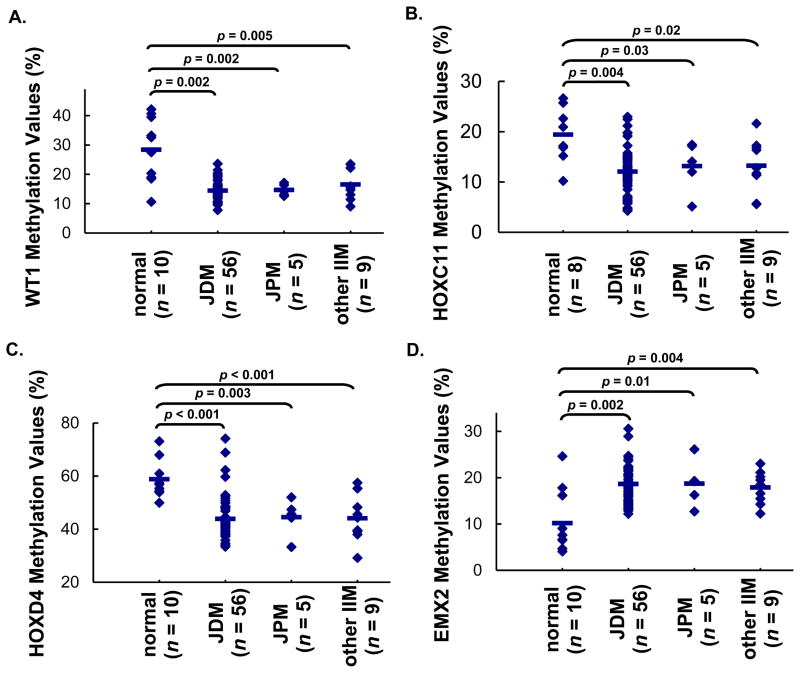
Comparison of genome-wide DNA methylation profiling between JDM and normal controls revealed 27 genes with significant methylation difference, enriched with transcription factors and cell cycle regulators, unrelated to duration of untreated disease. Six homeobox genes were among them: ALX4, HOXC11, HOXD3 and HOXD4 were hypomethylated; EMX2 and HOXB1 were hypermethylated. WT1 was significantly hypomethylated in JDM (Δβ = −0.41, p < 0.001). Bisulfite pyrosequencing verification in 56 JDM samples confirmed the methylation alterations of these genes. Similar methylation alterations were observed in Juvenile Polymyositis (JPM, n = 5) and other IIM (n = 9). Concordantly, WT1 protein was increased in JDM muscle, with average positive staining of 11.6%, but was undetectable in normal muscles (p < 0.05).
Conclusions
These results suggest that affected muscles of children with JDM and IIM have the capacity to repair, and that homeobox and WT1 genes are epigenetically marked to facilitate this repair process, potentially suggesting new avenues of therapeutic intervention.
Juvenile dermatomyositis (JDM) is a severe and often chronic childhood autoimmune disease. As a consequence of the immune system mediated vasculopathy, the children exhibit a characteristic rash often accompanied by symmetrical, proximal muscle weakness (1,2). In spite of decades of effort to improve the prognosis of JDM, at least 22% JDM patients have chronic damage (3) and there is variability in their recovery of full muscle strength and endurance. Although new data is beginning to emerge, there is a scarcity of information concerning a spectrum of long term outcomes (1,4,5). A major barrier to identifying and providing effective therapy for children with JDM is the lack of prognostic indicators of disease severity and the individual child’s ability to recover (2). Current medication for JDM utilizes immunosuppression to diminish inflammation, which may have different components in skin and muscle (6). The muscle’s ability to recover function has therefore drawn special and practical interests. The muscle repair process includes the postnatal muscle growth, regeneration of skeletal muscle and remodeling of post injury through the muscle stem cells, which are mainly satellite cells and mesenchymal stem cells (7,8).
In search for the accessible biomarkers for JDM, we performed a methylation profiling study of diagnostic muscle biopsies from untreated JDM children. As an important gene regulatory mechanism, DNA methylation is essential to normal development as well as to the evolution of a range of diseases (9–11). Our results show that several genes that have stem-cell related functions display methylation alterations in JDM, suggesting that the affected muscles of children with JDM may undergo self-renewal and repair processes involving stem cells by epigenetically marking a few critical genes.
