Abstract
Cell proliferation is a metabolically demanding process1,2. It requires active reprogramming of cellular bioenergetic pathways towards glucose metabolism to support anabolic growth1,2. NF-κB/Rel transcription factors coordinate many of the signals that drive proliferation during Immunity, inflammation and oncogenesis3, but whether NF-κB regulates the metabolic reprogramming required for cell division during these processes is unknown. Here, we report that NF-κB organizes energy metabolism networks by controlling the balance between the utilization of glycolysis and mitochondrial respiration. NF-κB inhibition causes cellular reprogramming to aerobic glycolysis under basal conditions and induces necrosis on glucose starvation. The metabolic reorganization that results from NF-κB inhibition overcomes the requirement for tumour suppressor mutation in oncogenic transformation and impairs metabolic adaptation in cancer in vivo. This NF-κB-dependent metabolic pathway involves stimulation of oxidative phosphorylation through upregulation of mitochondrial synthesis of cytochrome c oxidase 2 (SCO2; ref. 4). Our findings identify NF-κB as a physiological regulator of mitochondrial respiration and establish a role for NF-κB in metabolic adaptation in normal cells and cancer.
Transcription factors of the NF-κB/Rel family are central regulators of immune and inflammatory responses3. They also promote oncogenesis3. The best documented function of NF-κB (nuclear factor-κ–light-chain-enhancer of activated B cells) in these processes is the upregulation of a transcriptional program encoding inflammatory mediators, immunoregulators and inhibitors of apoptosis, as well as growth factors and factors stimulating cell migration and differentiation3.
Both immunity and tumorigenesis also involve a rapid rate of cell division. This presents substantial bioenergetic and biosynthetic challenges, which the cell meets by increasing glucose metabolism. This reliance on glucose under aerobic conditions (a phenomenon known in cancer as the Warburg effect2) enables dividing cells to fuel glycolysis and the pentose phosphate pathway to generate NADPH, macromolecules and ATP required for the doubling of biomass1,2. Glucose is therefore an essential nutrient for both cancer and normal proliferating cells1. When glucose is scarce, however, energy sensing by AMP-activated protein kinase (AMPK) arrests anabolic pathways, and metabolism is redirected to fatty acid oxidation and oxidative phosphorylation (OXPHOS), thus maximizing energy efficiency with the available resources1,5,6. Nevertheless, mitochondrial defects, partly due to altered expression and/or function of mitochondrial factors (for example, cytochrome c oxidase (COX) complex proteins)7,8, as well as oncogenic mutations can result in glucose addiction1,2,5,8,9. Whether the metabolic reprogramming required for cell proliferation is controlled by NF-κB, however, is unknown.
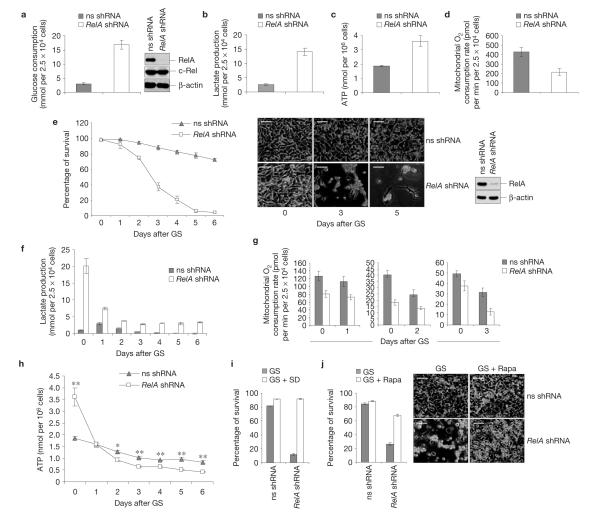
Strikingly, knock down of RelA, the dominant NF-κB transactivating subunit3, markedly enhanced glucose consumption and lactate production in mouse embryonic fibroblasts (MEFs), under normal culture conditions (Fig. 1a,b), thus recapitulating the Warburg effect. Similar results were obtained using early passage knockout MEFs (Supplementary Fig. S1a,b). RelA-deficient cells also exhibited increased ATP levels and decreased oxygen consumption levels (Fig. 1c,d and Supplementary Fig. S1c). Hence, basal NF-κB activity restrains reprogramming to aerobic glycolysis.
Figure 1.
