Abstract
In Escherichia coli, the molecular chaperones DnaK and DnaJ cooperate to assist the folding of newly synthesized or unfolded polypeptides. DnaK and DnaJ bind to hydrophobic motifs in these proteins and also each other to promote folding. This system is thought to be sufficiently versatile to act on the entire proteome, which creates interesting challenges in understanding the large-scale, ternary interactions between DnaK, DnaJ and their thousands of potential substrates. To address this question, we computationally predicted the number and frequency of DnaK- and DnaJ-binding motifs in the E. coli proteome, guided by free energy-based binding consensus motifs. This analysis revealed that nearly every protein is predicted to contain multiple DnaK- and DnaJ-binding sites, with the DnaJ sites occurring approximately twice as often. Further, we found that an overwhelming majority of the DnaK sites partially or completely overlapped with the DnaJ-binding motifs. It is well known that high concentrations of DnaJ inhibit DnaK-DnaJ-mediated refolding. The observed overlapping binding sites suggest that this phenomenon may be explained by an important balance in the relative stoichiometry of DnaK and DnaJ which determines whether they bind synergistically or competitively. To test this idea, we measured the chaperone-assisted folding of two denatured substrates and found that the distribution of predicted DnaK- and DnaJ-binding sites was indeed a good predictor of the optimal stoichiometry required for folding. These studies provide insight into how DnaK and DnaJ might cooperate to maintain global protein homeostasis.
Introduction
DnaK and DnaJ regulate protein homeostasis in E. coli by facilitating the folding of nascent polypeptides and the re-folding of damaged proteins 1–3. The DnaK-DnaJ system appears to be especially important under conditions of cellular stress because mutants lacking either chaperone are particularly sensitive to elevated temperatures, antibiotics and other insults 4, 5. Survival under these conditions requires that a wide range of vulnerable polypeptides be protected from aggregation and then refolded when conditions improve. How are DnaK and DnaJ able to act on such a wide number of possible substrates? In this study, we aimed to predict how DnaK and DnaJ might interact with their substrates across the E. coli proteome to better understand how they coordinate global protein homeostasis.
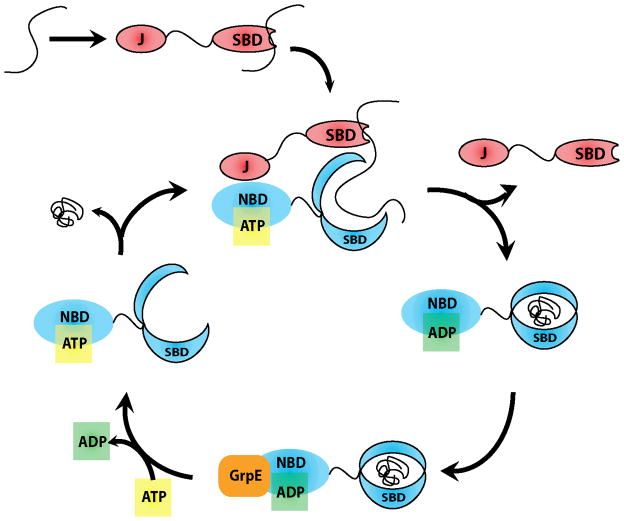
DnaK is a member of the widely expressed and highly conserved heat shock protein 70 (Hsp70) family. Like other Hsp70s, it is a 70 kDa chaperone that consists of two major domains: a nucleotide-binding domain (NBD) and a substrate-binding domain (SBD) 6–8. The NBD contains a nucleotide-binding cassette of the sugar kinase/Hsp70/actin superfamily, which has intrinsic ATPase activity. The SBD consists of a β-sandwich subdomain that has a hydrophobic cavity for binding to substrates 8, 9 and a C-terminal, α-helical subdomain that acts as a “lid”, opening and closing to accept and release these substrates. This two-domain architecture allows regulated control over substrate binding. Specifically, ATP hydrolysis in the NBD causes movements in the SBD that close the “lid” (Figure 1). The substrates of the SBD include extended linear, hydrophobic polypeptides with lengths of at least 7 to 8 amino acids 10, 11, while recent experiments suggest that more complex, partially-folded peptides may also be bound 12. Finally, interactions of these substrates with the SBD strongly stimulate ATPase activity, suggesting that allosteric communication is bi-directional 13–17.
FIGURE 1. DNAK ATPASE CATALYTIC CYCLE.
J-domain proteins recruit client proteins, dock to NBD of DnaK, and catalyze ATP hydrolysis. This converts DnaK from the ATP bound low affinity conformation to the ADP bound conformation with high affinity for client proteins. Nucleotide exchange factors (NEFs), such as GrpE, trigger ADP release. Binding of ATP completes the catalytic cycle by causing release of client protein and co-chaperones.
DnaJ is the founding member of the J domain-containing family of co-chaperones 18. DnaJ is a ~40 kDa protein that is composed of a conserved, N-terminal J domain, a glycine/phenylalanine (G/F)-rich motif, a central zinc finger region and a C-terminal domain. Through its J-domain, DnaJ binds to DnaK and allosterically promotes ATP hydrolysis 19–24. A recent NMR study has clarified that the J domain interacts with DnaK in a region between the NBD and SBD 25. The adjacent G/F motif has also been shown to help stimulate DnaK’s ATPase activity, while the zinc-finger domain is implicated in contributing to the binding of unfolded peptides 26, 27. Structural studies have suggested that short, hydrophobic peptide sequences bind in a shallow groove in the zinc finger region 28.
Earlier studies on model substrates had provided a model for how DnaK and DnaJ might cooperate to fold substrates 29–31. In this model, DnaJ is thought to bind first and then recruit DnaK through its J domain 23, 32. Stimulation of ATP turnover by the combined action of DnaJ and interactions with substrate would then enforce tight binding (Figure 1). This idea is supported by observations that model substrates containing DnaJ-binding motifs fused to consensus DnaK-binding regions are very effective at forming ternary complexes 33, 34. This synergy is required for the folding of denatured proteins, as evident from the strict dependence for including both DnaK and DnaJ to catalyze folding in vitro 35. Further, folding is blocked by deletion of the J domain, suggesting the interactions between the chaperones are required 24, 36, 37. Together, these observations have suggested that DnaK, DnaJ and substrates may need to form transient, ternary complexes to properly assist with the folding of polypeptides (Figure 1). However, DnaJ is able to prevent protein aggregation without the assistance of DnaK 27, 38, 39, suggesting that some chaperone functions do not have the same requirements. Also, DnaJ will inhibit refolding at elevated concentrations 27, 40, further suggesting that the relationships between DnaK and DnaJ in the refolding cycle are complex. Specifically, DnaJ oligomerization, DnaJ binding to DnaK’s substrate binding cleft, and competition for shared binding sites on unfolded client substrates are all possible explanations of this observed phenomenon 41, though the last theory has been most favored. Here, we show that DnaJ competes with DnaK by binding to shared target sites and therefore sequestering substrate from the larger chaperone.
