Summary
Genetic evidence indicates that Saccharomyces cerevisiae Sgs1, Top3, and Rmi1 resolve topologically linked intermediates arising from DNA replication and recombination. Using purified proteins, we show that Sgs1, Top3, Rmi1, and replication protein-A (RPA) coordinate catenation and decatenation of dsDNA through sequential passage of single strands of DNA, establishing a unique pathway for dsDNA decatenation in eukaryotic cells. Sgs1 is required for dsDNA unwinding and, unexpectedly, also has a structural role in DNA strand passage. RPA promotes DNA unwinding by Sgs1 by trapping ssDNA and it stimulates DNA strand passage by Top3. Paradoxically, Rmi1 has a unique regulatory capacity that slows DNA relaxation by Top3, but stimulates DNA decatenation. We establish that Rmi1 stabilizes the “open” Top3-DNA covalent complex formed as a transient intermediate of strand passage. This concerted activity of the Sgs1–Top3–Rmi1–RPA represents an important mechanism for disentangling structures resulting from the topological features of duplex DNA.
Keywords: BLM, Sgs1, Top3, Rmi1, DNA helicase, Holliday junction, recombination, topoisomerase, decatenation, hemicatenane, replication protein-A
Introduction
The RecQ family of DNA helicases functions to maintain genomic stability (Chu and Hickson, 2009). Mutations in three of the five human members of this helicase family, BLM, WRN, and RecQ4, result in chromosomal abnormalities that are associated with increased cancer risk and premature aging manifested by the Bloom, Werner, and Rothmund-Thomson syndromes, respectively. The budding yeast Saccharomyces cerevisiae possesses a single RecQ family member, Sgs1, which is most homologous to human BLM. This helicase was identified in a screen for slow growth suppressors of Topoisomerase III (Top3), a phenotype that gave Sgs1 its name (Gangloff et al., 1994). The association of RecQ helicases with a type IA topoisomerase is conserved from bacteria to humans, and it has become clear that many RecQ/Sgs1/BLM functions are carried out in concert with the cognate associated topoisomerase (Bennett et al., 2000; Harmon et al., 2003; Harmon et al., 1999; Wu et al., 2000). More recently, additional components of this complex were discovered, namely Rmi1 in yeast, and RMI1 and RMI2 in human cells (Chang et al., 2005; Mullen et al., 2005; Yin et al., 2005). Together, through direct physical interactions, the eukaryotic proteins form a functional unit termed the Sgs1–Top3–Rmi1 complex (Mankouri and Hickson, 2007).
The phenotypes of BLM−/− or sgs1 cells are complex, and reflect multiple and possibly overlapping functions. Mutants are characterized by both mitotic and meiotic hyper-recombination associated with increased DNA crossovers, leading to chromosomal translocations, rearrangements, and sister-chromatid exchanges. Slow growth, chromosome loss, and sensitivity to various DNA-damaging agents are also characteristic (Gangloff et al., 1994). Collectively, these findings suggest a function of the Sgs1–Top3–Rmi1 complex at the interface of DNA recombination and replication (for review, see (Mankouri and Hickson, 2007)).
RecQ, Sgs1, and BLM have several functions in homologous recombination (HR). Early in HR they provide the important helicase activity or structural function needed to produce resected dsDNA with a 3′-ssDNA overhang in cooperation with a specific nuclease: RecJ in E. coli (Handa et al., 2009); Dna2 in yeast (Gravel et al., 2008; Mimitou and Symington, 2008; Zhu et al., 2008); and either DNA2 or EXO1 in humans (Nimonkar et al., 2011; Nimonkar et al., 2008). After resection, the exposed 3′-ssDNA tail becomes a substrate for a DNA strand exchange protein, and both RecQ and BLM disrupt joint molecules in vitro, indicating a likely regulatory role of RecQ homologues in this process (Bugreev et al., 2007; Harmon and Kowalczykowski, 1998). Finally, both the Sgs1–Top3 and BLM–TopoIIIα complexes separate double Holliday junctions (dHJ), a late intermediate of HR, in a reaction that couples DNA unwinding by the helicase with unlinking by the type IA topoisomerase to dissolve these recombination intermediates without crossing over (Bzymek et al., 2010; Cejka et al., 2010b; Plank et al., 2006; Wu and Hickson, 2003).
The Sgs1–Top3–Rmi1 proteins also resolve structures arising from stalled or collapsed replication forks, which arise from lesions or adducts in the DNA. Although the exact mechanism remains largely unknown, BLM can regress model replication forks (Ralf et al., 2006) which, when coupled to limited DNA replication, can lead to lesion bypass. Alternatively, regression of replication forks leading to gap repair was proposed to produce, through a recombination and post-replicative repair-dependent process, X-shaped replication intermediates (also called sister chromatid junctions) (Bernstein et al., 2009; Carotenuto and Liberi, 2010; Liberi et al., 2005). These junctions could represent catenated DNA molecules that are template-switch intermediates. Interestingly, since they accumulate specifically in sgs1, top3, and rmi1 mutants, the Sgs1–Top3–Rmi1 complex is required for their resolution (Bernstein et al., 2009; Liberi et al., 2005).
The Sgs1–Top3–Rmi1 complex might also resolve late replication intermediates, structures arising when two replication forks nearly converge to leave a short region of unreplicated dsDNA (Wang, 1991). This topologically-linked intermediate can be resolved by coupling DNA unwinding by a RecQ helicase to unlinking by the respective topoisomerase, as shown in vitro for the bacterial proteins (Suski and Marians, 2008). The BLM–TopoIIIα–RMI1 complex was observed to localize on ultra-fine DNA bridges in mitosis in human cells (Chan et al., 2009). Thus, Sgs1–Top3–Rmi1 and homologues might also be required for efficient completion of DNA replication, and proper segregation of daughter chromosomes (Chu and Hickson, 2009). E. coli RecQ and Topo III were shown to catenate and decatenate dsDNA, showing that RecQ helicase generates ssDNA for the DNA strand passage capacity of Topo III (Harmon et al., 1999).
The phenotype of rmi1 null mutants strongly resembles that of top3 mutants, and is also suppressible by deletion of SGS1 (Chang et al., 2005; Mullen et al., 2005), suggesting that Rmi1 is required for the function of Top3. Little is known about the biochemical function of Rmi1/hRMI1/hRMI2. These proteins contain OB-folds (oligonucleotide/oligosaccharide binding), which could mediate DNA binding or protein-protein interactions (Chen and Brill, 2007; Mullen et al., 2005; Raynard et al., 2008; Wu et al., 2006; Xu et al., 2008). In agreement, both hRMI1 (Raynard et al., 2006; Wu et al., 2006) and hRMI2 (Singh et al., 2008), and yeast Rmi1 (Cejka et al., 2010b) stimulate the dissolution of an oligonucleotide-based dHJ.
Previously, we demonstrated that Rmi1 stimulated a late step in the dissolution of a double Holliday junction (dHJ) by Sgs1 and Top3, suggesting that Rmi1 promoted, minimally, the dissolution of a hemi-catenane, the final predicted intermediate of dHJ dissolution (Cejka et al., 2010b). In the present study, we show that the Sgs1–Top3–Rmi1 complex can promote the catenation and decatenation of dsDNA. Sgs1 catalytic function is needed to unwind the dsDNA to produce the ssDNA substrate for Top3, as expected, but unexpectedly, Sgs1 is also needed in a structural capacity for DNA strand passage. Also unanticipated, RPA promotes the reaction beyond simply stimulating DNA unwinding by Sgs1 and trapping ssDNA. Finally, we demonstrate an unanticipated function of Rmi1: it regulates the outcome of Top3 activity to increase the opportunity for DNA strand passage. Rmi1 achieves this control by the unique mechanism of stabilizing the lifetime of the Top3–DNA cleavage complex. Taken together, we show that the components of the Sgs1–Top3–Rmi1 complex and RPA act in a remarkably concerted manner to untangle a wide variety of DNA replication and recombination intermediates.
