SUMMARY
The fidelity of chromosome segregation depends on the spindle assembly checkpoint (SAC). In the presence of unattached kinetochores, anaphase is delayed when three SAC components (Mad2, Mad3/BubR1, and Bub3) inhibit Cdc20, the activating subunit of the Anaphase-Promoting Complex (APC/C). We analyzed the role of Cdc20 autoubiquitination in the SAC of budding yeast. Reconstitution with purified components revealed that a Mad3-Bub3 complex synergizes with Mad2 to lock Cdc20 on the APC/C and stimulate Cdc20 autoubiquitination, while inhibiting ubiquitination of substrates. SAC-dependent Cdc20 autoubiquitination required the Mnd2/Apc15 subunit of the APC/C. General inhibition of Cdc20 ubiquitination in vivo resulted in high Cdc20 levels and a failure to establish a SAC arrest, suggesting that SAC establishment depends on low Cdc20 levels. Specific inhibition of SAC-dependent ubiquitination, by deletion of Mnd2, allowed establishment of a SAC arrest but delayed release from the arrest, suggesting that Cdc20 ubiquitination is also required for SAC inactivation.
INTRODUCTION
The spindle assembly checkpoint (SAC) ensures accurate chromosome segregation by delaying the onset of anaphase until all sister chromatids are properly bioriented on the mitotic spindle (Musacchio and Salmon, 2007). The SAC is a signaling system that senses defects in sister chromatid attachments at the kinetochore and blocks anaphase by inhibiting a ubiquitin ligase called the Anaphase-Promoting Complex or Cyclosome (APC/C) (Barford, 2011).
The APC/C, together with its activator subunit Cdc20, normally initiates anaphase by targeting securin and mitotic cyclins for ubiquitination, leading to their destruction by the proteasome. Securin destruction unleashes separase, which cleaves cohesin to initiate sister chromatid separation; cyclin destruction inactivates cyclin-dependent kinases, allowing dephosphorylation of their substrates and thus the completion of mitosis. In late mitosis, Cdc20 is replaced by a related activator subunit called Cdh1, which maintains APC/C activity in G1.
Cdc20 and Cdh1 are substrate adaptor subunits that recruit substrates to the APC/C core for ubiquitination (Barford, 2011). These substrates generally contain ‘Destruction-box (D-box)’ or ‘KEN-box’ sequence motifs that bind a WD40 domain in the activator. Cdc20 and Cdh1 also contain multiple binding motifs, including an N-terminal ‘C-box’ motif and a C-terminal Isoleucine-Arginine ‘IR’ motif, which mediate a very high affinity interaction with the APC/C (Figure S1A).
Cdc20 levels decrease rapidly in late mitosis during a normal cell cycle, and various lines of evidence suggest that this decrease is due to a combination of two mechanisms. First, Cdc20 turnover in late mitosis and G1 is mediated in part by the alternate activator Cdh1, which interacts with a D-box at the N-terminus of Cdc20 and thereby targets Cdc20 for ubiquitination (Prinz et al., 1998). However, Cdc20 levels decrease in late mitosis even when its D-box is mutated or in cells lacking Cdh1 (Foe et al., 2011; Robbins and Cross, 2011). Mutation of the IR motif of Cdc20 leads to stabilization of the protein in yeast cells (Thornton et al., 2006), and recent studies indicate that Cdc20 autoubiquitinates at significant rates while bound as an activator (Foe et al., 2011). Cdc20 autoubiquitination is also likely an important mechanism for promoting rapid Cdc20 turnover during a SAC arrest (Ge et al., 2009; Mansfeld et al., 2011; Nilsson et al., 2008; Pan and Chen, 2004; Varetti et al., 2011).
The key components of the SAC include the proteins Mad1, Mad2, Mad3 (BubR1 in vertebrates), and Bub3 (Musacchio and Salmon, 2007). The SAC signal is initiated by formation of a stable Mad1-Mad2 complex at unattached kinetochores (Kulukian et al., 2009; Shah et al., 2004). Mad2 within this complex interacts with soluble Mad2 molecules, thereby catalyzing the formation of a complex between soluble Mad2 and Cdc20 (De Antoni et al., 2005; Hewitt et al., 2010; Maldonado and Kapoor, 2011; Simonetta et al., 2009). Cdc20 also interacts with Mad3 and its tightly bound partner Bub3 (Hardwick et al., 2000). The interdependencies of these binding events are not well understood, but the final output is an inhibitory complex known as the mitotic checkpoint complex (MCC), which contains Cdc20, Mad2, Mad3/BubR1, and Bub3 (Sudakin et al., 2001).
The mechanism of APC/C inhibition by these proteins remains unclear. While Mad2 and Mad3-Bub3 can each inhibit APC/C activity in vitro, together these proteins are much more potent inhibitors (Fang, 2002; Fang et al., 1998; Kulukian et al., 2009; Tang et al., 2001). Early evidence suggested that the MCC sequesters Cdc20, preventing its binding to the APC/C. However, in subsequent work all proteins of the MCC were found to associate with the APC/C and block its activity, probably by inhibiting substrate binding (Braunstein et al., 2007; Chao et al., 2012; Herzog et al., 2009). The contribution of different MCC components is unclear, although Mad3/BubR1 acts in part as a pseudosubstrate inhibitor of Cdc20 (Burton and Solomon, 2007; Chao et al., 2012; Lara-Gonzalez et al., 2011; Malureanu et al., 2009).
APC/C inhibition during a SAC arrest might also depend on Cdc20 autoubiquitination. In budding yeast, the instability of Cdc20 during a SAC arrest requires Mad2 and Mad3 (King et al., 2007; Pan and Chen, 2004). Cdc20 is also degraded rapidly in human cells during a SAC arrest, and stabilization of Cdc20 (by mutation of all lysines in the protein) causes cells to bypass the arrest (Ge et al., 2009; Nilsson et al., 2008). These studies led to the suggestion that destruction of Cdc20 is required for inhibition of APC/C function during a SAC arrest, perhaps because it reduces Cdc20 levels below some threshold required for APC/C activation.
On the other hand, evidence from studies in human mitotic extracts led to speculation that Cdc20 ubiquitination promotes disassembly of the MCC and thus checkpoint inactivation (Reddy et al., 2007; Stegmeier et al., 2007). Consistent with this idea, numerous recent studies in human cells and cell lysates suggest that MCC disassembly and checkpoint inactivation depend on APC/C-dependent ubiquitination, ATP hydrolysis, and proteasomal function (Braunstein et al., 2007; Garnett et al., 2009; Jia et al., 2011; Ma and Poon, 2011; Miniowitz-Shemtov et al., 2010; Teichner et al., 2011; Varetti et al., 2011; Visconti et al., 2010; Williamson et al., 2009; Zeng et al., 2010). In vertebrates, MCC disassembly is also promoted by the Mad2-binding protein p31comet (Fava et al., 2011; Habu et al., 2002; Hagan et al., 2011; Jia et al., 2011; Mapelli et al., 2006; Reddy et al., 2007; Teichner et al., 2011; Varetti et al., 2011; Westhorpe et al., 2011; Xia et al., 2004; Yang et al., 2007). Recent studies also suggest that the human APC/C subunit, Apc15, is required for efficient MCC turnover on the APC/C (Mansfeld et al., 2011). However, the biochemical mechanisms underlying MCC disassembly remain poorly understood, and the contributions of Cdc20 autoubiquitination to APC/C function in the SAC are not clear.
Here we explored the mechanism and function of Cdc20 ubiquitination in the SAC of budding yeast. Studies with purified proteins revealed that Mad2 and Mad3-Bub3 synergize to effectively inhibit securin ubiquitination, while at the same time promoting Cdc20 autoubiquitination. This activity required the APC/C subunit Mnd2/Apc15. To test the role of Cdc20 ubiquitination in SAC function, we analyzed mutants that reduce Cdc20 ubiquitination. Using a Cdc20 mutant that is poorly ubiquitinated throughout the cell cycle, we found that Cdc20 turnover is required for establishment of a SAC arrest. On the other hand, specific inhibition of MCC-dependent Cdc20 autoubiquitination, achieved by deletion of Mnd2, allowed a checkpoint arrest but delayed release from the arrest. Our results argue that Cdc20 ubiquitination has multiple functions: it suppresses Cdc20 levels to allow establishment of the SAC arrest and is also required for efficient release from the arrest.
RESULTS
Mad2 and Mad3 have opposing effects on autoubiquitination
Cdc20 is a highly unstable protein throughout the cell cycle. We showed previously that Cdc20 turnover in a normal cell cycle depends primarily on APC/C-dependent ubiquitination that is distinct from that of a canonical substrate (Foe et al., 2011). We dissected Cdc20 ubiquitination by reconstituting the reaction with purified proteins. By incubating radiolabeled Cdc20 with purified APC/C and other ubiquitination components, we found that Cdc20 is rapidly ubiquitinated while bound in its activator position on the APC/C. Autoubiquitination was not greatly affected by mutation of the D-box or by addition of unlabeled Cdc20, but was inhibited by mutation of the IR or C-box sequence motifs (Figure S1B, bottom panel) (Foe et al., 2011). We also performed Cdh1-mediated Cdc20 ubiquitination reactions in parallel and the resulting Cdc20 ubiquitination depended only on its D-box (Figure S1B, middle panel). For clarity and ease of quantitation, we prevented polyubiquitin chain formation on Cdc20 by using methylated ubiquitin and the E2 Ubc4, which is a poor catalyst of polyubiquitin assembly (Figure S1C) (Rodrigo-Brenni and Morgan, 2007).