Mutations in E. coli dnaK or dnaJ cause global problems in protein homeostasis, suggesting that this system acts on a relatively large percentage of the proteome 42. How can these chaperones be versatile enough to recognize thousands of different sequences in potential substrates? Several studies have explored this question 9, 28, 43, 44. The major conclusion of these studies is that both molecular chaperones show a strong preference for hydrophobic residues but with only weak ability to discriminate between amino acids. Consistent with this idea, DnaK is known to make contact with both side chains and the peptide backbone, while DnaJ only binds to side chains 44, providing a way for these chaperones to interact with hydrophobic sequences without strong preference for sequence. Such hydrophobic sequences are commonly found in the interior of folded proteins, allowing for chaperones to discriminate between folded and unfolded proteins. Together, these features appear to provide the necessary promiscuity that permits them to bind a wide range of substrates.
Although a great deal is known about how the DnaK-DnaJ system works, we were interested in the characterization of its ability to promote folding across the entire proteome. Are DnaK and DnaJ binding sites regularly distributed across all proteins? How many sites are there and what is the spacing between these sites? How does the distribution of DnaK- and DnaJ-binding sites influence the ability of the chaperones to refold each protein? Most studies towards these questions have been necessarily limited to a handful of well-known chaperone “clients”, such as luciferase, malate dehydrogenase and citrate synthase 45–49. Do these classic enzymes accurately represent the properties of the average chaperone substrate in vivo? The answers to these questions are critical to understanding how the DnaK-DnaJ system maintains global protein homeostasis in the E. coli cytosol.
Towards that goal, we sought a computational, sequence-based approach that would allow us to predict patterns of substrate binding in the E. coli proteome. This process relied on the known “consensus” binding motif for DnaK defined by the Bukau group using data obtained from peptide cellulose arrays covering the sequence of several known chaperone clients 9. Unfortunately, no such motif existed for DnaJ. Recently a binding sequence for a yeast J protein was proposed 43, but this analysis employed several restrictions that limit the results of such a study. Thus, in this report, we first developed a position-specific, consensus-binding motif for DnaJ using analogous methods as the Bukau group to analyze data from DnaJ binding to client peptide cellulose arrays 9, 44. Then, we employed both the DnaK-and DnaJ-binding preferences to reveal predicted sites across the E. coli proteome. As previously suggested 9, 44, we found that DnaK and DnaJ are predicted to bind frequently on nearly every protein. Unexpectedly, we also found that the majority of DnaK sites seem to be partially or completely overlapped by DnaJ sites. By studying the experimental folding of two denatured substrates, we also found that the number of shared DnaK-DnaJ sites was a good predictor of the optimal chaperone stoichiometry (e.g. [DnaK]:[DnaJ]) needed for refolding. Based on these findings, we propose a revised model of DnaK-DnaJ action in which the chaperones bind either synergistically or competitively, depending on the distribution of sites and the relative stoichiometry of the chaperones.
Experimental Procedures
Materials
Reagents were obtained from the following sources: Platinum Pfx DNA Polymerase (Invitrogen, Calsbad, CA), pMCSG7 plasmid (Midwest Center for Structural Genomics, Bethesda, MD), ATP-agarose column (Sigma, St. Louis, MO), firefly luciferase and Steady-Glo reagent (Promega, Madison, WI). All of the OD and luminescence measurements were performed using a SpectraMax M5 (Molecular Devices, Sunnyvale, CA).
DnaJ affinity matrix
Previously reported results that measured the binding of DnaJ to cellulose-bound peptide arrays 44 were used to generate amino acid frequencies for binding (Pb) and non-binding (Pn) peptides. Binding sites were identified using analysis in ImageJ. This frequency table was then used to estimate the Gibb’s free energy values for binding of DnaJ to each amino acid at each position in the 13mer peptides using the equation of Rudiger et al. 9: ΔΔGbinding = − RT ln(Pb/Pn).
Proteome-wide analyses
Proteomic libraries were obtained from UniProtKB databases. Using the DnaK and DnaJ affinity matrices, every possible 13mer for each sequence in the proteome was parsed, shifting one amino acid at a time, evaluating each 13mer for its ΔΔGbinding. Positive binding sites were those in which the ΔΔGbinding was calculated as < −5 kJ/mol (DnaK) or < −2 kJ/mol (DnaJ). These free energy values represent a binding probability of >80%, as previously described30. The algorithm used is readily available for public use upon request of the authors.
Plasmids and Protein Purification
The E. coli dnaK, dnaJ, and grpE genes were amplified by PCR using Platinum Pfx DNA Polymerase and inserted into the pMCSG7 plasmid through ligation-independent cloning, as previously described 50. His-tagged DnaK, DnaJ and GrpE were expressed and purified as described previously 51. Firefly luciferase and malate dehydrogenase (MDH) were purchased from Promega.
Refolding Assays
The ability of the DnaK-DnaJ system to refold denatured luciferase was evaluated as described 40, 52. Briefly, a concentrated stock of luciferase (8.2 μM) was denatured using 6 M guanidinium hydrochloride (GuHCl) in HEPES buffer (25 mM HEPES, 50 mM potassium acetate, 5 mM DTT, pH 7.2). This stock was incubated at room temperature for one hour, diluted to 0.2 μM in HEPES buffer lacking GuHCl, and then frozen into aliquots. Enzyme mix (10 μL) containing DnaK, DnaJ, GrpE, denatured firefly luciferase in 39 mM HEPES (170 mM potassium acetate, 1.7 mM magnesium acetate, 3 mM DTT, 12 mM creatine phosphate, 50 U/mL creatine kinase, pH 7.6) was first added into each well of a 96-well plate and then 4 μL of 3.5 mM ATP was added to start the reaction. The final concentrations were: DnaK (1 μM), GrpE (0.2 μM), denatured luciferase (8 nM), and ATP (1 mM). After one hour of incubation at 37 °C equilibrium was reached and 14 μL of 2% (v/v) SteadyGlo reagent in 50 mM glycine buffer (30 mM MgSO4, 10 mM ATP and 4 mM DTT, pH 7.8) was added into each well. For each experiment, the signal from a negative control containing all of the components except DnaK was subtracted.
The MDH refolding assay was based on a previous report 53. Briefly, MDH (0.2 μM) was denatured at 47 °C for 30 minutes in 20 mM MOPS buffer (2 mM magnesium acetate, 200 mM KCl, pH = 7.4)). Next, 15 μL of this mixture was transferred into each well of a transparent 96-well plate and the refolding reaction was started by adding 10 μL of enzyme mix containing DnaK (1 μM), DnaJ, GrpE (0.2 μM), DTT (5 mM), and ATP (2.5 mM) dissolved in 20 mM MOPS buffer with 25 mM creatine phosphate and 87.5 U/mL creatine kinase. After 1.5 hour incubation at 30 °C, 75 μL of MOPS buffer containing NADH (0.28 mM), OAA (0.5 mM), and BSA (1 mg/mL) was added to each well, and the decline of OD340 over 5 minutes was recorded. The slope of this data was obtained by linear regression in GraphPad Prism 4.0.