Materials and Methods
Patient Population
For all IIM and normal controls, age appropriate and parental consent were obtained under an IRB-approved protocol (Children’s Memorial Research Center IRB # 200813457). Muscle biopsy samples from children with definite or probable inflammatory myopathy (3 or 4 of Bohan and Peter criteria (12)) were obtained from an area positive on MRI examination (T-2 weighted image, fat suppressed), usually the vastus lateralis. Muscles of the trunk are commonly involved in JDM, resulting in decreased core strength, documented and validated by the Child Myositis Assessment Scale which is used internationally to assess muscle strength and endurance in children with inflammatory myopathy (13). Controls were non-inflammatory muscles, obtained from an anatomically similar source (trunk muscles) of age-gender-race matched otherwise healthy children undergoing an orthopedic procedure.
For the methylation profiling study, only patients who had not been given any immunosuppressive or other therapies before the diagnostic muscle biopsies were selected (n = 20). Of the 20 children, 15 (75%) were girls, 16 (80%) were White/Hispanic, the mean age at biopsy was 6.4 years ± 3.5 SD. Since the duration of untreated disease (DUD), as determined by the length of symptoms from first symptom (rash and/or weakness) to the time of biopsy, is a critical variable influencing both clinical (14) and gene expression profiling findings (15), we classified our JDM samples into short DUD (< 2 months of symptoms, n = 6) and long DUD (≥2 months of symptoms, n = 14). The average DUD for all 20 children was 6.2 ± 4.8 months. Four normal controls were used.
For the pyrosequencing confirmation study, 36 more children with JDM were included to make a total of 56 JDM samples. Of the 56 JDM children, 43 (77%) were girls, 33 (59%) were White/Hispanic, 14 (25%) had been given some type of medication before biopsies. The mean age at biopsy was 6.8 ± 3.5 years, the average DUD was 8.3 ± 10.2 months. In addition, children with a range of inflammatory myopathies were tested as well: JPM (n = 5) and other types of juvenile IIM (n = 9) with a spectrum of overlap syndromes and myositis-associated antibodies (U-1RNP, U-2RNP, Ro, Pm/Scl). Myositis specific and associated antibody studies were performed as part of the diagnostic evaluation in the University of Oklahoma Research Foundation Clinical Immunology Laboratory. JPM children had not been given any type of medication before biopsies, whereas 2 of the children with other IIM had been given some treatment. Ten normal controls were used.
Illumina Infinium BeadChip Assay
Genomic DNA was extracted with Qiagen DNeasy tissue kit (Qiagen, Valencia, CA). Methylation profiling was carried out in the Genomics Core Facilities of the Northwestern University, using Illumina Infinium HumanMethylation27 BeadChip and following protocols provided by Illumina (Illumina Inc., San Diego, CA). This array interrogates 27,578 CpG loci mostly selected from promoter regions covering more than 14,000 genes. Methylation levels for each CpG site, determined by the fluorescence signals for methylated and unmethylated alleles, were represented by beta values (β) ranging from 0 (completely unmethylated) to 1 (completely methylated).
DNA Methylation Data Analysis
Methylation data was analyzed with Illumina’s GenomeStudio software (GenomeStudio methylation module v1.0, 2008). CpG sites with poor detection signals (defined as Detection p-value > 0.05) were eliminated. Methylation signals were first normalized with background before further analysis. Significantly differentially methylated CpG sites were defined as Δβ > 0.17 with p < 0.05 after computing false discovery rate. Gene annotation was extracted with MetaCore software (GeneGo, Inc.) and DAVID (http://david.abcc.ncifcrf.gov/).
Pyrosequencing Verification
PCR reactions were carried out using the Hotstart Taq polymerase kit (Qiagen, Valencia, CA) in 25 μL total volume and with 50 pm of forward primer and reverse primer. For each PCR reaction, 50 ng of the bisulfite converted DNA in 1 μL was used as a template. After 5 min of initial denaturation at 95°C, the cycling conditions of 40 cycles consisted of denaturation at 95°C for 15 s, annealing at 58°C or 60°C for 30 s, and elongation at 72°C for 30 s. The PCR products were stored at 4°C until ready for pyrosequencing.
Pyrosequencing was performed using the PyroMark MD Pyrosequencing System (Biotage, Charlottesville, VA). In brief, the PCR product was bound onto streptavidin-Sepharose HP beads (GE Healthcare, Uppsala, Sweden). Beads containing the immobilized PCR product were denatured using a 0.2M NaOH solution and neutralized. Pyrosequencing primer at a concentration of 0.3 μM was annealed to the purified single-stranded PCR product at 28°C. Methylation quantification was performed using the manufacturer-provided software. The primers used in the PCR runs and pyrosequencing reactions are shown in Supplementary Table 1.
