Abstract
Mutations in the second EF-hand (D61N, D63N, D65N, E72A) of S100B were used to study its Ca2+-binding and dynamic properties in the absence and presence of abound target, TRTK-12. With D63NS100B as an exception (D63NKD = 50 ± 9 µM), Ca2+-binding to EF2-hand mutants were reduced by more than 8-fold in the absence of TRTK-12 (D61NKD = 412 ± 67 µM; D65NKD = 968 ± 171 µM; E72AKD = 471 ± 133 µM), when compared to wild-type protein (WTKD = 56 ± 9 µM). For the TRTK-12 complexes, the Ca2+-binding affinity to wild type (WT+TRTKKD = 12 ± 10 µM) and the EF2 mutants were increased by 5- to 19-fold versus in the absence of target (D61N+TRTKKD = 29 ± 1.2 µM; D63N+TRTKKD = 10 ± 2.2 µM; D65N+TRTKKD = 73 ± 4.4 µM; E72A+TRTKKD = 18 ± 3.7 µM). In addition, Rex, as measured using relaxation dispersion for side chain 15N resonances of Asn63 (D63NS100B) was reduced upon TRTK-12 binding when measured by nuclear magnetic resonance (NMR). Likewise, backbone motions on multiple time scales (ps-ms) throughout wild type, D61NS100B D63NS100B, and D65NS100B were lowered upon binding TRTK-12. However, the X-ray structures of Ca2+-bound (2.0 Å) and TRTK-bound (1.2 Å) D63NS100B showed no change in Ca2+ coordination, so these and analogous structural data for the wild-type protein could not be used to explain how target binding increased Ca2+-binding affinity in solution. Thus, a model for how S100B-TRTK12 complex formation increases Ca2+ binding is discussed, which considers changes in protein dynamics upon binding the target TRTK-12.
Keywords: NMR, relaxation dispersion, 15N relaxation, calcium-binding proteins, EF-hand, X-ray crystallography, S100 proteins, S100B
Introduction
S100 proteins (S100s) are mammalian Ca2+-binding proteins that were named based on their solubility in 100% saturated ammonium sulfate1. They have no inherent enzymatic activity and function by regulating biological pathways via specific Ca2+-dependent protein-protein interactions. S100s are distributed in a tissue-specific manner, a trend that is recapitulated in a large number of human cancers. Many S100s are clinical markers for cancer, and for S100B, elevated levels directly correlate with poor patient prognosis in malignant melanoma, glioblastoma, and anaplastic astrocytoma. Although the mechanism for how S100B contributes to cancer progression is not fully understood, elevated S100B contributes to lowering p53 protein levels and its tumor suppression activities, including those involving growth arrest and apoptosis. Thus, therapeutic strategies are underway to (i) target Ca2+-bound S100B with small molecule inhibitors; (ii) block the Ca2+-dependent S100B-p53, S100B-hdm2, and S100B-hdm4 interactions, and (iii) restore p53-dependent tumor suppressor activities in cancers with elevated S100B and wild-type p53, such as in malignant melanoma13. To help achieve this goal, it is necessary to characterize the structural, dynamic, and thermodynamic/kinetic properties associated with the formation of functionally important protein-protein interactions involving S100 proteins such as S100B. A feature of many EF-hand signaling proteins is that they do not bind Ca2+ with high affinity unless they are bound to a biological target(s). In other words, in the absence of target, the Ca2+-binding affinity for most S100 proteins is relatively low (i.e. in the µM range), but when bound to peptides (i.e. TRTK-12) or full-length proteins (i.e. S100A1 bound to full length RyR17), the Ca2+-binding affinity can be increased by 5- to 300-fold, respectively. This property is important physiologically because there are over six hundred EF-hand Ca2+-binding proteins in the human genome, yet Ca2+ homeostasis is maintained within each cells such that sufficient free Ca2+ ion concentrations is available at all times for proper signaling (i.e. 100 to 500 nM). Thus, as a physiological control mechanism, S100s and many other EF-hand proteins do not sequester significant amounts of free Ca2+ from the intracellular pool at any given time unless their functionally relevant molecular target is available. For drug design, this phenomenon is important to understand at the molecular level because an S100 inhibitor needs to fully mimic an EF-hand-target protein complex to be therapeutically effective with minimal side effects.
One possible mechanism for an increased Ca2+-binding affinity is that a structural change occurs upon target binding that provides more optimal Ca2+ coordination. For example, a high Ca2+-binding affinity is achieved for the EF-hand protein parvalbumin (KD = 400 nM) due to an oxygen ligand at the 9 coordination position from a glutamic acid residue20;21; where as, S100 proteins and most other EF-hand proteins bind Ca2+ with lower affinity and have an oxygen ligand contributed by a water molecule at this position. However, comparison of X-ray crystallographic structures demonstrated that Ca2+ coordination remains the same, with a H2O at position 9 for wild-type S100B (i.e. ± TRTK-12)14. Thus, the increased Ca2+ ion-binding affinity observed in the presence of TRTK-12 was not the result of a structural change involving a new ligand from the protein in the Ca2+-coordination sphere (i.e. from position 9).
An alternative mechanism is that dynamic properties throughout the protein contribute to lower Ca2+-binding affinities in the absence of target, and that stabilizing these motions via S100-target complex formation impact residues involved in binding Ca2+ within EF2. Interestingly, when the TRTK-12 peptide was bound to S100B, changes in 15N relaxation was observed for backbone and side chain amides throughout the protein. These results, as well as those of mutant constructs in EF2, are consistent with a model in which the Ca2+-bound S100B equilibrium is shifted from a dynamic ensemble of weaker Ca2+-binding states in the absence of TRTK-12, to a more structurally refined set of states with fewer dynamic properties.
Results
Metal ion and TRTK-12 binding to wild type and EF2-hand mutants of S100B
Mutations in the second EF-hand (EF2; residues 61–72) were engineered at the canonical positions one (D61NS100B), three (D63NS100B), five (D65NS100B), and twelve (E72AS100B) of S100B to determine which of these residue(s) are important for Ca2+-binding (± TRTK-12). The Ca2+-binding affinity (KD) was determined using Ca2+/Tb3+ competition experiments by monitoring Tb3+ luminescence at 37°C, as previously described for many EF-hand proteins (Table 1). Such experiments are highly accurate for measuring Ca2+ binding to S100B since only one Tb3+ ion binds to the EF2 motif of S100B with no Tb3+ binding to the pseudo EF-hand (EF1; residues 18–31) or to nonspecific sites under the conditions used, as previously reported.
Table 1.
Dissociation constants of Ca2+ and TRTK-12 from Ca2+-S100B or Ca2+-S100B-target complexesa
| Ca EF2KD (µM)b | Ca EF2 + TRTKKD (µM)c | TAMRA-TRTKKD (µM)d | TRTKKD (µM)e | |
|---|---|---|---|---|
| S100B | 56 ± 9 (5)f | 12 ± 10 (5)g | 1.2 ± 0.2 (2)g | 2.9 ± 0.5 (2)g |
| D61NS100B | 412 ± 67 (3) | 29 ± 1.2 (3) | 1.4 ± 0.2 (2) | 2.1 ± 0.7 (2) |
| D63NS100B | 50 ± 8.6 (3) | 10 ± 2.2 (4) | 0.47 ± 0.04 (2) | 3.7 ± 0.3 (2) |
| D65NS100B | 968 ± 171 (3) | 73 ± 4.4 (3) | 0.34 ± 0.16 (2) | 1.6 ± 0.4 (2) |
| E72AS100B | 471 ± 133 (4) | 18 ± 3.7 (4) | 0.33 ± 0.11 (2) | 4.8 ± 1.1 (2) |
Values in parenthesis are the number of experiments performed.
Dissociation constants of Ca2+ from the tight site (EF2) of S100B and S100B mutants; Ca EF2KD = [S100B][Ca2+]/[S100B-Ca2+]EF2
Dissociation constants of Ca2+ from the tight site (EF2) of S100B and D63NS100B in the presence of TRTK-12 peptide.
Dissociation constants of TAMRA-TRTK-12 from Ca2+-S100B and Ca2+- D63NS100B as measured directly from fluorescence polarization.
Dissociation constants of TRTK-12 from Ca2+-S100B as measured from competition experiments with TAMRA-TRTK-12.
Values reported by Rustandi et al.14
Values reported by Charpentier et al.7
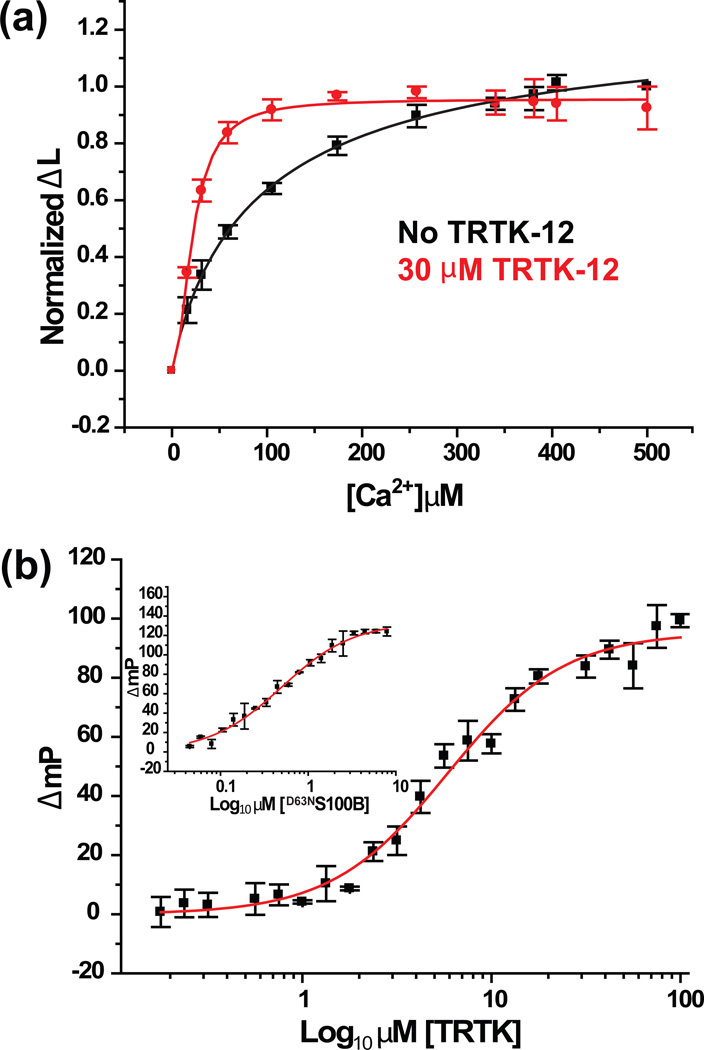
In the absence of target, the Ca2+/Tb3+ competition experiments showed reduced Ca2+-binding affinity for the D61NS100B and D65NS100B mutants by 8- to 19-fold, respectively, when compared to wild type protein (D61NKD = 412 ± 67 µM; D65NKD = 968 ± 171 µM). In many EF-hand containing proteins, the role of the bidentate ligand from the glutamate residue at position twelve is well established23. For S100B, position twelve is also important since the Ca2+-affinity of the E72AS100B was 9-fold lower than that found for wild type S100B (E72AKD = 471 ± 133). At position three, however, a single side chain oxygen atom (i.e. via Asn) is sufficient for binding Ca2+ (D63NKD = 50 ± 9 µM) since it binds Ca2+ with the same affinity as wild-type S100B (WTKD = 56 ± 9 µM; Table 2; Figs. 1a, S1–S3). Interestingly, seventeen S100 protein family members have an Asn residue at position 3 indicating that the third position of EF2 only requires a single side chain oxygen atom (i.e. via an Asn versus Asp residue) for several S100 family members, including S100B (Table 2).
Table 2.
Calcium-coordinating ligands in the pseudo EF-hand (EF1) and canonical EF-hand (EF2) of S100 proteinsa, b, c
| Position (EF1) | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S100B | S | G | R | E | G | D | K | H | K | L | K | K | S | E |
| S100A1 | S | G | K | E | G | D | K | Y | K | L | S | K | K | E |
| S100A2 | S | C | Q | E | G | D | K | F | K | L | S | K | G | E |
| S100A3 | A | G | R | C | G | D | K | Y | K | L | C | Q | A | E |
| S100A4 | S | G | K | E | G | D | K | F | K | L | N | K | S | E |
| S100A5 | S | G | R | E | G | S | K | L | T | L | S | R | K | E |
| Position (EF2) | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ||
| S100B | D | N | D | G | D | G | E | C | D | F | Q | E | ||
| S100A1 | D | E | N | G | D | G | E | V | D | F | Q | E | ||
| S100A2 | D | E | N | S | D | Q | Q | V | D | F | Q | E | ||
| S100A3 | D | T | N | K | D | C | E | V | D | F | V | E | ||
| S100A4 | D | S | N | R | D | N | E | V | D | F | Q | E | ||
| S100A5 | D | K | N | S | D | Q | E | V | D | F | K | E | ||
Shown in  are the amino acid residues that have their side chain carboxylate oxygen atoms coordinate Ca2+ in EF1and EF2.
are the amino acid residues that have their side chain carboxylate oxygen atoms coordinate Ca2+ in EF1and EF2.
Shown in  are the amino acid residues that have their backbone carbonyl oxygen atoms coordinate Ca2+ in EF1 and EF2.
are the amino acid residues that have their backbone carbonyl oxygen atoms coordinate Ca2+ in EF1 and EF2.
