SUMMARY
Heart development is exquisitely sensitive to the precise temporal regulation of thousands of genes that govern developmental decisions during differentiation. However, we currently lack a detailed understanding of how chromatin and gene expression patterns are coordinated during developmental transitions in the cardiac lineage. Here, we interrogated the transcriptome and several histone modifications across the genome during defined stages of cardiac differentiation. We find distinct chromatin patterns that are coordinated with stage-specific expression of functionally related genes, including many human disease-associated genes. Moreover, we discover a novel pre-activation chromatin pattern at the promoters of genes associated with heart development and cardiac function. We further identify stage-specific distal enhancer elements and find enriched DNA binding motifs within these regions that predict sets of transcription factors that orchestrate cardiac differentiation. Together, these findings form a basis for understanding developmentally regulated chromatin transitions during lineage commitment and the molecular etiology of congenital heart disease.
INTRODUCTION
Developmental decisions during lineage commitment are precisely coordinated at the genome level as broad gene expression programs are jointly activated or repressed (Davidson, 2010). Heart development requires the concurrent differentiation of cardiovascular cell types including endothelial cells, smooth muscle cells, and cardiomyocytes that must be organized into a complex organ. This process involves specification of pluripotent cells to mesodermal and cardiac precursors prior to terminal differentiation (Evans et al., 2010; Murry and Keller, 2008; Srivastava, 2006). Thus, heart development depends on precise temporal control of gene expression patterns, and disruption of transcriptional networks in heart development underlies congenital heart disease (CHD) (Bruneau, 2008; Evans et al., 2010; Srivastava, 2006). It is not known how groups of genes are co-regulated during lineage commitment in the cardiac lineage.
Chromatin regulation is fundamental in specifying different cell types during embryonic development and in generating cellular responses to the environment. Studies in mammalian cells have shown that histone modifications are correlated with active, repressed, and poised expression states and define cell state (Barski et al., 2007; Cui et al., 2009; Ernst et al., 2011; Guenther et al., 2007; Mikkelsen et al., 2007; Zhou et al., 2011). Histone marks also predict non-coding DNA elements, such as distal enhancers, that regulate tissue-specific gene expression (Creyghton et al., 2010; Ernst et al., 2011; Heintzman et al., 2009; Heintzman et al., 2007; Rada-Iglesias et al., 2011; Zentner et al., 2011). While we have considerable knowledge of the epigenetic landscape of specific cell types, how chromatin states are coordinated with gene expression during lineage commitment is poorly understood.
Emerging evidence indicates that faulty epigenetic regulation contributes to congenital heart disease (Chang and Bruneau, 2012). Mutations in the histone methyltransferase MLL2 in humans cause congenital heart defects in Kabuki syndrome (Ng et al., 2010). Transcription factors implicated in inherited congenital heart disease, such as Tbx5 and Nkx2–5, interact with histone modifying enzymes to regulate gene expression (Miller et al., 2008; Miller et al., 2010; Nimura et al., 2009), The H3K27 methyltransferase Ezh2 regulates gene expression programs that are important for heart development and homeostasis (Delgado-Olguin et al., 2012; He et al., 2012). In addition, epigenetic changes at cardiac-specific genes are observed during direct reprogramming of cardiac fibroblasts into cardiomyocytes (Ieda et al., 2010). Therefore, dissecting the dynamic chromatin and transcriptional landscapes during cardiomyocyte differentiation is critical for understanding heart development and will improve our ability to design stem cell-based therapies for cardiac-related diseases.
Here, we have defined the dynamic epigenetic and transcriptional landscapes during cardiac differentiation. We used a directed differentiation system representing the stepwise differentiation of mouse embryonic stem cells (ESCs) into cardiomyocytes (CM) that allows for isolation of developmental intermediates, including mesoderm (MES) and cardiac precursors (CP). We analyzed histone modifications at promoters to define chromatin states that accompany gene expression changes during cardiac differentiation. By using a dynamic model of lineage determination, we discovered previously unknown chromatin state transitions, including a pre-activation pattern associated with a set of genes with cardiac functions. We also used chromatin marks to discover thousands of stage-specific enhancers that may define new transcriptional networks deployed during cardiac differentiation. Our data illustrate the strengths of a differentiation time course in identifying gene regulatory networks and reveal a chromatin-level determination of cell fates in the earliest stages of differentiation that may be key to heart development.
RESULTS
Expression and chromatin states in cardiac differentiation
We investigated how global patterns of gene expression and chromatin organization are coordinated in the cardiac lineage. We used directed differentiation of mouse embryonic stem cells (ESCs) to cardiomyocytes as a model system (Figure S1). This approach reproduces normal cardiomyocyte differentiation (Kattman et al., 2011) and resulted in roughly 70% cardiac Troponin T (cTnT)-positive cardiomyocytes (Figure S1 and Supplementary Movie 1). Differentiating cultures were highly enriched at earlier stages for the cardiac transcription factors Nkx2–5 and Isl1, indicating that these cells progress efficiently through normal cardiac differentiation (Figure S1). Based on marker gene analysis, we selected four stages of differentiation that represent key cell types in the transition from pluripotent cells to cardiomyocytes (Figure 1A): undifferentiated embryonic stem cells (ESC) expressing pluripotency genes (Pou5f1/Oct4, Nanog), cells expressing mesodermal markers (Mesp1, Brachyury) (MES), cells expressing cardiac transcription factors (Nkx2–5, Tbx5, Isl1) but not yet beating (Cardiac Precursors, CP), and functional cardiomyocytes (CM) with cardiomyocyte-specific gene expression (Myh6, Myh7).
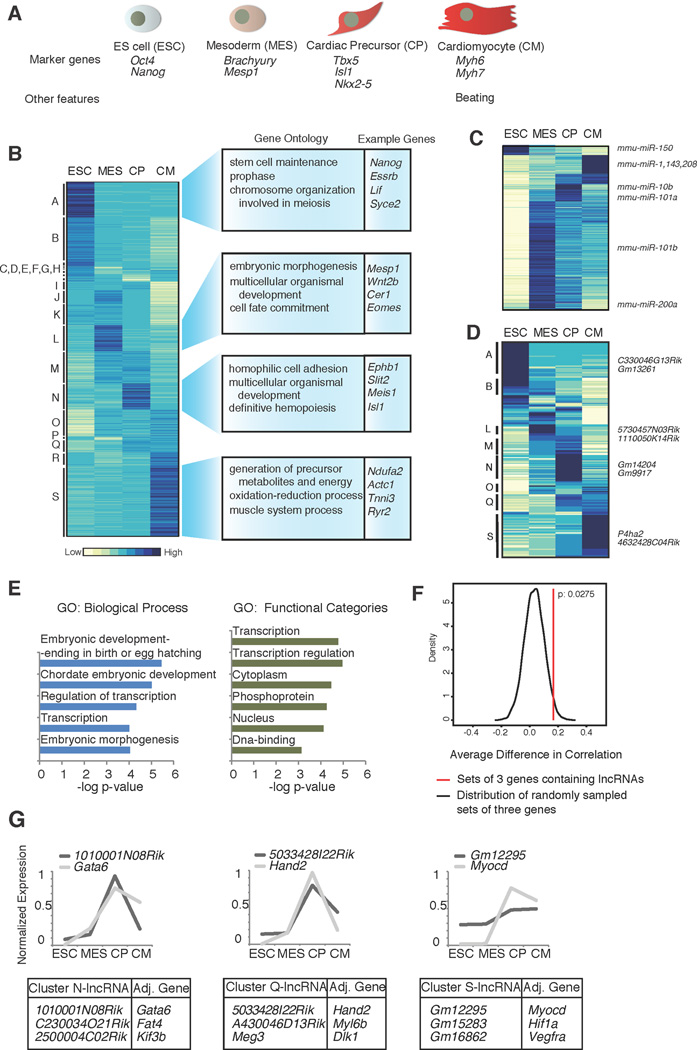
Figure 1. Transcriptional analysis of cardiac differentiation.