NF-κB counters reprogramming to aerobic glycolysis and promotes metabolic adaptation to nutrient starvation. (a–d) Glucose consumption (a, left), lactate production (b), ATP concentration (c) and oxygen consumption (d) in immortalized MEFs expressing non-specific (ns) or RelA-specific shRNAs, under normal culture conditions. (a), Right, western blots with the cells in a–d, showing levels of RelA (knockdown efficiency), c-Rel and β-actin (knockdown specificity). (e) Left, viability of immortalized MEFs expressing non-specific or RelA shRNAs after glucose starvation (GS). Middle, images of representative cells. Right, western blots with non-specific and RelA shRNA cells. Similar results were obtained using two additional non-specific shRNAs (ns2, shc003v), two additional RelA-specific shRNAs and eGFP-specific, luciferase-specific, laminA/C-specific and cyclophilinB-specific shRNAs. (f–h) Lactate production (f), oxygen consumption (g) and ATP concentration (h) in cells treated as in e. Lactate values in f after day 4 should be interpreted with caution, owing to massive necrosis in RelA-shRNA cells. (i,j) Survival of the cells in e after 4-day glucose starvation, either alone (GS) or together with serum deprivation (GS+SD; i) or rapamycin (GS Rapa; j, left). (j) Right, representative images. In a–j the values denote mean ± s.e.m. n = 3 (a–c,e,f,h–j); n = 4 (d); n = 10 (g). In h, *P < 0.05; **P < 0.01. ± Scale bars: 50 μm.
To determine whether this glycolytic switch represented a homeostatic response to increased proliferation, we examined the ability of RelA-deficient cells to turn to OXPHOS for energy provision on glucose starvation. Remarkably, RelA inhibition impaired adaptation to glucose starvation, causing cell death (Fig. 1e and Supplementary Fig. S1d). In contrast, the viability of wild-type cells was virtually unaffected. Similar results were obtained with inhibitors of the NF-κB-inducing kinase, IκB kinase (IKK)β (Supplementary Fig. S1f,g). RelA-deficient cells remained dependent on glycolysis for energy production, even during glucose starvation, shown by their high lactate production and decreased oxygen consumption levels (Fig. 1f,g and also Supplementary Fig. S1h). This resulted in a marked decrease of ATP levels in glucose-starved RelA-deficient cells, but not in starved wild-type cells (Fig. 1h and Supplementary Fig. S1e). RelA deficiency did not impair proliferation arrest following glucose starvation (Supplementary Fig. S2a,b), thereby excluding defects in glucose-starvation-induced cell-cycle checkpoints1 as the cause of death in mutant cells. Hence, NF-κB inhibition hinders the ability of cells to meet the metabolic demand during glucose starvation by impairing OXPHOS, thus causing a metabolic crisis and cell death. Accordingly, decreasing the metabolic demand2,5 through serum deprivation or blockade of protein biosynthesis with the mTOR inhibitor rapamycin1,6 rescued RelA-deficient cells from glucose-starvation-induced cell death (Fig. 1i,j and Supplementary Fig. S2c).
Survival following nutrient deprivation depends on macroautophagy, an energy-producing auto-digestive process5,10. Indeed, silencing Beclin1 or ATG7, two essential macroautophagy effectors10, or treatment with the macroautophagy inhibitors, 3-methyladenine and chloroquine10, induced toxicity in glucose-starved wild-type MEFs to an extent similar to RelA inactivation (Supplementary Fig. S3a–c). However, neither the induction of the macroautophagy effector, LC3 (ref. 10), nor the formation of macroautophagic vesicles10, was impaired by RelA inhibition during glucose starvation (Supplementary Fig. S3d,e). Instead, this inhibition basally induced autophagic flux, shown by the enhanced rates of autophagosome formation and degradation exhibited by untreated RelA small hairpin RNA (shRNA) cells relative to control cells (Supplementary Fig. S3d–f, 0 h; see the increased levels of LC3 and eGFP–LC3 punctate staining, and decreased levels of the autophagic cargo protein p62 in RelA-shRNA cells not exposed to bafilomycin A1, inhibiting lysosomal vacuolar ATPase). Hence, defective macroautophagy is not responsible for the demise of glucose-starved RelA-deficient cells. Similarly, RelA-deficient and control cells exhibited no obvious differences in glucose-starvation-induced caspase activation (Supplementary Fig. S4a), and blocking apoptosis using caspase-8-specific shRNAs or the pan-caspase inhibitor z-VADfmk (10) failed to rescue the RelA-deficient cells from programmed cell death (PCD) after metabolic challenge (ref. Supplementary Fig. S4b–d). Notably, RelA inactivation retained toxicity in glucose-starved Bax−/−/Bak−/− MEFs, which are refractory to apoptosis10 (Supplementary Fig. S4e,f). Starved RelA knocked-down cells also showed morphological signs of necrosis, lacked nucleosomal fragmentation, a hallmark of apoptosis, and their culture media contained elevated levels of the necrosis marker HMGB1 (ref. 10; Supplementary Fig. S4g; data not shown). Thus, NF-κB-afforded protection during glucose starvation involves suppression of a necrotic death pathway.