Shown in  is a water molecule that helps to coordinate calcium in the 9th position of EF2.
is a water molecule that helps to coordinate calcium in the 9th position of EF2.
Fig. 1.
Dissociation constants of Ca2+ and TRTK-12 from Ca2+-S100B or Ca2+-S100B-target complexes. (a) Competition of 2 µM Tb3+ bound to 2 µM D63NS100B (50 mM Hepes, pH 7.2 and 1 mM DTT) by CaCl2 as monitored by the change of Tb3+-luminesence (Δ L) at 37°C in the presence (red) and absence (black) of 30 µM TRTK-12. (b) Displacement of the TAMRA-TRTK-12 peptide from the Ca2+-D63NS100B-TAMRA-TRTK-12 complex by unlabeled TRTK-12 as monitored by fluorescence polarization. The solution contained 2 µM of D63NS100B, 50 nM of TAMRA-TRTK-12 peptide, and 10 mM CaCl2 in 50 mM Hepes, pH 7.2. (Inset) Binding of TAMRA-TRTK-12 to Ca2+-D63NS100B as monitored by fluorescence polarization. The solution contained 50 nM of TAMRA-TRTK-12 peptide and 10 mM CaCl2 in 50 mM Hepes, pH 7.2. The dissociation constants derived from these titrations are listed in Table 2.
Using fluorescence polarization competition assays24, the binding of the S100B target, TRTK-12, to Ca2+-loaded S100B and the Ca2+-liganding mutants (D61NS100B, D63NS100B, D65NS100B, and E72AS100B) was then examined (Fig. 1b, inset; Table 1). As an example, data are illustrated for TAMRA-labeled TRTK-12 binding to Ca2+-D63NS100B and fit to a single hyperbolic curve (TAMRA-TRTKKD = 0.47 ± 0.04 µM). The subsequent competition titration with unlabeled TRTK-12 is shown together with the appropriate fitting algorithm (TRTKKD = 3.7 ± 0.3 µM), which was indistinguishable from that determined previously for wild type S100B (TRTKKD = 2.9 ± 0.5; Table 2). Like D63NS100B, data for the other mutant constructs (D61NS100B, D65NS100B, and E72AS100B) did not hinder TRTK-12 binding despite their decreased Ca2+ ion binding affinity in the absence of peptide (Table 1). Next, Ca2+/Tb3+ competition data was collected to obtain the dissociation of Ca2+ from S100B when the target, TRTK12, is bound. Using exact same conditions and methods described previously for wild type S100B14, the EF2 mutants of S100B bound Ca2+ with a 5- to 17-fold higher affinity in the presence of TRTK-12 (D61N+TRTKKD = 29 ± 1.2 µM; D63N+TRTKKD = 10.4 ± 2.2 µM; D65N+TRTKKD = 73 ± 4.4 µM; E72A+TRTKKD = 18 ± 3.7 µM; Table 1; Figures S1, S3). In summary, mutations at positions one, five, and twelve (D61NS100B, D65NS100B, and E72AS100B) of the canonical EF-hand of S100B lowered Ca2+-binding in the absence of target, but wild type and all the mutant constructs bound TRTK-12 normally, and showed a >5-fold increase in their affinity for Ca2+ upon TRTK12 complex formation.
The X-ray structures of Ca2+-bound D63NS100B (± TRTK-12)
One possible mechanism for tightening Ca2+-binding upon binding TRTK-12 is via a structural change that provides a more optimal Ca2+-coordination. To ascertain whether this was the case for the D63NS100B, its three-dimensional structure was determined using X-ray crystallography in the absence (2.0 Å resolution) and presence (1.2 Å resolution) of TRTK-12. These structures were compared to each other and to the analogous structures solved previously for the wild type protein (Figure 2 and Figure 3; PDB IDs for Ca2+-S100B: 3IQO, TRTK-Ca2+-S100B: 3IQQ, Ca2+-D63NS100B: 3RLZ, TRTK-Ca2+-D63NS100B: 3RM1)14.
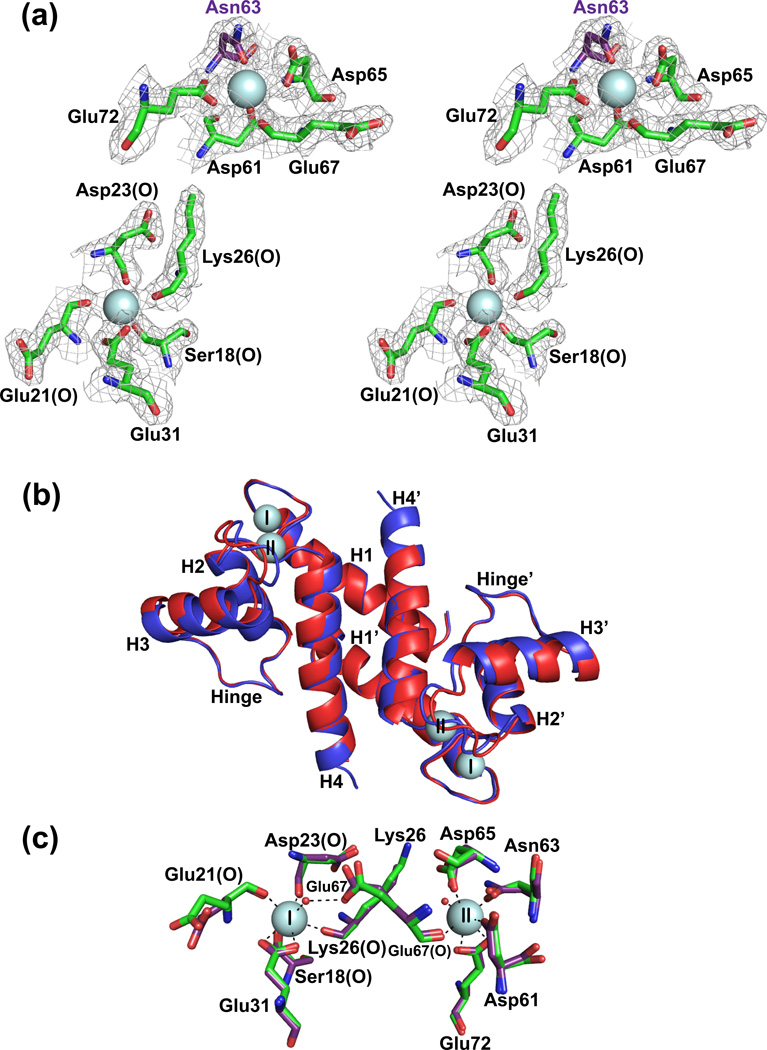
Fig. 2.
Comparison of X-ray crystallographic structures of Ca2+-S100B and Ca2+-D63NS100B in the absence of TRTK-12. (a) Stereo view of the S100-EF-hand (Ser18, Glu21, Asp23, Lys26, and Glu31) and the canonical EF-hand (Asp61, Asp/Asn63, Asp65, Glu67, and Glu72) of the D63NS100B mutant with the Asp63 to Asn63 mutation highlighted in purple. The electron density map was calculated with the 2mFo-DFc coefficients and contoured at 1.0σ for both Ca2+-binding sites. (b) Overlay of Ca2+-bound S100B (in red; PDB ID 3IQO) with Ca2+-bound D63NS100B (in blue; PDB ID 3RL). The most notable difference is that the Ca2+-boundD63NS100B structure extends to Glu89, where as S100B only has interpretable electron density to Phe88. (c) The positions of the side chains of the Ca2+ coordinating residues were compared for the pseudo EF-hand (Ser18, Glu21, Asp23, Lys26, and Glu31) and canonical EF-hand (Asp61, Asp/Asn63, Asp65, Glu67, and Glu72) Ca2+-binding sites for S100B and D63NS100B (purple).
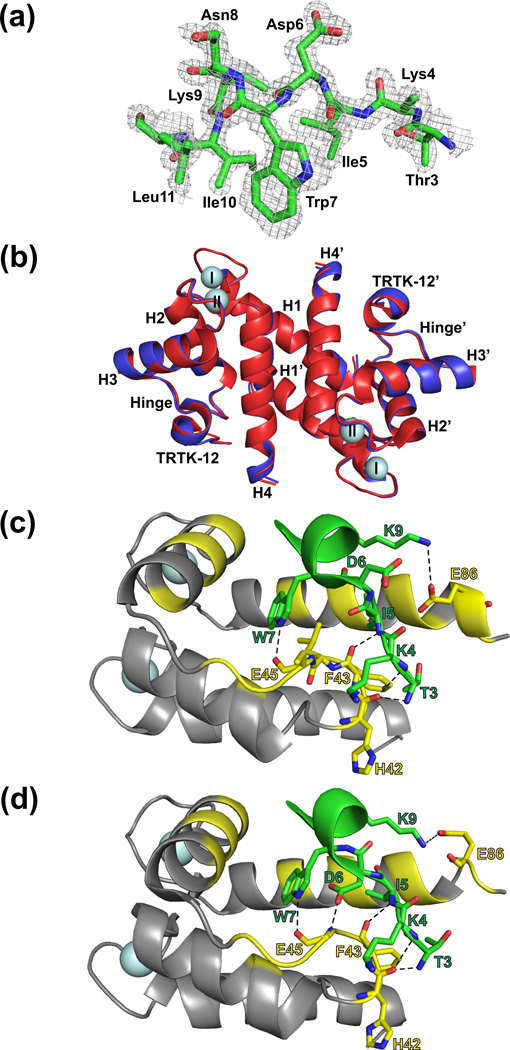
Fig. 3.
Comparison of X-ray crystallographic structures of Ca2+-S100B and Ca2+-D63NS100B bound to a peptide target, TRTK-12. (a) Electron density of the TRTK-12 peptide bound to Ca2+-bound D63NS100B at 1.2Å (contoured at 1.0σ). (b) Overlay of Ca2+-S100B (in red; PDB ID 3IQQ) with Ca2+-boundD63NS100B (in blue; PDB ID 3RM1). The most notable difference is that helix 4 in the TRTK-Ca2+-D63NS100B structure extends to Phe87; where as helix 4 in the TRTK-Ca2+-S100B terminates at His85. Close-up of the interactions between (c) Ca2+-bound S100B and TRTK-12; (d) Close-up of the interactions between Ca2+-bound D63NS100B and the TRTK-12 peptide. Shown in yellow are residues of Ca2+-S100B that contribute to hydrophobic interactions with the TRTK-12 peptide. The black dashed lines indicate hydrogen bonds between the peptide and S100B (yellow).
In the absence of TRTK-12, the final asymmetric unit of Ca2+-bound D63NS100B has two S100B subunits (model A and model B) as a symmetric dimer, with each subunit containing 90 residues (Met0-Glu89), and two Ca2+-ions per subunit. Initial phases were determined using molecular replacement, and the final refined model was one of high quality, as judged by both the refinement statistics (Table 3; Figure 2a) and Ramachandran analyses. The global fold of Ca2+-D63NS100B matched that of wild type Ca2+-S100B, with a pair wise RMSD for all residues of 0.728 Å2 (Figure 2b). As typically found for S100 proteins, each subunit of the D63NS100B dimer contained four α-helices (helix 1, M0-G19; helix 2, K28-L40; helix 3, E49-D61; helix 4, D69-F89), two β-strands, and an X-type four-helix bundle involving helices 1, 1’, 4, 4’ at the dimer interface. For Ca2+-bound D63NS100B, electron density was observed from Met 0 to Glu89, which was extended by one residue when compared to wild type Ca2+-S100B, which only had discernible electron density up to Phe8814. Despite the mutation in the 3rd position of the canonical EF-hand (D63N), no differences in Ca2+ coordination and few if any other changes were observed when compared to the analogous wild-type protein structure (RMSDall residues = 0.283 Å2; Fig. 2c; Table S1).
Table 3.
Diffraction and refinement statisticsa
| Ca2+- D63NS100B | TRTK-Ca2+- D63NS100B | |
|---|---|---|
| PDB Identification |
3RLZ | 3RM1 |
| Diffraction Statistics | ||
| Space Group | C2 | C2221 |
| Cell dimensions a, b, c (Å) | 89.8, 35.0, 57.6 | 35, 89.3, 59.7 |
| Cell angles α, β, γ (deg) | 90, 92.8, 90 | 90, 90, 90 |
| Resolution (Å) | 50.00-2.01 (2.08-2.01) | 44.63-1.24 (1.27-1.24) |
| No. of unique reflections | 11263 (751) | 25228 (1817) |
| Completeness (%) | 97.6 (89.5) | 98.6 (98.5) |
| Rsym | 0.079 (0.303) | 0.085 (0.559) |
| Average I/σ | 14.4 (3.8) | 18.0 (3.0) |
| Multiplicity | 1.9 (1.7) | 3.6 (3.3) |
| Refinement Statistics | ||
| Rcrys (%) | 20.8 (26.9) | 20.4 (31.2) |
| Rfree (%) | 25.8 (48.3) | 22.3 (37.4) |
| Protein Atoms | 1432 | 771 |
| Water Molecules | 83 | 121 |
| Non-Hydrogen Atoms | 1517 | 894 |
| RMSD | ||
| Bond Length (Å) | 0.0153 | 0.0134 |
| Bond Angles (Å) | 1.494 | 1.393 |
| Mean B values (Å2) | 34.98 | 18.78 |
| Ramachandran plot (%) | ||
| Most Favored | 100 | 100 |
| Additionally Allowed | 0.0 | 0.0 |
| Generously Allowed | 0.0 | 0.0 |
Numbers in parentheses represent the diffraction and refinement statistics for the last outer shell.