Peptide Microarrays
Glass microarrays were purchased from Jenrin Peptide Technologies (JPT, Berlin). The entire sequence of firefly luciferase was printed as 15 residue segments with overlaps of 10 residues. All peptides were arrayed in triplicate. Solutions (300 μL) containing His-tagged DnaJ (3 μM) or DnaK (5μM) in 25 mM HEPES (100 mM NaCl, 40 mM KCl, 8 mM MgCl2, 0.01% Tween 20, pH 7.4) were incubated in a sandwich apparatus (array slide and blank slide separated by 20μm plastic spacers) for 1 hour at room temperature in a humidified chamber. Arrays were then washed three times (5 minutes per wash) with TBS-T (50 mM Tris-HCl, 150 mM NaCl, 0.1% Tween 20, pH 7.4) at room temperature in Petri dishes. Excess buffer was removed by centrifugation prior to incubation of the slide with a TBS-T solution containing 1:1000 anti-His antibody conjugated with HiLyte-555 (Anaspec) for one hour at room temperature in a humidified chamber protected from light. Following three washes with TBS-T, two with water, and drying under a stream of nitrogen (all carried out in the dark), arrays were scanned with a 532 nm laser at 10 μm resolution on a GenePix 4100A Personal Scanner. Analyses were performed using GenePix Pro software and morphological background correction.
Results
DnaJ and DnaK Consensus Binding Sequence
To probe the E. coli proteome for molecular chaperone binding sites, we first needed a sequence-based affinity matrix for DnaK and DnaJ. Although there has been a published matrix of affinity values for DnaK 9, 54, an equivalent matrix for DnaJ has not yet been established. This was probably due to DnaJ’s propensity to bind some full-length proteins with a better affinity than smaller peptides 55. However, for the exclusive purposes of elucidating internal binding sites for DnaJ on fully unfolded proteins (such as might be found in nascent polypeptides), we sought to build such a matrix. For DnaK, Rudiger et al. examined every amino acid at each position of a predicted 13 residue binding site 9. Using a similar approach, we used previously published data on DnaJ binding to cellulose-bound peptide arrays derived from the sequences of several classic chaperone substrates 44 to estimate the affinities. Specifically, we constructed a matrix of ΔΔGbinding values for each amino acid at every position in a possible 13mer binding site (Supplemental Table 1) in a fashion similar to that done for DnaK 9. The total ΔΔGbinding for a 13mer peptide was calculated as the sum of each individual residue. It should be noted that although previous studies have shown a minimal preference of DnaJ for 7mer or 8mer substrates 28, 43, we have chosen to proceed with the longer, 13mer substrate sites because of their use in the cellulose-bound arrays from which this matrix is derived 44.
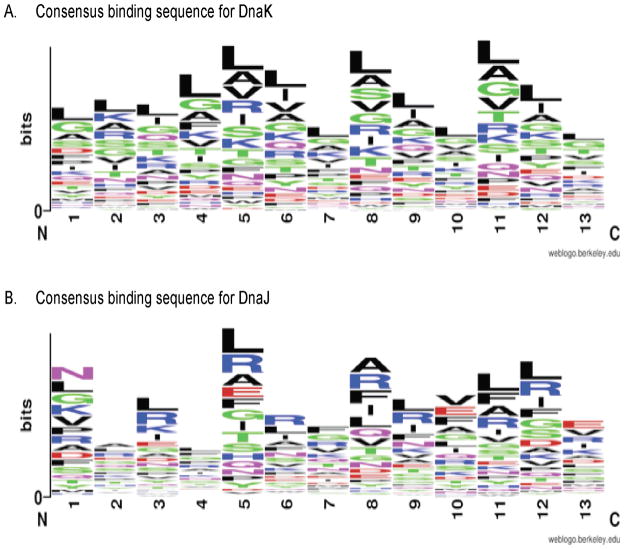
Using this data, we developed visual representations of the consensus binding sequences for DnaK and DnaJ (Figure 2, 56). In concurrence with previous reports 9, DnaK shows a preference for hydrophobic amino acids (Leu, Ile, Val, Ala, Gly), especially in the core positions 5 through 9. Additionally, DnaK has a preference for positively charged amino acids (Arg, Lys) over negatively charged amino acids (Asp, Glu). Similar to DnaK, we found that DnaJ also has a preference for hydrophobic amino acids, especially aromatic amino acids in positions 5 and 8–11. Although this DnaJ affinity matrix is derived from a relatively limited data set, it agrees well with previous suggestions about a possible consensus 28, 43, 44. Further, it is noteworthy that for both chaperones, certain amino acids are overrepresented at specific positions in the 13mer peptide, instead of displaying a site-independent preference. Together, these affinity matrices and consensus sequences positioned us to explore the binding sites in the entire E. coli proteome.
FIGURE 2. CONSENSUS BINDING SEQUENCES OF E. COLI MOECLULAR CHAPERONES.
(A) DnaK: Hydrophobic residues, e.g. Leu, Ile, Gly, Val, and basic residues (Arg, Lys) are highly preferred, while acidic residues are disfavored. (B) DnaJ: Hydrophobic and aromatic residues are clearly preferred, but in a position-specific manner. Figure made using WebLogo Tool (Crooks 2004).
Proteome-Wide DnaK and DnaJ Binding Sites
The DnaK affinity matrix has already been used to predict binding sites in individual proteins 9 and we wished to extend this search to a proteome-wide scale to understand whether classical chaperone substrates are unusual in their distribution or number of DnaK- and DnaJ-binding sites. To address this question, a tight-affinity DnaK binding site has been previously defined as ΔΔGbinding < −5 kJ/mol, which corresponds to a probability of binding of >80%. This value has been verified using experimental peptide-binding studies 30. Using a similar threshold of 80% binding probability, the cutoff for a tight-affinity DnaJ-binding site was determined to be a ΔΔGbinding < minus;2 kJ/mol. We next applied the DnaK and DnaJ affinity matrices to every 13mer for each protein in the E. coli proteome, shifting by a single amino acid with each iteration. Sites that met the free energy cutoffs described above were then annotated as predicted binding sites.
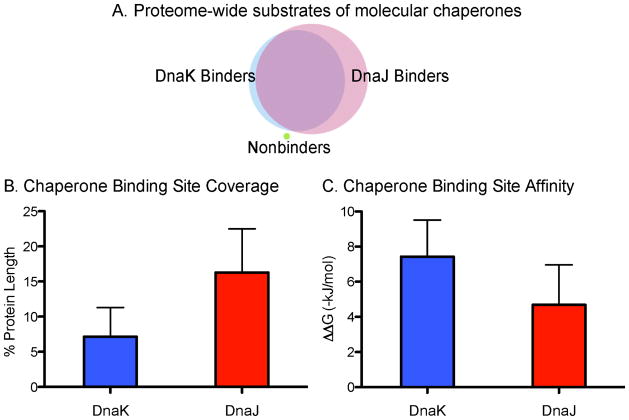
We found that DnaK was predicted to bind 4,215 proteins, or 98% of the total annotated E. coli proteome (Figure 3A, Supplemental Table 2). Further, DnaK binds each protein an average of 23 times, although there was a large variance (Supplemental Table 2). This is in good agreement with predictions that DnaK would bind once every 36 residues, a value based on a handful of classic substrates 9. Thus, we conclude that the model DnaK substrates (e.g. luciferase, citrate synthase, etc.) are relatively average in their number of predicted binding sites. As might be expected, one major factor in determining the absolute number of DnaK-binding sites in a protein was the length of the polypeptide. To normalize for differences in length, we calculated the contribution of DnaK binding sites to the total protein size and found that over 7% of an individual protein’s sequence consists of potential DnaK binding sites (Figure 3B, Supplemental Table 2). Additionally, DnaK’s binding affinity is relatively consistent from site to site, as the average ΔΔGbinding was −7.43 ± 2.08 kJ/mol (Figure 3C, Supplemental Table 2).