Results
Sgs1, Top3, and Rmi1 catalyze formation of DNA multimers in the presence of ssDNA-binding proteins
Previously, we showed that Rmi1 was nearly essential for decatenation of the multiply linked kinetoplast circular dsDNA by Sgs1 and Top3 into monomeric units (Cejka et al., 2010b). These results suggested that Rmi1 has a specific but undefined function in DNA strand passage mediated by Sgs1 and Top3. Depending on the direction of the strand passage event, two circular DNA molecules can be either separated (i.e., decatenated) or topologically linked (i.e., catenated). Therefore, catenation and decatenation should be mechanistically identical processes, and the directionality of the reaction would be determined by reaction component concentrations (see further below) (Harmon et al., 2003). Initially, we analyzed catenation of DNA by the Sgs1–Top3–Rmi1 complex (Figure 1A) because the experimental variables can be more easily defined.
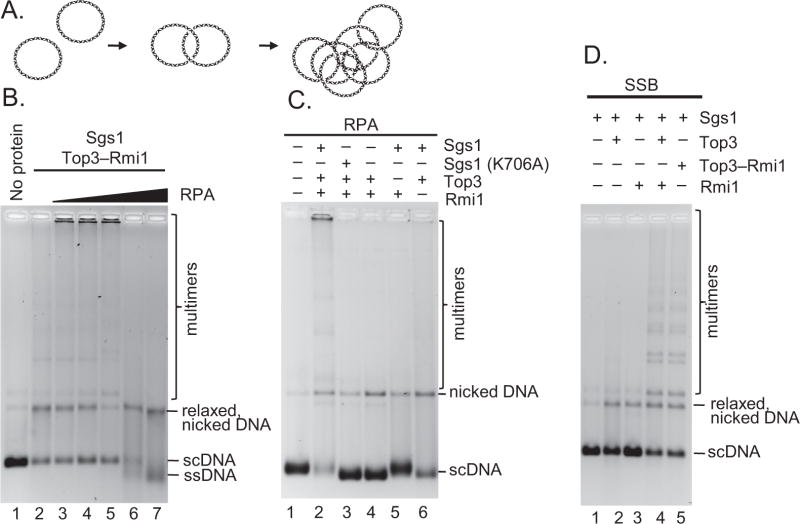
Figure 1. Sgs1, Rmi1, and RPA stimulate formation of DNA multimers by Top3.
(A) DNA catenation. (B) Catenation with Sgs1 (60 nM), Top3-Rmi1 (400 nM), and RPA (0.1, 0.33, 1, 3.1 and 6 μM). (C) Catenation with Sgs1 (60 nM), Sgs1(K706A) (60 nM), Top3 (400 nM), Rmi1 (400 nM), and RPA (1 μM). (D) Catenation with Sgs1 (100 nM), Top3-Rmi1 (300 nM), Top3 (300 nM), Rmi1 (300 nM), and SSB (3.8 μM).
As shown in Figure 1B, Sgs1, Top3, and Rmi1 proteins promote the formation of a modest amount of DNA multimers, when starting with monomeric negatively supercoiled DNA (scDNA). We observed that RPA increased the yield of DNA multimers, and resulted multimers that were often too large to enter the agarose gel during electrophoretic analysis, remaining trapped in the wells (Figure 1B, lanes 3–5). Formation of these large DNA complexes required catalytically active Sgs1, Top3, and was also largely dependent on Rmi1 (Figure 1C). Only a sub-stoichiometric amount of RPA was needed (~50% saturation of the potential ssDNA), and a further increase in RPA concentration inhibited catenation but led to the formation of a new fast-migrating DNA species (Figure 1B, lanes 6–7). Subsequent analysis of the fast-migrating species revealed that it is ssDNA produced from the complete unwinding and unlinking of the plasmid substrate into its component single-strands (Figure S1A). Thus, these proteins comprise a highly active DNA unwinding and strand passage complex.
RPA not only binds ssDNA, but also physically and functionally interacts with Sgs1 to promote DNA unwinding (Cejka et al., 2010b; Cobb et al., 2003). To separate the effect of binding to ssDNA from direct stimulation of Sgs1, we substituted RPA with E. coli SSB, which does not interact with Sgs1 (Cejka et al., 2010b; Cobb et al., 2003). In this case, the reactions produced a distinct pattern of DNA multimeric bands that extended above the monomer species, with only a small fraction of DNA remaining in or near the wells (Figure 1D). While utilization of the scDNA substrate was similar to the experiments with RPA, SSB stimulated the reactions to a lower extent, as measured by the amount of the larger multimeric complexes (compare Figure 1B, C and D). As was the case with RPA, Rmi1 greatly stimulated the reaction: without Rmi1, multimers were barely detectable (Figure S1B, lane 7); increasing the Rmi1 concentration led to a proportional increase in DNA multimers (Figure S1B, lanes 7–11). The formation of the multimeric product was dependent on time (Figure S1C); concentration of Top3-Rmi1 (Figure S1D); and barely detectable in the absence of Rmi1 (Figure S1E). The requirement for Sgs1, Top3, Rmi1, and RPA makes it unlikely that a contaminating eukaryotic type II topoisomerase is responsible for the catenation, but to eliminate that possibility, we added etoposide, which is a potent inhibitor of eukaryotic type II topoisomerases. The etoposide did not inhibit the multimerization reaction (Figure S2A) but did inhibit a type II topoisomerase control reaction (Figure S2B), confirming our conclusion that the catenation is specific to the type IA topoisomerase, Top3, and either requires or is greatly stimulated by Sgs1, Rmi1, and RPA.
The DNA multimers produced by Sgs1, Top3, and Rmi1 are catenanes
Next, we set out to identify the nature of the multimers produced by Sgs1–Top3–Rmi1. For these experiments, multimers produced by the protein complex in the presence of SSB were used initially, because the distinct ‘ladder’ formed with SSB facilitated analysis. The multimers were expected to fall into one of three classes of DNA species, or the combination thereof: (1) denatured and randomly annealed ssDNA molecules; (2) hemicatenanes; or (3) full catenanes (Figure 2A). When the products of an Sgs1–Top3–Rmi1 reaction were analyzed by denaturing gel electrophoresis, the slower-migrating multimeric species were maintained, demonstrating that the multimers are topologically linked (Figure 2B), either as hemi- or full catenanes, and are not annealed ssDNA.
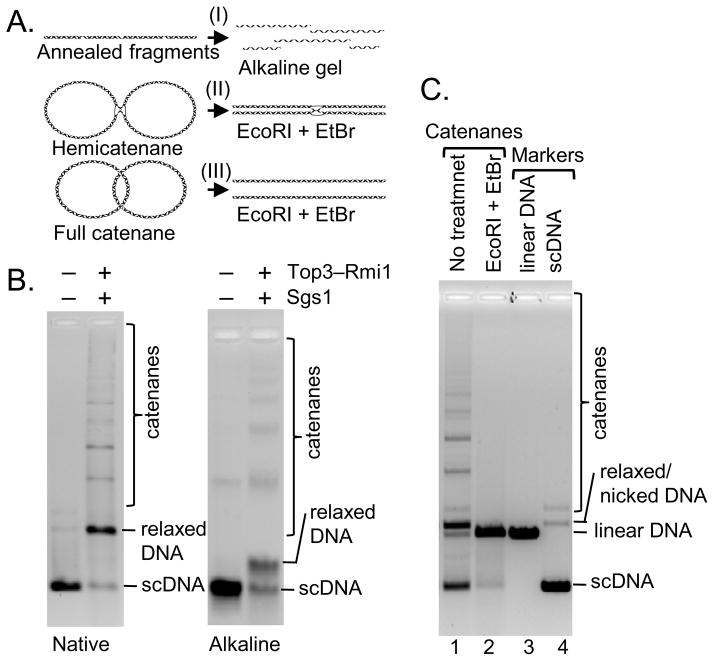
Figure 2. Sgs1, Top3, and Rmi1 promote formation of full dsDNA catenanes.