With TRTK-12 bound, the final asymmetric unit of the Ca2+-bound D63NS100B consisted of two S100B subunits as a symmetric dimer, with clear electron density for 89 residues per S100B subunit (Met0-Phe88), 9 residues for each TRTK-12 peptide (Thr3 to Leu12), and two Ca2+-ions. As in the absence of TRTK-12, analysis of the refinement and validation statistics indicated a high-quality structure determination (Table 3). The TRTK-bound S100B structures for wild type and the D63N mutant proteins were nearly identical, with an RMSDall residues value of 0.287 Å2. One small difference was that helix 4 was extended by two residues in D63NS100B, ending at Phe87 rather than at His85, as found in wild type S100B (Figure 3b); however, this difference is likely due to the higher resolution data collected for TRTK12-bound D63NS100B14. Like other S100 proteins, Ca2+-bound D63NS100B interacts with TRTK-12 via hydrophobic interactions and hydrogen bonds involving residues in the helical TRTK-12 peptide (I5, W7, I10, and L11) and numerous residues from Ca2+-D63NS100B including those from helix 2 (I36), loop 2 (hinge region: H42, F43, L44, E45, E46, I47), helix 3 (V52, K55, V56, T59), and helix 4 (F76, M79, I80, A83, C84, F87) (Figure 3c). These residues at the TRTK12-D63NS100B interface were identical to those found previously for the wild type S100B-TRTK12 complex14 and provide an explanation for why TRTK-12 binds to D63NS100B with a similar binding affinity as wild-type protein. Hydrogen bonds originating from loop 2, termed the hinge region, were observed for D63NS100B, including the specific residues shown in Figure 3c. These H-bonds were also found in the wild type complex; however, the orientation of Asp6 in TRTK-12 bound D63NS100B is not consistent with hydrogen bonding between the terminal carboxyl-oxygen of Asp6 and the backbone amide of Glu45 as in wild type S100B (Figure 3c and 3d). The distinct orientation of Asp6 in TRTK12-bound D63NS100B is likely directed by a crystal lattice contact observed between Asp6 and His42 from another subunit (3.38 Å), which is absent in the wild-type protein. Nonetheless, as with TRTK-12 in the wild-type S100B-TRTK12 complex, TRTK-12 in the mutant was also helical throughout this region of the peptide. Thus, the D63NS100B mutant is nearly identical to wild type S100B in the TRTK12 complex and explains why this mutation did not affect either Ca2+- or TRTK-12 binding (Table 1). Further, the insertion of a nitrogen atom in the EF2 side chain provides a very useful probe for measuring dynamic properties via NMR nearby the coordinating oxygen atom at this position in D63NS100B (Figure 2c).
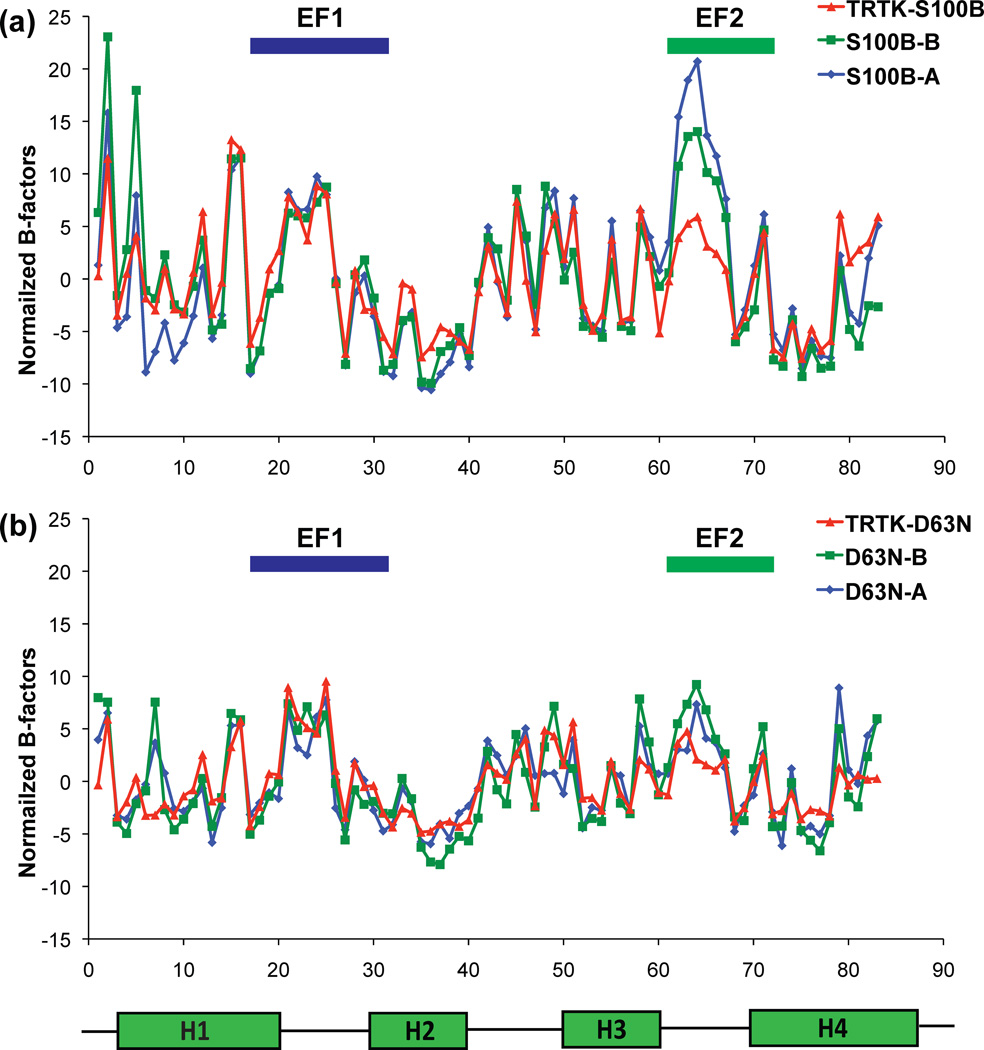
The B-factor values of most residues in wild type and D63NS100B were nearly the same (± TRTK-12) with the major exceptions being for residues in EF2 (i.e. S62, D63, G64, D65 and G66) where they were significantly higher in the absence of target. Such elevated B-factors were observed in both models A and B of the S100B dimer, thus excluding the possibility of model-specific variations (Fig. 4a)14. However, the magnitude of these differences was less for residues in the D63NS100B mutant than for the wild-type protein when the structures in the absence and presence of target peptide were compared (i.e. ± TRTK-12 bound; Fig. 4b). One simple explanation for this observation is that the two D63NS100B structures were obtained from higher quality crystals than the two wild type complexes, so the absolute value for all the B-factor values were lower, making their relative differences lower (Table 3); however, the more likely explanation is that differences in the crystal lattice contacts account for the relative magnitude of the B-factors within EF2. For wild type S100B, it was found that Lys48 forms a very weak lattice hydrogen bond with the backbone carbonyl oxygen of Asp65 (3.26 Å)14 in the absence of TRTK-12, and when TRTK-12 was bound, the crystal lattice contact changed such that an ionic interaction between Lys48 and the carboxylate oxygen atom of Glu67 occurred (2.79 Å)14. Where as, no crystal lattice contacts within 4 Å of any EF2 residue were observed for either Ca2+-bound D63NS100B or TRTK-Ca2+-D63NS100B. Despite these differences in lattice interactions, the B-factors were still significantly lower for several residues in EF2 when TRTK-12 was bound to both the wild type and the D63NS100B mutant giving some indication that motion exists within EF2 in the absence of TRTK-12, which could be stabilized upon binding the peptide target. However, it was necessary to examine directly the dynamic properties of S100B within EF2 (± TRTK-12 bound) via solution NMR methods to rigorously test this conclusion.
Fig. 4.
Graphs showing normalized B-factor values for each residue in Ca2+-S100B and Ca2+-D63NS100B in the absence and presence of bound TRTK-12. The normalized B-factor was calculated by first averaging all of an atoms’ total B-factors in a given amino acid residue. Next, the average of the total B-factors for each amino acid residue was averaged for the entire protein and subtracted from each averaged individual total B-factor. This was done for every model. Average of all atoms’ normalized B-factors per residue of model A (blue diamonds), model B (green squares), and TRTK-bound protein (red triangles) for (a) D63NS100B and (b) wild type S100B.
NMR 15N relaxation measurements for wild type and an EF2 mutant of S100B (± TRTK-12)
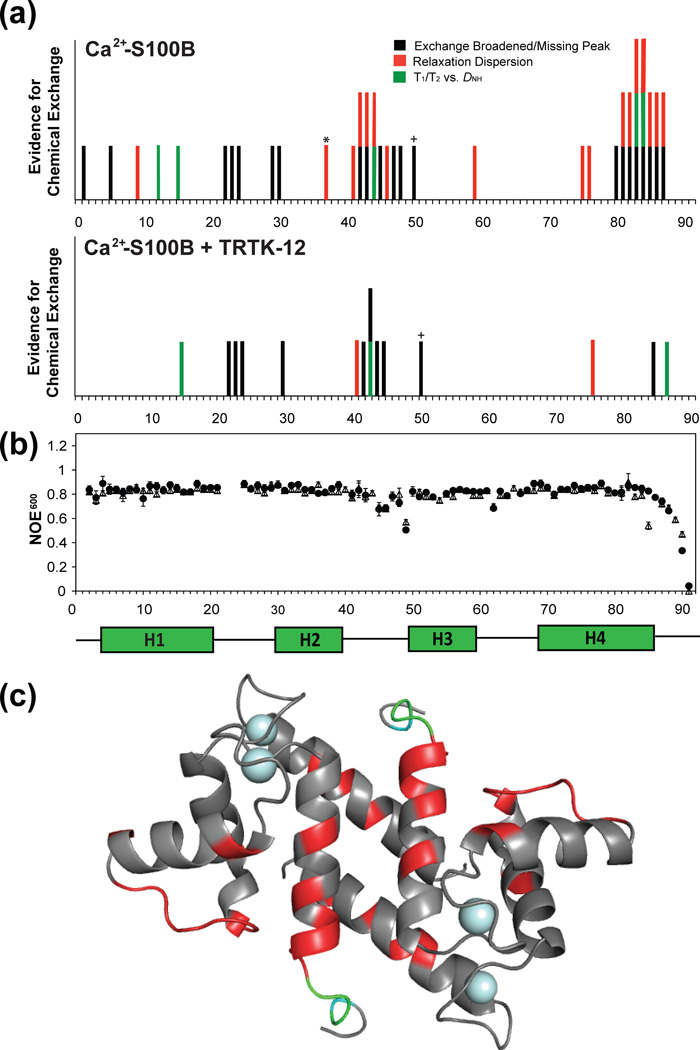
Backbone NMR 15N relaxation data were compared at two magnetic field strengths (14.4 T, 18.8 T) for wild type and the EF2 mutant, D63NS100B (Figures 5 and S4). In the Ca2+ bound state, data for 77 of the 91 amide resonances of D63NS100B were of high quality with the remaining 14 residues not evaluated due to spectral overlap or because of exchange broadening. The 10% trimmed mean values of R1 (1/T1), R2(1/T2), and the NOE ratios are summarized in Table 4. Residues that were exchange broadened, or identified as having Rexvia R2/R1 versus DNH data28, were located in helix 1 (D12, V13) the S100 EF-hand (G22, D23, K24), the hinge region (L41, H42, F43, L44, I47, K48, Q50), and the C-terminal tail (T81, T82, A83, C84, H85, E86, F87) of Ca2+-bound D63NS100B. These Rex values likely arise from motion in the C-terminal loop on the µs-ms time scale, as found previously in apo- and Ca2+-loaded forms of wild type S100B, since the only differences observed here for D63NS100B was the lack of convincing Rex for Lys5, His15, and Val80 (Figure 5a and Figure S4a). On faster time scales (ns-ps), hetero nuclear NOE values below 0.75 were observed in the Ca2+-bound state for D63NS100B and the wild-type proteins in the hinge region (E46, K48, E49), in the canonical EF-hand (E62), and in the C-terminus (E86–E91; Figures 5b and S4b). Although {1H}-15N NOE values below 0.75 were also observed for Glu45 and His85 of wild-type S100B, the data for these residues had a high level of uncertainty in both proteins, due to exchange broadening effects30. Thus, it was concluded that wild-type and D63NS100B have very similar backbone dynamic properties, on multiple time scales, in the Ca2+-bound state, and therefore, they were used next to examine the effect that TRTK-12 binding has on the dynamic properties of S100B.
Fig. 5.
Summary of fast and slow time-scale motion of 15N backbone amide relaxation rates for Ca2+-bound S100B in the absence and presence of the CapZ domain S100B binding domain, TRTK-12. (a) Summary chart of evidence for chemical exchange for the backbone amide of S100B with and with out TRTK-12. Residues that are missing or display exchange broadening in the NMR spectra are highlighted in black, backbone amides considered to be outliers (>2× the standard deviation) of residual dipolar couplings, DNH, plotted against 15N T1/T2ratios collected at 61 MHz are highlighted in green, and residues that were found to have chemical exchange through relaxation dispersion experiments are red. (b) {1H}- 15N heteronuclear NOE (η + 1) data are given for nitrogen Larmor frequencies of 61 MHz in black for Ca2+-S100B (open triangles) and TRTK- Ca2+-S100B (closed circles). (c) The NMR structure of Ca2+-bound S100B where residues that no longer showed evidence of chemical exchange (µs-ms) in the presence of the molecular target TRTK-12 are highlighted in red. In cyan are those residues that no longer had an NOE ratio below 0.75 when bound to target, which indicates movement in the fast time-scale (ns-ps) regime. Highlighted in green are those residues that no longer show evidence of both fast and slow time-scale mobility in the presence of peptide.
