Summary
Directed differentiation of human embryonic stem cells (ESCs) into cardiovascular cells provides a model for studying molecular mechanisms of human cardiovascular development. Though it is known that chromatin modification patterns in ESCs differ markedly from those in lineage-committed progenitors and differentiated cells, the temporal dynamics of chromatin alterations during differentiation along a defined lineage have not been studied. We show that differentiation of human ESCs into cardiovascular cells is accompanied by programmed temporal alterations in chromatin structure that distinguish key regulators of cardiovascular development from other genes. We used this temporal chromatin signature to identify regulators of cardiac development, including the homeobox gene MEIS2. We demonstrate using the zebrafish model that MEIS2 is critical for proper heart tube formation and subsequent cardiac looping. Temporal chromatin signatures should be broadly applicable to other models of stem cell differentiation to identify regulators and provide key insights into major developmental decisions.
Introduction
Cardiovascular cells derived from pluripotent stem cells in vitro have potential both as a cell-based therapy for cardiac regeneration and as tools to analyze basic developmental processes (Murry and Keller, 2008). Insights from model organisms have permitted harnessing of the signaling pathways controlling cardiovascular development, enabling the directed differentiation of mouse and human ESCs into the major definitive cell types of the heart, namely cardiomyocytes, smooth muscle cells and endothelial cells (Bu et al., 2009; Domian et al., 2009; Kattman et al., 2006; Kattman et al., 2011; Laflamme et al., 2007; Murry and Keller, 2008; Yang et al., 2008). In contrast to the relatively advanced knowledge of signaling pathways, the epigenetic alterations that accompany or potentiate cardiogenesis are largely unexplored.
Methylation of lysine residues on the tail of histone H3 accompanies many major genomic functional processes (Guenther et al., 2007; Ringrose and Paro, 2004; Schuettengruber et al., 2007). H3K4me3 and H3K36me3 are deposited by Trithorax group proteins and mark chromatin associated with, respectively, transcription initiation and elongation (Bannister et al., 2005; Li et al., 2007; Vakoc et al., 2006). H3K27me3 modifications result from activities within the Polycomb repressive complex 2 (PRC2) which includes SUZ12 and EZH2 (Boyer et al., 2006; Lee et al., 2006; Simon and Kingston, 2009). Studies of the distribution of H3K27me3 in both mouse (Bernstein et al., 2006) and human (Hawkins et al., 2010; Pan et al., 2007) ESCs have revealed that H3K27me3 deposition is preferentially enriched at promoters of regulatory genes (Boyer et al., 2006; Lee et al., 2006) controlling diverse developmental pathways including neuronal (Mohn et al., 2008a) and hematopoietic lineages (Mazzarella et al., 2011a), where frequently, in combination with H3K4me3, it may denote 'poising' of genes that are destined for rapid activation upon lineage commitment (Bernstein et al., 2006; Rada-Iglesias and Wysocka, 2011). Indeed, `bivalent' promoters marked by both H3K4me3 and H3K27me3 in pluripotent cells are found to have either the H3K4me3 or the H3K27me3 mark (but not both) in definitive cell types, implying that this bivalency resolves to either a transcriptionally active or silent state (Mikkelsen et al., 2007). It is currently unknown how Polycomb- and Trithorax- related histone modification patterns evolve during the transition from pluripotency to definitive cells during lineage differentiation. Moreover, because Polycomb-driven marking of key regulatory genes in ESCs is not specific to any given lineage, it is currently not possible to identify lineage-specific regulators simply from the chromatin state of pluripotent stem cells.
We sought to determine whether temporal patterning of histone modifications during differentiation along a defined lineage would clarify the relationships among key regulators of lineage-specific differentiation and further distinguish regulatory from lineage-specific structural genes. Specifically, we hypothesized that key regulators of differentiation would show stage-specific changes in H3K4me3 and H3K27me3, because inappropriate activation or silencing of these genes could alter cell fate by affecting many downstream target genes. The differentiation of human ESCs into cardiomyocytes has been extensively described (Kattman et al., 2011; Laflamme et al., 2007; Yang et al., 2008), and is of great potential therapeutic importance (Murry and Keller, 2008). We therefore asked how chromatin modifications evolve temporally along the cardiac differentiation axis, and whether the temporal patterning of histone modifications might provide a reliable signature that could discriminate key known regulators of cardiovascular development, as well as enable the identification of regulators.
Results
Directed differentiation of human ESCs into cardiomyocytes
Directed differentiation of H7 human ESCs to the cardiovascular lineage was achieved by allowing the cells to form embryoid bodies (EBs) in the presence of defined serum-free medium as previously described (Kattman et al., 2011; Yang et al., 2008). Mesoderm induction was accomplished using bone morphogenetic protein 4 (BMP4), activin A and basic fibroblast growth factor (bFGF). On day 5 (T5) of differentiation, a tripotential cardiovascular progenitor emerges, identified based on low expression of VEGFR2 (KDR) and PDGFRα (Kattman et al., 2011). Over time, this progenitor gives rise to cultures that contain predominantly cardiomyocytes, and also contain endothelial cells and smooth muscle cells, identified by flow cytometry for cardiac troponin T (cTnT), CD31/PECAM1 and smooth muscle alpha actin (SMA), respectively (Figure S1). For all genome-wide experiments, parallel samples were maintained in culture after the times of harvest for immunoprecipitation, and only runs that were >80% KDR+/PDGFRα+ at the T5 progenitor stage and >50% cTnT+ cardiomyocytes at the T14 definitive cardiovascular cell stage were used.
Chromatin states measured along the time course of differentiation
We used chromatin immunoprecipitation coupled to massively parallel sequencing (ChIP-seq) to map H3K4me3, H3K27me3 and H3K36me3 modifications genome-wide at five key developmental stages during cardiovascular directed differentiation including: pluripotent cells (T0), mesodermal progenitors (T2), specified tripotential cardiovascular progenitors (T5), committed cardiovascular cells (T9), and definitive cardiovascular cells (primarily cardiomyocytes, T14). We performed differentiation and ChIP-seq experiments in duplicate, with high reproducibility between biological replicates at each time point (average Pearson correlation 0.94). Figure S2A shows two biological replicates for chromatin modification at the TBX5 locus, a transcription factor (TF) implicated in generation of the first heart field (Takeuchi et al., 2003). Across all time points, all three histone modification patterns with respect to the transcription start site (TSS) showed the expected morphologies (Figure S2B), with H3K4me3 signal peaking within a narrow window around the TSS; H3K27me3 signal peaking in the vicinity of the TSS (though more broadly distributed than H3K4me3) with a prominent decrease over the TSS (consistent with reduced nucleosomal density and increased turnover in this region (Mobius and Gerland, 2010)) and H3K36me3 signal increasing rapidly from the TSS region to a plateau over the gene body, consistent with its deposition by the elongating polymerase complex (Pjanic et al., 2011).
