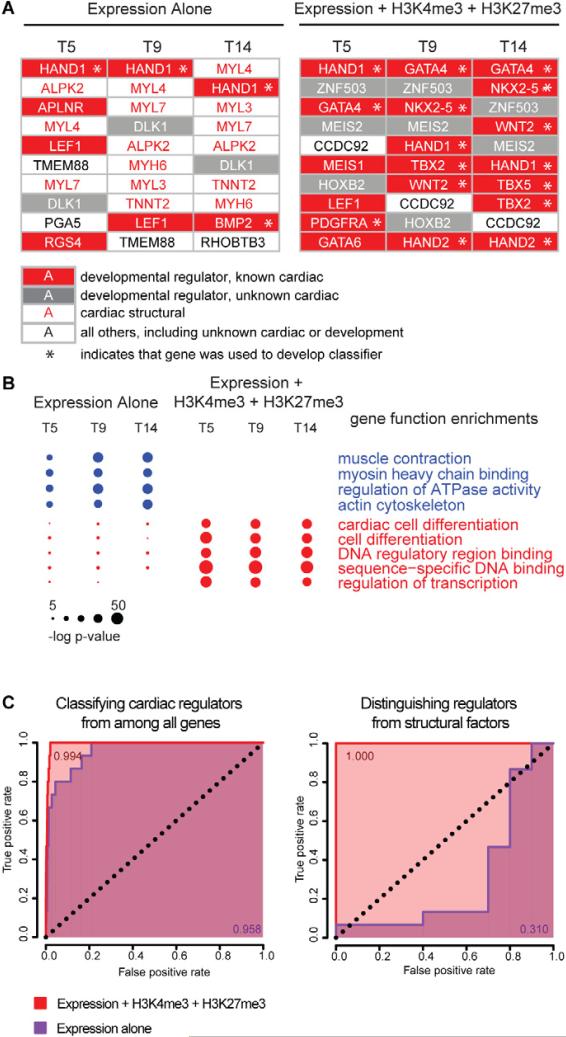
Figure 2. Accurate discrimination of key regulators of cardiac differentiation from other lineage-specific genes.
(A) All genes were ranked by two different methods in order to identify key regulators of cardiac differentiation. They were ranked at days 5, 9 and 14 by a formula using either (left) RNA expression alone, or (right) one that accounts for levels of H3K4me3, H3K27me3 and RNA expression. At each time point the top 10 candidate regulators are depicted for the respective methods. Developmental regulators with known roles in cardiac differentiation are shown in white text on red background, developmental regulators with no currently-appreciated role in cardiac differentiation are shown in white text on grey background, and genes whose function pertains to the structure and function of heart cells with no known regulatory roles are shown in red text on white background. All other genes are shown in black text on white background. Genes that were used in the training set for identifying the chromatin + expression regulator signature are indicated with an asterisk. (B) The top 100 candidates provided by each ranked list (see Figure S4) were analyzed to determine the degree to which the lists were enriched in 11 key gene ontology functional categories. The size of each circle is proportional to the significance of the enrichment of genes with the indicated functional role within the given list of 100 genes at each time point/ method of ranking genes. Ranking genes using H3K4m3, H3K27me3 and RNA expression yields lists that are not contaminated by structural factors and are more enriched with known regulators of cardiac differentiation. (C) Classification based on H3K4me3, H3K27me3 and expression (red) is more specific and sensitive than classification based on expression alone (purple). Shown are ROC curves for the identification of key regulators of cardiac differentiation from among all genes (left panel) or among all genes involved in heart development (right panel). The expression-only classifier systematically misclassifies structural factors that are involved in heart function but do not regulate cardiac development (right panel), leading to a lower area under the curve when classifying key regulators from among all genes (left panel). The genes used to generate true-positives for cardiac regulatory and structural genes are given in Figure 1.