Table 4.
The 10%-trimmed means of R1, R2, and hetNOE backbone measurements for S100B and D63NS100B (± TRTK-12) at 14.4 and 18.8 T
| Ca2+-S100B | Ca2+- D63NS100B | |||
|---|---|---|---|---|
| No TRTK-12c | TRTK-12 | No TRTK-12 | TRTK-12 | |
| R1 600a | 1.28 ± 0.04 | 1.18 ± 0.03 | 1.26 ± 0.04 | 1.18 ± 0.02 |
| R2 600a | 13.34 ± 0.17 | 13.86 ± 0.37 | 13.96 ± 0.41 | 13.34 ± 0.26 |
| 600NOE | 0.796 ± 0.02 | 0.825 ± 0.02 | 0.793 ± 0.02 | 0.833 ± 0.02 |
| R1 800b | 0.894 ± 0.04 | 0.878 ± 0.05 | 0.999 ± 0.04 | 0.889 ± 0.02 |
| R2 800b | 19.67 ± 0.90 | 20.36 ± 0.60 | 19.38 ± 0.71 | 19.83 ± 0.39 |
| 800NOE | 0.835 ± 0.02 | 0.841 ± 0.02 | 0.840 ± 0.02 | 0.851 ± 0.02 |
R1 = 1/T1 and R2 = 1/T2 at 600 MHz (14.4 T) both given in s−1
R1 = 1/T1 and R2 = 1/T2 at 800 MHz (18.8 T) both given in s−1
Values were published in Wright et al., 200827
Backbone 15N relaxation data for wild type (77 of the 91 residues) and D63NS100B (73 of the 91 residues) illustrated that TRTK-12 binding similarly affected the dynamics of these proteinson a residue-by-residue basis (Table 4). Upon TRTK-12 binding to wild-type S100B, Rex was diminished or lost forIle47, Lys48, and Gln50 in the hinge region (Figure 5a), and another stretch of residues in the C-terminus, Val80-Cys84; where as, residues in helix 1 (H15), the S100 EF-hand (G22, D23, K24), the hinge (H42, F43, L44, E45) and in the C-terminal loop (H85, F87) retained conformational exchange (i.e. Rex; Figure 5a). Likewise, TRTK-12 binding caused a decrease in µs-ms mobility for D63NS100B including for residues Leu41 and Ile47 in the hinge and for residues in the C-terminus, including Thr81, Thr82, Cys84, Glu86, and Phe87 (Figure S4). Like wild-type protein, residues in helix 1 (H15), the S100 EF-hand (G22, D23, K24, K29, S30), the hinge region (H42, F43, L44, E45, K48) and in the C-terminus (H85, F88) retained Rex with target peptide bound. As with slower time scale motions, TRTK-12 binding also diminished the fast-time scale dynamic properties of several residues in wild type (H85-F88) and D63NS100B (E86–F88; Figure S4b). Although, 15N NOE values below 0.75 were retained for residues in the hinge region (E45, E46, K48, E49), the canonical EF-hand (E62), and in the C-terminus (F88-E91) of wild-type TRTK12-Ca2+-S100B and for residues in the hinge region (E46, K48, E49), canonical EF-hand (E62), and the C-terminus (E89–E91) of TRTK12-Ca2+-D63NS100B. In summary, the global 10% trimmed mean values for the S100B complexes examined here showed little or no significant difference; however, TRTK-12 binding to wild-type and D63NS100B quenched dynamic properties, on multiple time scales, for several residues in loop 2, termed the hinge region, and in the C-terminal loop (Figures 5 and S4).
15N Relaxation dispersion data for wild type and D63NS100B (± TRTK-12)
To examine slow time scale motions (µs-ms; Rex) in more detail, Carr-Purcell-Meiboom-Gill (CPMG) relaxation dispersion experiments were collected for wild type and D63NS100B (± TRTK-12). Assuming two-site exchange, relaxation dispersion parameters were calculated including the exchange correlation time (τex), intrinsic relaxation rates , change in chemical shift (δω), and relative populations (pa and pb) undergoing chemical exchange.
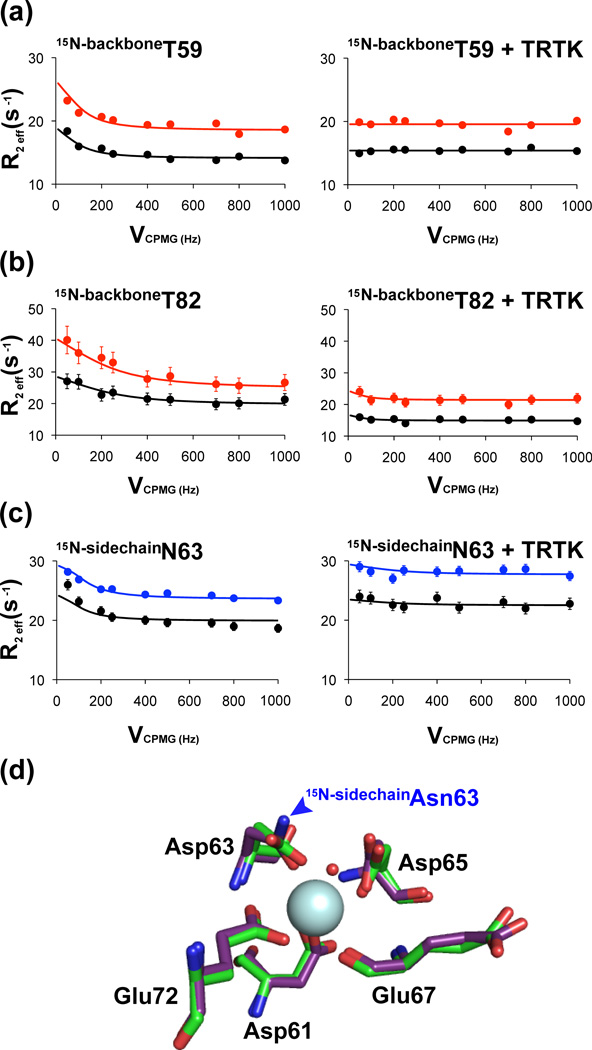
Data for 84 of the 91 backbone amide resonances of wild type S100B in the Ca2+-bound state were examined, with the remaining residues not being analyzed due to missing peaks or severe spectral overlap. As summarized in Figure 5, residues in helix 1 (A9), the hinge region (S41, H42, F43, L44, E46), helix-3 (T59), and helix-4 (A75, F76, M79, T81-E87) showed evidence for chemical exchange based on their relaxation dispersion profiles (Figure 6). The majority of these residues have exchange correlation times of between 0.7 and 1.5 ms, and when grouped together, global parameters for τex and pa were determined to be 0.880 ± 0.350 ms and 0.979 ± 0.001, respectively (Table 5). In the presence of the molecular target TRTK-12, only two residues in wild-type S100B retained conformational exchange including a residue in the hinge region (S41) and one in helix 4 (F76; Table 5). Thus, Rex was quenched for residues in helix 1 (S1, A9), helix 3 (T59), the hinge region (H42, F43, L44, E46) and helix 4 (A75, M79, T81-C84, E86, E87) upon binding TRTK12 (Figure 5c). Not surprisingly, residues in helix 3, helix 4, and the hinge, which lack conformational exchange in the TRTK12-bound complex, were located in a well-defined hydrophobic pocket that defines the peptide-S100B interface for the wild type protein (Figure 3d; Figure S5).
Fig. 6.
Selected relaxation dispersion curves of Ca2+-bound S100B (± TRTK-12). Dispersion profiles of the backbone amides of Thr59 (a) and Thr82 (b) in the absence and presence of TRTK-12 are shown with data collected at 800 (red) and 600 (black) MHz. The relaxation dispersion curve of one of the terminal amide correlation of Ca2+-boundD63NS100B (± TRTK-12) is shown in (c) with data collected at 800 (blue) and 600 (black) MHz. (d) Over lay of the canonical EF-hand of Ca2+-bound S100B (green) and Ca2+-boundD63NS100B (purple). The RMSD of all atoms between the wild type and D63NS100B mutant canonical EF-hand is 0.283Å2.
Table 5.
| Residues with Rex | δω (Hz) | χ2/N d | ||||
|---|---|---|---|---|---|---|
| Ca2+-S100B | ||||||
| 9 | 46.91 ± 12.9 | 14.90 ± 0.12 | 18.42 ± 0.46 | 1.52 | ||
| 41 | 49.61 ± 6.10 | 13.93 ± 1.09 | 16.78 ± 1.99 | 0.40 | ||
| 42 | 96.97 ± 38.6 | 15.42 ± 0.19 | 20.55 ± 0.36 | 0.37 | ||
| 43 | 70.29 ± 9.60 | 13.46 ± 0.20 | 17.42 ± 0.38 | 0.63 | ||
| 44 | 103.5 ± 5.60 | 14.78 ± 0.03 | 16.86 ± 0.15 | 0.61 | ||
| 46 | 72.99 ± 12.9 | 13.06 ± 0.04 | 17.29 ± 0.54 | 0.29 | ||
| 59 | 83.93 ± 9.30 | 13.73 ± 0.21 | 17.96 ± 0.47 | 1.32 | ||
| 75 | 70.53 ± 11.1 | 13.80 ± 0.51 | 17.00 ± 0.94 | 0.89 | ||
| 76 | 83.06 ± 2.20 | 13.07 ± 0.23 | 15.83 ± 0.63 | 0.52 | ||
| 81 | 117.4 ± 21.0 | 16.76 ± 0.75 | 19.73 ± 1.28 | 0.25 | ||
| 82 | 132.4 ± 31.1 | 19.78 ± 0.22 | 25.39 ± 0.15 | 0.14 | ||
| 83 | 86.79 ± 3.90 | 14.22 ± 0.10 | 18.95 ± 0.36 | 0.48 | ||
| 84 | 70.05 ± 10.0 | 15.45 ± 0.28 | 18.74 ± 0.55 | 0.39 | ||
| 86 | 66.37 ± 11.4 | 14.45 ± 0.19 | 21.52 ± 0.57 | 0.84 | ||
| 87 | 69.80 ± 2.10 | 13.48 ± 0.06 | 16.87 ± 0.18 | 0.34 | ||
| Ca2+-S100B + TRTK | ||||||
| 41 | 58.92 ± 13.4 | 13.96 ± 0.44 | 18.70 ± 0.02 | 0.45 | ||
| 76 | 115.1 ± 6.40 | 12.84 ± 1.12 | 19.03 ± 0.61 | 0.34 | ||
The values for δω and were independently obtained by holding the global parameters of τex and pa fixed to 0.880 ± 0.35 ms and 0.979 ± 0.001 for Ca2+-bound S100B.
The values for δω and were independently obtained by holding the global parameters of τex and pa fixed to 0.880 ± 0.14 ms and 0.986 ± 0.001 for Ca2+-loaded S100B + TRTK-12.
The uncertainties for each value was calculated from the Monte-Carlo simulations using the above τex and pa values as initial parameters.
The χ2 values listed here are normalized with respect to the number of degrees of freedom.
The relaxation dispersion data for D63NS100B were collected next in the absence and presence of TRTK-12. Specifically, data were analyzed for 84 of the 91 backbone amide resonances with chemical exchange identified in helix 1 (A9), the hinge region (S41, H42, F43, L44, E46), and helix-4 (A75, M79, T81-C84, E86, E87; Figure S4) in the absence of TRTK-12. Like wild type protein, the residues undergoing chemical exchange in the D63NS100B were found to have a correlation time for exchange of between 0.7 and 1.5 ms in the Ca2+-bound state when individually fit. Likewise, sub-groups of residues with similar exchange properties could not be distinguished for D63NS100B, so global values of τex and pa, 0.880 ± 0.30 and 0.986 ± 0.003, were calculated, respectively (Table S8). These values were essentially identical to those calculated for wild type S100B, in the absence of TRTK-12, with no significant difference between the changes in δω (p = 0.72), pa (p = 0.23), . An attempt was made to further divide residues into subgroups with similar chemical exchange rates in the fitting protocols; however, as for wild type protein, subgroups such as these could not be distinguished statistically for D63NS100B. In the presence of TRTK-12, a similar pattern was observed with D63NS100B, as found for wild type protein, such that elimination of slow time-scale movement was observed in helix 1 (A9) and for several backbone amides located in areas of the protein known to be critical for TRTK-binding (S41, E46, A75, M79, T81-C84, E86, E87). Thus, once TRTK-12 was bound, Rex was detected in only two residues of D63NS100B (H42, F88; Table S8). Therefore, in the presence of Ca2+, Rex observed by relaxation dispersion for several residues in the C-terminal loop and hinge regions of wild type and D63NS100B were quenched upon binding TRTK-12 (Figures 5, S4, and S5).