Identification of distinct chromatin signatures for different functional categories of cardiac factors
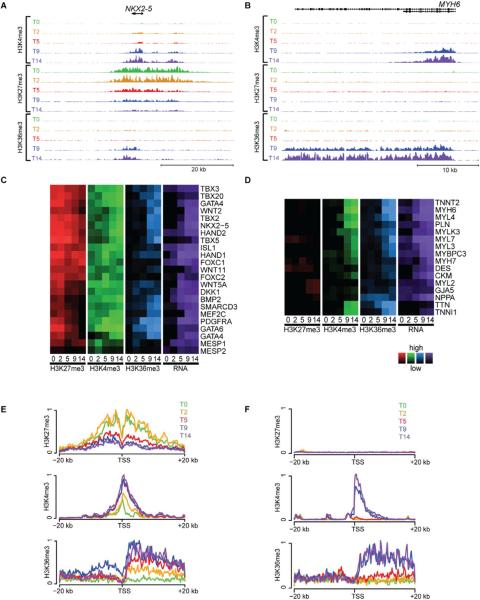
To gain insight into the pattern of sequential epigenetic alterations accompanying cardiovascular differentiation, we next examined the histone modification profiles and RNA expression of genes with established roles in heart development and function. These ranged from sequence-specific transcriptional regulators, such as NKX2.5 (Figure 1A) to cardiomyocyte contractile proteins, including alpha myosin heavy chain (MYH6) (Figure 1B), as well as genes expressed in cardiovascular progenitors, smooth muscle cells and endothelial cells (Figure S3A–C). A majority of cardiac transcription factors showed high levels of H3K27me3 during pluripotency that gradually decreased as differentiation progresses, paralleled by gradual increases in H3K4me3, H3K36me3 and RNA expression (Figure 1A, C, E). Similarly, many of the members of key signaling pathways involved in cardiac development, such as the TGFβ family, Wnt, Notch, Hedgehog, the FGF family, PDGF and VEGF showed stage-specific activation (H3K4me3, H3K36me3, RNA expression) and repression (H3K27me3), compatible with tight control over signaling pathways that direct cell fate (Figure S3D). In contrast, although genes encoding cardiomyocyte contractile proteins showed similar time-dependent increases in H3K4me3, H3K36me3 and RNA expression, these genes did not have appreciable levels of H3K27me3 deposition at any time (Figure 1B, D, F). As such, the temporal evolution of histone modifications provides a chromatin `signature' that differentiates key cardiac regulatory genes, including both transcription factors and soluble signals, from lineage-specific genes encoding proteins that regulate cardiac function and homeostasis.
Figure 1. Key regulators of cardiac differentiation share a temporal chromatin signature.
At five different time points of directed differentiation of human embryonic stem cells into cardiomyocytes (T0, 2, 5, 9 and 14), the levels of histone modifications H3K4me3 (activating), H3K27me3 (repressing) and H3K36me3 (transcribed) are shown within a ~50 kb region around (A) NKX2-5, a well-known regulator of cardiac differentiation (scales used: 1 to 250/150/50 tags per 150 bp, for H3K4me3/H3K27me3/H3K36me3) and (B) MYH6, a well-known structural component of cardiac cells (scales used: 1 to 500/100/50 tags per 150 bp, for H3K4me3/H3K27me3/H3K36me3). The relative levels of histone modifications (red, green and blue) and RNA expression (purple) are shown for (C) selected regulators of cardiac differentiation and (D) cardiac structural factors at all five time points. The averaged levels of epigenetic marks within ±20 kb of the TSS of (E) known regulators of cardiac differentiation and (F) known cardiac structural factors are plotted across all five time points (0, 2, 5, 9 14 = green, yellow, red, blue, purple). For key regulators of cardiac differentiation, levels of H3K4me3 and H3K36me3 increase during differentiation while H3K27me3 begins high and decreases, while H3K27 remains consistently low for cardiac structural factors. Note GATA4 is shown twice in panel (c) due to activation of two different promoters in our system. See Figure S3 for patterns found for other gene groups.
The temporal chromatin signature of cardiac regulators
To assess the cardiac lineage-specificity of the temporal chromatin signature for cardiac regulators, we compared this signature with the temporal chromatin patterns at genes involved in specification of non-cardiac mesoderm, neuroectoderm and endoderm fates (Figures S3E–I). Similar to cardiac transcription factors, most of the major transcription factor genes involved in hematopoiesis (e.g. TAL1, RUNX1, HHEX and LMO2) and skeletal muscle differentiation (e.g. MYOD1, MYF5 and MYF6) had low levels of H3K4me3 and high levels of H3K27me3 in the pluripotent state. However unlike cardiac TFs, non-cardiac factors do not activate in this system and generally resolve to only H3K27me3 by T5. Some lineage-selective factors such as GATA1, a key regulator of erythroid fate, showed negligible levels of any of the histone modifications at any time point. Genes encoding transcription factors involved in neuroectoderm differentiation, such as NEUROD1, show high levels of H3K27me3 (often in large domains (Guenther et al., 2010)) in combination with low H3K4me3 in the pluripotent state (Figure S3H). But whereas H3K27me3 levels at neuroectodermal genes remain high throughout differentiation, they evince a dramatic drop in H3K4me3 by T2. Notably, hematopoietic and skeletal muscle transcription factors also show declines in H3K4me3, but not until T5. This ordering recapitulates the later fate choice for mesoderm subtypes vs. the early choice of primary germ layer (mesoderm vs. ectoderm). Interestingly, several genes typically associated with endoderm formation are activated at T5, including FOXA2 and SOX17. This could be due to the presence of a small number of endodermal cells in our cultures or the fact that many of these genes have roles in both mesoderm and endoderm formation.