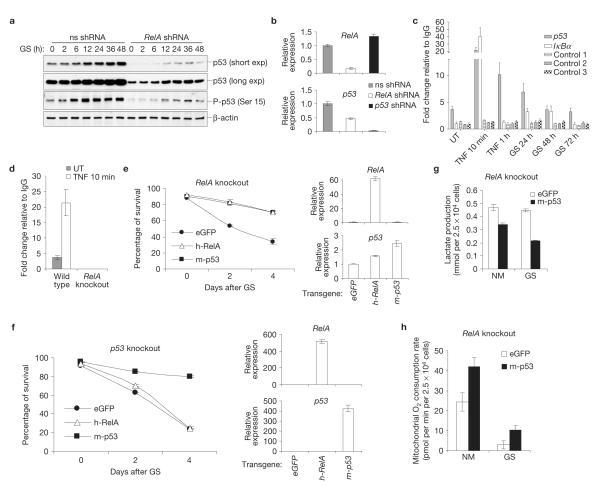
Adaptation to low glucose availability is orchestrated by the energy sensor AMPK, which suppresses anabolic pathways and induces cell-cycle arrest, thus dampening ATP consumption1,5,6. RelA deficiency, however, had no effect on glucose-starvation-induced activation of the Akt and mTOR pathways, promoting macromolecular biosynthesis1,5, or AMPK itself (Supplementary Fig. S5a). This deficiency nevertheless caused marked downregulation of p53 expression, in both unchallenged and glucose-starved cells (Fig. 2a).
Figure 2.
p53 mediates NF-κB-dependent protection against glucose starvation. (a) Western blots with antibodies against total or Ser15-phosphorylated (P) p53 (equivalent to mouse p53-Ser18) in non-specific (ns)- and RelA-shRNA-expressing immortalized MEFs after glucose starvation (GS). exp, exposure. (b) qRT-PCR with RelA- or p53-specific primers and RNAs from immortalized MEFs expressing the shRNAs shown. (c,d) Chromatin immunoprecipitation with antibodies against RelA, qPCR primers specific for B-containing regions of the p53 and IκBα promoters or control genomic regions (control 1–3) and extracts from untreated (UT), TNFα-treated or glucose-starved immortalized wild-type (c,d) and RelA−/− (RelA knockout; d) MEFs. (e,f) Survival of early passage RelA−/− (e) and p53−/− (p53 knockout; f) MEFs expressing exogenous eGFP, mouse (m)-p53 or human (h)-RelA before and after glucose starvation (left). qRT-PCR with RNAs from the same cells (0 h; right). (g,h) Lactate production and oxygen consumption in RelA−/− MEFs expressing exogenous eGFP or mouse p53 in the presence of normal medium (NM) and during glucose starvation (GS). In b–h the values denote mean ± s.e.m. n=3 (b–g); n 15 = (h). Uncropped images of blots are shown in Supplementary Fig. S8.
Basal p53 activity decreases the level of glycolysis and increases the level of OXPHOS, hence countering the Warburg effect1,5,7,11. Thus, we investigated whether p53 mediated any of the metabolic activities of NF-κB. Significantly, RelA inhibition downregulated basal messenger RNA levels of p53, as well as of the NF-κB target A20, consistent with a low basal NF-κB activity in MEFs (Fig. 2b and Supplementary Figs S4f and S5c; also Supplementary Figs S1g and S5d, phosphorylated I B and IKKα/β in untreated MEFs). RelA inhibition also impaired p53 mRNA induction by TNFα (Supplementary Fig. S5b). Conversely, p53 inactivation had no effect on RelA transcripts (Fig. 2b and Supplementary Fig. S4f). Chromatin immunoprecipitation showed that NF-κB/RelA complexes bound to a known κB element in the proximal p53 promoter region12, even under basal conditions (Fig. 2c). This binding was specific, as it was not detected with control genomic regions or RelA−/− cells (Fig. 2c,d). RelA binding to the p53 promoter increased after TNFα stimulation or glucose starvation, which induce NF-κB (ref. 3; Supplementary Fig. S5d), and was more stable at this promoter than at that of IκBα, another target of NF-κB (ref. 3; Fig. 2c). Thus, basal and induced p53 expression is under direct NF-κB-dependent transcriptional control. In contrast, NF-κB did not affect the relative glucose-starvation-dependent induction of total or Ser15-phosphorylated p53 (Fig. 2a), which is controlled by AMPK (refs 6,13), nor did it affect p53 protein stability11 (Supplementary Fig. S5e,f).