Side chain relaxation dispersion data for Asn and Gln were recorded in 50% D2O, so NHD selection could be achieved and proton-proton relaxation effects resulting from two terminal amide protons (NH2) were diminished. Using parameters summarized in Table 6 and S9, the dispersion profiles were recorded in the presence of Ca2+ for side chain 15N resonances of Asn and Gln residues upon binding TRTK-12 for wild-type S100B (Q16, Q50, Q71, N37 and N38). In addition, the Asp→Asn substitutions in the D61NS100B, D63NS100B, and D65NS100B mutants enabled Rex to also be measured at positions 1, 3, and 5 of EF2 in the absence and presence of TRTK-12 (see footnote 2).
Table 6.
Average R2 eff values with optimized correlation times (τex) and pa for select terminal Asn and Gln 15N side chain resonancesa, b, c, d
| No TRTK-12 | + TRTK-12 | |||||
|---|---|---|---|---|---|---|
| R2 eff AVE (s−1) |
pa | τex (10−3 s) |
R2 eff AVG (s−1) |
pa | τex (10−3 s) |
|
| S100B | ||||||
| Asn37 | 18.8 ± 0.48 | 0.97 ± 0.01 | 2.00 ± 0.65 | 21.1 ± 0.23 | NE | NE |
| Asn38 | 13.6 ± 1.33 | NE | NE | 16.6 ± 2.34 | NE | NE |
| Gln50 | 11.3 ± 0.16 | 0.97 ± 0.01 | 3.66 ± 1.51 | 13.6 ± 0.18 | 0.73 ± 0.20 | 73 ± 60 |
| D61NS100B | ||||||
| Asn37 | 11.7 ± 0.43 | 0.98 ± 0.02 | 29 ± 6.20 | 19.2 ± 2.06 | NE | NE |
| Asn38 | 9.76 ± 0.65 | NE | NE | 11.6 ± 1.56 | NE | NE |
| Gln50 | 5.85 ± 0.03 | - | - | 10.3 ± 0.17 | 0.98 ± 0.003 | 3.78 ± 0.29 |
| Asn61 | 11.6 ± 0.75 | 0.98 ± 0.01 | 4.69 ± 1.33 | 28.0 ± 1.81 | 0.97 ± 0.03 | 0.13 ± 0.09 |
| D63NS100B | ||||||
| Asn37 | 14.7 ± 0.09 | 0.97 ± 0.02 | 5.56 ± 2.53 | 17.8 ± 0.64 | NE | NE |
| Asn38 | 11.7 ± 0.27 | NE | NE | 15.3 ± 0.79 | NE | NE |
| Gln50 | 9.82 ± 0.07 | 0.94 ± 0.04 | 5.30 ± 4.42 | 10.8 ± 0.08 | 0.98 ± 0.01 | 62 ± 5.9 |
| Asn63 | 19.0 ± 1.88 | 0.76 ± 0.16 | 57.5 ± 39 | 21.4 ± 1.29 | NE | NE |
| D65NS100B | ||||||
| Asn37 | EB | EB | EB | 20.8 ± 0.71 | NE | NE |
| Asn38 | 16.0 ± 1.25 | 0.93 ± 0.06 | 2.06 ± 1.31 | 12.2 ± 1.79 | NE | NE |
| Gln50 | 13.6 ± 0.94 | 0.91 ± 0.03 | 36.5 ± 6.5 | 10.9 ± 0.08 | NE | NE |
| Asn65 | EB | EB | EB | EB | EB | EB |
The average (AVE) and RMSD values for R2 eff are given for 14.1 T.
τex and pa was obtained by independently fitting the R2 eff data from both proton amide resonances from the side chains of Asn and Gln using a two-site exchange model28. The average values of the exchange parameters are reported together with the standard error estimated from the values obtained from the two proton side chain correlations.
NE = no exchange detected and EB = exchange broadened.
Although the R2 eff AVE is not exclusively the result of 15N R2 relaxation since 1H Z-relaxation is also present (< 1%) during the CPMG period, R2 eff AVE is provided here to show an overall change in R2.
In the absence of TRTK-12, relatively large R2 eff values were recorded for the side chain amide resonances of Asn63 (position 3) in D63NS100B when compared to those of Gln16 and Gln71 (Table 6 and S9), consistent with Asn63 being at least partially coordinated to Ca2+ and not in rapid conformational exchange like the solvent exposed side chain amides, Gln16 and Gln71. When comparing EF2 residues, the side chain amide resonances of Asn61 (D61NS100B; position 1 of EF2) had an average R2 that is lower than for Asn63 in the Ca2+-bound state (D63NS100B; position 3), consistent with the Asn61 side chain having faster Rex values than that of Asn63 (Table 6 and S9; see footnote 2); whereas, the side chain resonances for Asn65 (at position 5) in the D65NS100B mutant were too exchanged broadened to collect meaningful relaxation dispersion data in the absence of TRTK-12 (Figure S3).
Upon binding TRTK-12, Rex values for several Asn and Gln side chain resonances throughout the proteins’ sequence (Q16, N37, N38, Q50, N61, N63, N65, Q71) were either significantly reduced (i.e. for Q50; τex > 10 ms) or eliminated altogether (N37, N38, N63; Table S9). For example, binding TRTK to either the wild type or the D63NS100B mutant proteins caused the correlation time for chemical exchange in Gln50 to be decreased by 20-fold. However, chemical exchange was still detected for side chain 15N resonances of the EF2 side chains, Asn61 and Asn65 (i.e. for D61NS100B and D65NS100B), with exchange broadening still being too problematic for collecting meaningful relaxation dispersion data for Asn65 (Table S9). On the other hand, the average R2 measured for side chain 15N resonances of Asn61 decreased significantly upon binding TRTK (Table 6) and displayed a more complex mode of exchange when compared to Ca2+-bound D61NS100B collected in the absence of target (Table S9). Importantly, no indication of chemical exchange remained for the side chain of Asn63 in EF2 when TRTK-12 was in a complex with Ca2+-loaded D63NS100B (Figure 6c). When bound to TRTK-12, the lack of exchange in the side chain of the Ca2+-coordinating residue, Asn63 (position 3), and the slower exchange rates observed for the other EF2 side chains could at least partially reflect the increase in Ca2+-affinity observed for S100B with TRTK-12 bound.
Discussion
In response to a calcium-signaling event, EF-hand containing proteins, such as S100B, bind Ca2+, undergo a conformational change, and bind specific targets as necessary to generate a biological response. However, the S100B calcium-signaling protein does not sequester appreciable amounts of Ca2+ due to its relatively low affinity unless it is bound to another protein target. To examine this phenomenon in more detail, mutations were engineered into the canonical EF-hand (EF2) of S100B and the structural, Ca2+-binding, and dynamic properties in the absence and presence of a bound target, TRTK-12, were examined. With mutation at position three (D63N) of EF2 as an exception (D63NKD = 50 ± 9 µM), the Ca2+ ion binding affinities of proteins mutated at positions one, five, and twelve were reduced by >8-fold in the absence of target (D61NKD = 412 ± 67 µM; D65NKD = 968 ± 171 µM; E72AKD = 471 ± 133 µM) as compared to wild-type protein (WTKD = 56 ± 9 µM). Like wild-type protein, (WT+TRTKKD = 12 ± 10 µM), however, the Ca2+-binding affinities were increased by >5-fold upon binding TRTK-12 for the mutant constructs (D61N+TRTKKD = 29 ± 1.2 µM; D63N+TRTKKD = 10 ± 2.2 µM; D65N+TRTKKD = 73 ± 4.4 µM; E72A+TRTKKD = 18 ± 3.7 µM). Thus, factor(s) other than side-chain oxygen atoms in EF2 are needed to understand how Ca2+-binding affinity is increased when S100B binds target. Three such possibilities were examined here in detail, which could be used to explain these results.
Model 1 – A discrete structural change induces a more optimal Ca2+-coordination
The most straight forward explanation for obtaining a higher affinity Ca2+ site upon binding a biological target is via a distinct structural change that provides a change from a weaker Ca2+ ion coordination geometry to a more optimal one. To examine this possibility directly, the X-ray crystal structures of wild type and D63NS100B were compared in the absence and presence of bound TRTK-12. Such an explanation was ruled out by these data, since as found for wild-type S100B, the structures of Ca2+-D63NS100B (2.0 Å) and TRTK-Ca2+-D63NS100B (1.2 Å) showed no detectable changes in Ca2+ coordination or coordination distances (<0.1 Å) upon binding TRTK-12 (Figure 2 and Figure 3). However, when examined closely, differences were observed in B-factor values, particularly for residues in EF2 (± TRTK-12; Figure 4). Residues in EF2 (i.e. S62, D63, G64, D65 and G66) displayed elevated B-factors in both models A and B of Ca2+-loaded S100B and D63NS100B in the absence of target, which were reduced significantly when TRTK-12 was bound. Elevated B-factors provide some initial indication that motion exists in EF2 of Ca2+-loaded S100B and Ca2+-D63NS100B, which could be stabilized upon binding TRTK-12. However, it was necessary to examine directly the dynamic properties via NMR to test this model more rigorously in solution.
Model 2 – A pre-equilibrium between the “open” and “closed” states
Another potential explanation for tightening of Ca2+-binding is that a pre-equilibrium between the “open” and “closed” states in S100B occurs prior to binding Ca2+, and that TRTK12-binding shifts this pre-equilibrium towards the “open” Ca2+-bound state. However, for wild type and all of the EF2 S100B mutants studied here, there was no evidence of TRTK-12 binding up to 20 mM target in the absence of Ca2+ (data not shown). Furthermore, 15N relaxation measurements with apo-S100B shows no Rex for residues in either EF2 or helix 3, so no conclusive evidence exists to support that a pre-equilibrium occurs between the “closed” and “open” states (where helix 3 swivels out approximately 90°) in the absence of Ca2+ (Figure S6)29. In such studies with apo-S100B constructs, conformational exchange was only detected in helix 1, the hinge (loop 2), and in the C-terminus of apo S100B (Figure S6b and c). More specifically, the chemical exchange observed in these regions could most readily be explained by a single movement of the disordered C-terminal loop within a protein cavity defined by loop 2 and helix 129. Nonetheless, relaxation dispersion measurements were carried out here for wild type and D63NS100B in the absence of Ca2+ (± TRTK12), as yet another method to identify whether or not residues in helix 3 and/or within EF2 in the “open” and “closed” states could be detected. As previously reported, there was no evidence for chemical exchange for any residues in helix 3 or EF2 that were indicative of S100B fluctuating from an open and closed state in the absence of Ca2+ (Figure S6). This included no measurable Rex for the backbone amide-proton correlation of Gly66 (± TRTK12), a residue found in the EF2 hand of S100B that undergoes a significant chemical shift perturbation upon the addition of Ca2+ and is a well-established indicator marking the structural transition of S100B from its “closed” to “open” state upon Ca2+-binding (Figure S6). These data confirmed that S100B does not undergo a major structural change in the absence of Ca2+ and that TRTK-12 does in fact only interact with S100B in a Ca2+-dependent manner and not via a pre-equilibrium of open/closed states. Such a Ca2+-dependent interaction with S100B and several other S100/target interactions have also been well documented in the literature.
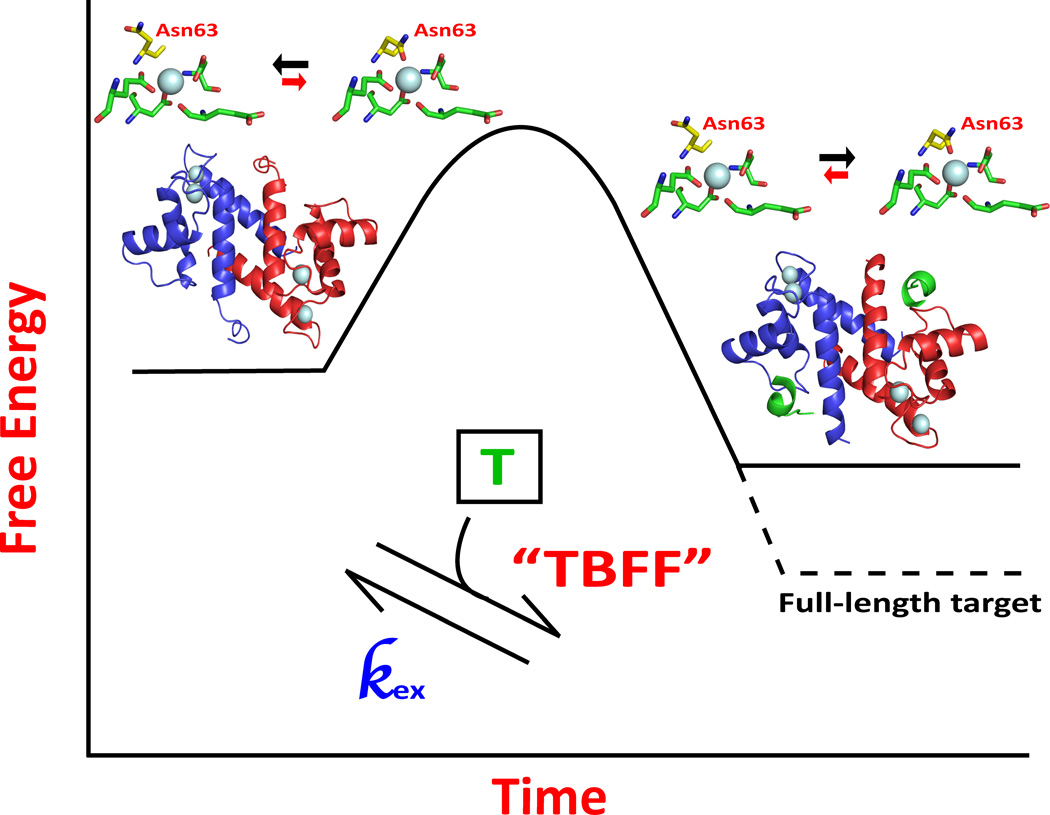
Model 3. The target binding and final mini-folding model (the TBFF model)
The third model considered was one in which protein dynamics contributed to a “less-defined” form(s) of the EF-hand protein that were stabilized upon binding target. In this model, a pre-equilibrium conformational averaging exists between weaker and higher affinity Ca2+-binding states in the absence of target, and that target binding shifts this equilibrium population from the dynamic and weakly bound states towards a higher affinity Ca2+-binding state(s) that has a narrower distribution of dynamic features throughout the protein. In energetic terms, such a model may also be thought of as a “binding and mini-folding” event that involves the entire protein sequence and thus can be highly specific for biological function(s) (Figure 7). Such a “target binding and functional folding” model is consistent with all of the experimental data presented here. Specifically, for D63NS100B, the 15N relaxation rate properties of the Asn63 side chain showed a significant decrease in Rex upon binding TRTK-12, as monitored using NMR relaxation dispersion methods. Likewise, loss of Rex occurred for 15N-labeled resonances throughout wild type, D61NS100B D63NS100B, and D65NS100B on fast and slow time scales upon S100B-TRTK-12 complex formation. These data are consistent with a model in which dynamic properties in the backbone and side chains throughout the S100B sequence contribute to weaker Ca2+-binding affinity prior to target binding. Consistent with this model, TRTK-12 binding leads to S100B-target complex with decreased conformational exchange, a more optimal Ca2+-coordination (i.e. N63 bound), and higher Ca2+-binding affinity, particularly for the wild type and D63NS100B mutant.