Identification of new regulators of human cardiac development
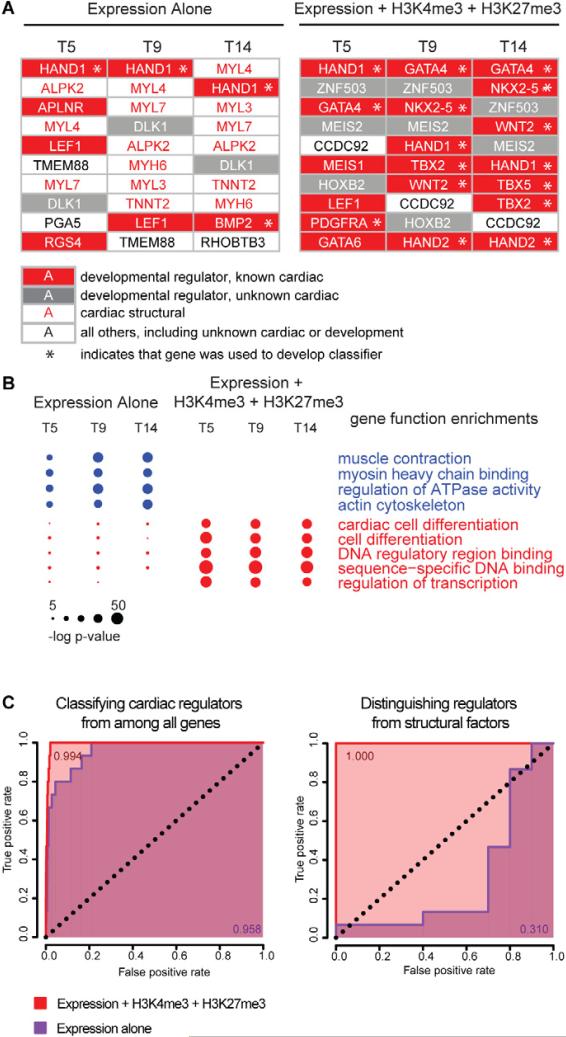
We next sought to determine if the temporal chromatin signature of cardiac regulators could be used to identify novel regulators of human cardiac development. Using a curated set of known cardiac regulators (Figure 1), we developed a classifier based on the concomitant induction of mRNA, loss of H3K27me3 marks, and anti-correlation coefficients of the H3K4me3 and H3K27me3 signals over the time course (see Methods). Next, we scored each gene against the classifier and rank-ordered each gene at T5, T9 and T14 by its classification score. For comparison, we performed a similar ranking using a classifier based on mRNA expression only. Figure 2A shows the top 10 genes at each time point using each algorithm, functionally annotated as cardiac developmental regulators, cardiac structural genes, developmental regulators with unknown cardiac roles, and all other genes (including unknown cardiac or developmental roles). A list of the top 100 genes identified using expression alone and chromatin + expression at each timepoint is shown in Supplemental Table 1. Using mRNA expression alone, 14 non-redundant genes comprised the top 10 list at the three time points. Of these, 6 (43%) encoded cardiac structural proteins, 5 (36%) encoded known cardiac developmental regulators, 1 (7%) encoded a developmental regulator with unknown cardiac function, and 2 (14%) were genes with unknown function. Using the chromatin + expression method, the top 10 lists across all time points contained 15 non-redundant genes. In contrast to the mRNA expression-only approach, 11 of these genes (73%) encode developmental regulators with known cardiac roles, 3 others (20%) encode developmental regulators with unknown cardiac roles, and 1 gene (7%) encodes a protein of unknown function. Interestingly, this gene (CCDC92) is predicted to contain a coiled-coil domain, which is found in many transcription factors. Notably, none of the top-scoring genes in the combined algorithm encode cardiac structural proteins.
Figure 2. Accurate discrimination of key regulators of cardiac differentiation from other lineage-specific genes.
(A) All genes were ranked by two different methods in order to identify key regulators of cardiac differentiation. They were ranked at days 5, 9 and 14 by a formula using either (left) RNA expression alone, or (right) one that accounts for levels of H3K4me3, H3K27me3 and RNA expression. At each time point the top 10 candidate regulators are depicted for the respective methods. Developmental regulators with known roles in cardiac differentiation are shown in white text on red background, developmental regulators with no currently-appreciated role in cardiac differentiation are shown in white text on grey background, and genes whose function pertains to the structure and function of heart cells with no known regulatory roles are shown in red text on white background. All other genes are shown in black text on white background. Genes that were used in the training set for identifying the chromatin + expression regulator signature are indicated with an asterisk. (B) The top 100 candidates provided by each ranked list (see Figure S4) were analyzed to determine the degree to which the lists were enriched in 11 key gene ontology functional categories. The size of each circle is proportional to the significance of the enrichment of genes with the indicated functional role within the given list of 100 genes at each time point/ method of ranking genes. Ranking genes using H3K4m3, H3K27me3 and RNA expression yields lists that are not contaminated by structural factors and are more enriched with known regulators of cardiac differentiation. (C) Classification based on H3K4me3, H3K27me3 and expression (red) is more specific and sensitive than classification based on expression alone (purple). Shown are ROC curves for the identification of key regulators of cardiac differentiation from among all genes (left panel) or among all genes involved in heart development (right panel). The expression-only classifier systematically misclassifies structural factors that are involved in heart function but do not regulate cardiac development (right panel), leading to a lower area under the curve when classifying key regulators from among all genes (left panel). The genes used to generate true-positives for cardiac regulatory and structural genes are given in Figure 1.
Beyond the top 10, the chromatin + expression algorithm identified many genes with less well-defined roles in cardiac development including transcription factors, cell surface proteins, signaling ligands, extracellular matrix proteins and enzymes. Gene ontology analysis demonstrated that the top 100 genes identified using chromatin + expression are highly enriched for regulation of transcription and differentiation functions (Figure 2B). To determine the accuracy with which each classifier could discriminate known cardiac regulators from all other genes, and specifically from cardiac structural genes, we computed a receiver operating characteristic (ROC) curve for each class using both the mRNA expression-only and the chromatin + expression classifier. These showed that the combined (histone modification + expression) classifier had extremely high sensitivity and specificity both for discrimination of cardiac regulators from all genes (ROC=0.994), and from cardiac structural proteins (ROC=1.000) (Figure 2C). By contrast, a classifier based on temporal mRNA patterns alone can reasonably discriminate genes with a cardiac role from all other genes, but cannot distinguish regulators from cardiomyocyte structural proteins involved in muscle contraction and energy production.