Immunohistochemistry Staining of WT1
Cold-acetone fixed frozen muscle tissue sections were blocked and incubated with monoclonal mouse anti-human WT1 antibody (Cat# M3561, Dako, Inc., CA, 1:100 dilution) overnight at 4°C. The sections were then incubated with goat anti-mouse biotinylated secondary antibody (Cat# 115-065-062, Jackson ImmunoResearch Laboratory, West Grove, PA) at room temperature for one hour. Avidin-biotin complex (ABC) and diaminobenzidine (DAB) reagent kits (Vector Laboratories, Burlingame, CA) were used to detect the secondary antibody. Counterstaining with hematoxylin and Scott’s bluing solution (Ricca Chemical, Texas) was performed. Mouse IgG was used as an isotype control (Cat#08-6599, Zymed, San Francisco, CA). Images of stained tissue sections were acquired with a Leica DMR-HC microscope (Leica Microsystems GmbH, Wetzlar, Germany) coupled to a Retiga 4000R camera using Openlab computer software 5.5.0 (Improvision Inc., Lexington, MA) and edited using Adobe Photoshop CS2 software. The tissue sections were evaluated by two independent observers blinded to the subject groups (JDM or normal control). Positive WT1 staining was detected in the cytoplasm of myofibers, and the percentage was defined as the ratio of myofibers expressing the protein at the cytoplasm level.
Statistical Analysis
Comparison of methylation levels in different groups was assessed with one-way ANOVA test. Computations were performed with the R statistical package (version 2.13.0, R Development Core Team, Vienna, Austria, http://www.R-project.org). For hierarchical clustering, Pearson’s correlation coefficient and average linkage were used as distance and linkage methods respectively.
Results
Methylation profiling distinguished JDM muscles from normal controls
In the first part of this study, with age-appropriate informed consent, 20 diagnostic muscle biopsies from untreated children with symptoms of active JDM together with 4 normal controls were tested using methylation arrays. For all 24 samples tested, CpG sites with poor detection signals (defined as detection p-value > 0.05) were eliminated, leaving 25,032 valid sites. By further comparing JDM with normal controls, a total of 27 genes were found to have significant methylation difference (Δβ > 0.17, p < 0.05 computing false discovery rate, Supplementary Table 2). The supervised hierarchical clustering using these 27 genes clearly distinguished JDM muscle samples from normal controls (Supplementary Figure 1). Conversely, the comparison between JDM with short DUD and JDM with long DUD did not yield any genes with significant methylation difference. The supervised hierarchical clustering with the 27 genes did not distinguish JDM with short DUD from JDM with long DUD either, indicating the methylation signature value of these genes with respect to the shared pathophysiology of any disease duration in JDM, but not the progressive process evolving over time.
Differentially methylated genes in JDM were enriched with transcription factors and cell cycle regulators
Among these 27 differentially methylated genes in JDM, a number of them are transcription factors and/or cell cycle regulators (Supplementary Table 2). These include 6 homeobox genes: ALX4, HOXC11, HOXD3, and HOXD4 were hypomethylated whereas EMX2 and HOXB1 were hypermethylated. The other known transcription factors include SIM2, BRDT, KLF1, ZNFN1A1, and WT1 (wilms tumor 1), whose functions comprise of neurogenesis (16), cell cycle control (17,18) and cell differentiation (19–22). Additional cell cycle regulators include SEPT9, CIDEB, and NUAK1. Although the methylation alterations of these genes were not directed in a uniform cell survival promoting pathway, or alternatively a cell death inducing route, changes in these genes would indeed lead to activation of a series of events in the muscle cells in response to damage. Previous gene expression studies of JDM have showed the alterations in expression of immune response genes, genes related to vascular remodeling, and genes involved in endoplasmic reticulum stress response pathway (15,23–25). These genes did not show significant methylation alterations in JDM, indicating the involvement of other regulatory mechanisms for these genes.
Pyrosequencing verification confirmed the methylation alterations of WT1 and homeobox genes in JDM as well as other types of juvenile IIM
In our study, WT1 gene showed the most significant methylation alteration in 20 untreated JDM MBx compared to muscle from controls (Δβ = −0.41). Represented on the array with reliable detection signals, a total of 12 CpG sites were located near the 5′-end of the WT1 gene. All these 12 CpG sites showed decrease in methylation values in JDM (Figure 1). To validate the methylation array data independently, DNA samples from muscle biopsies from 36 additional JDM children, together with the 20 samples used for methylation profiling study, were subjected to bisulfite treatment followed by pyrosequencing (n = 56). Additional normal muscles, together with those samples previously profiled, were used as controls (n = 10). Hypomethylation of WT1 in JDM was confirmed by this verification. We also included in this verification MBx from patient with JPM (n = 5) and other forms of IIM (n = 9). Compared to muscle from healthy children, hypomethylation of WT1 were consistently observed in all types of juvenile IIM tested (Figure 2A). To eliminate the effects of medication, the results were re-analyzed by removing samples that had been given any types of medication before diagnostic muscle biopsy. Hypomethylation of WT1 was still observed in all untreated samples, with similar p-values as those observed in all samples (Supplementary Figure 2A).