Fig. 7.
Model for how target binding to Ca2+-loaded S100B increases the affinity of the complex for Ca2+. In the absence of molecular target (T), Ca2+-bound S100B exists in a dynamic set of states where the overall affinity for Ca2+ is low. A molecular target, such as TRTK-12, binds to a subset of these conformers and pushes the equilibrium towards a narrower range of dynamic states, including within EF2, for which the overall affinity for Ca2+ is higher. However, in this model the “target binding and final mini-folding” (TBFF) event is only partially achieved with a target-peptide interaction and would need a full-length biologically relevant target to achieve a proper TBFF event that would further narrow the distribution of dynamic states within the protein as necessary for binding Ca2+ at physiologically relevant concentrations (i.e. 100 to 500 nM), as was observed previously for the S100A1-ryanodine receptor complex47.
It is important to recognize that the TBFF model for increasing Ca2+-binding affinity considers both protein dynamic properties and Ca2+-coordination geometry within EF2 of S100B. The nomenclature for Ca2+ coordination in calcium-binding proteins was originally defined as being an octahedral geometry with X, Y, Z, −X, −Y, −Z coordination; however, numerous structures by X-ray crystallography generally now recognize that Ca2+ coordinates seven ligands in a pentagonal bipyramidal geometry23. In a typical EF-hand such as EF2 of S100B (residues 61–72; EF2), the backbone carbonyl oxygen of the residue in the −Y position and side chains oxygen atoms at the Y and Z positions together with an invariant glutamate, which provides two coordinating oxygen atoms at the −Z, and −Z’, form an approximate coplanar pentagon, as was observed for wild type S100B and for D63NS100B (Figure 2). Likewise, the side chain oxygen atoms at positions X and −X are arranged at vertices of the pentagonal plane with the −X position usually having an intervening H2O molecule23. Specifically, the Ca2+ ion bound to EF2 of S100B has six oxygen atoms coordinated from the protein including the carboxylate oxygen atoms of Asp61 (position 1), Asp63 (position 3), Asp65 (position 5) at the X, Y, and Z sites, a backbone carbonyl oxygen from Glu67 (position 7) at the −Y site, and bidentate liganding from both carboxylate oxygen atoms of Glu72 (position 12) at the −Z and −Z’ sites. The final ligand is provided by a water molecule intervening between Asp-69 (position 9) and the −X site of coordination such that the overall geometry of the calcium coordination sphere is pentagonal bipyramidal as found for most other EF-hand containing proteins, and target binding (i.e. p53, TRTK12, etc.) does not change this overall coordination geometry (Figure 2). Thus, the use of coordination geometry from X-ray structures cannot be used alone to explain why S100B binds Ca2+ more tightly in the presence of a target, as was observed here and elsewhere. However, upon TRTK-12 binding, one change that was observed via X-ray crystallography was that elevated B-factor values observed for residues in EF2 (i.e. S62, D63, G64, D65 and G66) were lowered significantly for both wild type and D63NS100B (Figure 4). While these data were suggestive that dynamic properties were quenched upon target binding, it was necessary to rigorously examine this possibility via NMR.
Interestingly, several residues throughout the protein's sequence showed fewer dynamic properties upon binding TRTK-12, including for coordinating side chain moieties within EF2 (i.e. N63 of D63NS100B; position 3). Thus, it is concluded that while peptide targets can at least partially quench protein dynamic properties within the coordination sphere of EF2 and provide S100-target complexes with increased Ca2+-ion binding affinities, it is likely that only full-length and biologically relevant protein target(s) can fully achieve this feat, as was previously reported for S100A1 binding to the full-length ryanodine receptor at 100 nM free Ca2+ (Figure 7). In particular, it is likely that position 3 is the only Ca2+ ligand to be fully stabilized by peptide binding since TRTK-12 binding was sufficient to abolish Rex observed for this residue (i.e. versus at positions 1 and 5, which still had Rex with peptide bound). This is likely a different scenario from full-length targets, which could potentially stabilize Rex at several or all positions within EF2 of target-bound S100B to achieve high affinity binding at resting Ca2+ levels, as was observed for the S100A1-RyR1 complex. It is also possible that residues important for this “tightening” effect in S100s are different from those observed for calmodulin and other EF-hand containing proteins because the overall fold of S100 proteins is very different from that of CaM and other members of its protein family. Furthermore, the “Ca2+-switch” for S100s is very different from CaM family members because helix 3, the entering helix, is the one that rotates 90 degrees upon binding Ca2+ in S100 proteins rather than helix 4, the exiting helix, as found in most other EF-hand containing proteins, including CaM48.
Summary
S100 proteins (S100s) are unique Ca2+-activated switches among EF-hand proteins that are distributed cell-specifically in mammals. With rising Ca2+ levels, S100s bind Ca2+, change conformation, and interact with specific targets to regulate biological activities. As summarized in figures 5 and S4, NMR data show that a number of residues in the hinge, helix-3 and the C-terminus, have motion in both slow (µs-ms) and fast (ns) time scales with Ca2+-bound, but in the absence of target (i.e. TRTK-12). Upon binding TRTK-12, Rex and fast time scale dynamics are abolished for many residues throughout the protein's sequence, a trend that was also translated to Rex values for side chain 15N resonances (i.e. for Asn, Gln residues; Figure 6c). This included the side chain of Asn63, which has its oxygen atom as a direct Ca2+ ligand of S100B (Figure 2a). Likewise, these data provided a mechanistic explanation for how target protein binding to S100 proteins can increase their Ca2+-binding affinities since at least one coordinating residue in EF2 is less mobile on the chemical shift time scale in the S100B-TRTK12 complex (Figures 7 and S7). It is also important to realize that weak Ca2+-binding for S100s in the absence of target is biologically relevant (Figure 1a). Since most target-free S100 proteins have a low affinity for Ca2+, this allows numerous stable S100 proteins to be in at high concentrations within the cell (> 1 µM) without depleting [Ca2+]free levels and “short-circuiting” Ca2+ oscillations. Thus, as many as twenty highly stable S100s are "poised and ready" for when their specific biologically relevant target(s) are expressed, as necessary for them to regulate numerous functions in mammalian cells. Lastly, to engineer therapeutically effective small molecule S100B inhibitors, it is important that the compound(s) can mimic a biologically relevant target to induce this “final mini-folding event”. Due to the allosteric nature of this biophysical process, such a model provides a means to obtain highly specific S100B inhibitors since residues involved in this “Ca2+-tightening” and “final mini-folding” event are not limited to residues within the protein-protein interface or EF2, but require consideration of amino acid residues throughout the S100-target complex.
Materials and Methods
Materials
All chemicals and reagents were of ACS grade or higher and were typically purchased from Sigma-Aldrich unless otherwise indicated. 15NH4Cl, D2O, and D7-glucose were purchased from Cambridge Isotope Laboratories (Andover, MA). All buffers were passed through Chelex-100 resin to remove trace metals prior to use. All peptides were made using solid-state peptide synthesis and were >95% pure using HPLC and mass spectrometry (Biosynthesis Inc. Lewisville, TX). The TAMRA-TRTK-12-am (TAMRA-TRTKIDWNKILS-am) is an N-terminal 5-TAMRA labeled peptide derivative of the TRTK-12 peptide derived from the actin binding protein CapZ (residues 265–276) with an amidated c-terminus. The TAMRA-TRTK peptide was suspended in dH2O and the pH was immediately adjusted to 7.2 and stored in 50 µL aliquots at −20°C. The concentration of TAMRA–TRTK-12-am was determined at pH 7.2 in dH2O using the extinction coefficient for TAMRA, ε547 = 65, 000 cm−1 M−124.
Bacterial expression and purification of S100B and the EF2 mutants
S100B mutants from the second EF-hand of S100B (EF2) were engineered using wild-type rat S100B template DNA and the site-directed Qiagen mutagenesis kit with the appropriate DNA primers. The wild-type and mutant S100B proteins were then expressed and purified from Escherichia coli (HMS174 (DE3) strain) as described previously29. Yields for the D61NS100B, D63NS100B, D65NS100B, and E72AS100B mutants were typically 10–20 mg of purified protein per liter of defined bacterial culture and their concentrations and homogeneity determined quantitatively by amino acid analyses and mass spectroscopy. For NMR relaxation data collection, 2H, 15N-labeled proteins were prepared using minimal media with 15NH4Cl (>99%) as the only nitrogen source and grown in 100% D2O containing buffer.
Thermodynamic binding studies
Fluorescence polarization competition assays were performed in Corning 96-well, flat-bottom plates (Corning, NY) using a PolarStar fluorescent plate reader (BMG Labtech, Durham, NC) kept at 37°C with a final volume of 200 µL24. The S100B mutant titrations into 50 nM of TAMRA-TRTK-12 contained 0 – 25 µM of protein, 50 mM Hepes, pH 7.2, 15 mM NaCl, 100 mM KCl, 10 mM CaCl2, 1 mM DTT, and 0.1% Triton X-100. Using the same buffer conditions as the previous experiment, 50 nM of TAMRA-TRTK-12 peptide with 1 – 2 µM of each S100B EF2 mutant was displaced using 0 – 100 µM of unlabeled TRTK-12 to determine the KD of the unlabeled peptide in the presence of CaCl2. Polarization was read after excitation at 544 ± 10 nm using an emission wavelength of 590 ± 10 nm. The binding data were fit using a single-site binding model with Origin software (Origin Lab Corp., Northampton, MA), with one peptide bound per S100B subunit. The binding affinity for unlabeled TRTK was calculated using an equation derived from Nikolovska-Coleska et al. using the IC50 values as follows: KD = [I]50/([L]50/TAMRA-TRTK-12KD + [P]0/TAMRA-TRTK-12KD + 1), where [I]50 is the concentration of unlabeled TRTK-12 at 50% inhibition, [L]50 is the concentration of the free TAMRA-TRTK-12 at 50% inhibition, [P]0 is the concentration of the free protein at 0% inhibition, and TAMRA-TRTK-12KD is the dissociation constant of TAMRA-TRTK-12 from the S100B mutant-TAMRA-TRTK-12 complex14
Binding of Ca2+ to apo- S100B mutants in the presence and absence of TRTK-12 peptide was analyzed by measuring the changes in Tb3+-luminescence using a Cary Eclipse Fluorescence Spectrometer (VARIAN, Walnut Creek, CA) as described previously. Previous work done by Chaudhuri et al. has thoroughly characterized the stoichiometry of Tb3+ to S100B22 and with the experimental conditions chosen here, only the tight site is populated and the risk of non specific Tb3+-binding is vastly reduced18. Therefore, competition studies with Ca2+ accurately measures the binding affinity of the EF2 hand. All protein, peptide, and buffer solutions were chelexed, filtered and equilibrated to 37°C prior to titration. First, S100 protein was titrated into a solution containing 50 mM Hepes (pH 7.2), 1 mM DTT and 2 µM of Tb3+ in the presence and absence of 30 µM TRTK-12 to determine the binding affinity for Tb3+ ± TRTK-12 (TbKD). Next, the decrease in Tb3+ luminescence was monitored upon titration with 0.5–10 mM of CaCl2 to determine the Ca2+-binding affinity for the particular S100B mutant in the presence and absence of TRTK-12. The resulting titration curves were analyzed using Origin by MicroCal, fit using the Hill equation, and the binding affinity for Ca2+ was determined using the Cheng-Prusoff equation as follows: KD = ([IC]50/(1 + ([L]T/Tb(3+)KD))), where [IC]50 is the apparent binding affinity for the S100 protein to Ca2+, [L]T is the concentration of Tb3+ used and Tb(3+)KD is the binding affinity for the S100 protein to Tb3+.