Analysis of temporal chromatin dynamics identifies regulators of other cellular fates
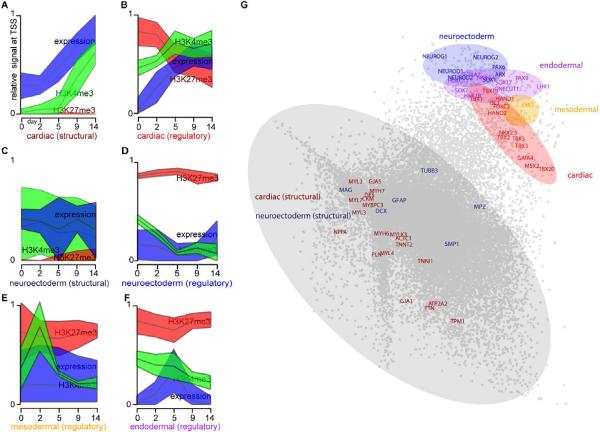
The selective discrimination of genes regulating cardiac differentiation by comparison of epigenetic profiles and expression led us to ask whether we could identify factors that regulate other lineage fates, not taken during the cardiovascular differentiation process. To explore this, we first examined a curated list of neuroectodermal regulatory genes and structurally-related genes. Remarkably, these showed distinct temporal chromatin signatures for developmental regulators vs. structural gene classes, even though the constituent genes were not significantly expressed at any time in our cardiac directed differentiation system (Figure 3A–F).
Figure 3. Temporal chromatin signatures enable cross-lineage identification of key regulatory factors.
(A–F) The median levels (middle line) and 95% confidence intervals (shaded regions) of H3K27me3 (red), H3K4me3 (green) and RNA expression (blue) at each time point are depicted for several categories of genes: (A) genes involved in cardiac structure and function, (C) genes involved in neuroectoderm structure/function, and regulators of differentiation for (B) cardiac, (D) neuroectoderm, (E) mesoodermal and (F) endodermal cells. (A–F) were identically normalized, such that the lowest and highest values for each individual mark across all time points and gene groups were plotted as 0 and 1, respectively. (G) Using principle component analysis, the 15 dimensional data for each gene (5 time points * 3 measurements of chromatin and mRNA) were reduced to two dimensions, and a scatterplot is shown depicted the relative locations of each gene in the reduced-dimensional space. Genes involved in structure and function of cells are contained within the largest cluster (grey) distinct from the cluster containing key regulators of cellular fate: cardiac (red), neuroectoderm (blue), endoderm (purple) and mesoderm (yellow). Non-annotated genes within each of the colored domains have a high probability of having unappreciated roles as key regulators of cellular fate.
To systematically identify candidate regulators of other cellular fates, we performed principal component analysis of the chromatin modification profiles (see Methods). This analysis revealed that genes with similar roles in cell fate determination clustered together (Figure 3G), remaining distinct from other lineage-specific genes. This result suggests that a rich layer of information on developmental programming can be mined from temporal analysis even of a single lineage, which may in turn provide insight into the mechanisms that determine numerous cell fates.
Functional validation of chromatin dynamics-based predictions
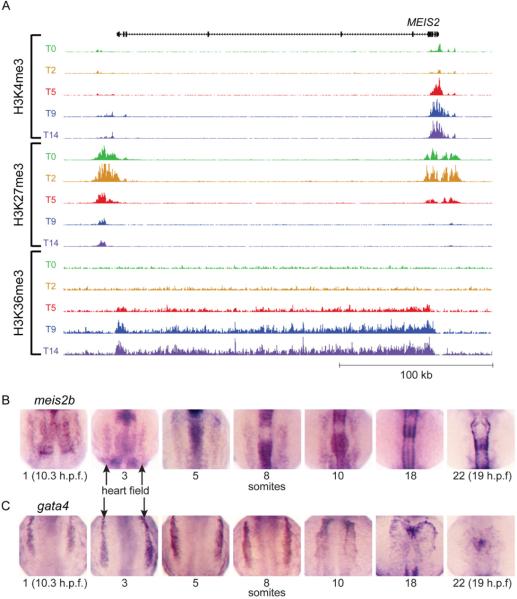
We next sought to confirm whether the temporal chromatin signature of cardiac regulators could identify novel bona fide cardiac regulators. Among the genes we identified without known roles in cardiac development was the homeodomain-containing transcription factor MEIS2 (Figure 4). MEIS transcription factors interact with HOX and PBX transcription factors to regulate downstream targets in multiple cellular processes, and previous studies have demonstrated a requirement for the related gene, Meis1, in mouse and zebrafish heart development (Maves et al., 2009; Minehata et al., 2008; Stankunas et al., 2008). As shown in Figure 4A, MEIS2 shares the histone modification pattern we have identified as a marker of cardiac regulatory genes. In zebrafish there are two orthologs of MEIS2: meis2a and meis2b. Interestingly, in situ time course experiments carried out in developing zebrafish embryos shows meis2b expression in the heart field closely resembles that of gata4, a known cardiac transcription factor (Figure 4B,C). In addition, meis2b expression is observed in the developing hindbrain and somites as previously described (Zerucha and Prince, 2001).
Figure 4. MEIS2 chromatin modifications during hESC differentiation and expression in developing zebrafish embryos resembles other regulators of cardiac development.
(A) The temporal pattern of epigenetic marks at the MEIS2 locus is similar to that of other regulators of cardiac development shown in Figure 1 (scales used: 1 to 250/150/25 tags per 150 bp, for H3K4me3/H3K27me3/H3K36me3). (B) Zebrafish meis2b expression are shown for developing embryos at the 1, 3, 5, 8, 10, 18 and 22 somite stages, showing (C) similar expression patterns within the bilateral heart fields (arrows) to gata4 through the 10 somite stage. By 18 somites, meis2b is no longer expressed in the cardiac mesoderm whereas gata4 maintains expression.