(A) Approaches used to distinguish various multimeric species; EtBr, ethidium bromide. (B) DNA substrate and catenation products using Sgs1 (100 nM), Top3 (400 nM), Rmi1 (800 nM), SSB (3.8 μM) separated by electrophoresis under native or denaturing conditions. (C) Catenation products using Sgs1 (60 nM), Top3-Rmi1 (400 nM), and SSB (3.8 μM), lane 1, were digested with EcoRI (lane 2) in the presence of 0.1 μg/ml ethidium bromide.
Hemicatenanes, which comprise another possibility for the multimeric species, are dsDNA molecules linked only by a single-strand of one duplex wrapped around a single-strand of another duplex; they can be distinguished from full catenanes by several complementary approaches. The first approach uses digestion with a restriction endonuclease that cuts once within the monomer (Harmon et al., 1999). Full catenanes will fall apart into monomeric linear dsDNA, whereas hemicatenanes will remain entangled (Figure 2A). Ethidium bromide was present in the reaction to limit thermal DNA branch migration of the hemicatenane off the end of the linearized DNA. Figure 2C (lane 2) and Figure S2C show that digestion with EcoRI caused the multimers to collapse into a band that comigrated with linear substrate DNA, suggesting that the DNA multimers are full catenanes. To verify this identification, we exploited the structure-selective endonuclease T7 Endonuclease I, which cleaves ssDNA, DNA mismatches, and branched DNA molecules including HJs (Figure S2D), but not dsDNA (de Massy et al., 1987) and is thus expected to cleave hemicatenanes to produce nicked or linear DNA monomers. As shown in Figure S2E (compare lanes 1 and 2), the DNA multimers were refractory to T7 Endonuclease I, suggesting they are neither hemicatenanes nor structures with any ssDNA character. Finally, we treated the catenated multimers with Drosophila Topo II. Topo II will decatenate full catenanes, but not hemicatenanes (Berger et al., 1996). When incubated with Topo II, all of the multimers formed by Sgs1–Top3–Rmi1 were decatenated into relaxed monomeric dsDNA (Figure S2E). Collectively, these experiments show that the majority, if not all, of the multimers produced by Sgs1–Top3–Rmi1 are full catenanes. Full catenanes were also the products of catenation by RecQ and Topo III (Harmon et al., 1999), implying that this is conserved activity of this RecQ–Topoisomerase pair.
Catenation is specific to the cognate complex of yeast proteins
To investigate whether the protein-protein interactions are important for catenation, we replaced the yeast protein components of this complex with homologous proteins from E. coli or humans. Neither BLM helicase, hTopo IIIα, EcTopo I, nor hRMI1 could replace their respective yeast homologues (Figure S3A). Therefore, the catenation reaction is specific to the yeast proteins, implying that direct protein-protein interactions are an integral part of the reaction mechanism.
Sgs1, Top3, and Rmi1 can decatenate topologically linked DNA
To this point, we have demonstrated that the yeast Sgs1–Top3–Rmi1 complex can efficiently catenate dsDNA molecules. We expected DNA strand passage to be reversible, and the catenation reaction to be in equilibrium with decatenation (Harmon et al., 2003). Consequently, we next tested whether Sgs1–Top3–Rmi1 could decatenate the multimeric catenanes. We prepared catenanes by incubating monomeric circular dsDNA with Sgs1–Top3–Rmi1–RPA, and deproteinized and purified the resulting DNA products. A significant proportion of the purified product comprised large catenanes that do not enter the agarose gel during electrophoresis (Figure 3, lane 1). This purified catenated DNA was the substrate for subsequent reactions, where we varied both the Sgs1 and the Top3–Rmi1 concentrations. In the presence of RPA, we found that the Sgs1–Top3–Rmi1 complex greatly reduced the amount of catenanes trapped in the wells, and converted a portion into monomeric circular dsDNA or into catenanes with a smaller number of units (Figure 3). The fraction of the large catenated complex that was converted to monomeric circles was inversely correlated with the concentration of the Sgs1–Top3–Rmi1 complex. A decrease in the Sgs1–Top3–Rmi1 concentration of approximately 10- to 20-fold favored decatenation over catenation (Figure 3). As expected, the Sgs1–Top3–Rmi1 complex exhibited a decatenation activity in the presence of SSB as well (Figure S3B). In summary, the yeast Sgs1–Top3–Rmi1 complex can both catenate and decatenate DNA, with the directionality of the reaction driven by the concentration of Sgs1, Top3, and Rmi1.
Figure 3. Sgs1, Top3, and Rmi1 decatenate DNA catenanes.
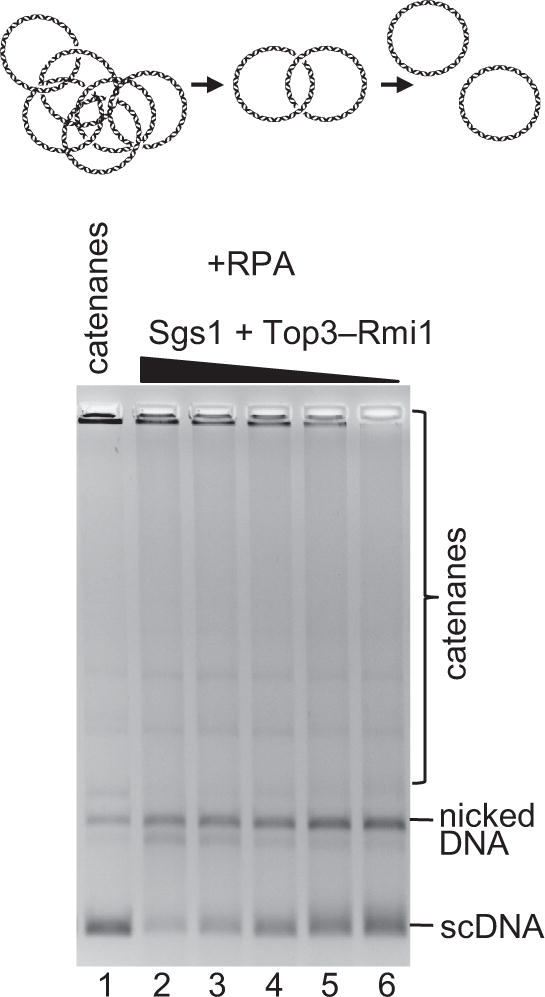
Catenation products using Sgs1 (60 nM), Top3-Rmi1 (400 nM), and RPA (1 μM), lane 1, were decatenated using Sgs1 (75, 60, 30, 10 and 5 nM), Top3-Rmi1 (300, 240, 120, 40 and 20 nM), and RPA (1 μM).