Cdc20 is turned over rapidly during a SAC arrest, and there is evidence that this turnover depends on Cdc20 autoubiquitination (Nilsson et al., 2008; Pan and Chen, 2004). Consistent with these results, we found that Cdc20 instability during a checkpoint arrest requires a functional APC/C and proteasome activity (Figure S2A), but is independent of the APC/C activator Cdh1 (Figure S2B) (Pan and Chen, 2004). In addition, checkpoint components are required for Cdc20 turnover (Figure S2C) (King et al., 2007; Pan and Chen, 2004).
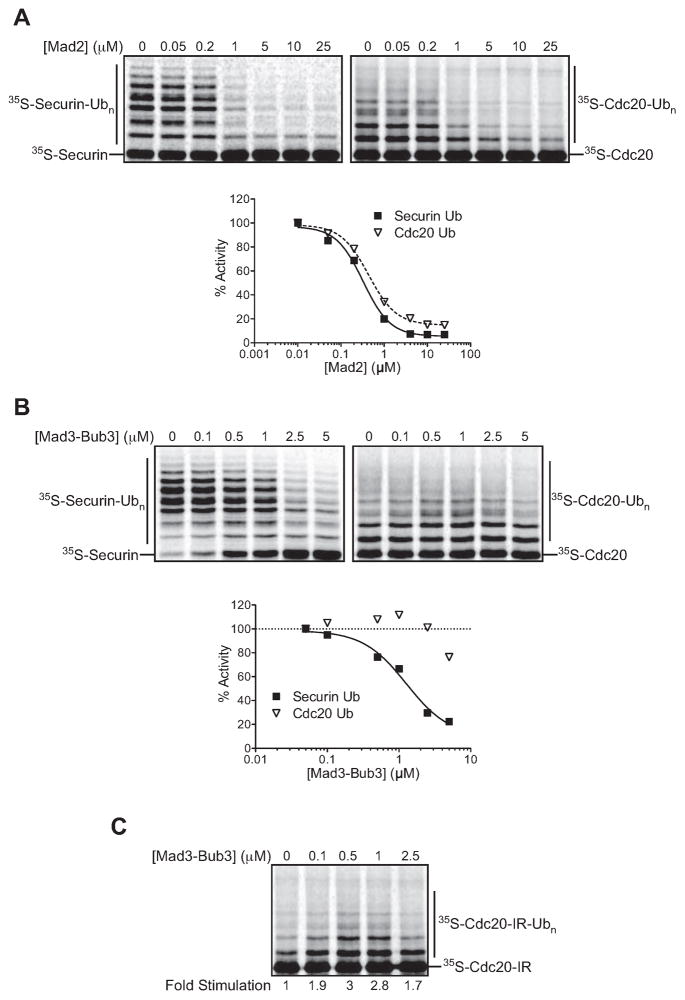
To explore the mechanisms underlying Cdc20 autoubiquitination in a SAC arrest, we used purified components to test the effects of checkpoint proteins on Cdc20 function and autoubiquitination. We first analyzed the role of Mad2 using recombinant protein purified from E. coli (Figure S3). As expected from previous studies, purified Mad2 inhibited Cdc20 activity toward securin (Figure 1A). Surprisingly, Mad2 had the same inhibitory effect on Cdc20 autoubiquitination (IC50 for both reactions was ~0.5 μM).
Figure 1. Mad2 and Mad3 have opposite effects on autoubiquitination.
(A) APC/C was immunopurified from lysates of CDC16-TAPcdh1Δ cells and used in ubiquitination reactions. Unlabeled Cdc20 (left) or 35S-Cdc20 (right), translated in and purifiedfrom rabbit reticulocyte lysates as ZZ-tagged proteins, was pre-incubated with the indicated Mad2 concentration before addition of APC/C (5 nM), E1/E2(Ubc4)/methyl-ubiquitin, and purified ZZ-tagged 35S-securin (left) or APC/C (5 nM) and E1/E2(Ubc4)/methyl-ubiquitin (right). Data were analyzed using Prism, with the zero concentration plotted on a log scale as 0.01 μM. Results are representative of two independent experiments.
(B) Purified unlabeled Cdc20 (left) or 35S-Cdc20 (right) was pre-incubated with the indicated Mad3-Bub3 concentration and APC/C (5 nM), as well as purified 35S-securin for the reactions at left. Reactions were started by the addition of E1/E2(Ubc4)/methyl-ubiquitin. The zero concentration was plotted on a log scale as 0.05 μM. Results are representative of two independent experiments.
(C) Purified 35S-Cdc20-IR (I609A, R610A) was pre-incubated with the indicated Mad3-Bub3 concentration and APC/C (5 nM). Reactions were started by the addition of E1/E2(Ubc4)/methyl-ubiquitin. Results are representative of two independent experiments.
Unlike Mad2, Mad3 expressed in E. coli was largely insoluble (Larsen et al., 2007). However, co-expression of Mad3 with Bub3 yielded a well-behaved complex (Figure S3). We found that this complex inhibited Cdc20-dependent securin ubiquitination (IC50 ~1 μM), but had little effect on autoubiquitination except at very high concentrations (Figure 1B).
To better characterize the Mad3-Bub3 effect on autoubiquitination, we used a sensitized Cdc20 mutant carrying a C-terminal IR motif mutation (Cdc20-IR) that reduces the affinity of Cdc20 for the APC/C and thereby reduces autoubiquitination (Figure S1B, bottom panel). The low level of Cdc20-IR autoubiquitination was stimulated by the Mad3-Bub3 complex (Figure 1C). Autoubiquitination was inhibited at the highest concentrations of Mad3-Bub3.
Checkpoint proteins regulate Cdc20 binding to the APC/C
The results in Figure 1 led us to hypothesize that the checkpoint proteins act, at least in part, by controlling the binding of Cdc20 to the APC/C. To test this possibility, we developed a quantitative Cdc20-APC/C binding assay, similar to a Cdh1-APC/C binding assay we described previously (Matyskiela and Morgan, 2009). In this assay, yeast APC/C is immunopurified on magnetic beads and incubated with radiolabeled Cdc20 prepared by translation in vitro, followed by washing to remove unbound Cdc20.
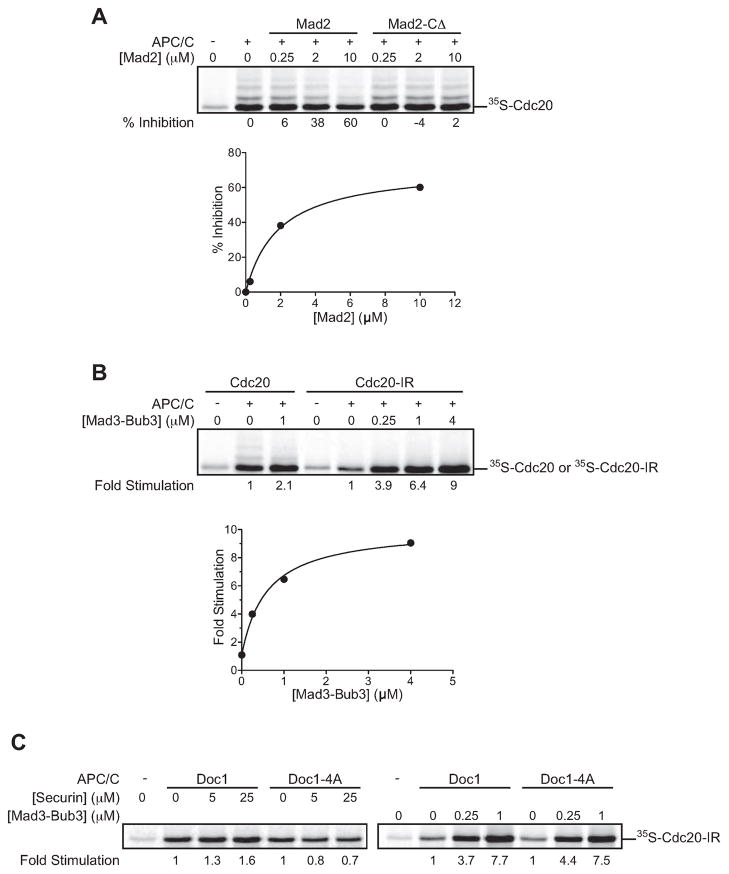
First, we tested the effect of Mad2 by pre-incubating Cdc20 with increasing concentrations of Mad2 or a Mad2 C-terminal deletion mutant (Mad2-CΔ) that is known to be defective in binding Cdc20 (Luo et al., 2000). Mad2 reduced Cdc20 binding to the APC/C, while Mad2-CΔ had no effect (Figure 2A). These results are likely to explain the ability of Mad2 to inhibit both securin ubiquitination and Cdc20 autoubiquitination (see Figure 1A).
Figure 2. Checkpoint proteins regulate Cdc20 binding to the APC/C.