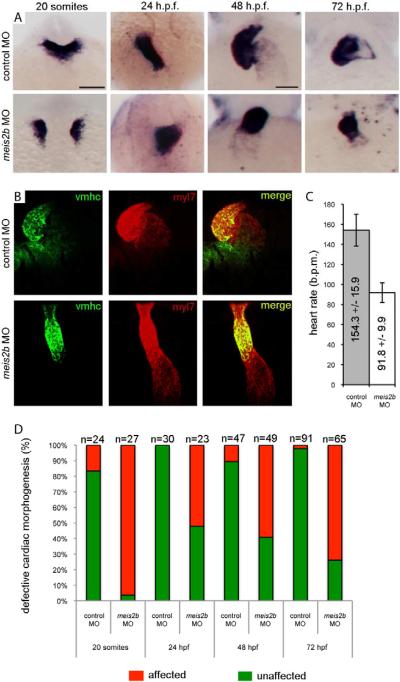
To validate the predicted role of MEIS2 in cardiac development based on its temporal chromatin signature, we utilized antisense morpholino oligonucleotide (MO) knockdown in developing zebrafish embryos (Figure 5). We found that delivery of a splice-blocking MO directed against meis2b (“meis2b-MO”) into fertilized zebrafish eggs results in defective cardiac morphogenesis. Defects are evident as early as 19 hours post fertilization (h.p.f.), when meis2b-MO embryos fail to fuse their bilateral heart fields into a linear heart tube at the midline (Figure 5A) (Glickman and Yelon, 2002). At 24 h.p.f. heart tubes have formed in meis2b-MO embryos, but they demonstrate an early looping defect as evidenced by the midline to right-sided location of the developing linear heart tube compared to the left-sided developing heart in control-MO embryos (Figure 5A) (Glickman and Yelon, 2002). By 48 and 72 h.p.f., control-MO embryos have normally looped hearts (Glickman and Yelon, 2002), while the meis2b-MO embryo hearts remain linear, displaying a persistent looping defect (Figure 5A–B). Furthermore, meis2b-MO embryos have a markedly reduced heart rate at 72 h.p.f. (92 +/− 10 beats per minute) compared with control-MO embryos (154 +/− 16 beats per minute), and they have pericardial edema indicative of cardiac failure (Figures 5C and S12). These findings indicate a requirement of meis2b for normal heart function (Figure 5D). Of note, these phenotypes are not attributable to a general developmental delay upon knock-down of meis2b as embryos were compared based on number of somites (developmental stage) as well as time post-fertilization. These phenotypes are also unlikely to be attributable to off-target effects as a separate, translation-blocking meis2b morpholino produces similar looping defects (Figure S5 and data not shown). These data establish meis2b as a regulator of cardiac development, confirming the chromatin signature-based prediction.
Figure 5. meis2b is required for cardiac morphogenesis.
(A) Expression of myl7 at 19 h.p.f., 24 h.p.f, 48 h.p.f. and 72 h.p.f. in control-MO (top row) versus meis2b-MO (bottom row) injected zebrafish embryos. Dorsal view, anterior is up in 20 somite and 24 h.p.f. embryos. Ventral view, anterior up in 48 h.p.f. and 72 h.p.f. embryos. At 19 h.p.f., meis2b-MO injected embryos display defects in fusion of the myl7+ cardiac progenitors at the midline compared with control-MO injected embryos. By 24 h.p.f., the heart tube has formed in meis2b morphants but displays aberrant cardiac morphogenesis and is either sitting at the midline or moving down the right side of the embryo, compared with the control-MO injected embryos where normal heart development proceeds with the heart tube emerging from under the head, down the left side of the embryo. At 48 and 72 h.p.f., control MO injected embryos display normal cardiac looping, while meis2b-MO injected embryos' hearts have not looped. (B) This failure of cardiac looping in meis2b-MO injected embryos is further evident in vmhc (green) and myl7 (red) double fluorescent in situs at 48 h.p.f.. (C) Heart rate is significantly reduced in meis2b-MO injected embryos compared with control-MO injected embryos at 72 h.p.f. (b.p.m. = beats per minute) Mean heart rate +/− s.d. is shown, n=10. P = 4 ×10−9 (Student's t-test, two-tailed). (D) Percentages of embryos displaying the depicted phenotypes. Scale bar in A (top left), 100 μm. Scale bar in A (top 3rd from the left), 50 μm. See also Figure S5.
Discussion
The chromatin landscape of both mouse and human ESCs has been intensively investigated. In the pluripotent state, many developmental loci are marked with both activating H3K4me3 and repressing H3K27me3, and are thus termed “bivalent” (Azuara et al., 2006; Bernstein et al., 2006; Pan et al., 2007). The notion is that these genes are simultaneously suppressed but poised for activation should the cell receive appropriate cues. Bivalent promoters have also been found at developmental loci in mouse embryos, both in the inner cell mass and trophectoderm, and also in zebrafish embryos (Dahl et al., 2010; Lindeman et al., 2010; Vastenhouw et al., 2010). Other studies have shown that bivalent promoters are present in progenitor and adult stem cell populations, including neural progenitors, mesenchymal stem cells and hematopoietic stem cells, and that these ultimately resolve to either active or inactive upon differentiation (Collas, 2010; Cui et al., 2009; Mazzarella et al., 2011b; Mohn et al., 2008b).
We sought to characterize the epigenetic changes that occur during cardiovascular differentiation from human ESCs by performing genome-wide mapping of three histone modifications, H3K4me3, H3K27me3 and H3K36me3, at five key developmental timepoints. Our study shows that the temporal trajectories of H3K4me3 and H3K27me3 during differentiation are more complex than a simple `resolution from bivalency' model. As an example of this, FGF19 and NODAL are highly transcribed in human ESCs with high levels of H3K4me3 and low levels of H3K27me3 (Figure SD). They subsequently lose H3K4me3 and gain H3K27me3 over time. If one were only to have taken time points T3 (sometime between T2 and T5) and T14, one might conclude that the genes began in a bivalent state and then resolved towards repression, while in reality the `bivalent' appearance was merely an artifact of the complete reversal from H3K4me3 to H3K27me3. Yet another example is the set of genes involved in mesodermal differentiation (Fig S7), which are highly expressed despite being heavily marked with H3K27me3. These and other examples point to a complex regulatory relationship than cannot be described by a simple `resolution from bivalency' model.
Transcription factors and signaling molecules known to play critical roles in cardiovascular development, such as NKX2.5, showed a unique chromatin signature that consisted of high enrichment for H3K27me3 in pluripotent ESCs that gradually decreased as H3K4me3, H3K36me3 and RNA expression increased over time. In contrast, structural proteins like alpha-myosin heavy chain (MYH6) demonstrated markedly increased H3K4me3 enrichment and RNA expression at later timepoints, without early H3K27me3 repression. The differences in chromatin markings between genes encoding developmental regulators and structural proteins are consistent with previous studies comparing pluripotent and differentiated cells. However, our study is the first to recognize that the complex temporal chromatin patterns over a time course of differentiation actually contain a far richer amount of information regarding the exact function of the genes they are marking.