These data indicate that NF-κB is required, besides AMPK, to engage the p53 pathway to direct the cellular response to metabolic stress. Consistently, p53 silencing recapitulated many of the metabolic effects of RelA inactivation, including the increase in basal lactate production, glucose consumption and ATP levels, and the decreased oxygen utilization level7,13,14 (Supplementary Fig. S6a–d and also Fig. 1a–d,f–h, and Supplementary Fig. S1a–c,e, RelA-deficient cells; Supplementary Fig. S6e, functional p53 status in immortalized MEFs). It also markedly increased glucose-starvation-induced PCD (Supplementary Fig. S6f,g and also Supplementary Fig. S4e), established previously6, with magnitude and kinetics similar to RelA inactivation (Fig. 1e and Supplementary Figs S1d and S4e). As with RelA deficiency, p53 inactivation did not impair cell-cycle arrest1 under the glucose starvation conditions used (Supplementary Fig. S2a), as reported previously6, and glucose-starvation-induced cell death of p53-deficient cells was rescued by serum deprivation (Supplementary Fig. S2d), indicating that these cells may undergo the same metabolic crisis faced by RelA-deficient cells. Remarkably, p53 expression rescued RelA-deficient cells from glucose-starvation-induced necrosis (Fig. 2e), whereas RelA expression afforded no protection to glucose-starved p53-deficient cells (Fig. 2f). p53 reconstitution also decreased the level of lactate production and increased the level of oxygen consumption in RelA−/− cells, thus reverting the metabolic effects of RelA loss, both basally and during glucose starvation (Fig. 2g,h). Hence, p53 is a crucial downstream mediator of NF-κB in the bioenergetic pathway controlling adaptation to metabolic stress.
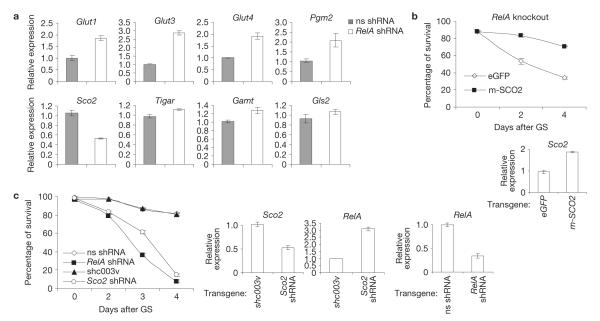
Reinforcing the notion that glucose addiction in NF-κB-deficient cells results from impaired p53 signalling, RelA inactivation upregulated basal expression of p53-repressed genes, including those encoding the glucose transporters GLUT1, GLUT3 and GLUT4 (refs 14–16), and the glycolytic enzyme phosphoglycerate mutase 2 (PGM2; ref.17; Fig. 3a). It also downregulated the p53 target SCO2 (Fig. 3a), a component of COX, the main cellular site of oxygen utilization4,7. Levels of metabolic enzymes controlled by p53 in a species- and/or tissue-specific manner13,18,19 were instead unaffected by NF-κB (Fig. 3a).
Figure 3.
SCO2 mediates NF-κB-dependent protection against glucose-starvation-induced PCD. (a) qRT-PCR with RNAs from non-specific (ns)- or RelA-shRNA-expressing immortalized MEFs under basal conditions. p53 metabolic targets referred to in the text, but not specified: TP53-induced glycolysis and apoptosis regulator18 (TIGAR), guanidinoacetate methyl transferase13 (GAMT ), glutaminase 2 (GLS2; ref. 19). (b) Top, survival of early passage RelA−/− MEFs expressing exogenous eGFP or mouse (m)-SCO2 after glucose starvation (GS). (c) Left, survival of immortalized MEFs expressing the indicated shRNAs after glucose starvation. (b) Bottom and (c) Right, qRT-PCR showing relative Sco2 and RelA expression in the same cells (0 h). In a–c the values denote mean±s.e.m. (n = 3).