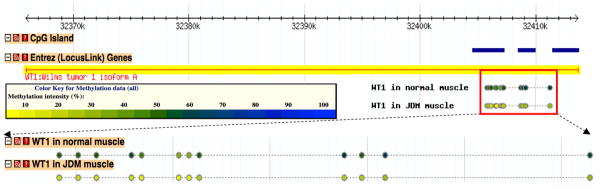
Figure 1. Genomic view of WT1 and its methylation patterns in both JDM and normal muscles.
Each dot represented one CpG site with valid methylation value. Methylation levels were represented by β values (0%-unmethylated, yellow; 100%-methylated, blue). Detailed view can be found at http://cmbteg.childrensmemorial.org/cgi-bin/gbrowse/btech/, by zooming in to WT1 gene (50).
Figure 2. Pyrosequencing verification of the methylation levels of specific genes in JDM and other types of juvenile IIM: WT1 (A), HOXC11 (B), HOXD4 (C), EMX2 (D).
Each dot in the results represented the methylation level of the specific gene in one individual sample and the line represented the average methylation levels for each group. The results confirmed the methylation levels as detected by methylation array. Comparisons among groups were performed by one way ANOVA, followed by post hoc Tukey HSD test. Significant differences were shown between groups: JDM vs. normal, JPM vs. normal, and other IIM vs. normal.
We also verified the methylation status of homeobox genes by bisulfite pyrosequencing: HOXC11 and HOXD4 were hypomethylated and EMX2 was hypermethylated, consistent with findings from the methylation array. Parallel to WT1 verification, we also included samples from JPM and other types of juvenile IIM. All verified homeobox genes displayed comparable methylation levels in all forms of juvenile IIM. These levels were significantly different from those displayed in normal controls (Figure 2B, C, and D). After removing samples that had been given any types of medication before biopsy, similar trends were observed, with p-values staying significant in all comparisons except for the case of hypomethylation of HOXC11 in other types of juvenile IIM (p = 0.06), although we anticipate that this would improve with increased sample size (Supplementary Figure 2B, C, D).
WT1 showed increased protein levels in JDM
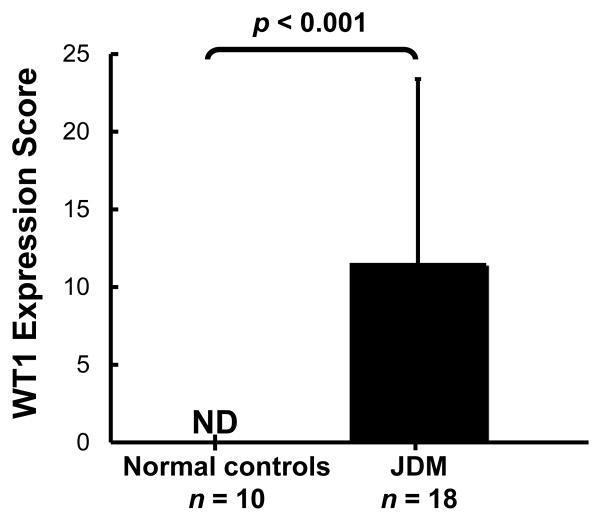
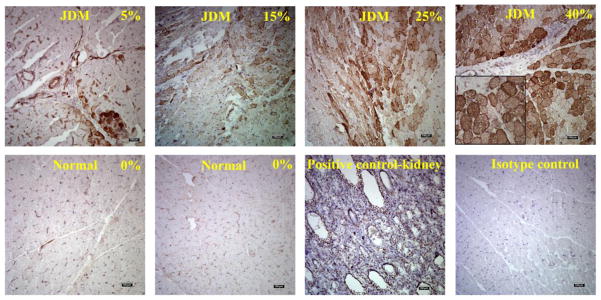
Based on the findings of WT1 hypomethylation in JDM, we further determined the expression levels of WT1 in JDM. As measured by immunohistochemistry, WT1 hypomethylation was in accord with increased WT1 protein expression. In JDM samples (n = 18), the average amount of myofibers positively stained of WT1 was 11.6%; whereas in all of the normal muscles (n = 10), WT1 expression was undetectable (Figure 3). This increased expression of WT1 in JDM muscle occurred in the cytoplasm of the myofibers (Figure 4).
Figure 3. Average protein levels of WT1 in JDM compared to normal muscles as detected by immunohistochemistry.
Comparisons between JDM and normal were performed by student’s t-test. ND: WT1 was not detectable in normal tissues.