A. The four stages of differentiation analyzed in this study. B. Hierarchical clustering of coding and non-coding polyA+ gene expression, across the four cell types. Enriched GO terms and example genes are shown to the right. C. Hierarchical clustering of miRNA expression (565 miRNAs included in NanoString probe set). D. Hierarchical clustering of lncRNA expression including 196 lncRNAs expressed at >1 RPKM in at least one time point. E. Enriched GO terms for the two nearest genes adjacent to the lncRNA genes expressed in the time course. F. Expression correlation between lncRNAs and-adjacent gene minus correlation of adjacent gene with the other neighbour in a three gene set in a 100 kb window, as compared to background model generated by randomly sampling similar sets of three genes. Distribution of the correlation differences in the background is plotted as the black curve. Difference in expression correlation for lncRNAs is significant (P= 0.0275, red line) relative to our background model. G. Example lncRNAs and highly correlated adjacent genes identified in expression clusters N, Q and S. Graphs display examples of genes with known roles in heart development (Gata6, Hand2 and Myocd) and expression pattern during the time course. See also Tables S1 and S2, Figures S1 and S2, and Movie S1.
We analyzed global expression patterns of polyadenylated transcripts and microRNAs (miRNAs) in the four cell types by RNA sequencing (RNA-Seq) and Nanostring, respectively. We identified over 13,500 genes expressed during the time course (reads per kilobase per million (RPKM) >1). Genes were clustered by expression pattern using HOPACH (Pollard and van der Laan, 2005), yielding distinct clusters (Figure 1B, Figure S2, Table S1), including groups of genes specifically expressed at each stage (e.g. Clusters A, L, N, S). The stage-specific clusters were enriched for expected Gene Ontology (GO) terms (Table S2). Using the Nanostring platform, we analyzed over 600 miRNAs and found that they also display dynamic and stage-specific expression (Figure 1C, Table S1). Our data confirmed expression of key ESC miRNAs, such as the miR290 cluster, as well as known cardiac miRNAs (including miR-1, miR-208, and miR-143) in the CM stage. These data also identify several other stage-specific miRNAs that represent potential new regulators of cardiac differentiation.
Long ncRNAs (lncRNAs) are non-coding polyadenylated transcripts with emerging roles in gene regulation (Pauli et al., 2011). LncRNAs are differentially expressed in mammalian cell types, suggesting roles in lineage commitment (Cabili et al., 2011). However, lncRNAs have not yet been implicated in heart development. Notably, we find that lncRNAs show striking stage-specific expression in our differentiation system (Figure 1D; Table S1). LncRNAs regulate gene expression in cis and trans and may also function as transcriptional enhancers (Orom et al., 2010). We expected that if lncRNAs function in cis to regulate lineage commitment, then their neighboring genes should have functions related to this process. To test this idea, we determined GO enrichment for the two nearest genes relative to lncRNAs expressed >1 RPKM in at least one stage (Figure 1D, E). Consistent with our hypothesis, we found enrichment of genes involved in development, morphogenesis, and transcriptional processes (Figure 1F).
We find that lncRNAs identified in our data are significantly correlated in expression with their neighboring genes compared to randomly selected neighboring protein coding genes (hypergeometric test p=4E-32). We tested the possibility that the observed correlations are attributable to coordinately regulated gene clusters, however, we find that lncRNA expression is more highly correlated with the nearest adjacent gene (p= 0.0275) relative to our background model (Figure 1F, Supplementary Methods). Our data suggest that some lncRNAs may regulate gene expression in cis during cardiomyocyte differentiation. Moreover, we find many correlated lncRNA-gene pairs are associated with known cardiac genes such as Gata6, Hand2 and Myocd (Figure 1G, Table S1). These data identify several potential non-coding regulators and will facilitate further study of lncRNAs in cardiogenesis.
Chromatin state dynamics during cardiac differentiation
Chromatin structure is key to transcriptional regulation, yet its role during differentiation is largely unknown. To this end, we performed chromatin immunoprecipitation and massively parallel sequencing (ChIP-Seq) for several histone modifications at the time points examined for gene expression (ESC, MES, CP, CM). Modifications included H3K27me3 and H3K4me3 (associated with inactive and active promoters respectively), and H3K4me1 and H3K27ac, (associated with promoters and enhancers) (Barski et al., 2007; Cui et al., 2009; Ernst et al., 2011; Mikkelsen et al., 2007; Zhou et al., 2011). We also determined binding of RNA polymerase II phosphorylated at serine 5 (RNAP), which is enriched at transcriptional start sites (TSS).
Given the dynamic nature of gene expression observed during cardiomyocyte differentiation, we initially focused on elucidating histone modification patterns at transcription start sites (TSSs). To identify gene promoters with similar patterns, we performed unsupervised clustering of ChIP signal 2kb around the TSS of each gene (Figure 2A, Table S1 and Supplementary Methods). Consistent with previous studies showing little variation of chromatin patterns at promoters across cell types (Heintzman et al., 2009), the largest cluster, Cluster 1, had high levels of H3K4me3, H3K27ac, and RNAP (all associated with active chromatin and transcription) across the time course. GO analysis showed that these genes are involved in fundamental cellular functions, such as metabolism and cell-cycle regulation (Table S3). Other clusters, however, revealed dynamic chromatin patterns, suggesting chromatin regulation is critical to cardiac gene expression and differentiation.
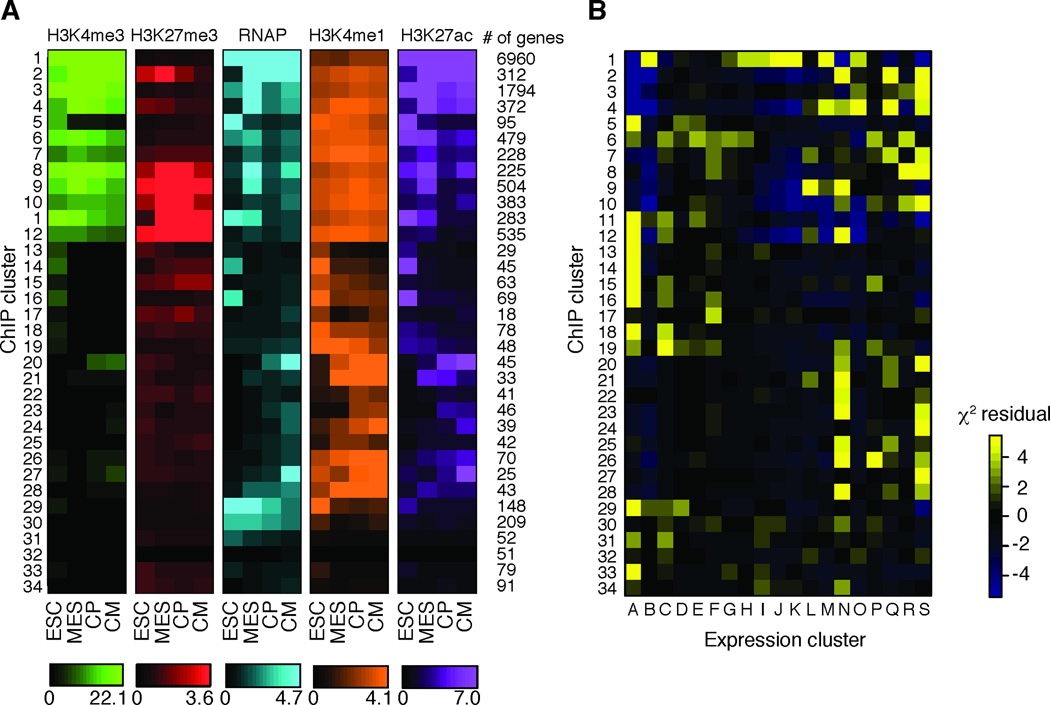
Figure 2. Chromatin state transitions during cardiac differentiation.