FIGURE 3. PROTEOME-WIDE MAPPING OF DNAK AND DNAJ BINDING SITES IN E. COLI.
(A) Venn diagram of DnaK and DnaJ-binders. The majority of the proteome is bound by both chaperones, with only one peptide binding neither (UniProtID: P60995). (B) Average binding affinities for DnaK and DnaJ across the proteome. Values given as −ΔΔGbinding. (C) Chaperones binding site coverage. Values are given as percent coverage of individual protein lengths, averaged over the entire proteome.
We then compared the DnaK and DnaJ binding sites. We were particularly interested in whether DnaJ might have more/fewer sites and whether the two types of sites were, on average, relatively close together or far apart in a linear peptide sequence. Our results showed that DnaJ was also predicted to interact with a large percentage of the E. coli proteome, binding 4,294 proteins or 99.8% of the total proteome (Figure 3A, Supplemental Table 3). Together, proteins with either a predicted DnaK- or DnaJ-binding site account for 99.99% of the proteome, with the only outlier being a single 16-mer His operon leader peptide (UniProtID: P60095). Interestingly, of the DnaK-binding proteins, all but 9 (0.2%) also were predicted to bind DnaJ. Conversely, 87 proteins (~2%) were predicted to bind DnaJ only (Figure 3A; Supplemental Tables 4 and 5).
To exclude the possibility that binding sites were arising from a skewed data population, we ran our algorithm against both the natural proteome and a randomized artificial proteome using our experimentally derived affinity matrices and various scrambled matrices. The controls using the artificial proteome or the scrambled matrices all gave a significantly decreased output in binding sites (data not shown).
While one might suspect that a relatively weaker free energy cutoff for DnaJ (−2 kJ/mol vs −5 kJ/mol) might yield sites with worse affinity, this was not the case. In fact, DnaJ was predicted to bind most proteins with good affinity (average ΔΔGbinding was −4.69 ± 2.27), only slightly weaker than DnaK (Figure 3C, Supplemental Table 3) and indeed statistically indifferent (p-value > 0.05). Next, we were curious as to how frequently DnaJ might bind each protein. Interestingly, despite an average affinity not significantly different from DnaK, DnaJ was predicted to bind over 52 times per protein, which is more than twice the frequency of DnaK. Normalization for protein length showed that DnaJ-binding sites accounted for over 16% of a protein sequence (Figure 3B, Supplemental Table 3). Since DnaK exhibits a symmetric affinity matrix, we also chose to examine how DnaJ’s substrates would change were it to bind similarly as its larger chaperone partner. By averaging the ΔΔGbinding values for positions 1 and 13, 2 and 12, 3 and 11, and so on, we were able to render the DnaJ affinity matrix symmetric. Interestingly, although the number of substrates for DnaJ did not decrease significantly, the number of times it bound a single protein, and the total number of binding sites decreased by over 30% (data not shown).
Binding Sites for DnaJ and DnaK Overlap in the E. coli Proteome
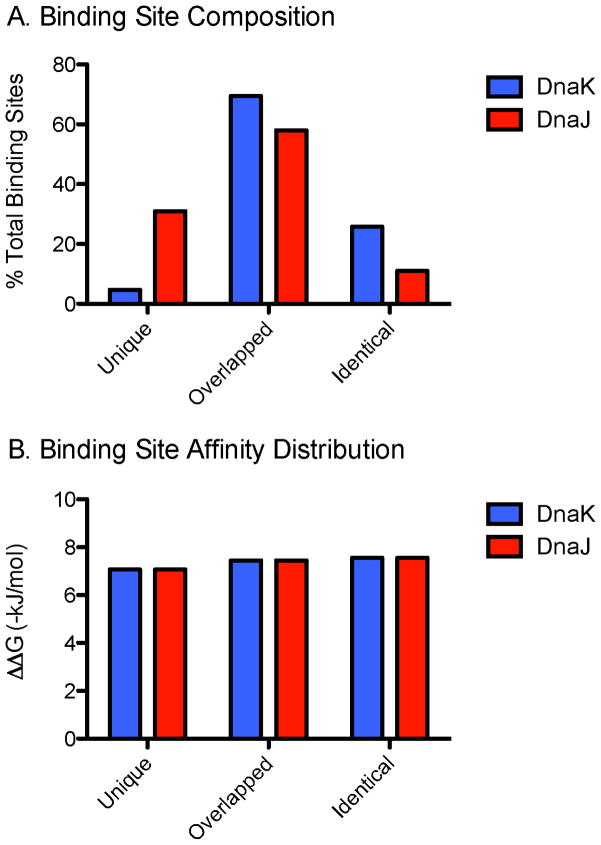
Previous studies on individual model proteins and peptides suggest that DnaK and DnaJ have similar binding sites that might partially or completely overlap in some cases 44–49. However, the significance and generality of this finding hasn’t been clear. To understand whether overlapping sites are common in a broader context, we first defined overlapping sites as those regions that contain a DnaJ-binding site within 13 residues of a DnaK site (or vice-versa). This distance was chosen because of structural work on substrate binding to both chaperones and predictions about steric exclusion 34, 57. We found that an astounding 95% of DnaK sites overlapped with DnaJ sites, with 26% being identical (Figure 4A). Conversely, only 69% of DnaJ binding sites were predicted to overlap with DnaK. This result is consistent with the promiscuity of DnaJ suggested both experimentally 44, 58 and computationally 43. Curiously, we found no difference in the apparent affinity of DnaK or DnaJ for their substrates regardless of whether the binding site was unique or overlapping (Figure 4B).
FIGURE 4. MOST DNAK SITES ARE ALSO DNAJ SITES.
(A) Chaperone binding site composition given as unique, overlapped (by corresponding chaperone), or identical, given as percent of total sites (DnaK: 95,558; DnaJ: 224,530). (B) Binding site affinity shows little to no variation across the three types of binding sites. Affinity values given as −ΔΔGbinding.
Comparison with GroEL-Dependent Substrates
GroEL-GroES is the other major arm of the bacterial protein folding system. GroEL forms a double-ring, cylindrical structure that is capped by GroES, forming a hollow interior chamber that promotes folding 59. Similar to the DnaK-DnaJ system, GroEL-GroES is thought to act on a relatively large number of protein substrates. Consistent with this idea, proteomic studies in a temperature-sensitive GroEL mutant strain revealed 302 proteins, or about 7% of the proteome, that are dependent on this chaperone for solubility 60. We wondered whether these specific substrates might have an unusual pattern of DnaK- or DnaJ-binding sites and applied our algorithm to this subset. However, we found that both DnaK and DnaJ bound nearly all of the GroEL substrates (DnaK: 98.3%, DnaJ: 99.7%) with no apparent differences in the predicted affinity or number of sites (data not shown). These findings suggest that the GroEL-dependent proteins also contain predicted DnaK-DnaJ sites; thus, some other feature must make these proteins particularly dependent on GroEL-GroES.