Figure 4.
Representative photographs of immunohistochemistry staining of WT1 in JDM muscle biopsies compared with normal controls. The photographs depicted a range of fibers positive for WT1 (5%, 25% and 40%, Scale = 100 μm). The inset picture was a regional enlargement (4x). Positive staining was indicated by brown color against the blue background. The % score represented the percentage of positive staining area for WT1 on the entire tissue section.
Discussion
Previous studies have documented that treatment with DNA methylation inhibitors caused fibroblasts to differentiate into myoblasts or myoblasts to differentiate into smooth muscle cells, providing strong evidence that DNA methylation plays an important role in establishing muscle cell lineage (26,27). In this epigenomic study of untreated muscle biopsies from patients with JDM, 27 genes were found to be differentially methylated. Amidst them are a number of transcription factors and/or cell cycle regulators, which include 6 homeobox genes (ALX4, HOXC11, HOXD3, HOXD4, EMX2, HOXB1), WT1, ZNFN1A1, KLF1, SIM2, SEPT9, CIDEB, NUAK1, and BRDT. ZNFN1A1, a zinc finger transcription factor, is associated with the pathogenesis of a similar autoimmune disease, systemic lupus erythematosus (28). KLF1 is another zinc finger transcription factor that inhibits cell growth and causes apoptosis. SIM2 has been shown to suppress hypoxia induced pro-cell death (17). Hypermethylation of SIM2 in JDM could alleviate this suppression and therefore promote cell death. Both SEPT9 and CIDEB are involved in cell cycle control and their regulation by methylation has been demonstrated (29,30). Since re-entry of the cell into the cell cycle is regarded as being associated with cell fate switch (31,32), our results appear to be consistent with an alteration of cell fate in damaged muscles in children with JDM.
Among the 6 homeobox genes, ALX4, HOXC11, HOXD3, and HOXD4 were hypomethylated whereas EMX2 and HOXB1 were hypermethylated. Homeobox genes have been commonly recognized as sequence-specific transcription factors involved in differentiation and limb development (33). Known as controlling the early development of specific compartments of the embryo (33), homeobox genes may also regulate muscle-specific genes in mature muscle tissues (34). Certain homeobox genes have been found to regulate a universal aspect of myogenesis, such as cell proliferation or subsequent differentiation during tissue remodeling or repair (35,36). Alterations in homeobox genes have been viewed as evidence of a “self-renewal” signature in stem cells (37,38). Of note, CpG dinucleotides, which are the exclusive sites of DNA methylation in mammalian somatic cells, are strongly preserved from mutational depletion in the coding regions of homeobox genes (39). This preservation does not exist in organisms that do not have DNA methylation (such as D. melanogaster and C. elegans), reinforcing the significance of DNA methylation in homeobox gene regulation (39). In our study, the methylation alterations of these genes in muscles from children with JDM and other IIM suggested that the homeobox regulatory network was altered after muscle damage, which could stimulate the stem cell pool in JDM muscles toward repair.
Furthermore, WT1 underwent significant hypomethylation in JDM. While the protein expression was undetectable in normal muscles, WT1 protein levels were increased in JDM muscles. WT1, first described in the development of the Wilms’ tumor of the kidney (40), has a spectrum of functions which are linked to normal development as well as to different disease states (41). One of the key functions of WT1 in mature tissues is to maintain the pluripotency of progenitor/stem cells (41). The role of WT1 in regulating muscle cell linage differentiation has been suggested in the development of primitive skeletal muscle in kidney with WT1 mutation (42) and the inhibition of myogenic differentiation by overexpressing WT1 in a murine myoblastic cell line (43). Although the latter evidence has been disputed by another study (44), the essential roles of WT1 in regeneration and repair of damaged hearts has been demonstrated (45–47). Re-expression of WT1 activated the progenitor cell populations in the adult heart after injury (46). Knockout of WT1 in cardiovascular epicardial progenitor cells reduced the numbers of mesenchymal progenitor cells and their derivatives (47). In our study, WT1 protein was re-expressed in JDM muscle in concordance with its hypomethylation. Of note, the methylation alterations of WT1 as well as homeobox genes were found not only in JDM, but also in JPM and other forms of juvenile IIM. Similar to JDM, JPM is also a type of IIM that often displays severe inflammation of the muscle, except that JPM does not manifest cutaneous inflammation and is much less common in children, with a ratio of 1:20 (48). Symmetrical proximal muscle weakness, however, is a hallmark of the majority of juvenile IIM (except for those who have a rash only without documented muscle weakness, who were not included in this study). Similar methylation alteration of WT1 and homeobox genes in all forms of IIM tested suggested a universal role of these genes in the damaged and inflamed muscle of juvenile IIM.