(A) 35S-Cdc20 in reticulocyte lysate was pre-incubated with the indicated Mad2 or Mad2-CΔ concentration and added to TAP-APC/C beads. Following a 30 min incubation, beads were washed and the bound Cdc20 was analyzed by SDS-PAGE and autoradiography. Results are representative of two independent experiments.
(B) 35S-Cdc20 or 35S-Cdc20-IR in reticulocyte lysate was incubated with the indicated Mad3-Bub3 concentration and TAP-APC/C beads, and analyzed as in panel (A). Similar results were obtained in three independent experiments.
(C) 35S-Cdc20-IR in reticulocyte lysate was incubated with the indicated concentration of securin fragment (aa 1–110) or Mad3-Bub3 and TAP-APC/C beads immunopurified from DOC1 or doc1-4A strains. Results are representative of two independent experiments.
Our observation that Mad3-Bub3 stimulated autoubiquitination (Figure 1B, C) suggested that Mad3 might promote the association of Cdc20 with the APC/C. Indeed, addition of 1 μM Mad3-Bub3 induced a small (~2-fold) but reproducible stimulation of binding (Figure 2B). This small effect is perhaps not surprising because the affinity of Cdc20 for the APC/C is likely to be very high. To sensitize this assay, we used the Cdc20-IR mutation to reduce Cdc20 affinity for the APC/C. Mad3-Bub3 dramatically stimulated Cdc20-IR binding to the APC/C (Figure 2B), consistent with its ability to promote autoubiquitination of this mutant (Figure 1C).
In previous work, we showed that APC/C substrates stimulate the association of Cdh1 with the APC/C through a bivalent interaction between the activator and the APC/C core (Matyskiela and Morgan, 2009). This stimulation requires the core subunit Doc1/Apc10, which may interact directly with substrate (Buschhorn et al., 2011; Carroll and Morgan, 2002; da Fonseca et al., 2011; Passmore et al., 2003). Mutation of four residues within Doc1 (Doc1-4A) inhibits substrate binding and blocks the ability of substrate to stimulate Cdh1 binding to the APC/C (Carroll et al., 2005; Matyskiela and Morgan, 2009). Similar stimulation and Doc1-dependence was observed for Cdc20 binding to the APC/C using a fragment of securin (Figure 2C, left panel) (Matyskiela and Morgan, 2009). To determine if Mad3-Bub3 promotes activator binding by a similar mechanism, we tested whether Doc1 was required for the stimulation of Cdc20 binding by Mad3-Bub3. Mad3-Bub3 had the same effect on Cdc20-IR binding to Doc1 and Doc1-4A APC/C (Figure 2C, right panel), suggesting that Mad3-Bub3 associates with the APC/C-Cdc20 complex through contacts not involving the substrate-binding site on Doc1.
Mad2 and Mad3-Bub3 synergize to inhibit securin ubiquitination and promote Cdc20 autoubiquitination
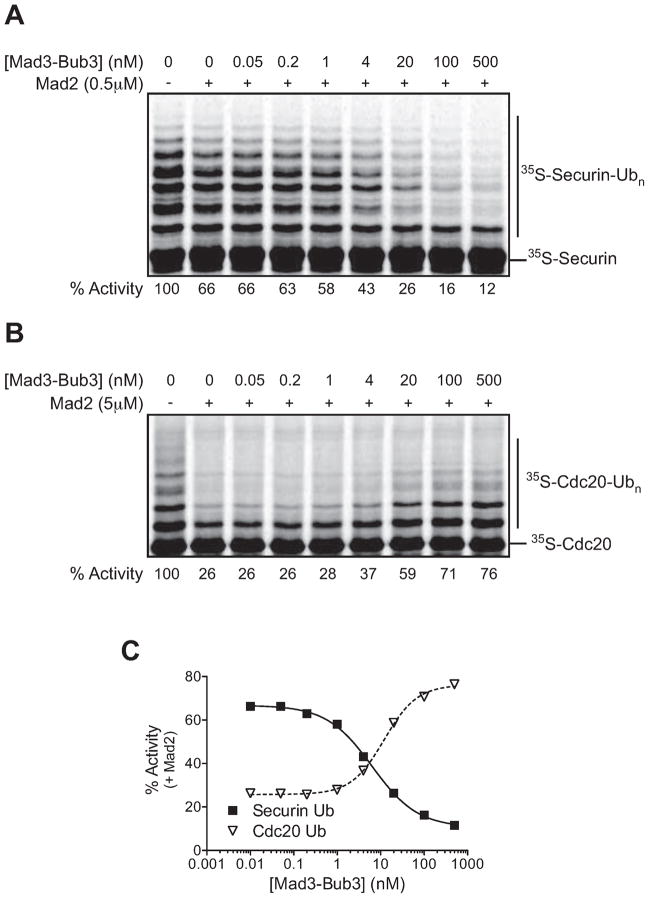
Mad2 and BubR1 (Mad3) are known to be poor inhibitors of vertebrate APC/C activity when tested individually, but inhibit at lower concentrations when added together (Fang, 2002). We tested this synergy using combinations of 0.5 μM Mad2 and increasing concentrations of the Mad3-Bub3 complex. The presence of Mad2 dramatically increased the potency of the Mad3-Bub3 complex, shifting the IC50 from ~1 μM to 6 nM (Figure 3A, C).
Figure 3. Mad2 and Mad3-Bub3 synergize to inhibit securin ubiquitination and allow autoubiquitination.
(A) Purified unlabeled Cdc20 was pre-incubated with 0.5 μM Mad2 and the indicated Mad3-Bub3 concentration. Reactions were started by the addition of purified 35S-securin, APC/C (5 nM), and E1/E2(Ubc4)/methyl-ubiquitin mix. Similar results were obtained in three independent experiments.
(B) Purified 35S-Cdc20 was pre-incubated with 5 μM Mad2 and the indicated Mad3-Bub3 concentration. Reactions were started by the addition of APC/C (5 nM) and E1/E2(Ubc4)/methyl-ubiquitin mix. Similar results were obtained in two independent experiments.
(C) The data from (A) and (B) were analyzed using Prism, and the zero concentration was plotted on a log scale as 0.01 nM.
Next we tested the combined effects of Mad2 and Mad3-Bub3 on Cdc20 autoubiquitination. Since we observed opposite effects of Mad2 and Mad3-Bub3 when they were tested individually (Figure 1), we were particularly interested in the possibility that Mad3-Bub3 might reverse the effect of Mad2. We used 5 μM Mad2, a concentration at which we observed clear inhibition of binding and autoubiquitination (Figure 1A and 2A). Strikingly, we found that Mad3-Bub3 reversed the inhibitory effect of Mad2 on Cdc20 autoubiquitination, and the dose response curve was the mirror image of the securin inhibition curve (Figure 3B, C): the IC50 of securin inhibition was similar to the EC50 for autoubiquitination (6 and 12 nM, respectively). We thus reconstituted the two major effects of the SAC – inhibition of securin ubiquitination and stimulation of Cdc20 autoubiquitination – at low concentrations of Mad3-Bub3. While Mad2 concentrations were higher, we suspect that the specific activity of our purified Mad2 preparations is low, and Mad2 is likely to be more active in vivo due to stimulation by the kinetochore-associated Mad1-Mad2 complex (De Antoni et al., 2005).
MCC-dependent Cdc20 autoubiquitination depends on the Mnd2 subunit of the APC/C
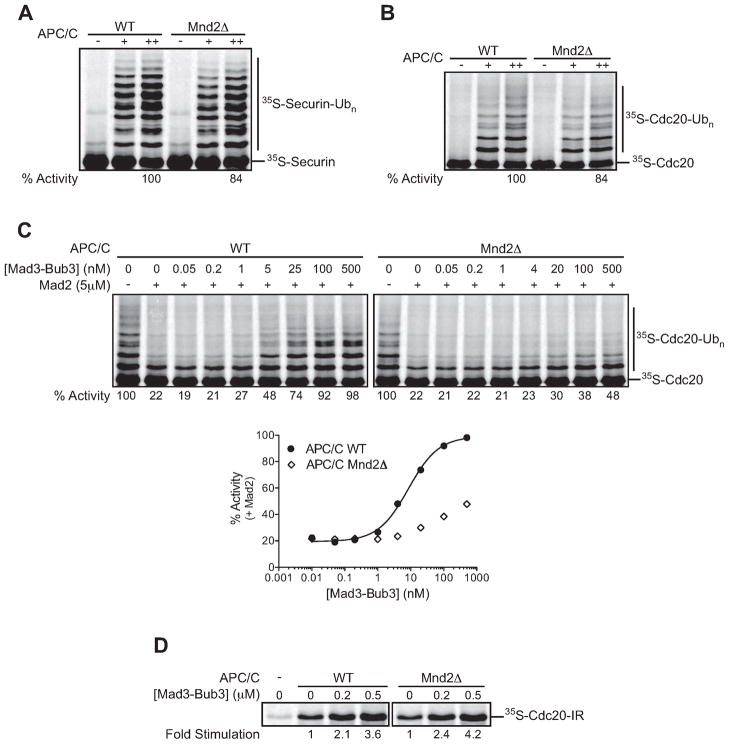
Recent studies in human cells suggest that Cdc20-MCC turnover during a SAC arrest depends on the nonessential APC/C subunit Apc15 (Mansfeld et al., 2011), but the effects of this subunit on Cdc20 autoubiquitination are not clear. The budding yeast homolog of Apc15 was suggested to be Mnd2 (Mansfeld et al., 2011). Mnd2 is known to be required for meiotic progression (Oelschlaegel et al., 2005; Penkner et al., 2005; Rabitsch et al., 2001) but is thought to have little, if any, effect in mitosis (Hall et al., 2003). To explore the roles of Mnd2 in yeast APC/C function and the SAC, we purified APC/C from an mnd2Δ strain. As previously shown, deletion of Mnd2 did not significantly affect the subunit composition of purified APC/C (Hall et al., 2003; Oelschlaegel et al., 2005). Also consistent with previous results, Mnd2Δ APC/C was similar to wild-type APC/C in its ability to target securin for ubiquitination in vitro, using Cdc20 as activator (Figure 4A) (Oelschlaegel et al., 2005). Mnd2Δ APC/C also displayed wild-type activity with the activator Cdh1 and with the E2 Ubc1 (J. Girard, unpublished results) (Oelschlaegel et al., 2005). Importantly, we found that Mnd2Δ APC/C also catalyzed Cdc20 autoubiquitination at wild-type rates in the absence of checkpoint components (Figure 4B).