Sgs1 unwinds covalently closed dsDNA
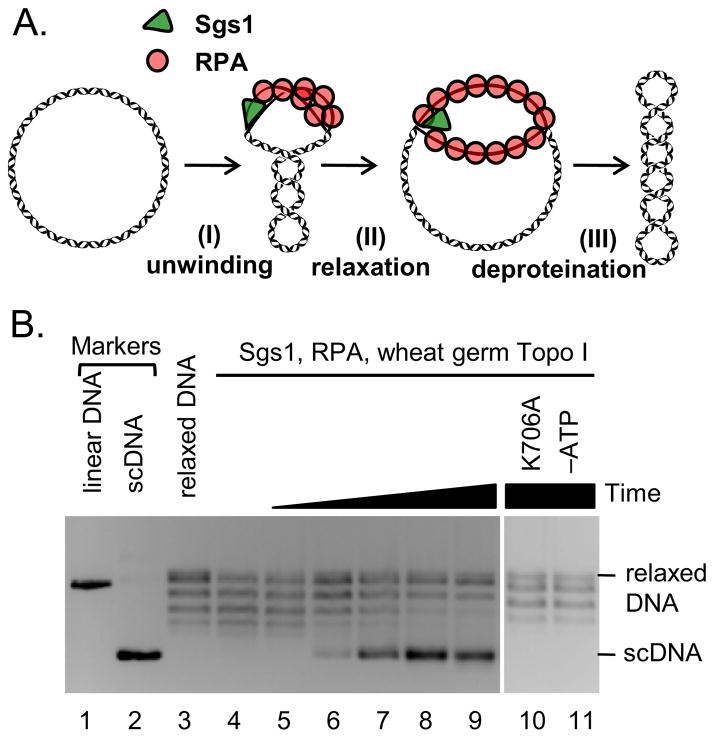
Because catenation by Sgs1–Top3–Rmi1 required the helicase activity of Sgs1 (Figure 1C), we expected that the unwinding activity of Sgs1 provides Top3 with ssDNA, which is the preferred substrate of this type IA topoisomerase. To detect DNA unwinding, we employed a Topo I-coupled assay used previously (Harmon et al., 1999). As illustrated in Figure 4A, helicase activity on covalently closed DNA in the presence of a ssDNA-binding protein results in the production of a denatured region, which leads to positive supercoiling within the remaining duplex region of the substrate. Wheat germ Topo I can rapidly remove this positive supercoiling. Upon deproteinization, removal of the ssDNA-binding protein results in renaturation of the denatured ‘bubble’, revealing negative scDNA (Figure 4A). Using this assay, we show that Sgs1 is indeed capable of unwinding covalently closed dsDNA (Figure 4B). This reaction is dependent on ATP hydrolysis indicating the activity is not a result of simple protein binding (Figure 4B). Clearly, Sgs1, similarly to E. coli RecQ helicase (Harmon et al., 1999), can unwind covalently closed dsDNA, and does not require a free end for loading onto DNA. The unwinding of dsDNA thus likely represents the first step in the dsDNA catenation/decatenation mechanism.
Figure 4. Sgs1 unwinds covalently closed dsDNA.
(A) Topo I-coupled unwinding assay. (B) Assay using Sgs1 (90 nM), RPA (1 μM), wheat germ Topo I (0.25 U/μl) for 5, 15, 30, 60 and 120 seconds, respectively; lane 10, Sgs1(K706A) catalytic mutant (90 nM) instead of wild-type Sgs1.
Sgs1 has a structural role in the catenation of DNA
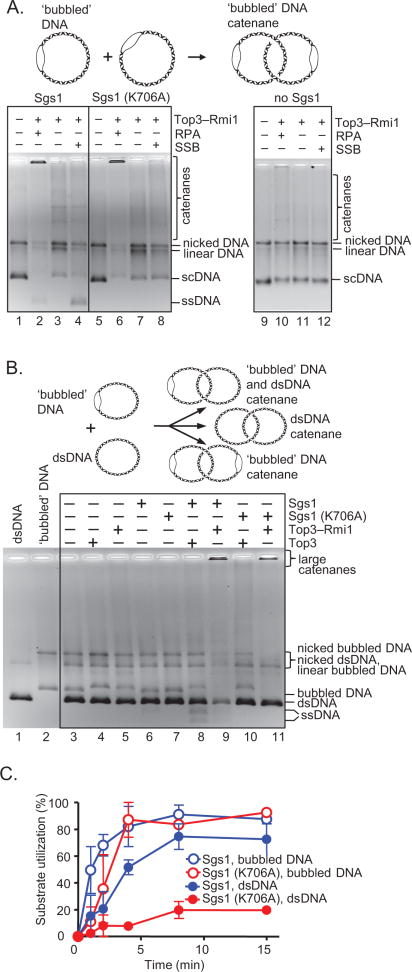
Thus far, we demonstrated that catalytically active Sgs1 is required, together with Top3 and Rmi1, for catenation. Therefore, the unwinding activity of Sgs1 likely generates ssDNA, which then becomes a substrate for the strand passage activity of Top3. To confirm this hypothesis, we utilized a covalently-closed dsDNA substrate containing a 450 nt region of unpaired ssDNA (Figure 5A). The Sgs1–Top3–Rmi1 complex, as expected, converted this substrate into multimers (Figure 5A, lane 2) as effectively as the fully duplex DNA. Surprisingly, when Sgs1 was omitted, there was little catenation by Top3–Rmi1 alone (Figure 5A, lane 10), despite the presence of a pre-existing region of ssDNA in all of the molecules. However, when the Sgs1 (K706A) helicase-dead mutant, which was ineffective in the standard reactions (Figure 1C) was added to Top3-Rmi1 and the bubbled DNA, formation of multimeric catenanes was restored to the same extent as seen with wild-type Sgs1 (Figure 5A, compare lanes 2 and 6). This result shows that Sgs1 has an unexpected structural role in promoting DNA catenation that is independent of its helicase activity. Sgs1, thus, has two functions in the catenation of fully duplex DNA: 1) the helicase activity of Sgs1 is required to unwind duplex DNA, and 2) the physical presence of Sgs1 protein is needed to promote DNA strand passage by the Top3–Rmi1 heterodimer.
Figure 5. Sgs1 and RPA play a structural role in DNA catenation.
(A) Catenation with ‘bubbled’ DNA (10 ng/μl), Sgs1 or Sgs1(K706A) (60 nM), Top3-Rmi1 (400 nM), and RPA (1 μM) or SSB (3.8 μM). Reactions were incubated with NaCl (500 mM, 5 min) before termination. (B) Catenation with both fully dsDNA and ‘bubbled’ DNA, and Sgs1 or Sgs1 (K706A), Top3 or Top3-Rmi1 and RPA as in panel A. (C) Kinetics of catenation for bubbled DNA versus dsDNA using Top3–Rmi1, RPA, and either Sgs1 or Sgs1(K706A); quantification is from two independent experiments ±SEM.
RPA promotes DNA strand passage by Top3–Rmi1 independently of its role in stimulating DNA unwinding by Sgs1
The catenation of dsDNA was stimulated by RPA, but also to a lower degree by the non-cognate SSB protein (Figure 1). The likely explanation is that SSB acts nonspecifically by trapping ssDNA created by Sgs1-catalyzed unwinding, whereas RPA, in addition to this nonspecific role, acts specifically via its direct interaction with and stimulation of Sgs1 (Cejka and Kowalczykowski, 2010; Cobb et al., 2003). However, because of the preexisting non-complementary ssDNA in the DNA bubble substrate, Sgs1 helicase function is not necessary, and neither RPA nor SSB should be needed. As we anticipated, SSB did not stimulate the catenation of the bubbled DNA substrate (Figure 5A, compare lanes 3 and 4). Unexpectedly, however, RPA clearly stimulated catenation of the bubbled DNA by Sgs1–Top3–Rmi1 (Figure 5A, compare lanes 2 and 4). The effect of RPA was largely dependent on the presence of Sgs1 (Figure 5A, compare lanes 2, and 10), but the helicase-dead (Sgs1 K706A) protein could also function with this bubbled DNA, but only in the presence of RPA, and not with SSB (Figure 5A, compare lanes 6 and 8). This shows that RPA not only promotes unwinding of dsDNA by Sgs1 and binds to the unwound products, but that it also specifically stimulates DNA strand passage by Top3–Rmi1 when complexed with Sgs1. This surprising result reveals an unanticipated species-specific function of RPA in dsDNA catenation that cannot be explained by specific stimulation of DNA unwinding by Sgs1; rather, RPA specifically affects DNA strand passage by Top3–Rmi1 via its interaction with Sgs1.
Catenation of dsDNA occurs primarily through the gating of single strands of DNA
Type IA topoisomerases such as Top3 act by transiently cleaving ssDNA, and it is believed that these enzymes then pass ssDNA through the open ‘gate’ (reviewed in (Baker et al., 2009)); however, the crystal structure of E. coli Topo III raised the possibility that these proteins can pass dsDNA through the open ssDNA gate (Changela et al., 2001). The catenation of dsDNA by Sgs1–Top3–Rmi1–RPA requires the passage of four DNA strands. This may occur either through four sequential ssDNA passages, which can take place when two ssDNA ‘bubbles’, formed by the helicase activity of Sgs1 on two separate DNA molecules, encounter one another. Alternatively, one ssDNA bubble on a single DNA molecule may be sufficient, provided that Top3 can pass a DNA duplex through the open gate, performing the catenation/decatenation reaction in only two steps.