To use the information contained within the temporal chromatin signatures to identify novel regulators, we developed a classifier to rank genes according to the likelihood that a given gene would modulate cardiogenesis. Interestingly, several genes that ranked highly were transcription factors that had not been previously studied in cardiac development. We therefore tested our hypothesis that these genes were in fact previously unappreciated key regulators of heart development by utilizing morpholino knock-down technology in zebrafish embryos. Indeed, knockdown of meis2b resulted in severe defects in heart looping in early stage zebrafish embryos. This provides in vivo evidence that our methodology of coupling stem cell differentiation with histone modification pattern identification can identify regulators of development. It is worth noting that several of these novel regulators could be identified using the rank list of the top 50 genes based on expression alone (Supplemental Table 1). However, these regulators usually rank much lower down the list, and could easily get lost amidst the noise of so many structural genes. For example, MEIS2 ranked 4th at T5, 4th at T9 and 5th at T14 using the chromatin signature ranking but 49th at T5, 37th at T9 and 48th at T14 based on expression alone. Our dataset will thus serve as a resource for scientists and clinicians across multiple fields. The list of novel cardiac regulators is extensive and each potential candidate gene warrants further investigation to flesh out its role in differentiation and/or morphogenesis. One can imagine that perturbations of these regulators could alter cardiogenesis during human development. Thus, screening for genetic etiologies of congenital heart disease could be expanded to include our list of regulators.
It is interesting to consider why the chromatin dynamics of genes involved in developmental regulation are so different from structural genes that regulate the function of differentiated cells. The principal difference in chromatin signatures is the high degree to which developmental regulators are repressed by H3K27me3 prior to their expression, while structural genes show no such modification. We propose that the consequence of inappropriate activation of developmental regulators is more deleterious, e.g. by inducing the wrong cell type, proliferative state or survival/death signals. These genes therefore require both loss of repression and gain of activation to be expressed. Conversely, inappropriate activation of a gene encoding contractile proteins, ion channels or metabolic enzymes may have less severe consequences for development, and chromatin regulation through activation mechanisms achieves sufficient fidelity.
The establishment of human ESC technology opened the doors to analyses that can provide insights into human development that have not been possible before. Directed differentiation of human ESCs into numerous cell types has been one of the major advances in the field over the past 10 years. In addition to the cardiovascular directed-differentiation model system we utilized in this study, similar protocols exist for the generation of other cell types that are particularly relevant to regenerative medicine, including neurons (Lee et al., 2007) and pancreatic beta-cells (Phillips et al., 2007). Thus, our approach of mapping chromatin states over the course of differentiation could easily be applied to other cell lineages, thereby facilitating the identification of novel regulators of differentiation and development of other organ systems.
Several limitations of the approach used in this study are important to note. First, while the directed differentiation model is very robust, it is not perfect. We are able to obtain highly enriched populations of cardiovascular cells; however, non-cardiac cells are also present in our cultures. Therefore, we cannot exclude the possibility that some of our chromatin patterns might be due to the presence of other cell types, such as non-cardiac mesoderm and endoderm. That said, the cell populations analyzed in this study contained at least 80% progenitors at T5 and 50% cardiomyocytes at T14, so the majority of the chromatin patterns are likely informative for cardiac development. Another issue is that the efficiency of directed differentiation is often dependent on the particular ESC cell line used, or even the batch of such cells used. Thus, it will be critical for these experiments to be repeated in other cell lines beyond the H7 ESC line used in our laboratory. Lastly, although human ESCs provide a platform to model human development in vitro, it is presently unclear how well this differentiation system mimics in vivo development in terms of expression patterns and epigenetic changes, such as histone modifications. These limitations are common to many directed differentiation systems, and reflect the current standard issues shared by stem cell biologists worldwide. On the other hand, there are currently no other ways to study early events in human development, and we were able to utilize our cardiovascular directed-differentiation system to identify novel regulators of heart development. Additionally, our study showed that the temporal chromatin profiles along cardiomyocyte differentiation contained enough information to identify genes with likely unappreciated roles in neurectodermal development, even though the vast majority of cells in the population split from that cellular-fate quite early in differentiation. As the chromatin states along other differentiation pathways are measured, the information from all of these model systems can be integrated using methods similar to those described in this study to provide even richer insights into all branches of developmental regulation.
Experimental Methods
Maintenance of Human Embryonic Stem Cells
H7 human ESCs were maintained on mouse embryonic fibroblasts (MEFs) in medium consisting of DMEM/F12 supplemented with 20% KnockOut serum replacement (Invitrogen), L-glutamine, non-essential amino acids, beta-mercaptoethanol and 8 ng/mL basic fibroblast growth factor (bFGF, Peprotech). Cells were passaged using collagenase IV and trypsin, as well as the ROCK inhibitor Y-27632 (10 μM, Tocris) to enhance cell survival. Prior to directed differentiation, human ESCs were passaged at least twice on Matrigel-coated plates to deplete the cultures of MEFs. For growth on Matrigel, cells were maintained in MEF conditioned medium (MEF-CM) supplemented with 8 ng/mL bFGF.
Cardiovascular Directed Differentiation
H7 human ESCs on Matrigel-coated plates were harvested using Collagenase IV and trypsin as during passaging. For embryoid body formation, cells were gently broken up into clusters of roughly 25–50 cells and plated into low-attachment plates in StemPro-34 medium (Invitrogen) supplemented with L-glutamine, ascorbic acid, transferrin and monothioglycerol (hereafter referred to as StemPro backbone) plus 0.5 ng/mL bone morphogenic protein 4 (BMP4, R+D). After 24 hours, embryoid bodies were gravity-settled and replated in fresh StemPro backbone plus 10 ng/mL BMP4, 6 ng/mL Activin A (R+D) and 5 ng/mL bFGF. After 3 days, the embryoid bodies were dissociated into single cells using trypsin and seeded onto Matrigel-coated plates at a density of 5 × 105 – 1 × 106 cells/cm2 in StemPro backbone. Medium was changed every 3–4 days thereafter until day 14 of differentiation.
Flow Cytometry
Cells were analyzed by flow cytometry at the cardiovascular progenitor (T5) and definitive cardiovascular cell (T14) stages of differentiation. Cultures were dissociated into single cells using trypsin enzymatic digestion. Progenitor stage cells were stained with mouse anti-human VEGFR2/KDR-PE (R+D) and mouse anti-human PDGFRα-APC (R+D). For intracellular staining, cells were fixed in 4% paraformaldehyde for 10 minutes followed by permeabilization with 0.75% saponin. Cardiomyocytes and smooth muscle cells were identified by staining with mouse anti-cardiac troponin T (LabVision/Neomarkers) and rabbit anti-smooth muscle actin (Abcam), followed by goat anti-mouse-PE (Jackson) and donkey anti-rabbit-APC (Jackson). Endothelial cells were identified using mouse anti-human CD31-PerCP-eFlour710 (eBioscience). Flow cytometric analysis was performed using a BD FACS Canto II machine.