Analysis of DnaK- and DnaJ-Binding Sites in Thermophilic Archaea
The DnaK-DnaJ system is highly conserved from bacteria to mammals. However, a recent study by the Kampinga group has identified a handful of archaea that lack an obvious DnaK or DnaJ ortholog 61. We are interested in whether the proteomes from these organisms might have distinct patterns of chaperone-binding sites. To this end, we explored the proteomes of Thermococcus kodakaraensis and Methanococcus janaschii, two archaea species lacking dnaJ and dnaK genes 61. As a control, we also examined the proteomes of two additional thermophilic archaea that have dnaJ and dnaK genes (Thermoplasma acidophilum and Methanosarcina mazei). Interestingly, we found that the proteomes of all four archaea contained thousands of predicted binding sites, with nearly complete coverage in every case (Table 1). This finding suggests that potential chaperone-binding sequences may have existed before the evolution of DnaK-DnaJ. In other words, the proteome within these organisms still folds and buries hydrophobic, aggregation-prone sequences, but lacks a molecular mechanism for aiding the folding of unfolded proteins or misfolded intermediates.
TABLE 1. EVOLUTIONARY CONSERVATION OF CHAPERONE BINDING SITES.
Analysis of archaeal organisms shows binding site coverage by DnaK and DnaJ of nearly the entire proteome.
| Organism | DnaK homolog | DnaJ homolog | DnaK | DnaJ |
|---|---|---|---|---|
| E coli | + | + | 95,558 sites 4,215 proteins (98.0%) |
224,530 sites 4,294 proteins (99.8%) |
| T. acidophilum | + | + | 33,849 sites 1,480 proteins (99.9%) |
89,326 sites 1,481 proteins (99.9%) |
| M. mazei | + | + | 62,680 sites 3,297 proteins (99.8%) |
169,210 sites 3,300 proteins (99.9%) |
| T. kodakaraensis | − | − | 132,343 sites 2,305 proteins (99.9%) |
191,386 sites 2,308 proteins (100%) |
| M. janaschii | − | − | 34,317 sites 1,785 proteins (99.9%) |
94,498 sites 1,786 proteins (99.9%) |
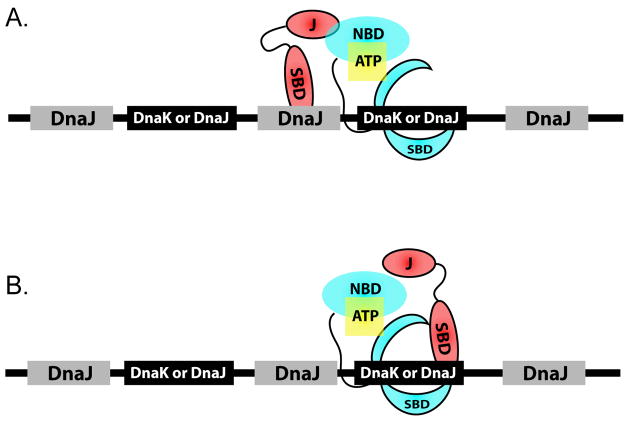
Revised Binding Model for the DnaK-DnaJ-Substrate Ternary Complex
Previous studies have converged on two possible models for how DnaK-DnaJ might cooperate to fold substrates: one in which DnaJ and DnaK separately bind polypeptide chains 33, 48 and a second in which the two chaperones compete for the same site and substrates are transferred from one chaperone to the other. The first model is often preferred based on data clearly showing enhanced folding by the DnaK-DnaJ system 33, 34, 44. However, it is also commonly observed that DnaJ will block folding at higher concentrations 27, 40, suggesting that competition can also take place.
Our data on predicted DnaK and DnaJ binding sites across the E. coli proteome suggest that both ideas may be valid. Because DnaJ sites outnumber and overlap nearly all DnaK sites, we suggest a more generalized model, incorporating both these ideas, in which unique DnaJ sites are interspersed between sites shared by both DnaK and DnaJ sites (Figure 5). This observation suggests that ternary complex formation will occur only if the shared DnaK/DnaJ sites are not saturated with DnaJ. Such a situation would allow DnaJ and DnaK to bind substrates synergistically, but only under conditions of permissive DnaK:DnaJ stoichiometry.
FIGURE 5. MODEL FOR THE ARRANGEMENT OF DNAK AND DNAJ BINDING SITES ON UNFOLDED SUBSTRATE PROTEINS.
Unique DnaJ sites are intersperced between common DnaK and DnaJ binding targets.
The Optimal DnaK:DnaJ Ratio for Substrate Refolding Correlates with the Composition of Computationally Predicted Binding Sites
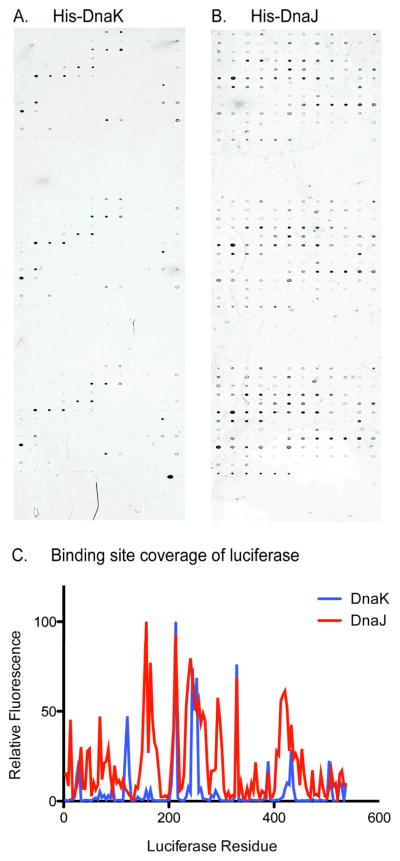
Before testing this hybrid model, we wanted to first understand whether our computationally predicted DnaK- and DnaJ-binding sites were consistent with experimentally determined binding regions. Towards that goal, we focused on firefly luciferase, a well-studied model chaperone substrate. We designed glass arrays spotted with peptides representing the entire luciferase protein; it contained 136 15mers with 10 residue overlaps in triplicate. Binding of His-tagged DnaK (5 μM) or DnaJ (3 μM) to these arrays was measured using fluorescent anti-His antibodies. For both DnaK and DnaJ, we found that the predicted binding sites matched remarkably well with the experimental results (Figures 6A, 6B, and 7B). Figure 6C gives a quantitative comparison of DnaK and DnaJ binding across the luciferase sequence. Further, this binding data appears to be conserved even in human chaperones (data not shown).
FIGURE 6. GLASS-SLIDE PEPTIDE MICROARRAYS CONFIRM COMPUTATIONAL PREDICTION OF CHAPERONE BINDING SITES ON LUCIFERASE.
(A) and (B). His-tagged DnaK and DnaJ,respectively, binding of luciferase confirm in silico predicitions (Figure 6). Results correlate well with previously published platforms (Rudiger 1997; Rudiger 2001). (C) Quantified binding site coverage for molecular chaperones on luciferase. Fluorescence is normalized to maximum values for each array.