Figure 4. MCC-dependent Cdc20 autoubiquitination depends on the Mnd2 subunit of the APC/C.
(A) APC/C was immunopurified from lysates of CDC16-TAPcdh1Δ (WT) or CDC16-TAPcdh1Δmnd2Δ cells and used in ubiquitination reactions with purified 35S-securin, APC/C (1 nM [+] or 5 nM [++]) or APC/C buffer (−) as a control, and E1/E2(Ubc4)/methyl-ubiquitin mix. Similar results were obtained in three independent experiments.
(B) Purified 35S-Cdc20 was incubated with wild-type (WT) or Mnd2ΔAP C/C (5 nM) and E1/E2(Ubc4)/methyl-ubiquitin mix. Similar results were obtained in three independent experiments.
(C) Purified 35S-Cdc20 was pre-incubated with 5 μM Mad2 and the indicated Mad3-Bub3 concentration. Reactions were started by the addition of wild-type (WT) or Mnd2Δ APC/C (5 nM) and E1/E2(Ubc4)/methyl-ubiquitin mix. Data were analyzed using Prism, and the zero concentration was plotted on a log scale as 0.01 nM. Similar results were obtained in two independent experiments.
(D) 35S-Cdc20-IR in reticulocyte lysate was incubated with the indicated concentration of Mad3-Bub3 and TAP-APC/C immunopurified from MND2 or mnd2Δ strains. Similar results were obtained in two independent experiments.
We also analyzed Cdc20 autoubiquitination in the presence of checkpoint proteins. We observed a striking defect in Cdc20 autoubiquitination with Mnd2Δ APC/C (Figure 4C), suggesting that the Mnd2 subunit is required for Cdc20 autoubiquitination in the context of the MCC, but not in its absence.
We next tested if the defect in Cdc20 autoubiquitination was due to a defect in Cdc20 binding to the APC/C in the context of the checkpoint proteins. Loss of the Mnd2 subunit had no detectable effect on the ability of Mad3-Bub3 to stimulate Cdc20 binding to the APC/C (Figure 4D), suggesting that MCC binding does not depend on Mnd2. Thus, reduced MCC-dependent Cdc20 autoubiquitination in the absence of Mnd2 does not result from poor Cdc20 binding but might instead result from a defect in the positioning of Cdc20 for autoubiquitination.
Stabilized Cdc20 allows bypass of the spindle assembly checkpoint
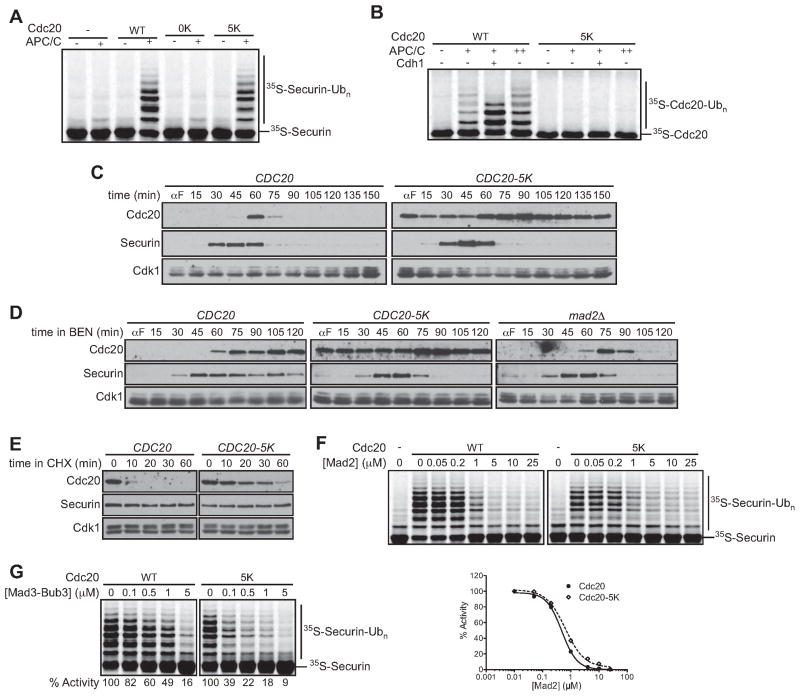
We next sought to explore the role of Cdc20 ubiquitination in vivo by creating mutations that prevent Cdc20 ubiquitination without affecting its activator function or interaction with checkpoint proteins. First, we generated a CDC20 mutant encoding a protein with all 39 lysines mutated to arginines (cdc20-0K mutant) (Figure S4A). Surprisingly, we found that this mutant rescued CDC20 function nearly as well as the wild-type gene at 25°C, implying that ubiquitination of Cdc20 is dispensable for viability (Figure S4B). However, the cdc20-0K mutant was temperature-sensitive and inviable at 37°C. Consistent with this sensitivity, we found that the Cdc20-0K protein was significantly less active toward securin in vitro (Figure 5A).
Figure 5. Stabilized Cdc20 allows bypass of the spindle assembly checkpoint.
(A) APC/C reactions were performed using purified unlabeled Cdc20 (WT), Cdc20-0K, Cdc20-5K, or a mock translation (−) incubated with APC/C (5 nM), 35S-securin and E1/E2(Ubc4)/methyl-ubiquitin.
(B) Reactions were performed using purified 35S-Cdc20 or 35S-Cdc20-5K, plus APC/C (1 nM [+] or 5 nM [++]). Recombinant His6-Cdh1 was purified from baculovirus-infected insect cells.
(C) Asynchronous log-phase cultures of strains carrying CDC20 or CDC20-5K at the endogenous locus were arrested in G1 with 1 μg/ml α-factor (αF) for 3 h. α-factor was washed out and cells were harvested at the indicated times. α-factor was re-added when a majority of the cells had budded. Samples were analyzed by western blotting with anti-Cdc20, anti-Myc (securin), and anti-Cdk1 (as a loading control). Similar results were observed by flow cytometry analysis of DNA content (data not shown).
(D) Asynchronous log-phase cultures of CDC20, CDC20-5K, or mad2Δ cells were arrested with α-factor and released into media containing 60 μg/ml benomyl. Cells were harvested at the indicated times and α-factor was re-added when a majority of the cells had budded. Samples were analyzed by western blotting.
(E) CDC20 or CDC20-5K strains, carrying a non-destructible securin mutant, PDS1- db, under the control of the GAL promoter, were arrested in benomyl in galactose-containing media to induce a metaphase arrest. 100 μg/ml Cycloheximide (CHX) was added and samples were analyzed by western blotting.
(F) Purified unlabeled Cdc20 (left) or Cdc20-5K (right) was pre-incubated with the indicated Mad2 concentration before addition of APC/C (5 nM), E1/E2(Ubc4)/methyl-ubiquitin, and purified 35S-securin. The (−) control represents background activity and was subtracted from activity in the presence of exogenous activator. The zero concentration was plotted on a log scale as 0.01 μM. Similar results were obtained in three independent experiments.
(G) Purified unlabeled Cdc20 (left) or Cdc20-5K (right) was pre-incubated with the indicated Mad3-Bub3 concentration and APC/C (5 nM). Reactions were started by the addition of 35S-securin and E1/E2(Ubc4)/methyl-ubiquitin. Results are representative of two independent experiments.
We hypothesized that specific lysines might be required for Cdc20 activity but not for ubiquitination. To identify these lysines, we carried out the analyses shown in Figure S4C and D. First, we generated three Cdc20 mutants, each with the lysines in one region (either N-terminal (N), middle (M), or C-terminal (C)) mutated to arginines (with the other regions remaining wild-type) (Figure S4A). Mutation of the nineteen C-terminal lysines (Cdc20-C mutant), but not mutation of lysines elsewhere in the protein, resulted in loss of activity towards securin in vitro (Figure S4C, left panel), suggesting that Cdc20 activity depends on lysines in the C-terminal region.