It has been reported that certain component from the endoplasmic reticulum (ER) stress response was present in the regenerating myofibers of patients with muscular dystrophy, but absent in non-regenerating myofibers (49). Concurrently, the involvement of the ER stress response pathway was well documented and laid the ground for muscle regeneration in autoimmune myositis (23). In our study, a number of transcription factors and cell cycle regulators have been found to be epigenetically altered in JDM, which implies the cascades of events occurring in the damaged muscles. Although ER stress response genes were not among the differentially methylated genes in JDM, the associations of these genes with ER stress response would merit further investigation. Moreover, the methylation alterations of WT1 and the homeobox genes in JDM and other IIM provide additional evidence that the damaged muscles in children have self-renewal capacity, and that critical genes are epigenetically altered in response to the muscle damage occurred during disease pathogenesis. Considering the autoimmune nature of JDM and other types of juvenile IIM, our findings may point to potential new therapeutic approaches in which the self-renewal ability of the damaged muscles in children may be enhanced to allow the muscle to repair by activating a viable and readily available resident source.
Supplementary Material
Acknowledgments
The authors thank Dr. John Sarwark and Teresa Philipp for their essential roles in recruiting the healthy pediatric donors to provide control muscle samples, and Peter Hendrickson, for his substantial contributions to the acquisition of data.
Supported in part by NIH/NINR, 1R01NR012692-01, CureJM Foundation, and Macy’s Miracle (to LMP).
References
- 1.Christen-Zaech S, Seshadri R, Sundberg J, Paller AS, Pachman LM. Persistent association of nailfold capillaroscopy changes and skin involvement over thirty-six months with duration of untreated disease in patients with juvenile dermatomyositis. Arthritis Rheum. 2008;58:571–6. doi: 10.1002/art.23299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feldman BM, Rider LG, Reed AM, Pachman LM. Juvenile dermatomyositis and other idiopathic inflammatory myopathies of childhood. Lancet. 2008;371:2201–12. doi: 10.1016/S0140-6736(08)60955-1. [DOI] [PubMed] [Google Scholar]
- 3.Schwartz T, Sanner H, Husebye T, Flato B, Sjaastad I. Cardiac dysfunction in juvenile dermatomyositis: a case-control study. Ann Rheum Dis. 2011;70:766–71. doi: 10.1136/ard.2010.137968. [DOI] [PubMed] [Google Scholar]
- 4.Huber AM, et al. Medium-and long-term functional outcomes in a multicenter cohort of children with juvenile dermatomyositis. Arthritis Rheum. 2000;43:541–9. doi: 10.1002/1529-0131(200003)43:3<541::AID-ANR9>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 5.Eimer MJ, et al. Clinical status and cardiovascular risk profile of adults with a history of juvenile dermatomyositis. J Pediatr. 2011;159:795–801. doi: 10.1016/j.jpeds.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shrestha S, Wershil B, Sarwark JF, Niewold TB, Philipp T, Pachman LM. Lesional and nonlesional skin from patients with untreated juvenile dermatomyositis displays increased numbers of mast cells and mature plasmacytoid dendritic cells. Arthritis Rheum. 2010;62:2813–22. doi: 10.1002/art.27529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aziz A, Sebastian S, Dilworth FJ. The Origin and Fate of Muscle Satellite Cells. Stem Cell Rev. 2012 doi: 10.1007/s12015-012-9352-0. [DOI] [PubMed] [Google Scholar]
- 8.De Bari C, Dell’Accio F, Vandenabeele F, Vermeesch JR, Raymackers JM, Luyten FP. Skeletal muscle repair by adult human mesenchymal stem cells from synovial membrane. J Cell Biol. 2003;160:909–18. doi: 10.1083/jcb.200212064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farthing CR, et al. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4:e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel DR, Richardson BC. Epigenetic mechanisms in lupus. Curr Opin Rheumatol. 2010;22:478–82. doi: 10.1097/BOR.0b013e32833ae915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie H, et al. Epigenomic analysis of Alu repeats in human ependymomas. Proc Natl Acad Sci U S A. 2010;107:6952–7. doi: 10.1073/pnas.0913836107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bohan A, Peter JB. Polymyositis and dermatomyositis (second of two parts) N Engl J Med. 1975;292:403–7. doi: 10.1056/NEJM197502202920807. [DOI] [PubMed] [Google Scholar]