To determine the mechanism of the DNA catenation by the Sgs1–Top3–Rmi1 complex, we performed the catenation reaction using a mixed population of bubbled and fully duplexed circular DNA monomers as substrate (Figure 5B). As expected, Sgs1–Top3–Rmi1 converted both substrates into multimeric catenanes. Importantly, however, when wild-type Sgs1 was replaced by the helicase-dead Sgs1 (K706A) mutant, the bubbled DNA was more efficiently converted into the catenane network than the fully complementary dsDNA (Figure 5B, lane 11). We further verified these findings by performing kinetic experiments (Figure 5C and Figure S4). These results demonstrate that Top3–Rmi1 in the physical presence of a helicase defective Sgs1 can gate dsDNA, although the rate is ~10-fold slower than for ssDNA. Thus, the catenation of dsDNA by Sgs1–Top3–Rmi1 occurs more rapidly through four sequential passages of single strands of DNA, but dsDNA can nonetheless be passed through the gate eliminating the obligatory need for two simultaneous ssDNA bubbles in the dsDNA partners.
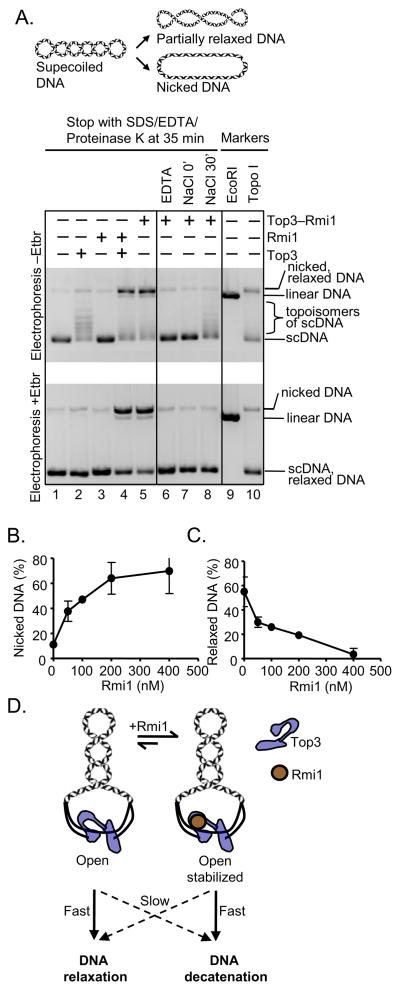
Rmi1 stalls Top3-mediated relaxation of supercoiled DNA
Top3 is a type IA DNA topoisomerase, the hallmark activity of which is relaxation of negative scDNA, which occurs by a three-step reaction mechanism (reviewed in (Viard and de la Tour, 2007). The type IA topoisomerases bind to one strand of DNA through either opportunistic binding of a pre-existing ssDNA bubble, or induced local denaturation of a dsDNA duplex upon binding. Once bound, the topoisomerase creates a transient ssDNA break in one of the DNA strands of the duplex. This break occurs by transesterification that creates a transient covalent intermediate between the topoisomerase and the 5′-terminated end of the break, while remaining bound to the other, 3′-terminated DNA end to preserve DNA integrity. This creates a protein-gate in the DNA backbone which allows passage of the intact ssDNA through the break; the topoisomerase then completes the reaction by bringing the two ends of the broken strand back together and reversing the transesterification reaction, sealing the break and dissociating from the DNA. In vitro, these assays are typically performed at elevated temperatures and in a reaction buffer containing high glycerol and low monovalent and divalent salt concentrations to facilitate the reaction by destabilizing the DNA duplex (Chen and Brill, 2007). Starting with negative scDNA, the expected products of these reactions are partially relaxed DNA and a small fraction of nicked DNA resulting from the trapping of the covalent Top3–DNA intermediate upon rapid deproteinization (see scheme atop Figure 6A, (Kung and Wang, 1977)). Because Rmi1 had a strong effect on catenation by Top3, we sought to determine its effect on relaxation by Top3.
Figure 6. Rmi1 slows Top3-dependent relaxation of negatively supercoiled DNA and increases the steady-state amount of Top3-DNA covalent intermediates.
(A) Relaxation of negative scDNA by Top3 and Rmi1 expressed individually or as a heterodimer (all 100 nM), lanes 1–5, plus either EDTA (25 mM, lane 6) or NaCl (0.5 M, lane 7); in lane 8, NaCl (0.5 M) was added at the end of a standard 30-minute incubation for 5 minutes. (B) Quantification of nicked dsDNA in reactions terminated with SDS/EDTA/proteinase K. (C) Quantification of relaxed DNA in reactions terminated with NaCl followed by SDS/EDTA/proteinase K. Quantification is from two independent experiments ±SEM. (D) Model for the effect of Rmi1 on the catalytic activity of Top3. By stabilizing the nicked “open” Top3–DNA intermediate, Rmi1 inhibits DNA relaxation, but promotes DNA catenation.
Top3 also has a vigorous relaxation activity, as indicated by the appearance of topoisomer bands with a reduced electrophoretic mobility relative to negative scDNA (Figure 6A). Rmi1, as expected, showed no relaxation activity (Figure 6A). Surprisingly, Top3 and Rmi1, purified either separately or together as a heterodimer, processed the supercoiled DNA to form a large fraction of apparently nicked DNA upon standard termination with SDS, EDTA, and proteinase K (Figure 6A, lanes 4–5). We verified the DNA species as nicked by analyzing the reaction products on an agarose gel containing ethidium bromide during electrophoresis, which can resolve nicked from fully relaxed DNA (Figure 6A, lanes 4–5; compare upper and lower panels). Neither Top3 nor Rmi1 showed significant nicking activity when incubated with substrate separately, showing that Top3-Rmi1 is responsible for this activity. To determine whether these nicks were the result of Top3 that was stalled by Rmi1 in a state that kinetically preserved the covalent intermediate, NaCl was added at the end of the 30 minute incubation period to 500 mM, and the reaction was incubated for an additional 5 minutes before inactivating the proteins with SDS, EDTA, and proteinase K. These conditions are known to allow type IA topoisomerases to complete the ligation of open complexes while preventing rebinding and subsequent strand scission (Kung and Wang, 1977). These termination conditions completely prevented nicking, resulting in partially relaxed DNA as the sole product (Figure 6, lane 8); in contrast, and as expected, addition of NaCl to 500 mM prior to initiation of the reaction completely inhibited relaxation (Figure 6, lane 7). No catenation of DNA was observed under these conditions (Figure S5A). Thus, Rmi1 was apparently trapping the covalently-linked, “open” intermediate formed between Top3 and DNA.
To further substantiate this conclusion, we examined the consequences of changing Rmi1 concentrations. Increasing the Rmi1 concentration increased the amount of DNA that was nicked (in the absence of NaCl treatment) (Figure 6B and Figure S5B). Interestingly, and consistent with our interpretations, when these same reactions were treated with NaCl to eliminate nicking, we found that increasing Rmi1 concentration reduced relaxation of scDNA by Top3 (Figure 6C and Figure S5B). In addition, a time-course of Top3 and Top3–Rmi1 reactions indicated that the Rmi1-dependent nicking occurred on the same time-scale as Top3-catalyzed relaxation (Figure S5C). Taken together, these data indicate that Rmi1 transiently slows Top3-catalyzed relaxation by kinetically stabilizing the Top3-DNA covalent intermediate. As a consequence, the steady-state concentration of the nicked Top3-DNA intermediate is increased, which is revealed upon rapid deproteinization of the complex (Figure 6B). We believe that this specific regulatory function of Rmi1 promotes the physiologically relevant reaction (i.e., decatenation), while inhibiting DNA relaxation (Figure 6D).
Discussion
Our experiments have revealed unexpected functions for the Sgs1, RPA, and Rmi1 proteins in regulating DNA strand passage by the topoisomerase, Top3. Top3 belongs to the type IA topoisomerase family. These enzymes catalyze the passage of one strand of DNA through a transient break in another strand (Viard and de la Tour, 2007). We showed that Sgs1, Top3, Rmi1, and RPA are all required for efficient conversion of circular dsDNA into topologically linked dsDNA rings, i.e., catenanes. The catenation reaction is reversible and in equilibrium with the physiologically more relevant decatenation reaction, because the same protein complex can separate interlinked dsDNA rings as well.