X-ray crystallography
As is the case for the wild-type protein, bovine D63NS100B mutant readily crystallized; therefore, this isoform of S100B was used to obtain the D63NS100B, Ca2+-D63NS100B and D63NS100B-Ca2+-TRTK structures (see footnote 1). Diffraction quality crystals were obtained by the sitting-drop vapor diffusion method at 22°C. Briefly, a 1:1 ratio of D63NS100B protein (3.7 mM D63NS100B, 7.5 mM CaCl2, ± 3.8 mM TRTK-12, and 20 mM cacodylate buffer, pH 7.2) was mixed with reservoir solution (7.5 mM CaCl2, 0.1 M cacodylate buffer, pH 7, and 25% PEG3350 for the protein alone or 0.1 M cacodylate buffer, pH 6.9, and 22% PEG3350 for the protein with TRTK-12) and allowed to equilibrate for 3–5 days. After crystal formation, the crystals were cryo protected with reservoir buffer containing 2% higher PEG3350 and 5% glycerol, flash-cooled and stored in liquid nitrogen prior to data collection.
X-ray data for Ca2+-D63NS100B were collected at 100 K using a MicroMax 7 S-ray generator (Rigaku/MSC, The Woodlands, TX) and a Raxis4++ image plate detector (Rigaku/MSC). X-ray data for D63NS100B-Ca2+-TRTK was collected remotely at the BL7-1 beamline of the Stanford Synchrotron Radiation Lightsource (Menlo Park, CA). The reflection intensities were integrated and scaled using the HKL2000 suite of computer programs51. The crystals of Ca2+-D63NS100B and D63NS100B-Ca2+-TRTK diffracted to 2.01 and 1.24 Å resolutions, respectively. Both structures were solved by molecular replacement using the structure of S100B-Ca2+ (PDB ID: 3IQO) with the ions removed as a search model and the computer program PHASER from the CCP4 suite52. Model building and refinement of the D63NS100B structure was completed using COOT and REFMAC5. The stereochemistry was checked with the programs WHATCHECK, PROCHEK, and MolProbity. Figures were generated using the program PyMol (http://www.pymol.org).
NMR spectroscopy
All NMR samples were prepared in 10 mM Hepes, pH 7.2, 15 mM NaCl, 10 mM CaCl2, 0.34 mM NaN3, 2 mM DTT, and protein that ranged from 350 µM – 400 µM for most NMR samples (see Footnote 2). For some side-chain relaxation dispersion NMR experiments, 1 – 2 mM protein was used to validate data collected at lower concentrations and the elevated protein concentration was shown to have no effect on the results. NMR spectra were collected at 37°C with a Bruker AVANCE III 600 NMR spectrometer (600.13 MHz for protons) and a Bruker AVANCE 800 NMR spectrometer (800.27 MHz for protons) equipped with four frequency channels and a triple-resonance z-axis gradient 5 mm cryoprobe. All NMR data were processed and analyzed with nmrPipe and nmrDraw Software58.
15N-labeled D63NS100B at 400 µM was used to collect backbone dynamic data in the presence and absence of 1.2 mM TRTK-12 target peptide. As reported previously29;30, backbone 15N R1 and R2 spectra were acquired with 32 scans per t1 point at both 600 and 800 MHz. A recycle delay of 3.0 s was used at both fields. R1 delay times of 40, 160 (2X), 320, 640, and 1280 ms were used for data collection at 600 and 800 MHz fields with and without target peptide TRTK-12. R2 delay times of 16, 32 (2X), 48 64, 96, 112, and 128 ms were used for data collection at 600 and 800 MHz fields with and without TRTK-12. {1H}- 15N NOE ratios were acquired in an interleaved fashion, with 56 scans at 600 and 800 MHz fields and with 256 t1 points. The NOE ratios were collected with a 3-s pre-saturation period and a 2-s saturation delay, while the control experiment had an equivalent 5-s delay.
Relaxation data were analyzed as described previously. Briefly, data were extended in the indirect (t1) dimension using linear prediction and apodization with a 5% shifted mixed Gaussian/exponential function was applied with weighting chosen to reproduce the natural line values. For the relaxation series, correlation peaks with S/N > 15 were selected and peak heights measured by fitting Gaussian surfaces to the transformed data and taking the maximum height of the fitted surface using the program NLINLS58. The computer program CURVEFIT [(A.G. Palmer, Columbia University] was used to fit the peak heights via the jackknife procedure with the Levenberg-Marquadrt nonlinear square algorithm. The heteronuclear NOE and reference experiments were also extracted in a similar manner as previously described.
For backbone 15N relaxation dispersion measurements, 350 µM of 2H, 15N protein in 10% D2O was used in the presence and absence of a 3-fold excess of TRTK-1232. Relaxation dispersion data were collected at two magnetic field strengths (61 and 81 MHz 15N resonance frequency) with the 15N-carrier frequency set at 117.1 ppm. For side chain measurements, a typical sample contained 50% D2O with 2 mM of 2H, 15N protein, with and with out a 3-fold excess of TRTK-12. Data collection was also done for samples prepared at lower concentrations with no changes in the results. The 15N-carrier frequency was set at 113 ppm, which is close to the 15N shifts of the terminal amide groups in Asn and Gln residues. A reference spectrum was acquired for all relaxation dispersion measurements, without a CPMG period, together with nine spectra containing a constant CPMG period, TCP in which the time between CPMG 180° pulses, 2Tcp, varied59. The effective field, νCPMG, is defined by 1/4Tcp. At each effective field, an R2 eff was calculated from the ratio of two signals ICP and I0, where I0 is the intensity of the peak in the reference spectrum and ICP is measured at the end of the TCP period. All spectra were recorded with TCP = 40 ms and νCPMG equal to 50, 100, 200, 250, 400, 500, 700, 800, and 1000 Hz. The 15N dispersion experiments were recorded with 512 and 1000–1280 complex points in F1 and F2 dimensions, respectively and with 8 scans per FID. The uncertainty in each R2 eff was calculated based on the random noise of the spectra31.
All dispersion profiles were fit using χ2 minimization by the Bloch-McConnell equation assuming a two-site exchange model as done previously31. First, the fits were performed for each amide 15N site using the datasets recorded at 61 and 81 MHz to optimize the intrinsic relaxation rate, , the difference in chemical shift between the exchanging species, δω, the relative population of the dominant exchanging species, ρa and the correlation time for exchange, τex. Uncertainties in the parameters were determined by the Monte-Carlo simulation31. For those residues undergoing chemical exchange, they were grouped together and a set of pa and τex was determined as global parameters for each domain by a grid search. At each grid point, the χ2-function was minimized with respect to the local parameters, δω, at 61 MHz, and at 81 MHz31. As described previously62, the uncertainties in τex and pa were obtained by graphically determining the confidence region, which is fitted to the normalized χ2 values one standard deviation larger than the minimum derived from the χ2 grid search (i.e. (χ2/N)MIN + 1.0). This region, often represented as an ellipsoid around a point (i.e. the lowest minimized χ2 value), can give the uncertainties associated with the two parameters calculated, τex and pa, with a confidence of approximately 95%. Uncertainties in the local parameters δω, at 61 MHz, and at 81 MHz, were independently determined by the Monte-Carlo simulation, using the optimized τex and pa as initial parameters. In order to identify residues that was undergoing chemical exchange, the fractional uncertainty, defined as (R2RMSD)/<R2>, where the r.m.s.d. of the set of R2 values measured in a relaxation dispersion profile is R2RMSD and <R2> is the average R2 of the dispersion profile, had to be one standard deviation greater than the R2 error31. For the side-chain dispersion measurements, there were two correlation peaks in the spectra for each Asn or Gln residue present in the S100B mutants and wild-type S100B. Each R2 measurement corresponding to a side chain correlation peak was individually fit using the Bloch-McConnell equations64. The exchange parameters extracted were then averaged for each pair of terminal amide resonances and the errors were estimated by comparing values obtained from fits of profiles derived from each of the two correlations for a given NHD group. The average R2 eff (R2 eff AVG) was reported, so a qualitative assessment of the internal dynamics could be used to quickly determine whether or not relaxation dispersion profiles could be fully analyzed (i.e. particularly for highly mobile 15N side chains).
Supplementary Material
S100B/EF-2 mutants bind Ca2+ tighter in the presence of TRTK.
The increase in Ca2+-binding is not due to a structural change in the EF2 of S100B
D63NS100B is a valid 15N probe to study side chain motion in the EF2 using NMR.
S100B and EF2 mutants show a decrease in fast/slow backbone time scale motions + TRTK.
The side chain dynamics (µs – ms) of Asn63 in D63NS100B quenches with TRTK bound.
Acknowledgements
This work was supported by the National Institutes of Health grants GM58888 (D.J.W.), CA107331 (D.J.W.) and CA144560-02 (M.A.L.). We thank the staff of the BL71 beamline of the Stanford Synchrotron Radiation Light source for their assistance in collecting X-ray diffraction data. The NMR spectrometers used in these studies were purchased, in part, with funds from shared instrumentation grants from the NIH (S10 RR10441; S10 RR15741; S10 RR16812; S10 RR23447 to D.J.W.) and from the National Science Foundation (DBI 1005795 to D.J.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers. The coordinates and structure factors for the Ca2+-boundD63NS100B and D63NS100B-Ca2+-TRTK X-ray structures were deposited in the PDB and assigned the accession numbers 3RLZ and 3RM1, respectively.
Attempts were made to crystallize and examine all of the EF2 mutant proteins, including D61NS100B, D65NS100B, and E72AS100B since their Ca2+-binding affinities were all increased by more than 5-fold with TRTK-12 bound; however, only the D63NS100B construct would crystallize in both the Ca2+- and TRTK12-bound forms. Attempts to crystallize these other mutant proteins in both complexes (i.e. Ca2+-bound; TRTK12-bound), if possible, will be the subject of future work.
For the more weakly binding EF2-mutants in the absence of target (CaKD> 400 µM), achieving occupancy>99.6% was not possible because the high levels of Ca2+ needed (>40 mM) caused aggregation of the protein, as previously described33. Therefore, the dispersion curves for these proteins in the absence of target may represent contributions of Rex from conformational exchange and/or from Cakoff. From the most conservative point of view, loss of Rex for these constructs upon TRTK12 binding may represent loss of conformational exchange and/or slower Cakoff values for Ca2+ release in solution. Therefore, discussion of the relaxation dispersion profiles is provided only for the backbone and side chain residues of wild-type S100B and D63NS100B constructs, which have occupancies exceeding 99.6% for Ca2+ bound to EF2 under the conditions used (+/− TRTK12). As stated in footnote 1, we are also searching conditions, so that occupancies for Ca2+-bound >99.6% can be achieved for the other mutant constructs without aggregation (i.e. at [Ca2+] > 40 mM). Such conditions could also impact our ability to crystallize these mutant constructs by avoiding protein aggregation.