Knocking down GLUT1, GLUT3 or PGM2 failed to protect RelA-deficient cells against glucose-starvation-induced PCD (Supplementary Fig. S7a), indicating that upregulation of the genes encoding these proteins cannot account for glucose addiction in these cells. Silencing them also promoted necrosis in glucose-starved wild-type cells. Hence, as reported previously20, increased glycolytic flux enhances, rather than impairs, cell tolerance to low glucose availability. Remarkably, this tolerance was restored in NF-κB-deficient cells by reconstitution with SCO2 (Fig. 3b), coincident with the decreased reliance of these cells on glucose metabolism (Supplementary Fig. S7b–d). In contrast, SCO2 downregulation to levels seen in RelA-deficient cells (Fig. 3a) induced PCD in glucose-starved wild-type cells to an extent comparable to RelA inactivation (Fig. 3c). SCO2 silencing also increased the levels of glucose consumption, lactate secretion and basal ATP production in untreated wild-type cells, and it decreased this production during glucose starvation7 (Supplementary Fig. S7e–g). It also caused a compensatory increase in the mRNA levels of p53-repressed glycolytic genes5 in RelA-deficient cells, but not in wild-type cells (Supplementary Fig. S7h), indicating involvement of a p53-dependent mechanism. Although SCO2 downregulation also increased RelA levels (Fig. 3c), Sco2-shRNA cells still succumbed to glucose starvation. Thus, SCO2 recapitulates many of the effects of NF-κB on energy homeostasis, metabolic adaptation and glycolytic gene expression. These data identify SCO2 as a downstream effector of the NF-κB–p53 bioenergetic pathway and underscore its importance in the protective activity of NF-κB during metabolic stress.
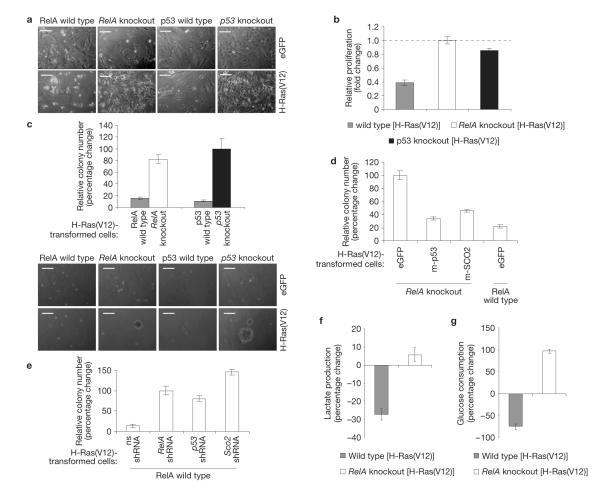
RelA or p53 disruption induces the Warburg effect, and deregulation of mitochondrial proteins (for example, COX components) and other metabolic factors is common in cancer1,2,5,7–9,19,20. We reasoned therefore that the metabolic reorganization resulting from RelA deficiency may overcome the requirement for tumour suppressor mutation in malignant transformation11. Indeed, on expression of oncogenic H-Ras(V12) (ref. 21), early passage RelA−/− MEFs acquired morphological features of transformed cells (Fig. 4a), and retained a proliferation rate comparable to their untransformed counterparts (Fig. 4b). Similarly treated wild-type cells underwent instead growth arrest and senescence, as expected (Fig. 4a,b). Foremost, unlike wild-type cells, H-Ras(V12)-expressing RelA−/− MEFs exhibited anchorage-independent growth (Fig. 4c), a hallmark of transformation14. These data demonstrate an ability of RelA-deficient cells to bypass oncogeneinduced senescence resulting in transformation even in the presence of wild-type p53.
Figure 4.