To identify specific C-terminal lysines required for activity, we analyzed Cdc20 proteins with single lysine-to-arginine mutations. Mutations at four lysines (K320, K431, K516, K550) each reduced Cdc20 activity toward securin over two-fold (Figure S4D). The K514R mutant also had a significant defect that became more pronounced when combined with the K516R mutation. These five residues fall within the predicted WD40 domain, and we suspect that these mutations affect the stability of this domain. To determine whether these five mutations caused the loss of function of the Cdc20-0K mutant, we generated a Cdc20-5K mutant in which these lysines were added back to the Cdc20-0K mutant. The Cdc20-5K protein exhibited near wild-type activity towards securin and other APC/CCdc20 substrates (Figure 5A, S4E), and the CDC20-5K mutant supported normal viability at all temperatures (Figure S4B).
We also analyzed ubiquitination of the Cdc20-N, M, and C mutants (Figure S4C, middle and right panels). Mutation of the 12 lysines in the N-terminal region (Cdc20-N mutant) resulted in significant loss of autoubiquitination, while only partially removing sites targeted by Cdh1. Autoubiquitination of human Cdc20 has also been mapped primarily to N-terminal lysines (Zeng and King, 2012). Autoubiquitination was also reduced in the Cdc20-C mutant, perhaps as a result of its general loss of stability (Figure S4C). Analysis of C-terminal lysine point mutants revealed that one mutation (K374R) reduced autoubiquitination despite having no apparent effect on activity (data not shown). Mutation of K374 reduced autoubiquitination in the Cdc20-N mutant, but two ubiquitination sites remained (Figure S4F).
Most importantly, we found that the Cdc20-5K protein was not detectably modified by either autoubiquitination or Cdh1-dependent ubiquitination in vitro(Figure 5B), indicating that this mutant lacks all major ubiquitination sites while retaining function as an APC/C activator (Figures 5A, S4B, S4E).
We constructed a strain in which the endogenous CDC20 was replaced with the CDC20-5K mutant. CDC20 and CDC20-5K strains were released from a G1 arrest into a single cell cycle. The Cdc20-5K protein was significantly stabilized, resulting in dampened oscillations with a peak mitotic level greater than that of the wild-type protein (Figure 5C). Oscillations in Cdc20-5K protein levels might be due to a low rate of ubiquitination in vivo on one or more of the five remaining lysines, which was not detectable in our ubiquitination studies in vitro. In addition, CDC20 transcription is known to peak in mitosis (Prinz et al., 1998). Despite the stabilization of the Cdc20-5K mutant protein, CDC20-5K cells progressed at wild-type rates through the cell cycle, undergoing DNA replication and mitosis at normal times (data not shown). Oscillations in securin levels appeared identical in the CDC20 and CDC20-5K strains (Figure 5C). Cdc20 ubiquitination and rapid turnover are therefore not required for normal cell-cycle progression.
We next tested the spindle checkpoint function of the CDC20-5K mutant by measuring sensitivity to the spindle poison benomyl. The CDC20-5K mutant showed a clear benomyl sensitivity that was similar to that of mad3Δ cells, but not as severe as that in the mad2Δ mutant (Figure S5).
We also arrested CDC20, CDC20-5K, mad2Δ, or mad3Δ strains in G1 and released the cells into high benomyl concentrations. In wild-type cells, the checkpoint arrest was accompanied by high levels of Cdc20 and securin (Figure 5D). However, the CDC20-5K, mad2Δ, and mad3Δ strains all failed to establish the SAC arrest, leading to destruction of securin with relatively normal timing (Figure 5D; mad3Δ data not shown). As in the normal cell cycle (Figure 5C), the Cdc20-5K protein was present at high levels in benomyl-treated mitotic cells. Consistent with this observation, Cdc20-5K was significantly stabilized in cells in which benomyl was added to cells arrested in metaphase by overexpression of a stabilized securin mutant (Figure 5E).
Studies with purified components revealed that the Cdc20-5K protein displayed wild-type sensitivity to Mad2 (Figure 5F). Mad3-Bub3 was also an effective inhibitor of securin ubiquitination by the mutant Cdc20-5K protein (Figure 5G). Thus, the checkpoint defect in CDC20-5K cells is not caused by a defect in the interaction of Cdc20-5K with checkpoint proteins. Instead, our results suggest that the stabilization of Cdc20 results in a defective spindle checkpoint, although they do not allow us to conclude whether this defect is in the establishment or maintenance of the checkpoint.
Mnd2 is required for efficient checkpoint release
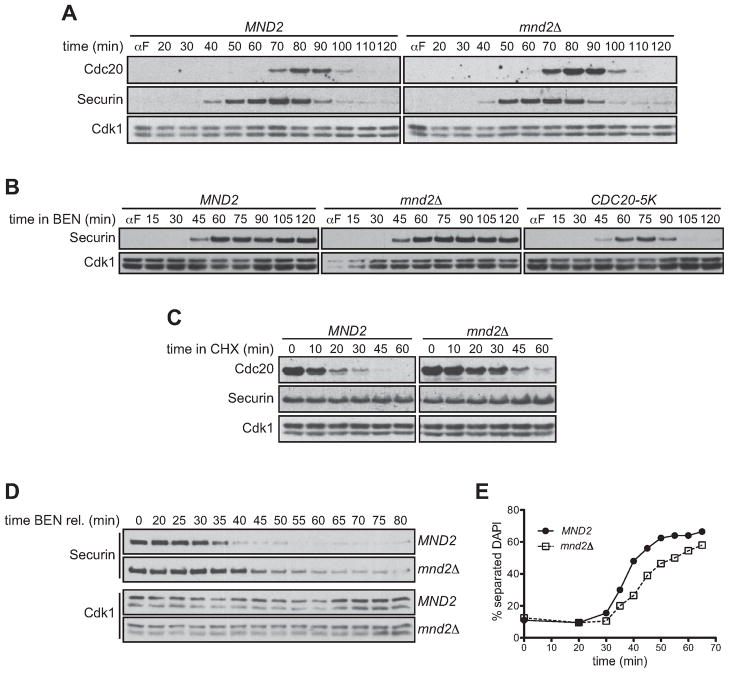
To further characterize the role of Cdc20 ubiquitination in SAC signaling, we sought a mutant that was specifically defective in Cdc20 ubiquitination during a checkpoint arrest, which would allow us to determine the importance of ubiquitination in maintenance of the arrest. The mnd2Δ strain seemed an ideal candidate for such a mutant, as our studies in Figure 4 had shown that Mnd2Δ APC/C is defective in Cdc20 autoubiquitination in the presence of checkpoint proteins but not in their absence.
Consistent with our evidence that removal of Mnd2 has no effect on APC/C activity in vitro (Figure 4A, B), we found that mnd2Δ cells released from G1 display no significant defects in progression through an unperturbed mitosis, with securin destruction occurring with normal timing (Figure 6A). Peak mitotic levels of Cdc20 were slightly elevated in mnd2Δ cells, suggesting that the nonessential SAC of budding yeast may be controlling Cdc20 levels in mitosis even in unperturbed cells. Importantly, the drop in Cdc20 levels after securin destruction was similar to that in wild-type cells, suggesting that Cdc20 turnover outside of the checkpoint was unaffected.
Figure 6. Mnd2 is required for efficient checkpoint release.
(A) Asynchronous log-phase cultures of MND2 or mnd2Δ cells were arrested in G1 with α-factor. α-factor was washed out and cells were harvested at the indicated times. αfactor was re-added when a majority of the cells had budded. Samples were analyzed by western blotting. Similar results were obtained by flow cytometry analysis of DNA content (data not shown).
(B) Asynchronous log-phase cultures of MND2, mnd2Δ, or CDC20-5K cells were arrested with α-factor and released into media containing 60 μg/ml benomyl. Cells were harvested at the indicated times and α-factor was re-added when a majority of the cells had budded. Samples were analyzed by western blotting.
(C) MND2 or mnd2Δ strains were arrested in benomyl before addition of cycloheximide. Samples were analyzed by western blotting.
(D) Asynchronous log-phase cultures of MND2 or mnd2Δ cells were arrested with αfactor and released into media containing 60 μg/ml benomyl. Cells were released from benomyl into α-factor and harvested at the indicated times. Samples were analyzed by western blotting.
(E) Cells from an experiment like that in panel (D) were analyzed for separation of DNA masses by DAPI staining. Two hundred cells were counted per time point. A similar delay was obtained by counting the percent of large budded cells and by flow cytometry analysis of DNA content (data not shown).
Interestingly, mnd2Δ cells displayed a normal SAC arrest when released from G1 into high benomyl concentrations (Figure 6B). In agreement with our studies showing that Mnd2 promotes SAC-dependent Cdc20 ubiquitination (Figure 4C), Cdc20 was stabilized in checkpoint-arrested mnd2Δ cells (Figure 6C), in contrast to the normal instability of Cdc20 in unperturbed cells (Figure 6A). We conclude that once the checkpoint is established, rapid Cdc20 turnover is not required to maintain the mitotic arrest.