How can a type IA topoisomerase, such as Top3, which requires ssDNA as a substrate, promote catenation and decatenation of dsDNA? We show that Sgs1 helicase activity is required when the DNA is fully duplex and that Sgs1 can unwind dsDNA at internal sites without the need for a free end. Thus, the first step in dsDNA linking and unlinking is formation of an ssDNA bubble by the helicase activity of Sgs1. This unwound region is trapped by RPA binding, which prevents its reannealing. RPA functions in a species-specific capacity to promote DNA unwinding by Sgs1 (Cejka and Kowalczykowski, 2010). Furthermore, DNA unwinding by Sgs1 is stimulated by both Top3 and Rmi1 via specific protein-protein interactions (Cejka et al., 2010a). The unwound ssDNA is a substrate for Top3 that, due to its tight association with Sgs1, permits delivery of Top3 to the ssDNA and the coordination of DNA unwinding and strand passage.
Unexpectedly, this species-specific interaction between Sgs1 and Top3 is essential even when the requirement for Sgs1 helicase function is bypassed through the use of DNA containing a non-complementary ssDNA bubble. In this case, Sgs1 protein is physically required, but catalytic function is not. Thus, Sgs1 plays a role both in DNA unwinding, where its helicase activity is essential, and in DNA strand transfer by Top3, where it serves a structural role. Similarly, and unexpectedly, RPA also has a structural role in the promotion of DNA strand passage by Top3 that is also revealed in the experiments with ssDNA bubbles. This structural function of RPA is also largely dependent on the physical presence of Sgs1, revealing the network of protein-protein interactions that are mostly centered on Sgs1.
DNA strand passage was strongly stimulated by Rmi1, a known interacting partner of both Sgs1 and Top3. We showed that Rmi1 stabilizes a kinetic intermediate of the Top3 reaction, where the topoisomerase remains covalently bound to a transient ssDNA break in a conformation that allows passage of the intact DNA strand through the open gate (Figure 6D). We hypothesize that by increasing the lifetime of this “open” intermediate, the Rmi1–Top3–DNA complex has a higher probability of a productive encounter with another strand of DNA in a different molecule (in trans) to permit its passage through the open gate and thereby promote DNA catenation/decatenation. We conclude that ssDNA molecules are primarily being passed through the open gate, although dsDNA can pass as well, albeit at a slower rate. We also established, as might be expected from this model for Rmi1 function, that Rmi1 inhibits DNA relaxation, which is the passage of DNA though a gate within the same DNA molecule (in cis). Thus, Rmi1 serves as an important discriminatory factor that inhibits DNA relaxation by Top3 and, instead, promotes the physiologically more relevant DNA decatenation.
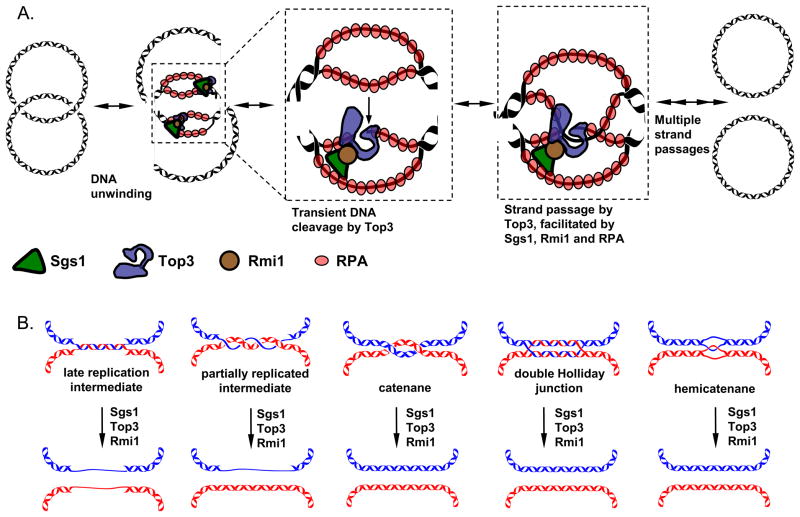
Collectively, these experiments revealed that the components of the Sgs1–Top3–Rmi1 and RPA complex have multiple functions, both catalytic and structural, to promote the activity of the other members of this complex. We propose the following model for dsDNA decatenation by the Sgs1–Top3–Rmi1 and RPA complex (Figure 7A). Sgs1 first unwinds dsDNA, and the resulting ssDNA is bound and stabilized by RPA. This enables Top3 to bind its preferred substrate, ssDNA, as a part of the Sgs1–RPA complex. Subsequently, Top3 catalyzes sequential rounds of DNA strand passage, which are both regulated and specifically stimulated by Sgs1, Rmi1, and RPA. DNA strand passage most often occurs via consecutive ssDNA transfers through the open gate. Double-stranded DNA can be passed through the ssDNA gate as well, albeit at a lower rate (~10-fold). We hypothesize that Sgs1 and RPA might also facilitate formation of a higher order complex of Top3-Rmi1, which could help coordinate multiple strand transfer reactions required in this model, and also to explain the observed structural role of Sgs1 and RPA in DNA catenation. Compatible with this idea, multimeric forms of the Sgs1 homologue, BLM, were observed by electron microscopy (Karow et al., 1999).
Figure 7. Model for the mechanism and function of DNA strand passage promoted by Top3, Sgs1, Rmi1, and RPA.
(A) Mechanism of DNA strand passage. Sgs1 unwinds dsDNA, and ssDNA is stabilized by RPA. Subsequently, Top3 catalyzes strand passages in steps stimulated by Rmi1, Sgs1, and RPA. (B) Schematic representation of various DNA replication and recombination intermediates that can be disentangled by the Sgs1–Top3–Rmi1 complex and its homologues.
What is the physiological relevance of the observed reactions? During various DNA metabolic processes such as replication or repair, it is essential to relieve superhelical twists and disentangle chromosomal DNA. These functions are carried out by the three topoisomerases, Top1, Top2, and Top3, in yeast (Wang, 1991). Topoisomerase I (Top1), is a type IB enzyme that relaxes dsDNA through a transient ssDNA break and rotational swiveling (Baker et al., 2009), while topoisomerase II (Top2) is a type II enzyme, which transports dsDNA through a transient dsDNA break (Berger et al., 1996). Top3 in conjunction with Sgs1 and Rmi1, and its homologs, can untangle various DNA molecules by transporting any DNA through a transient ssDNA break (Cejka et al., 2010b; Chan et al., 2007; Harmon et al., 1999; Plank and Hsieh, 2006; Suski and Marians, 2008; Wu and Hickson, 2003). In yeast, sgs1 top1 and top3 top1 double mutants show synthetic growth defects or are not viable (Lu et al., 1996; Tong et al., 2001), suggesting that Sgs1–Top3 functions in a parallel pathway to that of Top1, although the relevant DNA intermediates and functions are not known. The sgs1 helicase-dead (K706R) top1 double mutant has a faster growth rate than sgs1 top1, and this effect is dependent on direct interaction between the Sgs1 (K706R) mutant and Top3 (Weinstein and Rothstein, 2008). These genetic experiments are in agreement with the in vitro data presented here that defined a structural role for Sgs1 in promoting DNA catenation/decatenation by Top3.