RelA suppresses oncogenic transformation by regulating energy metabolism. (a) Images of representative early passage RelA−/−, p53−/− and wild-type MEFs infected with pWPT–eGFP or pWPT–H-Ras(V12), showing transformation features (that is, higher density, spindled morphology) in mutant cells, but not in wild-type cells. (b) Fold change in cell numbers in H-Ras(V12)-expressing MEFs relative to the respective eGFP-expressing controls after a 48 h culture. (c) Top, percentage change in colony numbers with H-Ras(V12)-transformed cells relative to H-Ras(V12)-infected p53−/− MEFs. Bottom, images of representative colonies. (d,e) Percentage change in colony numbers with H-Ras(V12)-transformed early passage RelA−/− MEFs expressing eGFP, mouse (m)-p53 or m-SCO2 and H-Ras(V12)-transformed early passage wild-type MEFs expressing eGFP relative to eGFP-expressing RelA−/− MEFs (d), and H-Ras(V12)-transformed early passage wild-type MEFs expressing non-specific (ns), RelA, p53 or Sco2 shRNAs relative to RelA-shRNA-expressing MEFs (e). (f,g) Percentage change in lactate production (f) and glucose consumption (g) in H-Ras(V12)-transformed MEFs relative to the respective eGFP-expressing controls (see also Supplementary Fig. S1a,b, absolute levels in the absence of H-Ras(V12)). The data in b,c,f and g are from the cells in a. In b–g the values denote mean±s.e.m. n=3 (b,f,g); n = 4 (c,e). Scale bars: 50 μm.
The effect of RelA loss on H-Ras(V12)-induced transformation was reverted by the ectopic expression of SCO2 or p53 (Fig. 4d). Conversely, SCO2 silencing had the same promoting effect on transformation as the silencing of RelA or p53 (Fig. 4e). Consistent with the role of glycolysis in proliferation and transformation, H-Ras(V12) expression further upregulated glycolytic flux in RelA−/− MEFs, and further decreased it in senescent wild-type cells, relative to untransformed cells (Fig. 4f,g). These data position the p53/SCO2-dependent metabolic pathway downstream of RelA in the suppression of oncogene-induced transformation (see also Supplementary Fig. S5g, p53 levels in H-Ras(V12)-expressing MEFs). They support a model whereby RelA suppresses transformation in part by upregulating a SCO2-mediated mechanism, which increases the rate of respiration and dampens the rate of glycolysis, thus limiting the Warburg effect. Accordingly, SCO2 inhibition induces the same reprogramming to glycolysis exhibited by p53-and RelA-deficient cells7 (Supplementary Fig. S7e–g), whereas SCO2 reconstitution reverts this reprogramming in RelA−/− MEFs (Supplementary Fig. S7b–d).
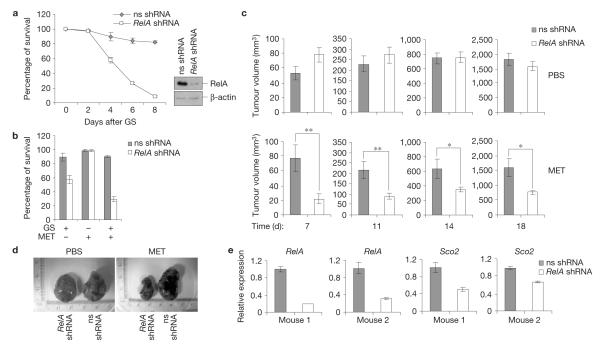
Our data indicate that the NF-κB-dependent increase in mitochondrial metabolism is a barrier for transformation. NF-κB, however, has been shown in many studies to promote tumorigenesis. Thus, we examined the relevance of the NF-κB metabolic function in colon carcinoma cells, a model system in which NF-κB plays this pro-tumorigenic role22,23. Following RelA silencing, CT-26 colon carcinoma cells became susceptible to metabolic challenge with glucose starvation, either alone or in combination with the anti-type-II diabetes drug, metformin, which inhibits mitochondrial complex I (ref. 24; Fig. 5a,b). Moreover, data in mice showed that, although having little or no effect on basal tumour survival and growth, NF-κB inhibition renders CT-26-derived tumours highly susceptible to mitochondrial stress in vivo, under conditions that do not affect NF-κB-proficient CT-26 tumours. This effect of RelA on systemic metformin treatment was especially pronounced at early times, when tumours expressed higher levels of RelA-targeting shRNAs and lower levels of Sco2 mRNAs (Fig. 5c,e). These findings indicate that suppressing mitochondrial metabolism in certain established cancer cells through inhibition of NF-κB and metformin diminishes tumorigenesis in vivo.
Figure 5.
NF-κB promotes metabolic adaptation in cancer in vivo. (a) Left, viability of CT-26 cells expressing non-specific (ns) or RelA shRNAs before and after glucose starvation (GS). Right, western blots showing RelA-knockdown efficiency. (b) Survival of the cells in a after a 4-day treatment with glucose starvation, metformin (MET) or glucose starvation plus metformin. (c) Growth of CT-26 tumours expressing non-specific or RelA shRNAs in nude mice treated with metformin or PBS. (d) Images of representative tumours from c. (e) qRT-PCR showing the relative RelA and Sco2 levels in tumours isolated from two representative metformin-treated mice at day 14. In a–c and e the values denote mean±s.e.m. n = 3 (a,b,e); n = 9 (c); *P < 0.05; **P < 0.01.