The mnd2Δ mutant allowed us to test the possibility that Cdc20 ubiquitination promotes release from the arrest in budding yeast, as suggested by previous studies in human cells and cell lysates (Jia et al., 2011; Mansfeld et al., 2011; Reddy et al., 2007; Stegmeier et al., 2007; Varetti et al., 2011). Benomyl-arrested wild-type and mnd2Δ cells were released from the arrest by removal of benomyl, and then held in the following G1 to prevent progression into the next cell cycle. Securin destruction and chromosome segregation were delayed by approximately ten minutes, and were less abrupt, in mnd2Δ cells relative to wild-type control cells (Figure 6D, E). These results are consistent with the notion that Mnd2-dependent Cdc20 autoubiquitination is required for efficient inactivation of the spindle checkpoint.
DISCUSSION
The regulation of Cdc20, like that of most key regulatory proteins, depends on its short lifetime in the cell. This instability arises primarily from self-inhibition by autoubiquitination, which targets the protein for destruction in the proteasome. In a normal cell cycle, autoubiquitination is the primary mechanism underlying the drop in Cdc20 levels that occurs in late mitosis (Foe et al., 2011). Surprisingly, however, we found that cells expressing the poorly ubiquitinated Cdc20-5K mutant are viable and display no significant cell cycle defects, despite the presence in these cells of high Cdc20 levels that decline only slightly outside of mitosis. Thus, rapid Cdc20 turnover is not essential in a normal cell cycle. The same is not true in a spindle checkpoint, however. The CDC20-5K strain failed to establish a spindle checkpoint arrest, to a similar extent as mutants lacking the checkpoint genes MAD2 or MAD3.
A lysine-free form of human Cdc20 also bypasses the SAC (Nilsson et al., 2008), but it has been suggested that this bypass might be due to a slight decrease in Mad2 affinity caused by mutation of two lysines in the Mad2-binding region (Varetti et al., 2011). However, budding yeast Cdc20 does not contain these lysines and instead contains one lysine N-terminal of the conserved interaction motif (Luo et al., 2002). Furthermore, we found that wild-type Cdc20 and Cdc20-5K interact equally well with checkpoint proteins in vitro (Figures 5F, G). We therefore conclude that the yeast Cdc20-5K mutant is not defective in its interaction with checkpoint proteins.
How, then, does the Cdc20-5K mutant allow progression through mitosis despite activation of the checkpoint? The most likely possibility, which is consistent with previous evidence that moderate CDC20 overexpression drives yeast cells through a checkpoint arrest (Pan and Chen, 2004), is that the higher levels of Cdc20-5K protein somehow provide resistance to activated checkpoint proteins. One simple possibility is that checkpoint proteins are limiting, allowing high levels of Cdc20 to outnumber Mad2 to allow formation of active APC/C-Cdc20 complexes despite the checkpoint activation. This seems unlikely, however, given the ability of mnd2Δ cells to arrest normally despite accumulating higher Cdc20 levels within the arrest (Figure 6C). A more appealing possibility stems from our observation that Mad2 does not inhibit the activity of preformed APC/C-Cdc20 complexes in vitro (data not shown), presumably because the binding affinity of Cdc20 is so high that Mad2 cannot gain access to its binding site on Cdc20. Thus, the downregulation of Cdc20 from late mitosis through S phase may be required to keep Cdc20 levels at a low level that prevents the premature formation of checkpoint-resistant APC/C-Cdc20 complexes. Thus, our results argue that the degradation of Cdc20 outside mitosis is required for efficient establishment of a checkpoint arrest.
Checkpoint proteins are required for rapid Cdc20 turnover during a checkpoint arrest (King et al., 2007; Pan and Chen, 2004) (Figure S2C). To explore the underlying mechanism, we reconstituted the effects of Mad2 and the Mad3-Bub3 complex with purified components. We found that Mad2 alone inhibited Cdc20 binding and autoubiquitination. The incomplete effect of Mad2, even at apparently saturating concentrations, suggests that the target of Mad2 may be just one of the multiple contact points that mediate Cdc20 binding to the APC/C. Given that the Mad2 binding motif in Cdc20 lies in the amino-terminal region near the C-box, a likely possibility is that Mad2 somehow interferes with C-box function.
Checkpoint proteins clearly synergize in forming the Cdc20-Mad2-Mad3-Bub3 complex (MCC). Although Mad2 inhibits Cdc20 binding to the APC/C, the Mad3-Bub3 complex reverses this effect and stimulates Cdc20 binding and autoubiquitination. We find that the three budding yeast checkpoint proteins are sufficient to promote robust Cdc20 autoubiquitination in the absence of additional components, in contrast to the dependence on p31comet in human cells and lysates (Reddy et al., 2007; Varetti et al., 2011). Yeast do not contain a clear homolog of p31comet, and so vertebrate cells may have evolved additional control mechanisms. The synergistic actions of checkpoint proteins are likely to depend on interactions between Mad2 and Mad3, as suggested by recent studies supporting a direct interaction between purified human Mad2 and BubR1 (Tipton et al., 2011). Recent structural analysis of fission yeast MCC components (Mad3, Mad2, and Cdc20) also revealed Mad2-Mad3 interactions that would explain the synergistic effect (Chao et al., 2012).
Interestingly, the full set of checkpoint proteins has opposite effects on two APC/C-dependent activities: inhibition of securin ubiquitination and stimulation of Cdc20 autoubiquitination. Thus, the MCC is not a global inhibitor of the APC/C, but inhibits only its substrate-targeting function. How is this possible? KEN boxes in Mad3 were proposed to function as pseudosubstrate inhibitor motifs that interfere with substrate binding to Cdc20 (Burton and Solomon, 2007), and recent structural data provides evidence for engagement of a Mad3 KEN box by the WD40 domain of Cdc20 (Chao et al., 2012). Interestingly, other studies suggest that the MCC causes a shift in the position of Cdc20 on the APC/C, away from the Doc1/Apc10 subunit that contributes to substrate binding (Herzog et al., 2009; Izawa and Pines, 2011); thus, the MCC might reduce substrate binding in part by separating Cdc20 from Doc1. We found that the majority of autoubiquitination sites lie within the Cdc20 N-terminal region, which is predicted to be unstructured and also contains the C-box motif, which recent studies suggest could interact with the Apc2 subunit at a location that is close to the site of E2 binding – and thus in a good position to attack the E2-ubiquitin conjugate (da Fonseca et al., 2011), as also suggested recently for human Cdc20 (Zeng and King, 2012). Perhaps MCC binding shifts Cdc20 to a position that reduces substrate interactions while favoring autoubiquitination.
The stimulation of Cdc20-APC/C binding by Mad3-Bub3 suggests that Mad3 (and/or Bub3) interacts with the APC/C core. Structural analysis of the APC/C-MCC complex suggests that the MCC could contact multiple subunits (Herzog et al., 2009). We found that Doc1/Apc10 is not required for the stimulation of Cdc20 binding by Mad3, suggesting that this subunit does not contribute to MCC binding – and consistent with the notion, mentioned above, that the MCC shifts Cdc20 away from Doc1. We also tested another nonessential subunit, Mnd2, based on recent evidence that a related human subunit, Apc15, is required for Cdc20-MCC turnover in the checkpoint (Mansfeld et al., 2011). Mnd2 has been suggested to interact with Apc1, Apc5, and Cdc23 (Hall et al., 2003), three subunits in the APC/C region where the MCC appears to bind (Chao et al., 2012; Herzog et al., 2009; Schreiber et al., 2011). Furthermore, Cdc23/Apc8 is particularly important in Cdc20 binding in the checkpoint (Izawa and Pines, 2011). Deletion of Mnd2 had a striking and specific effect: autoubiquitination and activity toward securin were largely unaffected in the absence of checkpoint proteins, but the loss of Mnd2 blocked the ability of Mad3-Bub3 to stimulate autoubiquitination in the presence of Mad2. Interestingly, we found that the loss of Mnd2 did not prevent the stimulation of Cdc20 binding to the APC by Mad3-Bub3. These results argue strongly that Mnd2 (and perhaps Apc15 in the human APC/C) is required for the MCC to shift the position of Cdc20 for checkpoint-induced autoubiquitination.
Surprisingly, despite the rapid turnover of Cdc20 that occurs in checkpoint-arrested cells, steady-state levels of Cdc20 appear constant (Figure 5D). Thus, a high rate of mitotic Cdc20 synthesis balances increased destruction. As recently proposed (Varetti et al., 2011), this constant flux of Cdc20 is likely to be important for reversing the effects of the checkpoint when all sister-chromatid pairs achieve correct spindle attachment. Cells lacking MND2 provided us with an effective approach to explore this possibility. In these cells, the lack of Mnd2 did not greatly affect Cdc20 oscillations in a normal cell cycle, and thus these cells do not have the general increase in Cdc20 levels that we observed in the CDC20-5K cells. Instead, mnd2Δ cells displayed a more specific defect in autoubiquitination and Cdc20 turnover in the presence of checkpoint proteins. These cells establish and maintain a checkpoint arrest, indicating that MCC-dependent autoubiquitination and rapid Cdc20 degradation are not required for the arrest. These results also support the notion, discussed above, that the Cdc20-5K mutant bypasses the arrest because of its high levels throughout the cell cycle.