A pair of converging replication forks can stall before completion, leaving a region of unreplicated DNA between the two forks. This unreplicated DNA (termed a late replication intermediate, Figure 7B) is topologically entangled (Suski and Marians, 2008; Wang, 1991). The activity of the Sgs1–Top3–Rmi1 complex and its homologs (Chan et al., 2007; Suski and Marians, 2008) can disentangle this structure. This occurs through decatenation of single strands of DNA, a property of Sgs1–Top3–Rmi1 and its homologues. The situation when only one DNA strand gets fully replicated before the other gives rise to another DNA intermediate with topologically linked single- and double-strands of DNA (Figure 7B). Based on the results presented here, the Sgs1–Top3–Rmi1 and RPA complex would be uniquely able to process this DNA structure. Full replication of both DNA strands would result in intertwined (catenated) dsDNA (Wang, 1991). This structure would be a good substrate for Top2, although our results presented here indicate that it could also be decatenated by the Sgs1–Top3–Rmi1 complex (Figure 7B). In this regard, it is relevant that human cells lacking the BLM helicase are specifically sensitive to ICRF 193 (Marple et al., 2006). This drug inhibits the ATPase activity of Topo II and, when used at lower concentrations, causes the accumulation of DNA catenanes associated with a transient G2 phase cell cycle arrest (Robinson et al., 2007). In contrast, BLM-deficient cells are not sensitive to etoposide (Marple et al., 2006), a drug that prevents DNA re-ligation by Topo II, resulting in DNA double strand breaks with the topoisomerase being covalently attached to the DNA ends, and presumably repair by recombination or NHEJ. These results suggest that BLM/Sgs1 could function in a backup pathway when dsDNA catenanes accumulate due to compromised dsDNA decatenation activity of Topo II.
The Sgs1–Top3–Rmi1 proteins and their homologues also clearly function in DNA repair, where they dissolve dHJs to complete HR, among other functions (Cejka et al., 2010a; Cejka et al., 2010b; Plank et al., 2006; Wu and Hickson, 2003). This occurs through convergent migration of the Holliday junctions by Sgs1–Top3 via unlinking of ssDNA (Cejka et al., 2010b). The last predicted intermediate in the dissolution pathway is a hemicatenane (Figure 7B). We showed previously that Rmi1 specifically promoted the latter unlinking steps of dissolution; in the absence of Top3 or Rmi1, a multiply-linked hemicatenane-like structure would persist. We have proposed that such structures may represent the stalled intermediates that form in top3 (or rmi1) mutants due to the essential function of these proteins in resolving catenanes linked by ssDNA. These intermediates may be responsible for the characteristic slow growth of top3 and rmi1 mutants, which would then be rescued by sgs1 mutation by preventing unwinding of dHJs into multiply-linked hemicatenanes (Liberi et al., 2005) and allowing alternative nucleolytic resolution pathways access to an interlinked pathological structure (Mankouri et al., 2011). The experiments presented here are compatible with this idea, in that they reveal the Sgs1–Top3–Rmi1 and RPA complex very efficiently unlinks hemicatenanes. Our results, implicating Sgs1-Top3-Rmi1 and RPA in the untangling of intermediates containing dsDNA, can now be used to relate the phenotypes associated with sgs1 mutations to their biochemical functions. Deletions in sgs1 reveal a genetic separation of recombination- and replication-associated functions (Bernstein et al., 2009); analysis of their biochemical activities would illuminate the complexities of Sgs1–Top3–Rmi1 and RPA function in the untangling of DNA as well as in other functions of these multi-faceted proteins.
In summary, we demonstrate here that the Sgs1–Top3–Rmi1 and RPA complex can decatenate a wide variety of topologically-linked DNA structures, both single- and double-stranded (Figure 7B). Due to the associated unwinding activity of Sgs1, and the regulatory roles of Rmi1 and RPA, the type IA topoisomerase, Top3, employs a distinctive mechanism to mediate and regulate topological transactions. By integrating these multiple activities, the Sgs1–Top3–Rmi1 and RPA complex can unlink 2-, 3-, and 4-stranded DNA intermediates that are generated in the course of repair and replication and, consequently, it possesses unique advantages relative to other topoisomerases in the broad spectrum of topologically linked molecules that it can disentangle and dissolve.
Experimental procedures
Catenation assay
Reactions (20 μl) contained 25 mM Tris-acetate (pH 7.5), 1 mM magnesium acetate, 37.5 mM NaCl, 0.1 mM dithiothreitol, 1 mM phosphoenolpyruvate, 100 μg/ml bovine serum albumin (New England Biolabs), 80 U/ml pyruvate kinase (Sigma), 1 mM ATP, 10 ng/μl pUC19 negative scDNA, and RPA or SSB, as indicated. Mixtures were assembled on ice, and Top3 and/or Rmi1 were added. Reactions were initiated by adding Sgs1 and shifting the temperature to 30 °C. After incubation for 30 minutes, unless noted otherwise, reactions were stopped with 6 μl STOP buffer (150 mM EDTA, 2% SDS, 30% glycerol, and 0.1% bromophenol blue) and 1 μl proteinase K (14–22 mg/ml, Roche) for 60 min at 37 °C. Where indicated, reactions were supplemented with 100 μM etoposide (Sigma). Products were analyzed by electrophoresis in 1% agarose TAE buffer (40 mM Tris-acetate (pH 8.0) and 1 mM EDTA) either with or without ethidium bromide (0.1 μg/ml). Where indicated, the deproteinized reaction products were purified by extraction with both phenol and phenol-chloroform and ethanol precipitation, and incubated with Sgs1, Top3, and Rmi1; T7 Endo I (New England Biolabs, in NEB buffer 2); or EcoRI (New England Biolabs, in NEB buffer 2), or DmTopo II (in 50 mM Tris-HCl (pH 7.5), 60 mM KCl, 10 mM MgCl2, 1 mM ATP, 0.1 mM EDTA, and 30 μg/ml bovine serum albumin).
Topo I-coupled unwinding of covalently closed plasmid DNA
Relaxed pUC19 dsDNA was prepared from negative scDNA using wheat germ Topo I (Promega). The DNA unwinding reactions were carried out and analyzed as described for catenation assays except 10 ng/μl pUC19 relaxed DNA, 2.3 μM RPA, 0.25 U/μl wheat germ Topo I and indicated Sgs1 were used, and NaCl was omitted.
Relaxation of negatively supercoiled DNA
Reactions contained 25 mM sodium HEPES (pH 7.0), 5 mM sodium acetate, 100 μg/ml bovine serum albumin, 0.2 mM EDTA, 0.3 mM magnesium acetate, 32% glycerol, 10 ng/μl pDHJS AN+ negative scDNA (Plank and Hsieh, 2006), and proteins as indicated. The reactions were assembled on ice and incubated at 42 °C for 30 minutes. Where indicated, the reactions were supplemented with 2 μl 5 M NaCl at the end of the 30 minutes incubation period for 5 minutes at 42 °C. The reactions were then stopped and analyzed as described above.
Supplementary Material
Highlights.
Sgs1, Rmi1, and RPA stimulate the type IA topoisomerase, Top3, to decatenate dsDNA.
Sgs1 and RPA have specific structural roles in DNA strand passage promoted by Top3.
Rmi1stabilizes the Top3-DNA covalent complex, and biases Top3 toward decatenation.
Concerted strand passage is a versatile biological pathway to disentangle linked DNA.