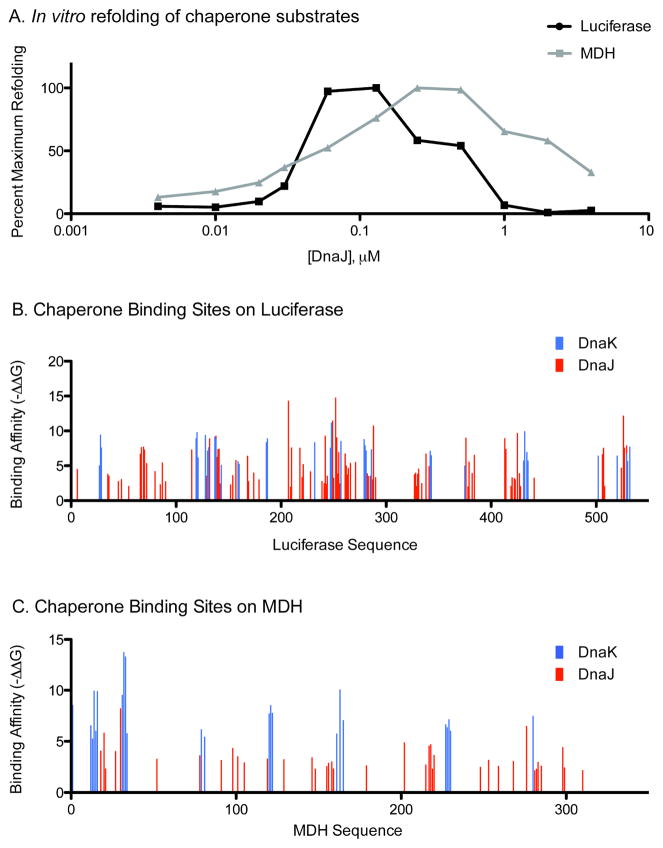
FIGURE 7. DNAJ DIFFERENTIALLY INHIBITS LUCIFERASE AND MDH REFOLDING.
(A) In vitro refolding assay shows DnaJ inhibiting refolding of luciferase at lower concentrations than for MDH. (B) and (C) Computational prediction of chaperone binding sites. Note luciferase contains many more DnaJ sites that bind at higher affinity than MDH.
Next, we wanted to understand how the proportion of unique and shared DnaK- and DnaJ-binding sites might contribute to the optimum stoichiometry between DnaK and DnaJ needed to achieve folding. In this task, we compared the chaperone-assisted folding of luciferase with another model substrate, malate dehydrogenase (MDH). These two substrates are particularly well suited for this question because luciferase is predicted to have 134 possible DnaJ binding sites, of which a remarkable ~81% are shared with DnaK, while MDH has 61 total DnaJ binding sites and only ~72% are predicted to be shared with DnaK. Thus, these two substrates differ in their total number of sites and their percentage of unique DnaJ binding sites (Table 2). Further, in vitro methods for the chaperone-mediated refolding of these substrates are well established 40. Thus, although analyses of linear, extended sequences are clearly an over-simplification of the complexity of protein folding reactions, we thought that these substrates might serve as useful tools for probing the potential relevance of the predicted binding sites. Towards that goal, we denatured recombinant luciferase and MDH, diluted them to the final concentration previously reported to be best for refolding 53, and then monitored the refolding reactions by monitoring the recovery of enzymatic activity (light production or malate conversion to oxaloacetate, respectively). To identify the optimal ratio between DnaK and DnaJ needed to achieve optimal refolding, we fixed the concentration of DnaK and varied the levels of DnaJ. This approach revealed that the optimal ratio between DnaK and DnaJ needed to refold luciferase was approximately 5:1. This result is consistent with previous reports suggesting luciferase refolding only occurs under conditions of substoichiometric DnaJ 21. Interestingly, the stoichiometry of these chaperones in the E. coli cytosol is a similar 10:1 62. Next, we performed parallel experiments using denatured MDH and, interestingly, found that the optimal DnaK:DnaJ ratio was only 2:1 (Figure 7A). As expected, higher concentrations of DnaJ suppressed folding of both substrates. The suppression of folding by DnaJ was more dramatic in the case of luciferase than MDH, but we hesitate to interpret this particular observation because luciferase appears to be more prone to off-pathway, irreversible aggregation.
TABLE 2. PREDICTED DNAK AND DNAJ BINDING SITES ON LUCIFERASE AND MDH.
Luciferase contains over 10% more shared binding sites relative to MDH and is predicted to bind DnaJ significantly tighter than MDH.
| Luciferase | MDH | |
|---|---|---|
| Unique DnaK sites | 0 | 1 (1.6%) |
| Unique DnaJ sites | 25(18.7%) | 17(27.8%) |
| Shared Binding Sites | 109(81.3%) | 43 (70.5%) |
| Total Sites | 134 | 61 |
| Average ΔΔGbinding for DnaK (kJ/mol); p-value > 0.05 | −7.51 ± 1.58 | −7.87 ± 1.32 |
| Average ΔΔGbinding for DnaJ (kJ/mol); p-value = 7.86e-5 | −5.27 ± 2.78 | −3.43 ± 1.27 |
Comparing the results of the refolding assays to the predicted number and composition of DnaK and DnaJ binding sites (Figures 7B and 7C) suggest some potentially interesting connections. As discussed above, luciferase is predicted to have a relatively higher percentage of shared binding sites, when compared to MDH. Additionally, DnaJ seems to bind its sites on luciferase with a higher affinity than on MDH (Table 2). What does this mean for the refolding reactions? If DnaJ randomly “selects” binding sites, as was suggested by the relatively narrow differences between free energies, then DnaJ would be expected to compete with DnaK binding sites more often in luciferase. This might be why luciferase becomes “saturated” with DnaJ at a relatively lower concentration. Alternatively, DnaJ’s higher affinity binding of luciferase compared to MDH might lead to a longer duration of interaction, preventing DnaK association, thus resulting in refolding inhibition. Although this hypothesis needs further validation using a wider range of substrates, these observations provide an intriguing initial link between computationally predicted binding sites and their possible impact on chaperone-mediated folding in vitro.
Discussion and Conclusions
There has been great interest in understanding how the molecular chaperones DnaK and DnaJ interact with substrates to assist their folding. Pioneering work by the Bukau group and others has established consensus-binding sequences for DnaK and provided substantial insight into this bimolecular interaction on a limited number of substrates 9, 28, 30, 44–49. In our study, we sought to expand this analysis to understand how DnaK and its important co-chaperone, DnaJ, might interact with the entire proteome. In doing so, we developed a similar affinity matrix for DnaJ (Supplemental Table 1) and probed the large-scale proteome. Our results indicate that the two chaperones are predicted to interact with 99.99% of the E. coli proteome (Figure 3A). While this number exceeds that of previous predictions 63, it is likely that the potential to bind DnaK or DnaJ is advantageous to a client, particularly during stress conditions when these chaperones are most active 4, 5. The frequency of DnaK sites is in good agreement with the frequency observed using classic chaperone substrates 9, suggesting at a proteome wide level that these classic chaperone substrates serve as a good representation of DnaK and DnaJ interactions with globular proteins. Furthermore, we found that both chaperones bound each substrate multiple times, with DnaJ binding sites taking up nearly twice as much space on the linear peptide sequences as sites for DnaK (16% vs. 7%). Unexpectedly, we found that more than 95% of predicted DnaK sites overlap with a DnaJ site (see Figure 4A). Moreover, given the lack of substantial differences in the affinities for the different binding sites, it is unlikely that DnaK and DnaJ strongly prefer one site to another in the absence of secondary structure or other factors.