A. Hierarchal clustering of genes based on enrichment of histone modifications and RNA Polymerase (serine 5 phosphorylated) within 2 kb of the TSS. Color represents median enrichment for each cluster of genes. Number of genes within each cluster is shown on the right. B. The overlap of genes between chromatin clusters (vertical axis) and expression clusters (horizontal axis). Color represents the Pearson residuals. Yellow represents significant overlap between the genes within chromatin cluster and expression cluster.. See also Table S3.
Dynamic chromatin states correlate with distinct expression patterns
We considered that genes with similar temporal expression patterns would share a common chromatin pattern. Conversely, common expression patterns may be represented by multiple different chromatin patterns. To test this, we tabulated the number of genes shared between each chromatin and expression cluster and determined statistical enrichment (Figure 2B). We found that the mesoderm-specific expression cluster L is primarily associated with chromatin cluster 9. However, the ESC-specific expression cluster A, comprising genes rapidly silenced upon differentiation, correlated with several chromatin patterns, including chromatin cluster 5 where active marks are lost without gain of additional marks tested, and cluster 11 where active marks are gradually lost while gaining the repressive H3K27me3 modification (Figure 2A). Regulators of stem cell state fell into more than one co-cluster (A11 Pou5f1/Oct4, A5 Nanog), suggesting expression of pluripotency regulators is controlled by multiple epigenetic mechanisms.
We next examined how chromatin and expression patterns were coordinated at each stage of differentiation. We find that genes with similar expression patterns showed considerable variation in chromatin states during differentiation. For instance, genes in co-cluster A11 include active genes with highly correlated chromatin states at the ESC stage (Figure 3A). Upon differentiation, these genes were down regulated at the MES stage. However, this initial change in expression did not correlate with changes in chromatin until later in differentiation at the CP stage. Conversely, genes in the mesoderm-specific expression Cluster L9 (Figure 3B), which are expressed at the MES stage and repressed at the CP stage, correspondingly have highly-correlated active MES and silent CP chromatin states.
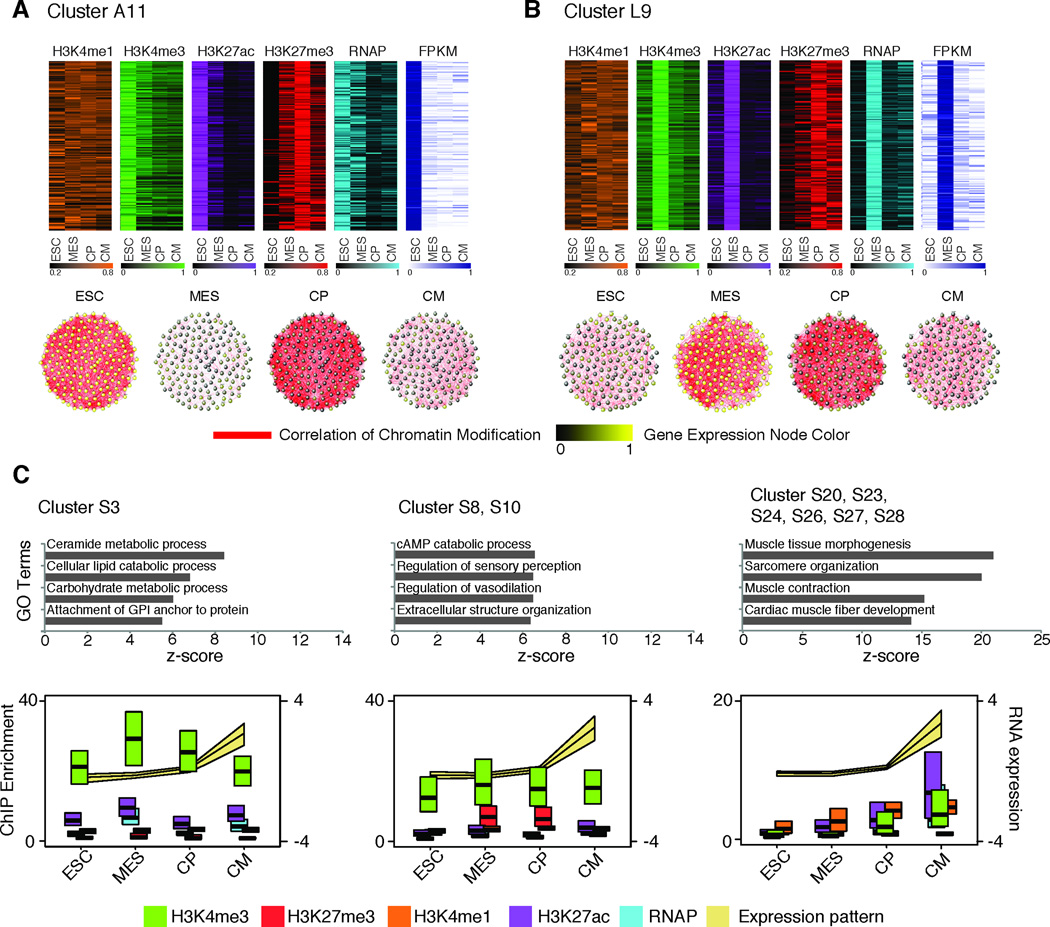
Figure 3. Dynamic and highly correlated chromatin and gene expression patterns during cardiomyocyte differentiation.
A. Heat maps (top) of magnitude transformed, chromatin fold enrichment values and gene expression values, for co-cluster A11. Co-cluster A11 correlation network (bottom), where nodes represent genes in each module and edges (red lines) represent Pearson correlations of chromatin marks, calculated with the magnitude transformed values with a threshold of 0.9. Node color corresponds to gene expression state; yellow and black indicates up- and down-regulated expression, respectively. B. Co-cluster L9, analysed in the same manner as co-cluster A11. C. Sub-groups of expression cluster S based on chromatin pattern segregate genes with distinct gene ontology. Chromatin and gene expression values are represented as median +/− interquartile range among all genes graphed. Expression values were normalized by interquartile range within each gene.
Our analysis also revealed that chromatin patterns could distinguish functionally distinct genes with a similar expression pattern (Figure 3C). For example, expression cluster S comprises genes expressed at the CM stage that are associated with diverse chromatin patterns, such as H3K4me3 and no H3K27me3 (S3), H3K4me3 and H3K27me3 (S8 and S10), and gain of low levels of H3K4me3 during differentiation without H3K27me3 enrichment (S20, S23, S24, S26, S27, and S28). However, each subgroup includes genes involved in distinct processes, including metabolism (S3), signalling (S8 and S10), and muscle contraction (S20, S23, S24, S26, S27, and S28). This indicates that despite similar expression patterns, groups of functionally related genes can be distinguished at the chromatin level.