Chromatin Immunoprecipitation Followed by Massive Parallel Sequencing
H7 human ESCs at different cardiovascular stages were crosslinked with 1% formaldehyde (Sigma) and sheared by Diagenode bioruptor. The antibodies used in ChIP assays were 9751 for histone H3 tri-methyl lysine 4 (Cell Signaling), 07–449 for histone H3 tri-methyl lysine 27 (Millipore), and ab9050 for histone H3 tri-methyl lysine 36 (Abcam). For each IP, Dynabeads (M-280, sheep anti-rabbit IgG, Invitrogen) were incubated with antibodies for 6 hours at 4°C and then incubated overnight with ~100 μg sheared chromatin. The complexes were rinsed sequentially with IP wash buffer I (50 mMTris-HCl pH 8.0, 150 mM NaCl, 1 mM EDTA pH8.0, 0.1 % SDS, 1 % Triton X-100, 0.1 % sodium deoxycholate), high salt buffer (50 mMTris-HCl pH8.0, 0.5 M NaCl, 1 mM EDTA pH8.0, 0.1 % SDS, 1 % Triton X-100, 0.1 % sodium deoxycholate), IP wash buffer II (50 mMTris-HCl pH8.0, 1 mM EDTA pH8.0, 1 % NP-40, 0.7 % sodium deoxycholate, 0.5 M LiCl) and TE buffer (10 mMTris-HCl pH8.0, 1 mM EDTA pH8.0). The complexes were incubated with elution buffer (10 mMTris-HCl pH 8.0, 0.3 M NaCl, 5 mM EDTA pH8.0, 0.5 % SDS) supplemented with RNase A (Ambion) at 65°C overnight. After separation, the DNA was treated with Proteinase K and purified by PCR purification column (QIAGEN). The sequencing libraries were prepared following a standard protocol using PE adapters (Illumina). For each library, an Illumina Genome Analyzer was used to generate 36 base pair sequence reads, yielding an average of ~18 million tags that mapped uniquely to the human genome (hg18).
Affymetrix RNA Expression Array
At the five time points indicated, an aliquot of cells were harvested in RNAlater (Ambion) and stored at −20°C. Total RNA was isolated using RNeasy kits (Qiagen) according to the manufacturer's instructions followed by quantification and quality assessment using a Bioanalyzer (Agilent). Approximately 3 μg RNA was subjected to in vitro transcription and labeling followed by hybridization to Affymetrix Human Exon 1.0 ST arrays (Affymetrix) according to the manufacturer's protocols.
Data Analysis
ChIP-seq tag density, the number of uniquely mapping sequenced tags within +/−75 bp, was calculated in 20 bp bins across the (hg18) reference human genome. Using annotated transcription start sites, the ChIP densities for three chromatin modifications (H3K4me3, H3K27me3 and H3K36me3) within 20 kb of each transcription start sites were collected at 20 bp resolution.
To determine the temporal chromatin profiles for different classes of genes TSS-centered profiles of chromatin changes were assembled. For H3K27me3 and H3K4me3 data, this was accomplished by `folding' the data within 2 kb of each TSS in half and performing a linear regression. The slope and intercept of the resulting line were directly proportional to the intensity of those signals and were used to assign a single number to each gene at each time point. For H3K36me3, the average signal intensity over the gene body was used as the signal intensity measure.
Two methods were used to rank genes according to potential roles as regulators of cardiac differentiation. The formulas for ranking genes according to these measures follow:
where `a' is the measure based on expression alone, and b is the measure based on expression (x), H3K4me3 (k4) and H3K27me3 (k27). The measure was calculated for each gene (i), using data from each of n=5 time points (j), and also using information specific to a given time point of interest `t'. The function `ρ' is the Pearson product moment correlation, and `min()' yields the minimum value from a set of numbers.
Principal component analysis was performed in R using 20 data points per gene, including 5 time points for each of 4 measurement types: H3K4me3, H3K27me3, H3K36me3 and RNA expression. The overview for all genes was plotted in two-dimensional space according to the top three principal components.
Zebrafish Morpholino Injections and In Situ Hybridization
The strain Tg(acta1a:GFP) (Higashijima et al., 1997) was used as a wild-type zebrafish strain and was maintained, crossed, injected, raised and staged as described (James et al., 2009) and in accordance with IACUC approved procedures. Heart rate was assessed in 10 randomly selected embryos from each group (control-MO versus meis2b-MO) by visualizing the heart beat in living embryos and counting for 1 minute. myl7 and vmhc probes were synthesized from published constructs (Yelon et al., 1999). Stock morpholinos (MO; Genetools, Philomath, OR) were dissolved in ddH2O to a concentration of 30 ng/nL, diluted to injection strengths [10 ng/nl for splice blocking E3I3 meis2b-MO (injected 4nl of 2.5ng/nl solution), 6 ng/nl for ATG blocking meis2b-MO], and 5 ng/nl (1–2 nl injected) Standard control-MO and injected into one-cell stage zebrafish embryos. The sequences for the morpholinos are as follows:
Splice-blocking E3I3 meis2b-MO: ACCGAAATCAATAACTTGCCTGTTT
ATG blocking meis2b-MO: 5–-CTTCGTACCGTTGAGCCATCAGCAT
Standard control-MO: 5'-CCTCTTACCTCAGTTACAATTTATA
RNA in situ hybridizations were performed as previously described (Maves et al., 2009; Talbot et al., 2010). Phenotype was scored “affected” if the heart displayed a defect as pictured versus “unaffected” if the heart appeared normal, compared with controls.
Supplementary Material
Highlights
Genome-wide chromatin modification dynamics during lineage-specific differentiation.
Chromatin modification dynamics distinguish structural from regulatory genes
A temporal chromatin signature predicts regulators of cardiac development.
Temporal chromatin signatures can identify regulators of other cell fates.