- 13.Lovell DJ, et al. Development of validated disease activity and damage indices for the juvenile idiopathic inflammatory myopathies. II. The Childhood Myositis Assessment Scale (CMAS): a quantitative tool for the evaluation of muscle function. The Juvenile Dermatomyositis Disease Activity Collaborative Study Group. Arthritis Rheum. 1999;42:2213–9. doi: 10.1002/1529-0131(199910)42:10<2213::AID-ANR25>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 14.Pachman LM, et al. Duration of illness is an important variable for untreated children with juvenile dermatomyositis. J Pediatr. 2006;148:247–53. doi: 10.1016/j.jpeds.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 15.Chen YW, Shi R, Geraci N, Shrestha S, Gordish-Dressman H, Pachman LM. Duration of chronic inflammation alters gene expression in muscle from untreated girls with juvenile dermatomyositis. BMC Immunol. 2008;9:43. doi: 10.1186/1471-2172-9-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rachidi M, et al. Spatial and temporal localization during embryonic and fetal human development of the transcription factor SIM2 in brain regions altered in Down syndrome. Int J Dev Neurosci. 2005;23:475–84. doi: 10.1016/j.ijdevneu.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Farrall AL, Whitelaw ML. The HIF1alpha-inducible pro-cell death gene BNIP3 is a novel target of SIM2s repression through cross-talk on the hypoxia response element. Oncogene. 2009;28:3671–80. doi: 10.1038/onc.2009.228. [DOI] [PubMed] [Google Scholar]
- 18.He LC, et al. Ikaros is degraded by proteasome-dependent mechanism in the early phase of apoptosis induction. Biochem Biophys Res Commun. 2011;406:430–4. doi: 10.1016/j.bbrc.2011.02.062. [DOI] [PubMed] [Google Scholar]
- 19.Arnaud L, et al. A dominant mutation in the geneencoding the erythroid transcription factor KLF1 causes a congenital dyserythropoietic anemia. Am J Hum Genet. 2010;87:721–7. doi: 10.1016/j.ajhg.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauer DE, Orkin SH. Update on fetal hemoglobin gene regulation in hemoglobinopathies. Curr Opin Pediatr. 2011;23:1–8. doi: 10.1097/MOP.0b013e3283420fd0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodfellow SJ, et al. WT1 and its transcriptional cofactor BASP1 redirect the differentiation pathway of an established blood cell line. Biochem J. 2011;435:113–25. doi: 10.1042/BJ20101734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuiper RP, et al. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia. 2007;21:1258–66. doi: 10.1038/sj.leu.2404691. [DOI] [PubMed] [Google Scholar]
- 23.Nagaraju K, et al. Activation of the endoplasmic reticulum stress response in autoimmune myositis: potential rolein muscle fiber damage and dysfunction. Arthritis Rheum. 2005;52:1824–35. doi: 10.1002/art.21103. [DOI] [PubMed] [Google Scholar]
- 24.Sallum AM, et al. MHC class I and II expression in juvenile dermatomyositis skeletal muscle. Clin Exp Rheumatol. 2009;27:519–26. [PubMed] [Google Scholar]
- 25.Baechler EC, Bilgic H, Reed AM. Type Iinterferon pathway in adult and juvenile dermatomyositis. Arthritis Res Ther. 2011;13:249. doi: 10.1186/ar3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lassar AB, Paterson BM, Weintraub H. Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell. 1986;47:649–56. doi: 10.1016/0092-8674(86)90507-6. [DOI] [PubMed] [Google Scholar]
- 27.Lee WJ, Kim HJ. Inhibition of DNA methylation is involved in transdifferentiation of myoblasts into smooth muscle cells. Mol Cells. 2007;24:441–4. [PubMed] [Google Scholar]
- 28.Cunninghame Graham DS, et al. Association of NCF2, IKZF1, IRF8, IFIH1, and TYK2 with systemic lupus erythematosus. PLoS Genet. 2011;7:e1002341. doi: 10.1371/journal.pgen.1002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toth K, et al. The influence of methylated septin 9 gene on RNA and protein level in colorectal cancer. Pathol Oncol Res. 2011;17:503–9. doi: 10.1007/s12253-010-9338-7. [DOI] [PubMed] [Google Scholar]
- 30.Da L, Li D, Yokoyama KK, Li T, Zhao M. Dual promoters control the cell-specific expression of the human cell death-inducing DFF45-like effector B gene. Biochem J. 2006;393:779–88. doi: 10.1042/BJ20051027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee N, Maurange C, Ringrose L, Paro R. Suppression of Polycomb group proteins by JNK signalling induces transdetermination in Drosophila imaginal discs. Nature. 2005;438:234–7. doi: 10.1038/nature04120. [DOI] [PubMed] [Google Scholar]