A novel chromatin state transition during CM differentiation
Our promoter clustering revealed a group of genes that showed enrichment for H3K4me1 enrichment prior to enrichment of H3K4me3 and RNA Pol II and transcriptional activation. As H3K4me1 has largely been associated with open chromatin at distal enhancers prior to activation (Creyghton et al., 2010; Ernst et al., 2011; Heintzman et al., 2009; Rada-Iglesias et al., 2011; Zentner et al., 2011), we hypothesized that H3K4me1 may mark a similar promoter state. While we found H3K4me3 and H3K4me1 were often enriched at the same TSSs, a fraction (15–20%) of genes marked by H3K4me1 was not H3K4me3 enriched (Figure 4A) and was poorly expressed (Figure 4B). These included many contractile protein genes, such as Actc1, for which H3K4me1 was present at the MES stage prior to transcriptional activation at the CP and CM stages (Figure 4C). Notably, H3K27me3 was not enriched at these promoters, suggesting that this group of genes was not repressed by Polycomb during cardiomyocyte differentiation.
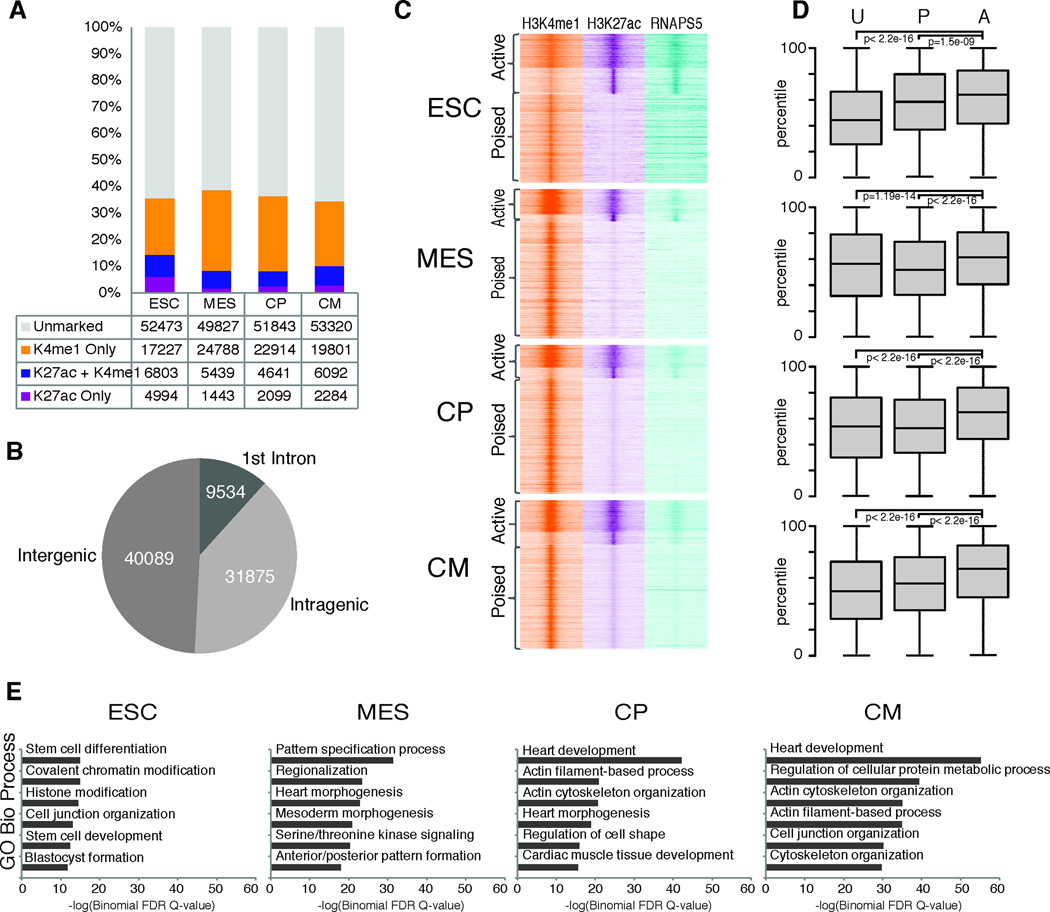
Figure 4. H3K4me1 marks cardiac contractile genes prior to gene activation.
A. Fraction of H3K4me1-marked genes that overlap with H3K4me3. An enrichment value at the TSS of 3 was used as the threshold to distinguish marked from unmarked genes. B. Average expression (RPKM) of genes marked with H3K4me1, H3K4me3, both H3K4me1 and H3K4me3, or neither modification for each stage of differentiation. C. Example of a pre-activated gene, Actc1. ChIP-Seq (H3K4me1, H3K27ac, H3K4me3, y-axis reads/million unique mapped reads) and RNA-Seq (FPKM) genome tracks (mm9) are shown. Scale for each modification is constant throughout the time course. D. Classification of genes based on gain of H3K4me1 and H3K4me3 enrichment at the TSS. Enrichment for genes with MGI cardiovascular expression was calculated using a Pearson residual. E. Example genes for each group. Left axis represents mean normalized chromatin enrichment values at the TSS. Right axis represents RPKM expression value.
To gain broader insights, we classified genes based on the pattern of H3K4me1 and H3K4me3 at their TSS. We identified three gene groups that showed interesting patterns of these modifications (Figure 4D,E). Group 1 gained H3K4me1 prior to H3K4me3 enrichment and transcriptional activation and was enriched exclusively for cardiovascular genes including those encoding contractile proteins associated with terminal differentiation and cardiomyocyte function. H3K4me1 was often maintained at these TSSs upon H3K4me3 enrichment and gene activation. Group II gained H3K4me1 over time, but failed to gain H3K4me3 or robust expression and included muscle lineage genes such as Ckm, Ckmt2, and Tcap, whose expression is associated with cardiomyocyte maturation. Group III genes transiently gained H3K4me1 at specific stages, but showed no H3K4me3 enrichment during differentiation. These genes were not expressed above background levels throughout differentiation and function in non-cardiac lineages. Group II and III genes did not acquire H3K27me3 suggesting Polycomb-independent silencing. While the functional role of the H3K4me1 pre-activation pattern is unknown, chromatin remodelling at early stages of cardiac differentiation may be necessary for gene activation during terminal differentiation.
Enhancer activity correlates with cardiac specific programs
While regulation at promoters is important for gene regulation, distal enhancers are key regulators of tissue-specific gene expression patterns during lineage commitment. In addition to their enrichment at TSSs, H3K4me1 and H3K27ac demarcate enhancer elements in a wide range of cell types (Creyghton et al., 2010; Ernst et al., 2011; Rada-Iglesias et al., 2011; Zentner et al., 2011). Using these modifications, we identified 81,497 putative distal enhancer regions during cardiac differentiation (Figure 5A,B, Figure S3A–C, Table S4 and Supplementary Methods). H3K4me1 marks most elements at each stage, whereas H3K27ac is enriched at a subset of these regions (Figure 5A). The broad enrichment of H3K4me1 is consistent with the idea that it represents a general mark of enhancers and open chromatin (Creyghton et al., 2010; Ernst et al., 2011; Heintzman et al., 2009; Rada-Iglesias et al., 2011; Zentner et al., 2011). Comparing our data to H3K4me1 and H3K27ac profiles from neural precursors, liver, and pro B cells (Creyghton et al., 2010) revealed that our enhancers are largely unique to the cardiac lineage (Figure S3D).