We have identified NF-κB as a physiological regulator of mitochondrial respiration, and have established that this function of NF-κB suppresses the Warburg effect and oncogenic transformation, and prevents necrosis on nutrient starvation. We also identified a role for NF-κB in metabolic adaptation in cancer in vivo. This metabolic function of NF-κB involves the p53-dependent upregulation of SCO2, which increases OXPHOS, thereby decreasing glycolytic flux. Our findings position NF-κB at a nodal checkpoint tethering cell activation and proliferation to energy sensing and metabolic homeostasis.
NF-κB–p53 crosstalk has been previously investigated during genotoxic stress and inflammation, with different outcomes, depending on tissue type, levels of induced nuclear activities and the nature of post-translational modifications25. We show here, however, that under basal conditions, as well as in response to certain stimuli, NF-κB and p53 cooperate to direct cellular bioenergetics. Consistently, RelA deficiency phenocopies some of the basal effects of p53 inactivation, including defective DNA repair and genomic instability, both in culture and mice11,26,27. Although NF-κB was reported to activate the p53 promoter in CAT/luciferase reporter assays12, the fundamental roles of NF-κB in the regulation of oxidative metabolism and metabolic adaptation had not been investigated. Of note, our unpublished data indicate that NF-κB may control metabolism also through p53-independent mechanisms (C.M., S.C.L. & G.F., unpublished data), underscoring the concept that it governs global energy metabolism networks, beyond p53. Further studies will determine the significance of these p53-independent functions of NF-κB. Nevertheless, here we identify NF-κB as a central regulator of energy homeostasis and metabolic adaptation.
The role of NF-κB in Ras-induced transformation, however, remains controversial14,15,21,28,29. A recent study indicated that RelA can promote this transformation by upregulating GLUT3 (ref. 14). The status of p53, however, is a confounding issue in this and other studies, as the systems used were either immortalized fibroblasts harbouring a defective p53 pathway or p53−/− models14,15,21,29. By using early passage p53 wild-type MEFs, we show instead that the NF-κB-imposed restraint of glucose metabolism and transformation is mediated by endogenous p53, which seemingly overrides other p53-independent glycolysis-associated activities of NF-κB. Consistently, NF-κB functions as an inhibitor of proliferation and a tumour suppressor in some tissues3,28,30.
Notwithstanding, NF-κB more often promotes progression and survival in cancer3,29,31. In this regard, our data indicate that NF-κB plays this pro-tumorigenic role in part by enabling cancer growth during metabolic stress, potentially permitting adaptation to hypoxic and hypoglycaemic environments. These findings are consistent with previous studies showing that mitochondrial metabolism is required for tumorigenesis32,33. Interestingly, the dichotomy we report for the pro- and anti-tumorigenic metabolic activities of NF-κB has been noted previously for many other oncogenes and tumour suppressors, including p53 (refs 1,11,19,24). However, NF-κB is often upregulated in cancer, and p53 is often mutated. Our data, nevertheless, do not exclude that in some tumours NF-κB may exercise its metabolic and pro-tumorigenic functions also through mutated p53. Although further studies are required to address this issue, our findings establish the physiological significance of the NF-κB-dependent control of energy metabolism to metabolic adaptation in cancer in vivo, and they define a bioenergetic pathway by which NF-κB can promote tumour progression and survival, independently of the regulation of apoptosis or inflammation. These findings have therapeutic implications, as they indicate that NF-κB inhibitors may enhance the anti-cancer efficacy of metabolic drugs.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturecellbiology
METHODS
Antibodies and reagents