- 32.Sustar A, Bonvin M, Schubiger M, Schubiger G. Drosophila twin spot clones reveal cell division dynamics in regenerating imaginal discs. Dev Biol. 2011;356:576–87. doi: 10.1016/j.ydbio.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fjose A, McGinnis WJ, Gehring WJ. Isolation of a homoeo box-containing gene from the engrailed region of Drosophila and the spatial distribution of its transcripts. Nature. 1985;313:284–9. doi: 10.1038/313284a0. [DOI] [PubMed] [Google Scholar]
- 34.Capovilla M, Kambris Z, Botas J. Direct regulation of the muscle-identity gene apterous by a Hox protein in the somatic mesoderm. Development. 2001;128:1221–30. doi: 10.1242/dev.128.8.1221. [DOI] [PubMed] [Google Scholar]
- 35.Goff DJ, Tabin CJ. Analysis of Hoxd-13 and Hoxd-11 misexpression in chick limb buds reveals that Hox genes affect both bone condensation and growth. Development. 1997;124:627–36. doi: 10.1242/dev.124.3.627. [DOI] [PubMed] [Google Scholar]
- 36.Houghton L, Rosenthal N. Regulation of a muscle-specific transgene by persistent expression of Hox genes in postnatal murine limb muscle. Dev Dyn. 1999;216:385–97. doi: 10.1002/(SICI)1097-0177(199912)216:4/5<385::AID-DVDY7>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 37.Costa BM, et al. Reversing HOXA9 oncogene activation by PI3K inhibition: epigenetic mechanism and prognostic significance in human glioblastoma. Cancer Res. 2010;70:453–62. doi: 10.1158/0008-5472.CAN-09-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaspar N, et al. MGMT-independent temozolomide resistance in pediatric glioblastoma cells associated with a PI3-kinase-mediated HOX/stem cell gene signature. Cancer Res. 70:9243–52. doi: 10.1158/0008-5472.CAN-10-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Branciamore S, Chen ZX, Riggs AD, Rodin SN. CpG island clusters and pro-epigenetic selection for CpGs in protein-coding exons of HOX and other transcription factors. Proc Natl Acad Sci U S A. 2010;107:15485–90. doi: 10.1073/pnas.1010506107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haber DA, et al. An internal deletion within an 11p13 zinc finger gene contributes to the development of Wilms’ tumor. Cell. 1990;61:1257–69. doi: 10.1016/0092-8674(90)90690-g. [DOI] [PubMed] [Google Scholar]
- 41.Huff V. Wilms’ tumours: about tumour suppressor genes, an oncogene and a chameleon gene. Nat Rev Cancer. 11:111–21. doi: 10.1038/nrc3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schumacher V, et al. Correlation of germ-line mutations and two-hit inactivation of the WT1 gene with Wilms tumors of stromal-predominant histology. Proc Natl Acad Sci U S A. 1997;94:3972–7. doi: 10.1073/pnas.94.8.3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyagawa K, et al. Loss of WT1 function leads to ectopic myogenesis in Wilms’ tumour. Nat Genet. 1998;18:15–7. doi: 10.1038/ng0198-15. [DOI] [PubMed] [Google Scholar]
- 44.Tiffin N, Williams RD, Robertson D, Hill S, Shipley J, Pritchard-Jones K. WT1 expression does not disrupt myogenic differentiation in C2C12 murine myoblasts or in human rhabdomyosarcoma. Exp Cell Res. 2003;287:155–65. doi: 10.1016/s0014-4827(03)00131-9. [DOI] [PubMed] [Google Scholar]
- 45.Wagner KD, et al. The Wilms’ tumor suppressor Wt1 is expressed in the coronary vasculature after myocardial infarction. Faseb J. 2002;16:1117–9. doi: 10.1096/fj.01-0986fje. [DOI] [PubMed] [Google Scholar]
- 46.Smart N, et al. De novo cardiomyocytes from within the activated adult heart after injury. Nature. 2011;474:640–4. doi: 10.1038/nature10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinez-Estrada OM, et al. Wt1 is required for cardiovascular progenitor cell formation through transcriptional control of Snail and E-cadherin. Nat Genet. 2010;42:89–93. doi: 10.1038/ng.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rider LG, Pachman LM, Miller FW, Bollar H. Myositis and you: A guide to juvenile dermatomyositis for patients, families, and healthcare providers. Alexandria, VA: The Myositis Association; 2007. [Google Scholar]
- 49.Nakanishi K, Sudo T, Morishima N. Endoplasmic reticulum stress signaling transmitted by ATF6 mediates apoptosis during muscle development. J Cell Biol. 2005;169:555–60. doi: 10.1083/jcb.200412024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang M, et al. BTECH: A Platform to Integrate Genomic, Transcriptomic and Epigenomic Alterations in Brain Tumors. Neuroinformatics. 2011;9:59–67. doi: 10.1007/s12021-010-9091-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.