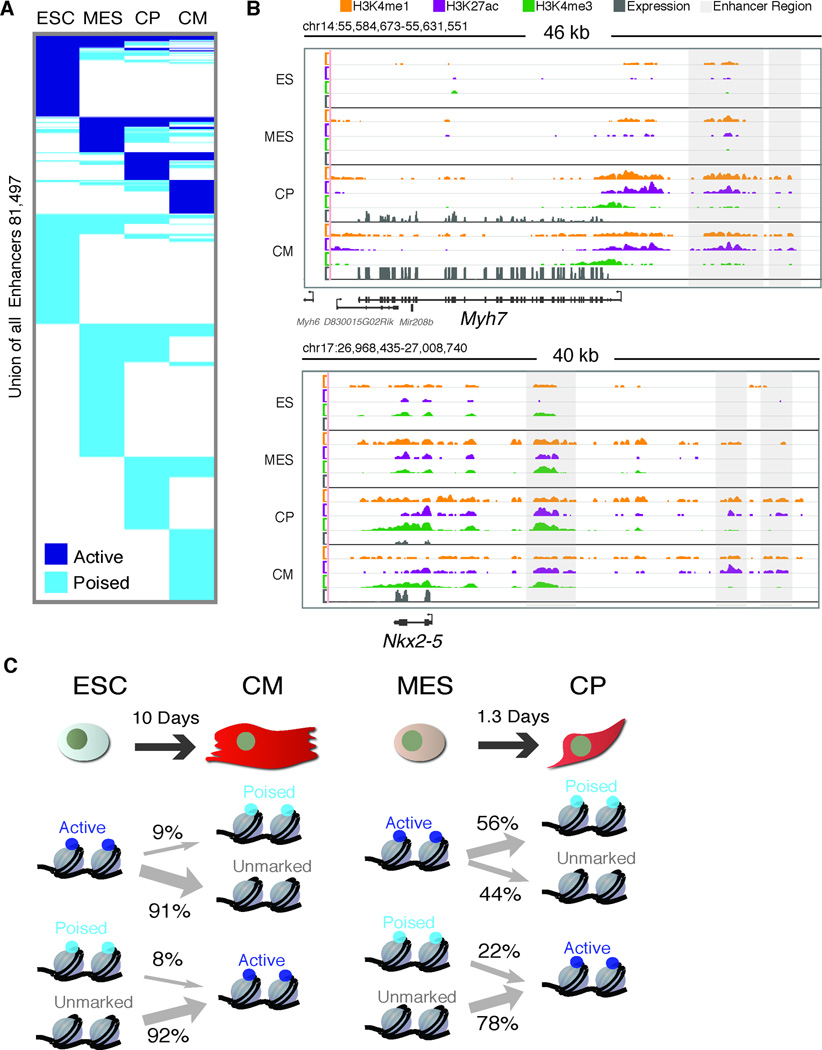
Figure 5. Identification of enhancer elements during cardiac differentiation.
A. Total distal enhancers identified in ESC, MES, CP, and CM categorized by H3K27ac and H3K4me1 status at each stage. B. Distribution of enhancers across the genome. C. Density of ChIP-Seq reads +/− 4kb relative to the midpoint of enriched regions for H3K4me1, H3K27ac and RNA Polymerase (serine 5 phosphorylated form). D. Boxplots of log2 transformed (FPKM) gene expression values for single nearest gene associated with unmarked (U), poised (P), and active (A) enhancer groups. p-values determined by Wilcoxon rank sum test with continuity correction. Boxplots show interquartile ranges (IQR) with whiskers extending to the furthest data point that was no further than 1.5 times the IQR from the interquartile boundaries. E. −Log(Binomial FDR Q-value) scores for GO Biological Process enriched in single nearest gene associated with active enhancers.
Our predicted set of enhancers significantly overlaps the smaller sets identified by p300/CBP binding in fetal mouse heart, fetal or adult human hearts, or by the binding of multiple transcription factors in the HL1 cardiomyocyte cell line (Blow et al., 2010; He et al., 2011; May et al., 2012), which supports our enhancer predictions (Figure S3D,E). Consistent with evidence indicating that cardiac enhancers are conserved over a limited phylogenetic distance (Blow et al., 2010; May et al., 2012), we find our predicted enhancers have low overlap with highly conserved (>600) Phastcons elements, which increases significantly when considering only highly conserved elements in placental mammals (Figure S3F,G). Thus, our analysis identified many putative heart enhancers that likely function in heart development, including novel enhancers that may regulate the transition from pluripotency to a functionally differentiated state.
Histone modification patterns can distinguish active enhancers (which correlate with tissue-specific expression) from poised enhancers (which correlate with potential gene expression later in development) (Creyghton et al., 2010; Ernst et al., 2011; Rada-Iglesias et al., 2011; Zentner et al., 2011). We classified our enhancers as active (H3K27ac+, H3K4me1+/−) or poised (H3K4me1+ only) at each stage of differentiation (Figure 5C). We find most enhancers at a given stage are poised, whereas a smaller subset is active. We then compared enrichment patterns of several other histone modifications and RNAP with these enhancer regions. We find that RNAP is highly enriched at active enhancers (Figure 5C), consistent with transcription initiation at these regulatory elements (Kim et al., 2010). Conversely, although H3K27me3 has been shown to demarcate poised enhancers elements (Rada-Iglesias et al., 2011; Zentner et al., 2011), this mark had a minimal overlap with our enhancer regions (Figure S4). Thus, while poised and active enhancers can be broadly defined by a limited set of histone modifications, there likely exist many other sub-states that comprise functionally distinct enhancer states.
To test whether active enhancers correlate with stage specific gene expression, we assigned active enhancers to their single nearest gene (Table S4) and found that genes associated with active enhancers are expressed at significantly higher levels than genes associated with unmarked or poised enhancers at each stage (Figure 5D). GO analysis of genes associated with active enhancers revealed enriched categories that progressively become cardiomyocyte-specific (Figure 5E, Figure S5A, Table S5). For example, enhancer-associated genes at the MES stage function in mesoderm and embryonic pattern specification, while at the CP stage they function in heart morphogenesis and cardiac tissue development. In CMs, we observe a transition to genes involved in cardiomyocyte structure and function. Many of these genes have important roles in heart development and their dysregulation is associated with heart defects and cardiovascular disease (Bruneau, 2008; Srivastava, 2006). Thus, we identified many new putative enhancers that correlate with genes involved in cardiac specification during embryonic development.
Enhancer transitions during CM differentiation
To dissect enhancer state transitions that govern cardiac gene expression programs, we clustered enhancers according to their states (unmarked, poised, or active) at each stage (Figure 6A). The set of active and poised enhancers is largely unique at each stage, indicating that enhancers are highly cell type-specific even between closely related cell types. A subset of enhancers showed poised-to-active state transitions concomitant with activation of the proximal gene (e.g. Myh7 and Nkx2–5) (Figure 6B). However, most poised enhancers failed to acquire an active state during cardiomyocyte differentiation. These initially poised enhancers may be required to specify cell fates in other lineages during early development, suggesting that cells retain significant plasticity during lineage commitment.
Figure 6. Transitioning enhancer states during cardiac differentiation.
A. Union set of enhancers combined from all 4 time points during cardiomyocyte differentiation clustered based on Unmarked, Poised (H3K4me1+) or Active (H3K27ac+;H3K4me1+/−) enhancers. B. Example ChIP-Seq (H3K4me1, H3K27ac, H3K4me3, y-axis reads/million unique mapped reads) and RNA-Seq (RPKM) genome tracks (mm9). Scale for each modification is constant throughout the time course. C. Enhancer state transitions in similar cell types relative to distant cell types.
The dynamic cell-type specificity of enhancer usage suggested that transitions between poised and active enhancer states occur rapidly between stages. Consistent with this, comparing enhancer states between each stage of cardiac differentiation showed that the fraction of active enhancers that transition through a poised state is largest during the MES to CP and CP to CM transition and lowest between unrelated cell types (Figure 6C and Figure S5). While enhancer transitions between ESCs and the differentiated cell types remain below ~5%, they comprise 22% to 56% during the MES-CP transition, respectively (Figure S5). The cell type specificity and rapid state transitions of enhancers suggests dynamic enhancer usage is an important regulatory mechanism for coordinating tissue specific gene expression.