Acknowledgements
The authors thank Nina Tan, Mark Saiget, James Fugate, Kristen Lee, Rajinder Kaul and Stanley Kim for technical expertise. This work was supported by NIH grants P01 GM081719, U01 HL100405, P01 HL094374, R01 HL084642, R01 HL64387, R03 AR057477 and UW ENCODE Center (U54HG004592). SP was supported through NIH F30 HL095343. HW was supported by R90HG001452. RTM is an investigator of the HHMI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bu L, Jiang X, Martin-Puig S, Caron L, Zhu S, Shao Y, Roberts DJ, Huang PL, Domian IJ, Chien KR. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009;460:113–117. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
- Collas P. Programming differentiation potential in mesenchymal stem cells. Epigenetics. 2010;5 doi: 10.4161/epi.5.6.12517. [DOI] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JA, Reiner AH, Klungland A, Wakayama T, Collas P. Histone H3 lysine 27 methylation asymmetry on developmentally-regulated promoters distinguish the first two lineages in mouse preimplantation embryos. PLoS One. 2010;5:e9150. doi: 10.1371/journal.pone.0009150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domian IJ, Chiravuri M, van der Meer P, Feinberg AW, Shi X, Shao Y, Wu SM, Parker KK, Chien KR. Generation of functional ventricular heart muscle from mouse ventricular progenitor cells. Science. 2009;326:426–429. doi: 10.1126/science.1177350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman NS, Yelon D. Cardiac development in zebrafish: coordination of form and function. Semin Cell Dev Biol. 2002;13:507–513. doi: 10.1016/s1084952102001040. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Frampton GM, Soldner F, Hockemeyer D, Mitalipova M, Jaenisch R, Young RA. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell. 2010;7:249–257. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima S, Okamoto H, Ueno N, Hotta Y, Eguchi G. High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev Biol. 1997;192:289–299. doi: 10.1006/dbio.1997.8779. [DOI] [PubMed] [Google Scholar]
- James RG, Biechele TL, Conrad WH, Camp ND, Fass DM, Major MB, Sommer K, Yi X, Roberts BS, Cleary MA, et al. Bruton's tyrosine kinase revealed as a negative regulator of Wnt-beta-catenin signaling. Sci Signal. 2009;2:ra25. doi: 10.1126/scisignal.2000230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-Specific Optimization of Activin/Nodal and BMP Signaling Promotes Cardiac Differentiation of Mouse and Human Pluripotent Stem Cell Lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- Lee H, Shamy GA, Elkabetz Y, Schofield CM, Harrsion NL, Panagiotakos G, Socci ND, Tabar V, Studer L. Directed differentiation and transplantation of human embryonic stem cell-derived motoneurons. Stem Cells. 2007;25:1931–1939. doi: 10.1634/stemcells.2007-0097. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lindeman LC, Winata CL, Aanes H, Mathavan S, Alestrom P, Collas P. Chromatin states of developmentally-regulated genes revealed by DNA and histone methylation patterns in zebrafish embryos. Int J Dev Biol. 2010;54:803–813. doi: 10.1387/ijdb.103081ll. [DOI] [PubMed] [Google Scholar]
- Maves L, Tyler A, Moens CB, Tapscott SJ. Pbx acts with Hand2 in early myocardial differentiation. Dev Biol. 2009;333:409–418. doi: 10.1016/j.ydbio.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzarella L, Jorgensen HF, Soza-Ried J, Terry AV, Pearson S, Lacaud G, Kouskoff V, Merkenschlager M, Fisher AG. Embryonic stem cell-derived hemangioblasts remain epigenetically plastic and require PRC1 to prevent neural gene expression. Blood. 2011a;117:83–87. doi: 10.1182/blood-2010-03-273128. [DOI] [PubMed] [Google Scholar]
- Mazzarella L, Jørgensen HF, Soza-Ried J, Terry AV, Pearson S, Lacaud G, Kouskoff V, Merkenschlager M, Fisher AG. Embryonic stem cell-derived hemangioblasts remain epigenetically plastic and require PRC1 to prevent neural gene expression. Blood. 2011b;117:83–87. doi: 10.1182/blood-2010-03-273128. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O'Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, Murry CE. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted hearts. Nature Biotechnology. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- Minehata K, Kawahara A, Suzuki T. meis1 regulates the development of endothelial cells in zebrafish. Biochem Biophys Res Commun. 2008;374:647–652. doi: 10.1016/j.bbrc.2008.07.075. [DOI] [PubMed] [Google Scholar]
- Mobius W, Gerland U. Quantitative test of the barrier nucleosome model for statistical positioning of nucleosomes up- and downstream of transcription start sites. PLoS Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1000891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008a;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schübeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008b;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Phillips BW, Hentze H, Rust WL, Chen QP, Chipperfield H, Tan EK, Abraham S, Sadasivam A, Soong PL, Wang ST, et al. Directed differentiation of human embryonic stem cells into the pancreatic endocrine lineage. Stem Cells Dev. 2007;16:561–578. doi: 10.1089/scd.2007.0029. [DOI] [PubMed] [Google Scholar]
- Pjanic M, Pjanic P, Schmid C, Ambrosini G, Gaussin A, Plasari G, Mazza C, Bucher P, Mermod N. Nuclear factor I revealed as family of promoter binding transcription activators. BMC Genomics. 2011;12:181. doi: 10.1186/1471-2164-12-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Wysocka J. Epigenomics of human embryonic stem cells and induced pluripotent stem cells: insights into pluripotency and implications for disease. Genome Med. 2011;3:36. doi: 10.1186/gm252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Stankunas K, Shang C, Twu KY, Kao SC, Jenkins NA, Copeland NG, Sanyal M, Selleri L, Cleary ML, Chang CP. Pbx/Meis deficiencies demonstrate multigenetic origins of congenital heart disease. Circ Res. 2008;103:702–709. doi: 10.1161/CIRCRESAHA.108.175489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Ohgi M, Koshiba-Takeuchi K, Shiratori H, Sakaki I, Ogura K, Saijoh Y, Ogura T. Tbx5 specifies the left/right ventricles and ventricular septum position during cardiogenesis. Development. 2003;130:5953–5964. doi: 10.1242/dev.00797. [DOI] [PubMed] [Google Scholar]
- Talbot JC, Johnson SL, Kimmel CB. hand2 and Dlx genes specify dorsal, intermediate and ventral domains within zebrafish pharyngeal arches. Development. 2010;137:2507–2517. doi: 10.1242/dev.049700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol. 2006;26:9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464:922–926. doi: 10.1038/nature08866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, et al. Human cardiovascular progenitor cells develop from a KDR(+) embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- Yelon D, Horne SA, Stainier DY. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev Biol. 1999;214:23–37. doi: 10.1006/dbio.1999.9406. [DOI] [PubMed] [Google Scholar]
- Zerucha T, Prince VE. Cloning and developmental expression of a zebrafish meis2 homeobox gene. Mech Dev. 2001;102:247–50. doi: 10.1016/s0925-4773(01)00299-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.