Next, we applied our algorithm to the proteomes of several archea, including two recently identified as lacking genes for homologs of DnaK or DnaJ 61. The results indicated that all sets of proteomes contained numerous chaperone target sites, even in those without actual chaperones (Table 1). This data supports a novel idea that chaperones may have emerged downstream of their clients in the evolutionary pathway. As the binding sites for yeast and human chaperones are elucidated, it will be most interesting to apply our algorithm to the corresponding proteomes for a more detailed analysis.
Finally, these studies have led us to propose a “hybrid” model for how DnaK and DnaJ bind to their peptide substrates. In this model, unique DnaJ sites are interspersed between sites that are shared by both DnaK and DnaJ (Figure 5). This model includes aspects consistent with previous ideas, which feature either simultaneous binding of substrate by both chaperones (e.g. formation of a ternary complex) or sequential transfer of client proteins from DnaJ to DnaK 23, 44. The hybrid model accommodates both these theories, and argues that the individual substrate may determine which mechanism is dominant. For example, the outcome of interactions between the DnaK-DnaJ system and its various substrates are predicted to depend on: (a) the relative stoichiometry of DnaJ and DnaK, (b) the proportion of shared sites and (c) the number of unique DnaJ sites. In contrast, the individual affinities of DnaJ and DnaK for their substrates are predicted to be relatively minor contributing factor, though can factor in on occasion (such as with luciferase compared to MDH, Table 2). The issue of stoichiometry, though, appears to be a particularly critical component in this model because of potential competition between DnaJ and DnaK on shared sites. In this way, our model agrees with published data regarding the optimal stoichiometry of DnaK and DnaJ for proper substrate folding 27, 40. Specifically, at substoichiometric levels, DnaJ is known to accelerate substrate refolding, likely due to its help in recruiting client to DnaK and stimulating ATP turnover. However, (as shown in Figure 7 and Table 1), our data suggests that the DnaJ-inhibition of refolding observed at is due to DnaJ competing with DnaK for shared binding sites. While this a thermodynamic computation analysis of binding sites, interestingly we have reached a similar conclusion to that postulated from a kinetic computational model of refolding. In 2006, Hu et al. used kinetic data regarding the on and off rates of DnaK, DnaJ, and substrate binding interactions to model refolding outcomes 41. They found that while DnaJ oligomerization or binding to DnaK’s substrate binding site would likely not explain DnaJ-mediated inhibition of refolding, indeed competition for substrate was a more plausible model. Taken together, the work of Hu and the work presented herein provide strong support for the proposed model. Indeed, high levels of DnaJ might compete with DnaK for shared binding sites, or alternatively block refolding by staying on the high-affinity unique sites for “too long”, as substrates are unlikely to fold if chaperones remain bound to their hydrophobic regions. Though individual substrates may indeed follow either mechanism, the latter idea – at least for luciferase and MDH – appears to be very important given the difference in their binding affinities for DnaJ. Thus, the reversibility of chaperone interactions is important, which could be an advantage of having a high percentage of shared DnaK- and DnaJ-binding sites.
Supplementary Material
Acknowledgments
We thank Dr. Stefan Rüdiger for supplying the DnaK affinity matrix and Drs. Bernd Bukau and Liu Kang for advice. This work was supported by NSF grant MCB-0844512 and NIH grants 5T32GM007863-32 (MSTP) and NS059690.
References
- 1.Feldman DE, Frydman J. Curr Opin Struct Biol. 2000;10:26–33. doi: 10.1016/s0959-440x(99)00044-5. [DOI] [PubMed] [Google Scholar]
- 2.Genevaux P, Georgopoulos C, Kelley WL. Mol Microbiol. 2007;66:840–857. doi: 10.1111/j.1365-2958.2007.05961.x. [DOI] [PubMed] [Google Scholar]
- 3.Georgopoulos C. Genetics. 2006;174:1699–1707. doi: 10.1534/genetics.104.68262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hansen S, Lewis K, Vulic M. Antimicrob Agents Chemother. 2008;52:2718–2726. doi: 10.1128/AAC.00144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu A, Tran L, Becket E, Lee K, Chinn L, Park E, Tran K, Miller JH. Antimicrob Agents Chemother. 2010;54:1393–1403. doi: 10.1128/AAC.00906-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ER. Proc Natl Acad Sci U S A. 2009;106:8471–8476. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blond-Elguindi S, Cwirla SE, Dower WJ, Lipshutz RJ, Sprang SR, Sambrook JF, Gething MJ. Cell. 1993;75:717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- 8.Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rudiger S, Germeroth L, Schneider-Mergener J, Bukau B. EMBO J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flynn GC, Pohl J, Flocco MT, Rothman JE. Nature. 1991;353:726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- 11.Gragerov A, Zeng L, Zhao X, Burkholder W, Gottesman ME. J Mol Biol. 1994;235:848–854. doi: 10.1006/jmbi.1994.1043. [DOI] [PubMed] [Google Scholar]
- 12.Schlecht R, Erbse AH, Bukau B, Mayer MP. Nat Struct Mol Biol. 2011;18:345–351. doi: 10.1038/nsmb.2006. [DOI] [PubMed] [Google Scholar]
- 13.Jiang J, Maes EG, Taylor AB, Wang L, Hinck AP, Lafer EM, Sousa R. Mol Cell. 2007;28:422–433. doi: 10.1016/j.molcel.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moro F, Fernandez-Saiz V, Muga A. Protein Sci. 2006;15:223–233. doi: 10.1110/ps.051732706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pellecchia M, Montgomery DL, Stevens SY, Vander Kooi CW, Feng HP, Gierasch LM, Zuiderweg ER. Nat Struct Biol. 2000;7:298–303. doi: 10.1038/74062. [DOI] [PubMed] [Google Scholar]
- 16.Slepenkov SV, Witt SN. FEBS Lett. 2003;539:100–104. doi: 10.1016/s0014-5793(03)00207-2. [DOI] [PubMed] [Google Scholar]
- 17.Swain JF, Dinler G, Sivendran R, Montgomery DL, Stotz M, Gierasch LM. Mol Cell. 2007;26:27–39. doi: 10.1016/j.molcel.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kampinga HH, Craig EA. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karzai AW, McMacken R. J Biol Chem. 1996;271:11236–11246. doi: 10.1074/jbc.271.19.11236. [DOI] [PubMed] [Google Scholar]