Integrating enhancers into gene networks
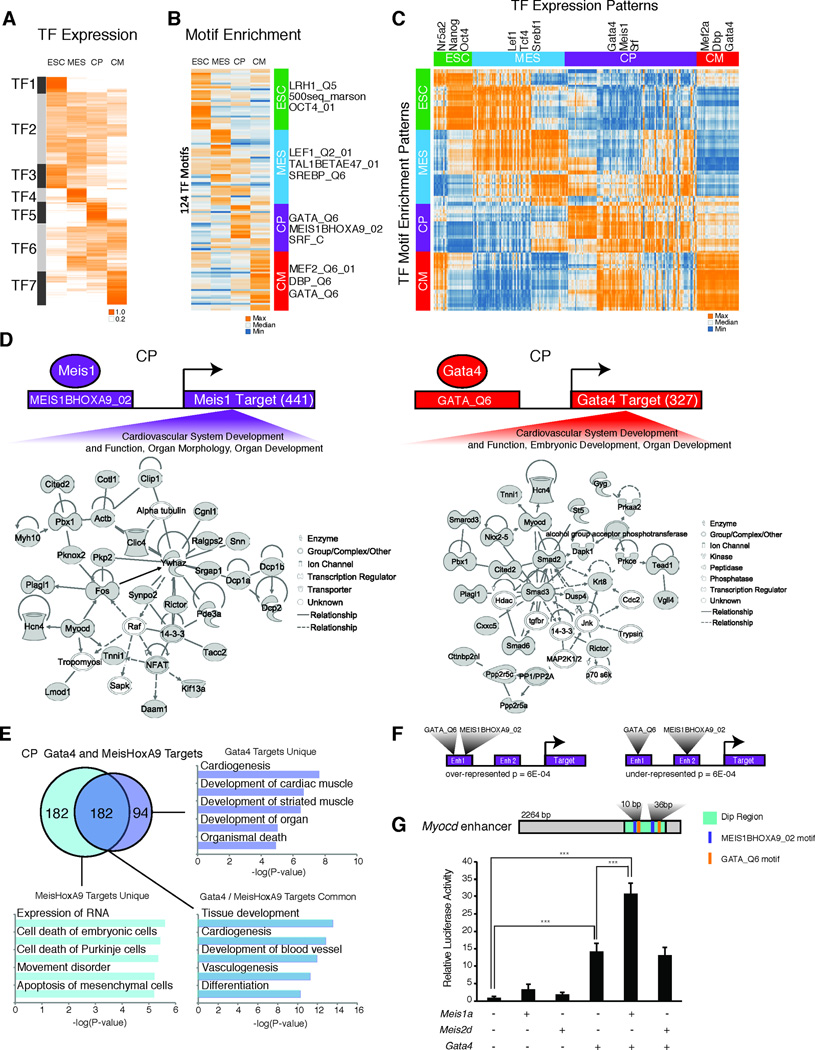
While enhancers regulate global developmental gene expression patterns, integrating these genomic elements into the core transcriptional regulatory circuitry is challenging. Transcription factors (TFs) can act as master regulators of gene expression programs by binding to specific motifs within cis-regulatory elements. Given the stage-specific expression of TFs in our time course (Figure 7A, Figure S2), we hypothesized that motifs for TFs that drive cardiac development would be enriched in active enhancers. Reasoning that TFs bind open chromatin regions (He et al., 2010; Verzi et al., 2010), we developed an algorithm to find depressions in the H3K27ac chromatin profile at active enhancer regions and used these regions to search for motifs. We found over-represented motifs at each stage, including those for TFs that regulate the ESC state (OCT4_01, LRH1_Q5, and 500seq_marson) and cardiac development (GATA_Q6, MEF_Q6_01, MEIS1BHOXA9_02, SRF_C) (Figure 7B). We compiled a list of TFs known to bind these highly conserved motifs and found strong correlations between TF expression and motif enrichment at each stage (Figure 7C). To address whether these predictions represented binding events, we analyzed ChIP-Seq data for OCT4 and SOX2 in ESCs (Marson et al., 2008) and GATA4 in the HL-1 cardiomyocyte cell line (He et al., 2011). OCT4 and SOX2 bound regions substantially overlapped the OCT4_01 and SOX9_B1 motifs at enhancers in ESCs (P = 2e10−74 and 3e10−26) (Figure S5D). GATA4 bound regions strongly correlated with enhancers that have a GATA_Q6 motif (P = 3e10−74 and 2e10−30 respectively) at the CP and CM stage.
Figure 7. Enhancer gene networks critical for heart development.
A. Hierarchical clustering of magnitude normalized FPKM values for transcription factors expressed during cardiac differentiation, subdivided into 7 groups (TF1–TF7). B. Clustering of magnitude normalized density based motif enrichment scores (−log(p-value)) shows stage specific enrichment of highly conserved TF motifs. C. Pearson correlation matrix between enriched TF motifs and the expression pattern of TFs known to bind the list of highly conserved motifs. D. Examples of predicted target gene networks. Grey nodes represent genes identified via motif enrichment analysis. E. Venn diagram shows overlap between MEISHOXA9 and GATA6_Q6 motif containing target genes, with associated GO terms for unique and common targets. F. Graphical representation of the preference for MEISHOXA9 and GATA6_Q6 motifs to occupy the same enhancer versus separate enhancers at common gene targets. G. Meis1a and Gata4 synergistically activate the Myocd enhancer. A graphical representation of the candidate MEIS and GATA sites within the enhancer dip are shown. The graph shows relative luciferase reporter activity normalized to reporter construct alone. Data are represented as mean + SEM. N=4; ***P<0.001 by ANOVA.
To construct gene regulatory networks connected to specific TF-motif pairs, we selected target genes positively correlated with TF expression. We find that many of these pairs were associated with genes that function in common pathways. For example, at the CP stage, MEIS, GATA, and NFATC enhancer-associated genes comprise networks implicated in cardiovascular development and function (Figure 7D, Figure S7). While Meis1 and Meis2 have been implicated in heart development (Crowley et al., 2010; Pfeufer et al., 2010; Stankunas et al., 2008), their targets in cardiac differentiation are unknown. Our data suggest MEIS1/2 regulate a subset of genes important in cardiac morphogenesis. Moreover, GATA factors potentially bind enhancers that may regulate important cardiac genes, including Nkx2–5, Mef2c, and Gata4 itself (Bruneau, 2008; Srivastava, 2006).
We further tested our enhancer predictions by analyzing the effects of loss of function of particular TFs on gene expression. Genes in the predicted OCT4-regulated network were highly correlated with genes affected by Oct4 knockdown (P=3e−44) (Loh et al., 2006). Despite considerable redundancy among GATA factors (Zhou et al., 2012), we found the GATA4 network also predicted many genes differentially regulated in Gata4 knockdown HL-1 cells (He et al., 2011) (P=9e−12 at CP stage, P=9e−15 at CM stage). These data indicate that we have identified a set of TFs that may regulate specialized gene expression networks by binding to distinct sets of enhancers during cardiomyocyte differentiation.