References
- 1.Moore BW. A soluble protein characteristic of the nervous system. Biochem Biophys Res Commun. 1965;19:739–744. doi: 10.1016/0006-291x(65)90320-7. [DOI] [PubMed] [Google Scholar]
- 2.Donato R. Perspectives in S-100 protein biology. Review article. Cell Calcium. 1991;12:713–726. doi: 10.1016/0143-4160(91)90040-l. [DOI] [PubMed] [Google Scholar]
- 3.Kligman D, Hilt DC. The S100 protein family. Trends Biochem Sci. 1988;13:437–443. doi: 10.1016/0968-0004(88)90218-6. [DOI] [PubMed] [Google Scholar]
- 4.Baudier J, Glasser N, Gerard D. Ions binding to S100 proteins. I. Calcium- and zinc-binding properties of bovine brain S100 alpha alpha, S100a (alpha beta), and S100b (beta beta) protein: Zn2+ regulates Ca2+ binding on S100b protein. J Biol Chem. 1986;261:8192–8203. [PubMed] [Google Scholar]
- 5.Harpio R, Einarsson R. S100 proteins as cancer biomarkers with focus on S100B in malignant melanoma. Clin Biochem. 2004;37:512–518. doi: 10.1016/j.clinbiochem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 6.Loppin M, Quillien V, Adamski H, Ollivier I, Garlantezec R, Chevrant-Breton J. Protein S100 beta and Melanoma Inhibitory Activity (MIA): a prospective study of their clinical value for the early detection of metastasis in malignant melanoma. Ann Dermatol Venereol. 2007;134:535–540. doi: 10.1016/s0151-9638(07)89264-7. [DOI] [PubMed] [Google Scholar]
- 7.Salama I, Malone PS, Mihaimeed F, Jones JL. A review of the S100 proteins in cancer. Eur J Surg Oncol. 2008;34:357–364. doi: 10.1016/j.ejso.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 8.Camby I, Lefranc F, Titeca G, Neuci S, Fastrez M, Dedecken L, Schafer BW, Brotchi J, Heizmann CW, Pochet R, Salmon I, Kiss R, Decaestecker C. Differential expression of S100 calcium-binding proteins characterizes distinct clinical entities in both WHO grade II and III astrocytic tumours. Neuropathol Appl Neurobiol. 2000;26:76–90. doi: 10.1046/j.1365-2990.2000.00223.x. [DOI] [PubMed] [Google Scholar]
- 9.Rustandi RR, Drohat AC, Baldisseri DM, Wilder PT, Weber DJ. The Ca(2+)-dependent interaction of S100B (beta beta) with a peptide derived from p53. Biochemistry. 1998;37:1951–1960. doi: 10.1021/bi972701n. [DOI] [PubMed] [Google Scholar]
- 10.Lin J, Blake M, Tang C, Zimmer D, Rustandi RR, Weber DJ, Carrier F. Inhibition of p53 transcriptional activity by the S100B calcium-binding protein. J Biol Chem. 2001;276:35037–35041. doi: 10.1074/jbc.M104379200. [DOI] [PubMed] [Google Scholar]
- 11.Lin J, Yang Q, Yan Z, Markowitz J, Wilder PT, Carrier F, Weber DJ. Inhibiting S100B restores p53 levels in primary malignant melanoma cancer cells. J Biol Chem. 2004;279:34071–34077. doi: 10.1074/jbc.M405419200. [DOI] [PubMed] [Google Scholar]
- 12.Lin J, Yang Q, Wilder PT, Carrier F, Weber DJ. The calcium-binding protein S100B down-regulates p53 and apoptosis in malignant melanoma. J Biol Chem. 285:27487–27498. doi: 10.1074/jbc.M110.155382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Markowitz J, Mackerell AD, Jr, Carrier F, Charpentier TH, Weber DJ. Design of Inhibitors for S100B. Curr Top Med Chem. 2005;5:1093–1108. doi: 10.2174/156802605774370865. [DOI] [PubMed] [Google Scholar]
- 14.Charpentier TH, Thompson LE, Liriano MA, Varney KM, Wilder PT, Pozharski E, Toth EA, Weber DJ. The effects of CapZ peptide (TRTK-12) binding to S100B-Ca2+ as examined by NMR and X-ray crystallography. J Mol Biol. 2010;396:1227–1243. doi: 10.1016/j.jmb.2009.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malashkevich VN, Varney KM, Garrett SC, Wilder PT, Knight D, Charpentier TH, Ramagopal UA, Almo SC, Weber DJ, Bresnick AR. Structure of Ca2+-bound S100A4 and its interaction with peptides derived from nonmuscle myosin-IIA. Biochemistry. 2008;47:5111–5126. doi: 10.1021/bi702537s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wright NT, Cannon BR, Wilder PT, Morgan MT, Varney KM, Zimmer DB, Weber DJ. Solution structure of S100A1 bound to the CapZ peptide (TRTK12) J Mol Biol. 2009;386:1265–1277. doi: 10.1016/j.jmb.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright NT, Prosser BL, Varney KM, Zimmer DB, Schneider MF, Weber DJ. S100A1 and Calmodulin Compete for the Same Binding Site on Ryanodine Receptor. J Biol Chem. 2008;283:26676–26683. doi: 10.1074/jbc.M804432200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markowitz J, Rustandi RR, Varney KM, Wilder PT, Udan R, Wu SL, Horrocks WD, Weber DJ. Calcium-binding properties of wild-type and EF-hand mutants of S100B in the presence and absence of a peptide derived from the C-terminal negative regulatory domain of p53. Biochemistry. 2005;44:7305–7314. doi: 10.1021/bi050321t. [DOI] [PubMed] [Google Scholar]
- 19.Rustandi RR, Baldisseri DM, Weber DJ. Structure of the negative regulatory domain of p53 bound to S100B (beta beta) Nat Struct Biol. 2000;7:570–574. doi: 10.1038/76797. [DOI] [PubMed] [Google Scholar]
- 20.Moews PC, Kretsinger RH. Refinement of the structure of carp muscle calcium-binding parvalbumin by model building and difference Fourier analysis. J Mol Biol. 1975;91:201–225. doi: 10.1016/0022-2836(75)90160-6. [DOI] [PubMed] [Google Scholar]
- 21.Moeschler HJ, Schaer JJ, Cox JA. A thermodynamic analysis of the binding of calcium and magnesium ions to parvalbumin. Eur J Biochem. 1980;111:73–78. doi: 10.1111/j.1432-1033.1980.tb06076.x. [DOI] [PubMed] [Google Scholar]
- 22.Chaudhuri D, Horrocks WD, Jr, Amburgey JC, Weber DJ. Characterization of lanthanide ion binding to the EF-hand protein S100 beta by luminescence spectroscopy. Biochemistry. 1997;36:9674–9680. doi: 10.1021/bi9704358. [DOI] [PubMed] [Google Scholar]
- 23.Strynadka NC, James MN. Crystal structures of the helix-loop-helix calcium-binding proteins. Annu Rev Biochem. 1989;58:951–998. doi: 10.1146/annurev.bi.58.070189.004511. [DOI] [PubMed] [Google Scholar]
- 24.Wilder PT, Charpentier TH, Liriano MA, Gianni K, Varney KM, Pozharski E, Coop A, Toth EA, Mackerell AD, Weber DJ. In vitro screening and structural characterization of inhibitors of the S100B-p53 interaction. Int J High Throughput Screen. 2010;2010:109–126. doi: 10.2147/IJHTS.S8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilder PT, Lin J, Bair CL, Charpentier TH, Yang D, Liriano M, Varney KM, Lee A, Oppenheim AB, Adhya S, Carrier F, Weber DJ. Recognition of the tumor suppressor protein p53 and other protein targets by the calcium-binding protein S100B. Biochim Biophys Acta. 2006;1763:1284–1297. doi: 10.1016/j.bbamcr.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 26.Zimmer DB, Wright Sadosky P, Weber DJ. Molecular mechanisms of S100-target protein interactions. Microsc Res Tech. 2003;60:552–559. doi: 10.1002/jemt.10297. [DOI] [PubMed] [Google Scholar]
- 27.Inman KG, Yang R, Rustandi RR, Miller KE, Baldisseri DM, Weber DJ. Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12. J Mol Biol. 2002;324:1003–1014. doi: 10.1016/s0022-2836(02)01152-x. [DOI] [PubMed] [Google Scholar]
- 28.de Alba E, Baber JL, et al. The use of residual dipolar coupling in concert with backbone relaxation rates to identify conformational exchange by NMR. J Am Chem Soc. 1999;121:4282–4283. [Google Scholar]
- 29.Inman KG, Baldisseri DM, Miller KE, Weber DJ. Backbone dynamics of the calcium-signaling protein apo-S100B as determined by 15N NMR relaxation. Biochemistry. 2001;40:3439–3448. doi: 10.1021/bi0027478. [DOI] [PubMed] [Google Scholar]
- 30.Wright NT, Inman KG, Levine JA, Cannon BR, Varney KM, Weber DJ. Refinement of the solution structure and dynamic properties of Ca(2+)-bound rat S100B. J Biomol NMR. 2008;42:279–286. doi: 10.1007/s10858-008-9282-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishima R, Torchia DA. Error estimation and global fitting in transverse-relaxation dispersion experiments to determine chemical-exchange parameters. J Biomol NMR. 2005;32:41–54. doi: 10.1007/s10858-005-3593-z. [DOI] [PubMed] [Google Scholar]
- 32.Ishima R, Torchia DA. Extending the range of amide proton relaxation dispersion experiments in proteins using a constant-time relaxation-compensated CPMG approach. J Biomol NMR. 2003;25:243–248. doi: 10.1023/a:1022851228405. [DOI] [PubMed] [Google Scholar]
- 33.Drohat AC, Baldisseri DM, Rustandi RR, Weber DJ. Solution structure of calcium-bound rat S100B (beta beta) as determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1998;37:2729–2740. doi: 10.1021/bi972635p. [DOI] [PubMed] [Google Scholar]
- 34.Drohat AC, Amburgey JC, Abildgaard F, Starich MR, Baldisseri D, Weber DJ. Solution structure of rat apo-S100B (beta beta) as determined by NMR spectroscopy. Biochemistry. 1996;35:11577–11588. doi: 10.1021/bi9612226. [DOI] [PubMed] [Google Scholar]
- 35.Zimmer DB, Weber DJ. The Calcium-Dependent Interaction of S100B with Its Protein Targets. Cardiovasc Psychiatry Neurol. 2010;2010 doi: 10.1155/2010/728052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kursula P, Tikkanen G, Lehto VP, Nishikimi M, Heape AM. Calcium-dependent interaction between the large myelin-associated glycoprotein and S100beta. J Neurochem. 1999;73:1724–1732. doi: 10.1046/j.1471-4159.1999.731724.x. [DOI] [PubMed] [Google Scholar]
- 37.Delphin C, Ronjat M, Deloulme JC, Garin G, Debussche L, Higashimoto Y, Sakaguchi K, Baudier J. Calcium-dependent interaction of S100B with the C-terminal domain of the tumor suppressor p53. J Biol Chem. 1999;274:10539–10544. doi: 10.1074/jbc.274.15.10539. [DOI] [PubMed] [Google Scholar]
- 38.Baudier J, Delphin C, Grunwald D, Khochbin S, Lawrence JJ. Characterization of the tumor suppressor protein p53 as a protein kinase C substrate and a S100b-binding protein. Proc Natl Acad Sci U S A. 1992;89:11627–11631. doi: 10.1073/pnas.89.23.11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santamaria-Kisiel L, Rintala-Dempsey AC, Shaw GS. Calcium-dependent and -independent interactions of the S100 protein family. Biochem J. 2006;396:201–214. doi: 10.1042/BJ20060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ivanenkov VV, Jamieson GA, Jr, Gruenstein E, Dimlich RV. Characterization of S-100b binding epitopes. Identification of a novel target, the actin capping protein, CapZ. J Biol Chem. 1995;270:14651–14658. doi: 10.1074/jbc.270.24.14651. [DOI] [PubMed] [Google Scholar]
- 41.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marlow MS, Dogan J, Frederick KK, Valentine KG, Wand AJ. The role of conformational entropy in molecular recognition by calmodulin. Nat Chem Biol. 2010;6:352–358. doi: 10.1038/nchembio.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai CJ, Ma B, Nussinov R. Folding and binding cascades: shifts in energy landscapes. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9970–9972. doi: 10.1073/pnas.96.18.9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 45.Dyson HJ, Wright PE. Elucidation of the protein folding landscape by NMR. Methods in enzymology. 2005;394:299–321. doi: 10.1016/S0076-6879(05)94011-1. [DOI] [PubMed] [Google Scholar]
- 46.Matsumura H, Shiba T, Inoue T, Harada S, Kai Y. A novel mode of target recognition suggested by the 2.0 A structure of holo S100B from bovine brain. Structure. 1998;6:233–241. doi: 10.1016/s0969-2126(98)00024-0. [DOI] [PubMed] [Google Scholar]
- 47.Prosser BL, Wright NT, Hernandez-Ochoa EO, Varney KM, Liu Y, Olojo RO, Zimmer DB, Weber DJ, Schneider MF. S100A1 binds to the calmodulin-binding site of ryanodine receptor and modulates skeletal muscle excitation-contraction coupling. J Biol Chem. 2008;283:5046–5057. doi: 10.1074/jbc.M709231200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yap KL, Ames JB, Swindells MB, Ikura M. Diversity of conformational states and changes within the EF-hand protein superfamily. Proteins. 1999;37:499–507. doi: 10.1002/(sici)1097-0134(19991115)37:3<499::aid-prot17>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 49.Donato R. S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int J Biochem Cell Biol. 2001;33:637–668. doi: 10.1016/s1357-2725(01)00046-2. [DOI] [PubMed] [Google Scholar]
- 50.Marenholz I, Heizmann CW, Fritz G. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature) Biochem Biophys Res Commun. 2004;322:1111–1122. doi: 10.1016/j.bbrc.2004.07.096. [DOI] [PubMed] [Google Scholar]
- 51.Otwinowski ZM, W Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 52.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 55.Laskowski RAM, M W, Thornton JM. PROCHECK: a program to check the steriochemical quality of protein structures. J Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 56.Laskowski RA, MacArthur MW, Thornton JM. Validation of protein models derived from experiment. Curr. Opin. Struct. Biol. 1998;8:631–639. doi: 10.1016/s0959-440x(98)80156-5. [DOI] [PubMed] [Google Scholar]
- 57.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 59.Tollinger M, Skrynnikov NR, Mulder FA, Forman-Kay JD, Kay LE. Slow dynamics in folded and unfolded states of an SH3 domain. J Am Chem Soc. 2001;123:11341–11352. doi: 10.1021/ja011300z. [DOI] [PubMed] [Google Scholar]
- 60.Skrynnikov NR, Mulder FA, Hon B, Dahlquist FW, Kay LE. Probing slow time scale dynamics at methyl-containing side chains in proteins by relaxation dispersion NMR measurements: application to methionine residues in a cavity mutant of T4 lysozyme. J Am Chem Soc. 2001;123:4556–4566. doi: 10.1021/ja004179p. [DOI] [PubMed] [Google Scholar]
- 61.Mulder FA, Skrynnikov NR, Hon B, Dahlquist FW, Kay LE. Measurement of slow (micros-ms) time scale dynamics in protein side chains by (15) N relaxation dispersion NMR spectroscopy: application to Asn and Gln residues in a cavity mutant of T4 lysozyme. J Am Chem Soc. 2001;123:967–975. doi: 10.1021/ja003447g. [DOI] [PubMed] [Google Scholar]
- 62.Press WHSAT, Vetterling WT, Flannery BP. Numerical Recipes in C: The Art of Scientific Computing. 2nd ed. Cambridge UK: Cambridge University Press; 1988. [Google Scholar]
- 63.Motulsky HJ, R L. Fitting curves to data using nonlinear regression: a practical and nonmathematical review. FASEB J. 1987;1:365–374. [PubMed] [Google Scholar]
- 64.McConnell HM. J. Chem. Phys. 1958;28:430–431. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.