Although mnd2Δ cells arrest in the checkpoint, they are less efficient than wild-type cells in inactivation of the checkpoint when spindle poisons are removed, as observed in human cells depleted of Apc15 (Mansfeld et al., 2011). Thus, inactivation of the checkpoint might depend, at least in part, on MCC-dependent Cdc20 autoubiquitination. How does ubiquitination promote checkpoint inactivation? In purified reactions, we have not seen evidence that polyubiquitination causes dissociation of Cdc20 from the checkpoint complex and APC/C (Figure S6). Instead, we suspect that MCC removal is an active process mediated in part by the proteasome and other factors, as suggested by recent evidence for the involvement of ATP hydrolysis and proteolysis (Ma and Poon, 2011; Miniowitz-Shemtov et al., 2010; Teichner et al., 2011; Visconti et al., 2010; Zeng et al., 2010). In vertebrates, checkpoint inactivation also depends on the protein p31comet, which has been proposed to provide a functionally redundant mechanism for driving MCC disassembly (Jia et al., 2011). We speculate that when the SAC signal is extinguished, the ATP-dependent removal of polyubiquitinated Cdc20, together with its checkpoint partners, helps allow newly synthesized Cdc20 to reactivate the APC/C. In this way, the dynamic features of the checkpoint – balanced high rates of Cdc20 synthesis and destruction – allow cells to rapidly initiate anaphase upon checkpoint inactivation.
EXPERIMENTAL PROCEDURES
Yeast Methods
See Table S1 for yeast strains. All strains were derivatives of W303. Synchronization and analysis of yeast cultures is described in Supplemental Experimental Procedures.
APC/C Assays
APC/C was purified from a TAP-CDC16cdh1Δ strain. E1, E2 (Ubc4 or Ubc1), APC/C, and Cdh1 were expressed and purified as previously described (Carroll and Morgan, 2005; Rodrigo-Brenni and Morgan, 2007). Cdc20, securin, Acm1, Dbf4 (amino acids 1–236), and Clb5 (with a C-terminal 3HA-tag) were cloned into plasmids containing a T7 promoter and an N-terminal or C-terminal ZZ tag. ZZ-tagged proteins were generated in vitro with TnT T7 Quick Coupled Transcription/Translation Systems (Promega) either in the presence of 35S-methionine or unlabeled methionine. ZZ-tagged proteins were purified from the reticulocyte lysate using IgG-coupled Dynabeads (Invitrogen) and cleaved using TEV protease. E2 charging was performed in the presence of E1 (Uba1, 300 nM), E2 (Ubc4 or Ubc1, 50 μM), ubiquitin (wild-type, K48R, or methyl-ubiquitin; Boston Biochem, 150 μM), and ATP (1 mM) for 20 min. Reactions were initiated by the addition of charged E2, APC/C (1–5 nM), purified Cdc20, and purified securin. Reactions were performed at 23°C for the indicated time. For reactions containing Mad2, Cdc20 was pre-incubated with the indicated final concentration of Mad2 before addition of other components. For Mad3 reactions, Cdc20 was pre-incubated with the APC/C and the indicated final concentration of Mad3-Bub3 before addition of other components. All reactions were stopped by the addition of SDS sample buffer, separated by SDS-PAGE, and visualized and quantified with a Molecular Dynamics PhosphorImager and ImageQuant (Amersham Biosciences/GE Healthcare).
Cdc20-APC/C binding assays
APC/C was immunopurified from TAP-CDC16cdh1Δ strains using IgG beads. APC/C-bound beads or beads incubated with untagged lysate were incubated with in vitro translated 35S-Cdc20. Immunoprecipitates were washed two times to remove unbound proteins and bound proteins were eluted with SDS sample buffer. For Mad2 experiments, Cdc20 was pre-incubated with the indicated final concentration of Mad2 or Mad2-CΔ before addition of beads. The securin fragment was generated as previously described (Matyskiela and Morgan, 2009).
Supplementary Material
Highlights.
Spindle checkpoint proteins synergize to stimulate Cdc20 autoubiquitination.
Mnd2/Apc15 is required for Cdc20 autoubiquitination in the checkpoint.
Cdc20 stabilization throughout the cycle prevents spindle checkpoint establishment.
Mnd2 delete cells maintain a checkpoint arrest, but delay checkpoint inactivation.
Acknowledgments
We thank H. Eshelman, M. Galli, J. Girard, L. Holt, M. Matyskiela, M. Lopez, D. Lu, N. Lyons, S. Naylor, M. Rodrigo-Brenni, J. Schaefer, V. Van Voorhis, and G. Yaakov for discussions; M. Lopez and M. Matyskiela for comments on the manuscript; M. Matyskiela for help with assay development and optimization; and D. Lu for help with data processing. This work was supported by funding from the National Institute of General Medical Sciences (GM053270).
Footnotes
Supplemental data include 6 figures, a table, and supplemental experimental procedures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barford D. Structure, function and mechanism of the anaphase promoting complex (APC/C) Q Rev Biophys. 2011;44:153–190. doi: 10.1017/S0033583510000259. [DOI] [PubMed] [Google Scholar]
- Braunstein I, Miniowitz S, Moshe Y, Hershko A. Inhibitory factors associated with anaphase-promoting complex/cyclosome in mitotic checkpoint. Proc Natl Acad Sci U S A. 2007;104:4870–4875. doi: 10.1073/pnas.0700523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JL, Solomon MJ. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev. 2007;21:655–667. doi: 10.1101/gad.1511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschhorn BA, Petzold G, Galova M, Dube P, Kraft C, Herzog F, Stark H, Peters JM. Substrate binding on the APC/C occurs between the coactivator Cdh1 and the processivity factor Doc1. Nat Struct Mol Biol. 2011;18:6–13. doi: 10.1038/nsmb.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Enquist-Newman M, Morgan DO. The APC subunit Doc1 promotes recognition of the substrate destruction box. Curr Biol. 2005;15:11–18. doi: 10.1016/j.cub.2004.12.066. [DOI] [PubMed] [Google Scholar]
- Carroll CW, Morgan DO. The Doc1 subunit is a processivity factor for the anaphase-promoting complex. Nat Cell Biol. 2002;4:880–887. doi: 10.1038/ncb871. [DOI] [PubMed] [Google Scholar]
- Carroll CW, Morgan DO. Enzymology of the Anaphase-Promoting Complex. Meth Enzymol. 2005;398:219–230. doi: 10.1016/S0076-6879(05)98018-X. [DOI] [PubMed] [Google Scholar]
- Chao WC, Kulkarni K, Zhang Z, Kong EH, Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484:208–213. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- da Fonseca PC, Kong EH, Zhang Z, Schreiber A, Williams MA, Morris EP, Barford D. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2011;470:274–278. doi: 10.1038/nature09625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Antoni A, Pearson CG, Cimini D, Canman JC, Sala V, Nezi L, Mapelli M, Sironi L, Faretta M, Salmon ED, et al. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol. 2005;15:214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- Fang G. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol Biol Cell. 2002;13:755–766. doi: 10.1091/mbc.01-09-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fava LL, Kaulich M, Nigg EA, Santamaria A. Probing the in vivo function of Mad1:C-Mad2 in the spindle assembly checkpoint. EMBO J. 2011;30:3322–3336. doi: 10.1038/emboj.2011.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe IT, Foster SA, Cheung SK, Deluca SZ, Morgan DO, Toczyski DP. Ubiquitination of Cdc20 by the APC Occurs through an Intramolecular Mechanism. Curr Biol. 2011;21:1870–1877. doi: 10.1016/j.cub.2011.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett MJ, Mansfeld J, Godwin C, Matsusaka T, Wu J, Russell P, Pines J, Venkitaraman AR. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat Cell Biol. 2009;11:1363–1369. doi: 10.1038/ncb1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S, Skaar JR, Pagano M. APC/C- and Mad2-mediated degradation of Cdc20 during spindle checkpoint activation. Cell Cycle. 2009;8:167–171. doi: 10.4161/cc.8.1.7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habu T, Kim SH, Weinstein J, Matsumoto T. Identification of a MAD2-binding protein, CMT2, and its role in mitosis. EMBO J. 2002;21:6419–6428. doi: 10.1093/emboj/cdf659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan RS, Manak MS, Buch HK, Meier MG, Meraldi P, Shah JV, Sorger PK. p31(comet) acts to ensure timely spindle checkpoint silencing subsequent to kinetochore attachment. Mol Biol Cell. 2011;22:4236–4246. doi: 10.1091/mbc.E11-03-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MC, Torres MP, Schroeder GK, Borchers CH. Mnd2 and Swm1 are core subunits of the Saccharomyces cerevisiae anaphase-promoting complex. J Biol Chem. 2003;278:16698–16705. doi: 10.1074/jbc.M213109200. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Johnston RC, Smith DL, Murray AW. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J Cell Biol. 2000;148:871–882. doi: 10.1083/jcb.148.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog F, Primorac I, Dube P, Lenart P, Sander B, Mechtler K, Stark H, Peters JM. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 2009;323:1477–1481. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt L, Tighe A, Santaguida S, White AM, Jones CD, Musacchio A, Green S, Taylor SS. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J Cell Biol. 2010;190:25–34. doi: 10.1083/jcb.201002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa D, Pines J. How APC/C-Cdc20 changes its substrate specificity in mitosis. Nat Cell Biol. 2011;13:223–233. doi: 10.1038/ncb2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Li B, Warrington RT, Hao X, Wang S, Yu H. Defining pathways of spindle checkpoint silencing: functional redundancy between Cdc20 ubiquitination and p31(comet) Mol Biol Cell. 2011;22:4227–4235. doi: 10.1091/mbc.E11-05-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King EM, van der Sar SJ, Hardwick KG. Mad3 KEN boxes mediate both Cdc20 and Mad3 turnover, and are critical for the spindle checkpoint. PLoS One. 2007;2:e342. doi: 10.1371/journal.pone.0000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulukian A, Han JS, Cleveland DW. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev Cell. 2009;16:105–117. doi: 10.1016/j.devcel.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Gonzalez P, Scott MI, Diez M, Sen O, Taylor SS. BubR1 blocks substrate recruitment to the APC/C in a KEN-box-dependent manner. J Cell Sci. 2011;124:4332–4345. doi: 10.1242/jcs.094763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen NA, Al-Bassam J, Wei RR, Harrison SC. Structural analysis of Bub3 interactions in the mitotic spindle checkpoint. Proc Natl Acad Sci U S A. 2007;104:1201–1206. doi: 10.1073/pnas.0610358104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Fang G, Coldiron M, Lin Y, Yu H, Kirschner MW, Wagner G. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat Struct Biol. 2000;7:224–229. doi: 10.1038/73338. [DOI] [PubMed] [Google Scholar]
- Luo X, Tang Z, Rizo J, Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol Cell. 2002;9:59–71. doi: 10.1016/s1097-2765(01)00435-x. [DOI] [PubMed] [Google Scholar]
- Ma HT, Poon RY. Orderly inactivation of the key checkpoint protein mitotic arrest deficient 2 (MAD2) during mitotic progression. J Biol Chem. 2011;286:13052–13059. doi: 10.1074/jbc.M110.201897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado M, Kapoor TM. Constitutive Mad1 targeting to kinetochores uncouples checkpoint signalling from chromosome biorientation. Nat Cell Biol. 2011;13:475–482. doi: 10.1038/ncb2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malureanu LA, Jeganathan KB, Hamada M, Wasilewski L, Davenport J, van Deursen JM. BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in interphase. Dev Cell. 2009;16:118–131. doi: 10.1016/j.devcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfeld J, Collin P, Collins MO, Choudhary JS, Pines J. APC15 drives the turnover of MCC-CDC20 to make the spindle assembly checkpoint responsive to kinetochore attachment. Nat Cell Biol. 2011;13:1234–1243. doi: 10.1038/ncb2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapelli M, Filipp FV, Rancati G, Massimiliano L, Nezi L, Stier G, Hagan RS, Confalonieri S, Piatti S, Sattler M, et al. Determinants of conformational dimerization of Mad2 and its inhibition by p31comet. EMBO J. 2006;25:1273–1284. doi: 10.1038/sj.emboj.7601033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyskiela ME, Morgan DO. Analysis of activator-binding sites on the APC/C supports a cooperative substrate-binding mechanism. Mol Cell. 2009;34:68–80. doi: 10.1016/j.molcel.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miniowitz-Shemtov S, Teichner A, Sitry-Shevah D, Hershko A. ATP is required for the release of the anaphase-promoting complex/cyclosome from inhibition by the mitotic checkpoint. Proc Natl Acad Sci U S A. 2010;107:5351–5356. doi: 10.1073/pnas.1001875107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Nilsson J, Yekezare M, Minshull J, Pines J. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat Cell Biol. 2008;10:1411–1420. doi: 10.1038/ncb1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelschlaegel T, Schwickart M, Matos J, Bogdanova A, Camasses A, Havlis J, Shevchenko A, Zachariae W. The yeast APC/C subunit Mnd2 prevents premature sister chromatid separation triggered by the meiosis-specific APC/C-Ama1. Cell. 2005;120:773–788. doi: 10.1016/j.cell.2005.01.032. [DOI] [PubMed] [Google Scholar]
- Pan J, Chen RH. Spindle checkpoint regulates Cdc20p stability in Saccharomyces cerevisiae. Genes Dev. 2004;18:1439–1451. doi: 10.1101/gad.1184204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, McCormack EA, Au SW, Paul A, Willison KR, Harper JW, Barford D. Doc1 mediates the activity of the anaphase-promoting complex by contributing to substrate recognition. Embo J. 2003;22:786–796. doi: 10.1093/emboj/cdg084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkner AM, Prinz S, Ferscha S, Klein F. Mnd2, an essential antagonist of the anaphase-promoting complex during meiotic prophase. Cell. 2005;120:789–801. doi: 10.1016/j.cell.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Prinz S, Hwang ES, Visintin R, Amon A. The regulation of Cdc20 proteolysis reveals a role for the APC components Cdc23 and Cdc27 during S phase and early mitosis. Curr Biol. 1998;8:750–760. doi: 10.1016/s0960-9822(98)70298-2. [DOI] [PubMed] [Google Scholar]
- Rabitsch KP, Toth A, Galova M, Schleiffer A, Schaffner G, Aigner E, Rupp C, Penkner AM, Moreno-Borchart AC, Primig M, et al. A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr Biol. 2001;11:1001–1009. doi: 10.1016/s0960-9822(01)00274-3. [DOI] [PubMed] [Google Scholar]
- Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- Robbins JA, Cross FR. Regulated degradation of the APC coactivator Cdc20. Cell Div. 2011;5:23. doi: 10.1186/1747-1028-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Brenni M, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–139. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Schreiber A, Stengel F, Zhang Z, Enchev RI, Kong EH, Morris EP, Robinson CV, da Fonseca PC, Barford D. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature. 2011;470:227–232. doi: 10.1038/nature09756. [DOI] [PubMed] [Google Scholar]
- Shah JV, Botvinick E, Bonday Z, Furnari F, Berns M, Cleveland DW. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol. 2004;14:942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- Simonetta M, Manzoni R, Mosca R, Mapelli M, Massimiliano L, Vink M, Novak B, Musacchio A, Ciliberto A. The influence of catalysis on mad2 activation dynamics. PLoS Biol. 2009;7:e10. doi: 10.1371/journal.pbio.1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Rape M, Draviam VM, Nalepa G, Sowa ME, Ang XL, McDonald ER, 3rd, Li MZ, Hannon GJ, Sorger PK, et al. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446:876–881. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Bharadwaj R, Li B, Yu H. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev Cell. 2001;1:227–237. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- Teichner A, Eytan E, Sitry-Shevah D, Miniowitz-Shemtov S, Dumin E, Gromis J, Hershko A. p31comet Promotes disassembly of the mitotic checkpoint complex in an ATP-dependent process. Proc Natl Acad Sci U S A. 2011;108:3187–3192. doi: 10.1073/pnas.1100023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton BR, Ng TM, Matyskiela ME, Carroll CW, Morgan DO, Toczyski DP. An architectural map of the anaphase-promoting complex. Genes Dev. 2006;20:449–460. doi: 10.1101/gad.1396906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton AR, Wang K, Link L, Bellizzi JJ, Huang H, Yen T, Liu ST. BUBR1 and closed MAD2 (C-MAD2) interact directly to assemble a functional mitotic checkpoint complex (MCC) J Biol Chem. 2011;286:21173–21179. doi: 10.1074/jbc.M111.238543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varetti G, Guida C, Santaguida S, Chiroli E, Musacchio A. Homeostatic control of mitotic arrest. Mol Cell. 2011;44:710–720. doi: 10.1016/j.molcel.2011.11.014. [DOI] [PubMed] [Google Scholar]
- Visconti R, Palazzo L, Grieco D. Requirement for proteolysis in spindle assembly checkpoint silencing. Cell Cycle. 2010;9:564–569. doi: 10.4161/cc.9.3.10581. [DOI] [PubMed] [Google Scholar]
- Westhorpe FG, Tighe A, Lara-Gonzalez P, Taylor SS. p31comet-mediated extraction of Mad2 from the MCC promotes efficient mitotic exit. J Cell Sci. 2011;124:3905–3916. doi: 10.1242/jcs.093286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc Natl Acad Sci U S A. 2009;106:18213–18218. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia G, Luo X, Habu T, Rizo J, Matsumoto T, Yu H. Conformation-specific binding of p31(comet) antagonizes the function of Mad2 in the spindle checkpoint. EMBO J. 2004;23:3133–3143. doi: 10.1038/sj.emboj.7600322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Li B, Tomchick DR, Machius M, Rizo J, Yu H, Luo X. p31comet blocks Mad2 activation through structural mimicry. Cell. 2007;131:744–755. doi: 10.1016/j.cell.2007.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, King RW. An APC/C inhibitor stabilizes cyclin B1 by prematurely terminating ubiquitination. Nat Chem Biol. 2012;8:383–392. doi: 10.1038/nchembio.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Sigoillot F, Gaur S, Choi S, Pfaff KL, Oh DC, Hathaway N, Dimova N, Cuny GD, King RW. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell. 2010;18:382–395. doi: 10.1016/j.ccr.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.