Combinatorial interactions among transcription factors can increase the diversity of regulatory modules governed by a particular factor.. We observed a significant overlap among target genes associated with enhancers containing MEIS1BHOXA9_02 and GATA_Q6 motifs at the CP stage (Figure 7E, Table S6). Upon further analysis, we identified groups of developmentally important genes regulated by only GATA, only MEIS, or MEIS and GATA together suggesting independent as well as co-regulation by MEIS/HOX and GATA factors. Enhancers co-enriched for both MEIS and GATA motifs are associated with genes important for cardiac development, such as Gata5, Irx4, Myocd, Zfpm2, Wnt2, and Smarcd3, and with genes that influence conduction system function (Hcn4), consistent with an association of MEIS1 with conduction parameters (Pfeufer et al., 2010). Notably, we find MEIS/GATA motifs are often enriched in the same enhancer (Figure 7F), suggesting a functional relationship between these factors.
We used luciferase reporter activation assays to test for co-regulation by MEIS and GATA factors. We tested 5 enhancers with motifs for MEIS and GATA, including a Myocd enhancer active in the developing heart (Creemers et al., 2006). Co-transfection of the Myocd reporters with combinations of expression constructs for GATA4, MEIS1A, or MEIS2D, showed that this enhancer responded to GATA4 and was synergistically activated by the combination of GATA4 and MEIS1A (Figure 7G). This appeared to be specific to MEIS1A, as co-transfection of MEIS2D with GATA4 did not lead synergistic activation. Furthermore, most (4 of 5) enhancers tested were synergistically activated by the combination of MEIS1A and GATA4 (Figure S7C). Thus GATA4 and MEIS1A can function together to activate certain cardiac enhancers.
Collectively, our work reveals a detailed picture of how gene expression programs may be coordinated during lineage commitment and provides novel insights into the key principles that underpin heart development and disease.
Discussion
We have defined chromatin state transitions during cardiac differentiation that provide new insights into the dynamic regulation of cellular differentiation and the coordinated regulation of gene expression programs. Our results show that there are complex but distinct chromatin patterns that accompany lineage decisions.
Dynamic epigenetic transitions in differentiation
The rapid loss of expression of pluripotency-associated genes upon differentiation can be achieved by at least 9 different chromatin patterns. These patterns comprise broad groups that include loss of active marks (e.g. Nanog), or gradual loss of active marks with the simultaneous acquisition of repressive marks (e.g. Oct4/Pou5f1). Conversely, during cardiac differentiation, we observed striking coherence among most mesoderm-specific genes, which share a specific chromatin pattern. On the other hand, genes expressed in later development can be classified by multiple distinct chromatin regulatory patterns that may precisely coordinate precursor and differentiated cardiomyocyte gene expression programs.
Our data also reveal that chromatin patterns can predict sets of functionally related genes. For example, genes associated with metabolic function share a similar chromatin pattern, while those involved in contractile function and sarcomere structure have a distinct pattern, although they share a similar expression profile. This implies that functionally-related and co-expressed genes have specific modes of regulation. Distinct modes of epigenetic regulation may exist to ensure that functional gene modules are synchronized, thus ensuring robust and coordinated expression of key processes that may be critical for cell function and importantly for adaptation to stress.
A novel dynamic pattern of histone modifications
Analysis of chromatin states during cardiomyocyte differentiation has led to the identification of novel patterns that are highly informative to understand developmental regulatory programs. In particular, we have identified a pattern of H3K4me1 deposition at the TSS that precedes the transcriptional activation and acquisition of H3K4me3 and recruitment of RNAP. This pre-activation pattern is consistent with the idea that molecular events in early lineage commitment mark genes for subsequent activation. A similar pattern has been observed in T-cell differentiation, in which H3K4me2 precedes transcriptional activation for a select group of genes (Zhang et al., 2012). This pre-activation pattern is likely important for genes that are not regulated by Polycomb complexes. The presence of H3K4me1 in anticipation of transcriptional activation is reminiscent of its presence at poised enhancers, for which only a minority show H3K27me3 enrichment. It is possible that H3K4me1 may mark regulatory elements such as proximal enhancers that may function to poise the TSS for activation. These data suggest that diverse mechanisms can poise specific classes of TSSs and enhancers for subsequent activation. We propose that early deposition of H3K4me1 at specific cardiac genes is a regulated step that facilitates later activation of these genes. It will be important to identify the chromatin regulator that is necessary to catalyse this modification as well as the factors required for its recruitment and whether the early deposition of this mark is required for proper activation of these genes.
Identification of transcriptional networks based on enhancer predictions
Chromatin marks at genomic regions distal to the TSS provides a means to identify candidate enhancers elements (Creyghton et al., 2010; Rada-Iglesias et al., 2011). We have identified a large number of enhancers that show stage-specific activation. This rich data set has allowed us to discover transcription factor motifs that predict novel enhancer-driven transcriptional regulatory networks during cardiomyocyte differentiation. For example, we identified networks for the GATA family of transcription factors that include GATA4, GATA5, and GATA6 in CP and CM stages. The GATA factors are known to orchestrate many developmental processes. In the developing heart, these transcription factors have broad roles in early differentiation, morphogenesis, and postnatal physiology (Zhao et al., 2008; Zhou et al, 2012). Mutations in GATA4 lead to congenital heart defects (Garg et al., 2003), highlighting the importance of elucidating how these factors regulate gene expression programs in heart development.
We also discovered potential new regulators of cardiac development. We identified enrichment for the Meis1HOXA9 motif, which predicts the binding of Meis factors, perhaps along with a partner Hox factor, at a subset of enhancers at the CP stage. Meis1 has been implicated in heart development because Meis1 null mice display congenital heart defects (Stankunas et al., 2008) and because MEIS1 has been identified in Genome Wide Association Studies (GWAS) of human arrhythmias (Pfeufer et al., 2010). Chromosomal deletions that include MEIS2 have also been identified in patients with congenital heart defects (Crowley et al., 2010). Our expression data show that both Meis1 and Meis2 are robustly and transiently activated at the CP stage, consistent with a role in cardiac progenitors. Thus, we have identified a novel network potentially under control of Meis factors. Finally, we observed a striking overlap between GATA- and the MEISHOXA9-binding sites at enhancers, and show that these enhancers can respond to GATA4 and Meis1/2 in reporter assays. Thus we have uncovered a previously unknown functional relationship between GATA and Meis TFs in heart development.
Together, our study establishes a platform to understand the process of cardiomyocyte differentiation and provides an opportunity to identify mechanisms of complex disease loci by comparison with GWAS. Moreover, our data lay the foundation for understanding how the epigenetic landscape of cardiac differentiation integrates transcriptional inputs during normal development. These insights will be valuable to develop improved cardiac reprogramming strategies (Ieda et al., 2010; Qian et al., 2012; Song et al., 2012; Takeuchi and Bruneau, 2009) and to elucidate how disruption of these diverse regulatory modules contributes to congenital heart disease.
Methods
Detailed experimental and analysis methods can be found in Supplementary.
Cardiomyocyte differentiation and analysis
E14 Tg(Nkx2–5-EmGFP) mouse ES cells (Hsiao et al., 2008) were cultured in feeder-free conditions using standard techniques. Directed differentiations and analyses were performed essentially as described in (Kattman et al., 2011).
RNA-Seq and analysis pipeline
Total RNA was isolated from 5×106 cells using TRIzol® Reagent. Sequencing libraries were prepared according to Illumina RNA-Seq library kit with minor modifications. Paired-end RNA-Seq 36 base pair reads were aligned to mm9 (Mus musculus assembly July 2007). DESeq (Anders and Huber, 2010) was used to normalize raw read counts and analyze differential gene expression. USeq 7.0 (Nix et al., 2008) was then used to generate gene-level read counts and estimate RPKM (reads per kilobase of exon per million reads mapped). Only genes with expression values >1 RPKM in at least one cell type were considered for subsequent analysis. Expression was normalized to the interquartile range across the time course; interquartile numbers were used for clustering using a cosine angle distance metric and the Hopach clustering package (http://www.bioconductor.org/packages/2.1/bioc/html/hopach.html).