Acknowledgments
We would like to thank to Ian Hickson, Pavel Janscak, Amitabh Nimonkar, Jason Bell and Behzad Rad for purified proteins, the members of the Kowalczykowski laboratory for helpful discussions and critical reading of the manuscript. This work was supported by the Swiss National Science Foundation Fellowship PA00A-115375 (to P.C.), National Cancer Institute Award T32CA108459 (to J.L.P.), and C.C.D.), and by US National Institutes of Health grant GM-41347 (to S.C.K.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker NM, Rajan R, Mondragon A. Structural studies of type I topoisomerases. Nucleic Acids Res. 2009;37:693–701. doi: 10.1093/nar/gkn1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, Noirot-Gros MF, Wang JC. Interaction between yeast sgs1 helicase and DNA topoisomerase III. J Biol Chem. 2000;275:26898–26905. doi: 10.1074/jbc.M003137200. [DOI] [PubMed] [Google Scholar]
- Berger JM, Gamblin SJ, Harrison SC, Wang JC. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- Bernstein KA, Shor E, Sunjevaric I, Fumasoni M, Burgess RC, Foiani M, Branzei D, Rothstein R. Sgs1 function in the repair of DNA replication intermediates is separable from its role in homologous recombinational repair. EMBO J. 2009;28:915–925. doi: 10.1038/emboj.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzymek M, Thayer NH, Oh SD, Kleckner N, Hunter N. Double Holliday junctions are intermediates of DNA break repair. Nature. 2010;464:937–941. doi: 10.1038/nature08868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carotenuto W, Liberi G. Mitotic inter-homologue junctions accumulate at damaged DNA replication forks in recQ mutants. DNA Repair (Amst) 2010;9:661–669. doi: 10.1016/j.dnarep.2010.02.017. [DOI] [PubMed] [Google Scholar]
- Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010a;467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Kowalczykowski SC. The full-length Saccharomyces cerevisiae Sgs1 protein is a vigorous DNA helicase that preferentially unwinds Holliday junctions. J Biol Chem. 2010;285:8290–8301. doi: 10.1074/jbc.M109.083196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Plank JL, Bachrati CZ, Hickson ID, Kowalczykowski SC. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat Struct Mol Biol. 2010b;17:1377–1382. doi: 10.1038/nsmb.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26:3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- Chang M, Bellaoui M, Zhang C, Desai R, Morozov P, Delgado-Cruzata L, Rothstein R, Freyer GA, Boone C, Brown GW. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. EMBO J. 2005;24:2024–2033. doi: 10.1038/sj.emboj.7600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changela A, DiGate RJ, Mondragon A. Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule. Nature. 2001;411:1077–1081. doi: 10.1038/35082615. [DOI] [PubMed] [Google Scholar]
- Chen CF, Brill SJ. Binding and activation of DNA topoisomerase III by the Rmi1 subunit. J Biol Chem. 2007;282:28971–28979. doi: 10.1074/jbc.M705427200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 2003;22:4325–4336. doi: 10.1093/emboj/cdg391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B, Weisberg RA, Studier FW. Gene 3 endonuclease of bacteriophage T7 resolves conformationally branched structures in double-stranded DNA. J Mol Biol. 1987;193:359–376. doi: 10.1016/0022-2836(87)90224-5. [DOI] [PubMed] [Google Scholar]
- Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa N, Morimatsu K, Lovett ST, Kowalczykowski SC. Reconstitution of initial steps of dsDNA break repair by the RecF pathway of E. coli. Genes Dev. 2009;23:1234–1245. doi: 10.1101/gad.1780709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon FG, Brockman JP, Kowalczykowski SC. RecQ helicase stimulates both DNA catenation and changes in DNA topology by topoisomerase III. J Biol Chem. 2003;278:42668–42678. doi: 10.1074/jbc.M302994200. [DOI] [PubMed] [Google Scholar]
- Harmon FG, DiGate RJ, Kowalczykowski SC. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: a conserved mechanism for control of DNA recombination. Mol Cell. 1999;3:611–620. doi: 10.1016/s1097-2765(00)80354-8. [DOI] [PubMed] [Google Scholar]
- Harmon FG, Kowalczykowski SC. RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev. 1998;12:1134–1144. doi: 10.1101/gad.12.8.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow JK, Newman RH, Freemont PS, Hickson ID. Oligomeric ring structure of the Bloom’s syndrome helicase. Curr Biol. 1999;9:597–600. doi: 10.1016/s0960-9822(99)80264-4. [DOI] [PubMed] [Google Scholar]
- Kung VT, Wang JC. Purification and characterization of an omega protein from Micrococcus luteus. J Biol Chem. 1977;252:5398–5402. [PubMed] [Google Scholar]
- Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Mullen JR, Brill SJ, Kleff S, Romeo AM, Sternglanz R. Human homologues of yeast helicase. Nature. 1996;383:678–679. doi: 10.1038/383678a0. [DOI] [PubMed] [Google Scholar]
- Mankouri HW, Ashton TM, Hickson ID. Holliday junction-containing DNA structures persist in cells lacking Sgs1 or Top3 following exposure to DNA damage. Proc Natl Acad Sci U S A. 2011;108:4944–4949. doi: 10.1073/pnas.1014240108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri HW, Hickson ID. The RecQ helicase-topoisomerase III-Rmi1 complex: a DNA structure-specific ‘dissolvasome’? Trends Biochem Sci. 2007;32:538–546. doi: 10.1016/j.tibs.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Marple T, Kim TM, Hasty P. Embryonic stem cells deficient for Brca2 or Blm exhibit divergent genotoxic profiles that support opposing activities during homologous recombination. Mutat Res. 2006;602:110–120. doi: 10.1016/j.mrfmmm.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Nallaseth FS, Lan YQ, Slagle CE, Brill SJ. Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Mol Cell Biol. 2005;25:4476–4487. doi: 10.1128/MCB.25.11.4476-4487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci U S A. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plank JL, Hsieh TS. A novel, topologically constrained DNA molecule containing a double Holliday junction: design, synthesis, and initial biochemical characterization. J Biol Chem. 2006;281:17510–17516. doi: 10.1074/jbc.M602933200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plank JL, Wu J, Hsieh TS. Topoisomerase IIIα and Bloom’s helicase can resolve a mobile double Holliday junction substrate through convergent branch migration. Proc Natl Acad Sci U S A. 2006;103:11118–11123. doi: 10.1073/pnas.0604873103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralf C, Hickson ID, Wu L. The Bloom’s syndrome helicase can promote the regression of a model replication fork. J Biol Chem. 2006;281:22839–22846. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- Raynard S, Bussen W, Sung P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75. J Biol Chem. 2006;281:13861–13864. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- Raynard S, Zhao W, Bussen W, Lu L, Ding YY, Busygina V, Meetei AR, Sung P. Functional role of BLAP75 in BLM-topoisomerase IIIalpha-dependent Holliday junction processing. J Biol Chem. 2008;283:15701–15708. doi: 10.1074/jbc.M802127200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson HM, Bratlie-Thoresen S, Brown R, Gillespie DA. Chk1 is required for G2/M checkpoint response induced by the catalytic topoisomerase II inhibitor ICRF-193. Cell Cycle. 2007;6:1265–1267. doi: 10.4161/cc.6.10.4225. [DOI] [PubMed] [Google Scholar]
- Singh TR, Ali AM, Busygina V, Raynard S, Fan Q, Du CH, Andreassen PR, Sung P, Meetei AR. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev. 2008;22:2856–2868. doi: 10.1101/gad.1725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suski C, Marians KJ. Resolution of converging replication forks by RecQ and topoisomerase III. Mol Cell. 2008;30:779–789. doi: 10.1016/j.molcel.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Viard T, de la Tour CB. Type IA topoisomerases: a simple puzzle? Biochimie. 2007;89:456–467. doi: 10.1016/j.biochi.2006.10.013. [DOI] [PubMed] [Google Scholar]
- Wang JC. DNA topoisomerases: why so many? J Biol Chem. 1991;266:6659–6662. [PubMed] [Google Scholar]
- Weinstein J, Rothstein R. The genetic consequences of ablating helicase activity and the Top3 interaction domain of Sgs1. DNA Repair (Amst) 2008;7:558–571. doi: 10.1016/j.dnarep.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Bachrati CZ, Ou J, Xu C, Yin J, Chang M, Wang W, Li L, Brown GW, Hickson ID. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci U S A. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Davies SL, North PS, Goulaouic H, Riou JF, Turley H, Gatter KC, Hickson ID. The Bloom’s syndrome gene product interacts with topoisomerase III. J Biol Chem. 2000;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Xu D, Guo R, Sobeck A, Bachrati CZ, Yang J, Enomoto T, Brown GW, Hoatlin ME, Hickson ID, Wang W. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev. 2008;22:2843–2855. doi: 10.1101/gad.1708608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Sobeck A, Xu C, Meetei AR, Hoatlin M, Li L, Wang W. BLAP75, an essential component of Bloom’s syndrome protein complexes that maintain genome integrity. EMBO J. 2005;24:1465–1476. doi: 10.1038/sj.emboj.7600622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.