- 20.Kelley WL. Trends Biochem Sci. 1998;23:222–227. doi: 10.1016/s0968-0004(98)01215-8. [DOI] [PubMed] [Google Scholar]
- 21.Laufen T, Mayer MP, Beisel C, Klostermeier D, Mogk A, Reinstein J, Bukau B. Proc Natl Acad Sci U S A. 1999;96:5452–5457. doi: 10.1073/pnas.96.10.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Proc Natl Acad Sci U S A. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Misselwitz B, Staeck O, Rapoport TA. Mol Cell. 1998;2:593–603. doi: 10.1016/s1097-2765(00)80158-6. [DOI] [PubMed] [Google Scholar]
- 24.Wall D, Zylicz M, Georgopoulos C. J Biol Chem. 1994;269:5446–5451. [PubMed] [Google Scholar]
- 25.Ahmad A, Bhattacharya A, McDonald RA, Cordes M, Ellington B, Bertelsen EB, Zuiderweg ER. Proc Natl Acad Sci U S A. 2011;108:18966–18971. doi: 10.1073/pnas.1111220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banecki B, Liberek K, Wall D, Wawrzynow A, Georgopoulos C, Bertoli E, Tanfani F, Zylicz M. J Biol Chem. 1996;271:14840–14848. doi: 10.1074/jbc.271.25.14840. [DOI] [PubMed] [Google Scholar]
- 27.Szabo A, Korszun R, Hartl FU, Flanagan J. EMBO J. 1996;15:408–417. [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Qian X, Sha B. Structure. 2003;11:1475–1483. doi: 10.1016/j.str.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 29.Hartl FU. Nat Med. 2011;17:1206–1210. doi: 10.1038/nm.2467. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez F, Arsene-Ploetze F, Rist W, Rudiger S, Schneider-Mergener J, Mayer MP, Bukau B. Mol Cell. 2008;32:347–358. doi: 10.1016/j.molcel.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 31.Summers DW, Douglas PM, Ramos CH, Cyr DM. Trends Biochem Sci. 2009;34:230–233. doi: 10.1016/j.tibs.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pierpaoli EV, Sandmeier E, Schonfeld HJ, Christen P. J Biol Chem. 1998;273:6643–6649. doi: 10.1074/jbc.273.12.6643. [DOI] [PubMed] [Google Scholar]
- 33.Han W, Christen P. J Biol Chem. 2003;278:19038–19043. doi: 10.1074/jbc.M300756200. [DOI] [PubMed] [Google Scholar]
- 34.Han W, Christen P. FEBS Lett. 2004;563:146–150. doi: 10.1016/S0014-5793(04)00290-X. [DOI] [PubMed] [Google Scholar]
- 35.Szabo A, Langer T, Schroder H, Flanagan J, Bukau B, Hartl FU. Proc Natl Acad Sci U S A. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Linke K, Wolfram T, Bussemer J, Jakob U. J Biol Chem. 2003;278:44457–44466. doi: 10.1074/jbc.M307491200. [DOI] [PubMed] [Google Scholar]
- 37.Perales-Calvo J, Muga A, Moro F. J Biol Chem. 2010;285:34231–34239. doi: 10.1074/jbc.M110.144642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan CY, Lee S, Cyr DM. Cell Stress Chaperones. 2003;8:309–316. doi: 10.1379/1466-1268(2003)008<0309:mfrohf>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu Z, Cyr DM. J Biol Chem. 1998;273:27824–27830. doi: 10.1074/jbc.273.43.27824. [DOI] [PubMed] [Google Scholar]
- 40.Chang L, Thompson AD, Ung P, Carlson HA, Gestwicki JE. J Biol Chem. 2010;285:21282–21291. doi: 10.1074/jbc.M110.124149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu B, Mayer MP, Tomita M. Biophys J. 2006;91:496–507. doi: 10.1529/biophysj.106.083394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCarty JS, Buchberger A, Reinstein J, Bukau B. J Mol Biol. 1995;249:126–137. doi: 10.1006/jmbi.1995.0284. [DOI] [PubMed] [Google Scholar]
- 43.Kota P, Summers DW, Ren HY, Cyr DM, Dokholyan NV. Proc Natl Acad Sci U S A. 2009;106:11073–11078. doi: 10.1073/pnas.0900746106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rudiger S, Schneider-Mergener J, Bukau B. EMBO J. 2001;20:1042–1050. doi: 10.1093/emboj/20.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frydman J, Hartl FU. Science. 1996;272:1497–1502. doi: 10.1126/science.272.5267.1497. [DOI] [PubMed] [Google Scholar]
- 46.Gamer J, Bujard H, Bukau B. Cell. 1992;69:833–842. doi: 10.1016/0092-8674(92)90294-m. [DOI] [PubMed] [Google Scholar]
- 47.Gamer J, Multhaup G, Tomoyasu T, McCarty JS, Rudiger S, Schonfeld HJ, Schirra C, Bujard H, Bukau B. EMBO J. 1996;15:607–617. [PMC free article] [PubMed] [Google Scholar]
- 48.Kim SY, Sharma S, Hoskins JR, Wickner S. J Biol Chem. 2002;277:44778–44783. doi: 10.1074/jbc.M206176200. [DOI] [PubMed] [Google Scholar]
- 49.Wawrzynow A, Zylicz M. J Biol Chem. 1995;270:19300–19306. doi: 10.1074/jbc.270.33.19300. [DOI] [PubMed] [Google Scholar]
- 50.Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. Protein Expr Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 51.Chang L, Miyata Y, Ung PM, Bertelsen EB, McQuade TJ, Carlson HA, Zuiderweg ER, Gestwicki JE. Chem Biol. 2011;18:210–221. doi: 10.1016/j.chembiol.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wisen S, Gestwicki JE. Anal Biochem. 2008;374:371–377. doi: 10.1016/j.ab.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 53.Diamant S, Goloubinoff P. Biochemistry. 1998;37:9688–9694. doi: 10.1021/bi980338u. [DOI] [PubMed] [Google Scholar]
- 54.de Crouy-Chanel A, Kohiyama M, Richarme G. J Biol Chem. 1996;271:15486–15490. doi: 10.1074/jbc.271.26.15486. [DOI] [PubMed] [Google Scholar]
- 55.Feifel B, Schonfeld HJ, Christen P. J Biol Chem. 1998;273:11999–12002. doi: 10.1074/jbc.273.20.11999. [DOI] [PubMed] [Google Scholar]
- 56.Crooks GE, Hon G, Chandonia JM, Brenner SE. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mayer MP, Rudiger S, Bukau B. Biol Chem. 2000;381:877–885. doi: 10.1515/BC.2000.109. [DOI] [PubMed] [Google Scholar]
- 58.Li J, Sha B. Biochem J. 2005;386:453–460. doi: 10.1042/BJ20041050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horwich AL, Farr GW, Fenton WA. Chem Rev. 2006;106:1917–1930. doi: 10.1021/cr040435v. [DOI] [PubMed] [Google Scholar]
- 60.Chapman E, Farr GW, Usaite R, Furtak K, Fenton WA, Chaudhuri TK, Hondorp ER, Matthews RG, Wolf SG, Yates JR, Pypaert M, Horwich AL. Proc Natl Acad Sci U S A. 2006;103:15800–15805. doi: 10.1073/pnas.0607534103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hageman J, van Waarde MA, Zylicz A, Walerych D, Kampinga HH. Biochem J. 2011;435:127–142. doi: 10.1042/BJ20101247. [DOI] [PubMed] [Google Scholar]
- 62.Bardwell JC, Tilly K, Craig E, King J, Zylicz M, Georgopoulos C. J Biol Chem. 1986;261:1782–1785. [PubMed] [Google Scholar]
- 63.Hartl FU, Hayer-Hartl M. Nat Struct Mol Biol. 2009;16:574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.