ChIP-Seq and analysis pipeline
Genome-wide localization of histone modifications (H3K4me3, H3K4me1, H3K27me3 and H3K27ac) and the serine 5 phosphorylated form of RNA Pol II for each stage was determined via chromatin immunoprecipitation followed by high throughput sequencing. Chromatin immunoprecipitations were performed according to the Young lab protocol (Lee et al., 2006) with minor modification. Details on analysis pipeline can be found in Supplementary.
Transient transactivation assays for enhancer validation
Candidate enhancers were cloned into a modified pGL3 luciferase reporter construct. Expression constructs for Gata4, Meis1a, and Meis2d (250ng each) were co-transfected with each reporter construct (500 ng each) in 10T1/2 cells. Luciferase activity was assessed 40 hours later and normalized to Renilla luciferase activity. Conditions were analyzed for statistical differences using a one-way repeated measures ANOVA followed by Tukey’s Multiple Comparison Test.
Supplementary Material
Article Highlights.
-
-
Chromatin signatures at promoters predict co-regulated genes
-
-
Identify novel pre-activation chromatin pattern at cardiac genes
-
-
Predict transcription factors that bind and activate cardiac specific enhancers
-
-
Dynamic chromatin and expression landscape reveals new clues into heart development
Acknowledgements
We thank T. Sukonnik for cloning enhancer fragments; S. Thomas (Gladstone Bioinformatics Core) for help with Figure 3; J. Wythe and P. Devine for the modified pGL3 constructs; M. Cleary, E.N. Olson, and D. Wotton for constructs; and Bruneau, Boyer, and Pollard lab members for helpful discussions. This work was supported by the NHLBI Bench to Bassinet Program (U01HL098179, to B.G.B., L.A.B., K.S.P., and D.S.), NIH F32-HL104913 (J.A.W.), the Lawrence J. and Florence A. DeGeorge Charitable Trust/American Heart Association Established Investigator Award (B.G.B.), by William H. Younger, Jr. (B.G.B.) and Massachusetts Life Sciences Center (L.A.B.). L.A.B. is a Pew Scholar in the Biomedical Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Deposition. All sequencing files have been deposited and can be found at https://b2b.hci.utah.edu/gnomex/.
References
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-Seq identification of weakly conserved heart enhancers. Nature genetics. 2010;42:806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943–948. doi: 10.1038/nature06801. [DOI] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & development. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CP, Bruneau BG. Epigenetics and Cardiovascular Development. Annu Rev Physiol. 2012;74:13.11–13.28. doi: 10.1146/annurev-physiol-020911-153242. [DOI] [PubMed] [Google Scholar]
- Creemers EE, Sutherland LB, McAnally J, Richardson JA, Olson EN. Myocardin is a direct transcriptional target of Mef2, Tead and Foxo proteins during cardiovascular development. Development. 2006;133:4245–4256. doi: 10.1242/dev.02610. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley MA, Conlin LK, Zackai EH, Deardorff MA, Thiel BD, Spinner NB. Further evidence for the possible role of MEIS2 in the development of cleft palate and cardiac septum. American journal of medical genetics Part A. 2010;152A:1326–1327. doi: 10.1002/ajmg.a.33375. [DOI] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468:911–920. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Olguin P, Huang Y, Li X, Christodoulou DC, Seidman CE, Seidman JG, Tarakhovsky A, Bruneau BG. Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nat Genet. 2012 doi: 10.1038/ng.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SM, Yelon D, Conlon FL, Kirby ML. Myocardial lineage development. Circulation research. 2010;107:1428–1444. doi: 10.1161/CIRCRESAHA.110.227405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, Matsuoka R, Cohen JC, Srivastava D. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003 Jul 24;424(6947):443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A, Kong SW, Ma Q, Pu WT. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5632–5637. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A, Ma Q, Cao J, von Gise A, Zhou P, Xie H, Zhang B, Hsing M, Christodoulou DC, Cahan P, et al. Polycomb repressive complex 2 regulates normal development of the mouse heart. Circ Res. 2012 doi: 10.1161/CIRCRESAHA.111.252205. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al. Nucleosome dynamics define transcriptional enhancers. Nature genetics. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature genetics. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Heng JC, Feng B, Han J, Jiang J, Kraus P, Ng JH, Orlov YL, Huss M, Yang L, Lufkin T, et al. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell stem cell. 2010;6:167–174. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Hsiao EC, Yoshinaga Y, Nguyen TD, Musone SL, Kim JE, Swinton P, Espineda I, Manalac C, deJong PJ, Conklin BR. Marking embryonic stem cells with a 2A self-cleaving peptide: a NKX2–5 emerald GFP BAC reporter. PLoS ONE. 2008;3:e2532. doi: 10.1371/journal.pone.0002532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-Specific Optimization of Activin/Nodal and BMP Signaling Promotes Cardiac Differentiation of Mouse and Human Pluripotent Stem Cell Lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May D, Blow MJ, Kaplan T, McCulley DJ, Jensen BC, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, et al. Large-scale discovery of enhancers from human heart tissue. Nat Genet. 2012;44:89–93. doi: 10.1038/ng.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Huang AC, Miazgowicz MM, Brassil MM, Weinmann AS. Coordinated but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression. Genes Dev. 2008;22:2980–2993. doi: 10.1101/gad.1689708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Molecular cell. 2010;40:594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nature genetics. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, Kaneda Y. A histone H3 lysine 36 trimethyltransferase links Nkx2–5 to Wolf-Hirschhorn syndrome. Nature. 2009;460:287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- Nix DA, Courdy SJ, Boucher KM. Empirical methods for controlling false positives and estimating confidence in ChIP-Seq peaks. BMC bioinformatics. 2008;9:523. doi: 10.1186/1471-2105-9-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nature reviews Genetics. 2011;12:136–149. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeufer A, van Noord C, Marciante KD, Arking DE, Larson MG, Smith AV, Tarasov KV, Muller M, Sotoodehnia N, Sinner MF, et al. Genome-wide association study of PR interval. Nature genetics. 2010;42:153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, van der Laan MJ. Cluster Analysis of Genomic Data. In: GR, CV, HW, IR, DS, editors. Bioinformatics and Computational Biology Using R and Bioconductor. Springer; 2005. [Google Scholar]
- Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012 doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Nam Y, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012 doi: 10.1038/nature11139. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Stankunas K, Shang C, Twu KY, Kao SC, Jenkins NA, Copeland NG, Sanyal M, Selleri L, Cleary ML, Chang CP. Pbx/Meis deficiencies demonstrate multigenetic origins of congenital heart disease. Circulation research. 2008;103:702–709. doi: 10.1161/CIRCRESAHA.108.175489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi MP, Shin H, He HH, Sulahian R, Meyer CA, Montgomery RK, Fleet JC, Brown M, Liu XS, Shivdasani RA. Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Developmental cell. 2010;19:713–726. doi: 10.1016/j.devcel.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome research. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. Dynamic Transformations of Genome-wide Epigenetic Marking and Transcriptional Control Establish T Cell Identity. Cell. 2012;149:467–482. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Watt AJ, Battle MA, Li J, Bondow BJ, Duncan SA. Loss of both GATA4 and GATA6 blocks cardiac myocyte diffferentiation and results in acardia in mice. Dev Biol. 2008;317:614–619. doi: 10.1016/j.ydbio.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nature reviews Genetics. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- Zhou P, He A, Pu WT. Regulation of GATA4 transcriptional activity in cardiovascular development and disease. Curr Top Dev Biol. 2012;100:143–169. doi: 10.1016/B978-0-12-387786-4.00005-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.