The antibodies used for western blots were: anti-RelA, (1:10,000; Assay Design); anti-caspase-8 (1:1,000; Alexis); anti-PARP1 (1:100; Calbiochem); anti-p53 (1:2,000; Oncogene); anti-beclin1 (1:1,000), anti-p21 (1:1,000; BD Biosciences); anti-HMGB1 (1:500; Abcam); anti-LC3 (1:100; Abgent); anti-ATG7 (1:200), anti-TSC2 (1:1,000), anti-P-TSC2 (1:1,000), anti-ACC (1:1,000), anti-P-ACC (1:1,000), anti-Akt (1:1,000), anti-P-Akt (1:1,000), anti-P-p53(Ser 15) (1:1,000), anti-AMPKα (1:1,000), anti-P-AMPKα (1:1,000), anti-p70-S6K (1:1,000), anti-P-p70-S6K (1:2,000), anti-IKKβ (1:1,000), anti-P-IKKα/β (1:1,000), anti-caspase-3 (1:1,000), anti-caspase-9 (1:1,000), anti-P-IκBα (1:1,000), anti-c-Rel (1:1,000), anti-p38 (1:1,000), anti-P-p38 (1:1,000), anti-ERK1/2 (1:1,000), anti-P-ERK1/2 (1:1,000; Cell Signaling); anti-MDM2 (1:1,000), anti-β-actin (1:3,000; Santa Cruz Biotechnology); anti-p62 (1:1,000; Sigma-Aldrich); anti-RelA (1:1,000) and rabbit control IgG (1:1,000) for chromatin immunoprecipitation were from Santa Cruz Biotechnology. Anti-BrdU (1:20) for cell-cycle analyses was from eBioscience. The reagents used were: mouse TNFα (1,000 U ml—1; Peprotech); the IKKβ inhibitor SC-514 (20 or 100 μM; Calbiochem); z-VADfmk (50 μM; Alexis); MG132 (0.5 or 50 μM), 3-methyladenine (5 mM), chloroquine (5 μM), metformin (2 mM), bafilomycin A1 (100 nM), daunorubicin (5 μM), cycloheximide (0.1 μg ml–1 or 10 μg ml–1; Sigma-Aldrich); rapamycin (100 nM; Calbiochem); BrdU (Roche); 7-AAD (BD Biosciences). Antimycin (1 μM) and rotenone (1 μM) used for mitochondrial oxygen consumption analyses were from Sigma-Aldrich.
Cell culture and lentiviral infections
Early passage RelA−/−, RelA+/+, p53−/− and p53+/+ MEFs were derived from 14-day-old mouse embryos using standard methods and used at passage (p)1 to p5. Immortalized wild-type and RelA−/− MEFs were described previously34. Immortalized Bax−/−/Bak−/− MEFs were provided by C.B. Thompson. The CT-26 wild-type mouse colon carcinoma cell line was purchased from ATCC. Cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; with glutamine, without sodium pyruvate) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich), antibiotics (100 μg ml−1 penicillin and 100 μg ml−1 streptomycin) and 1 mM glutamine. For glucose starvation, early passage MEFs were cultured in DMEM with no glucose (with glutamine, without sodium pyruvate), supplemented with 10% dialysed FBS (Sigma-Aldrich), antibiotics and 2 mM sodium pyruvate. Glucose starvation with immortalized MEFs was carried out in the same DMEM with no glucose, supplemented with 10% FBS, antibiotics and 2 mM sodium pyruvate, in the presence of 6 mM d-glucose, unless otherwise specified. Glucose starvation with CT-26 wild-type cells was carried out as for immortalized MEFs, in the presence of 14 mM d-glucose. For serum deprivation, cells were cultured in DMEM with no glucose, supplemented with antibiotics and 2 mM sodium pyruvate, without FBS. Production of high-titre lentiviral preparations in HEK293T cells and lentiviral infections were carried out as described previously34,35.
Lentiviral vectors
The targeting shRNAs used are listed in Supplementary Table SI. The DNA sequences encoding shRNAs specific for eGFP, firefly luciferase and mouse RelA (RelA shRNA), caspase-8, beclin1, Atg7, p53 (p53 shRNA, p53-2 shRNA), laminA/C and cyclophilinB, as well as the control non-specific sequences, ns and ns2, were introduced between the BamHI and HpaI restriction sites of the lentiviral vector pLentiLox3.7 (ref. 35). A variant of pLentiLox3.7 (pLL–red), expressing enhanced red fluorescent protein (eRFP), was used to deliver non-specific and RelA-specific shRNAs to immortalized MEFs in Supplementary Fig. S3e. The DNA-coding sequences of the shRNAs specific for mouse Glut1 (also known as Slc2a1), Glut3 (also known as Slc2a3), Pgm2 or Sco2, and the additional RelA-specific sequences, RelA-2 shRNA and RelA-3 shRNA, as well as the control sequence, shc003v, in the pLKO.1 lentiviral vector were purchased from Sigma-Aldrich. The pWPT lentiviral vector, expressing eGFP, has been described previously36. The full-length complementary DNAs of human H-Ras(V12), human RelA and mouse p53 were introduced between the BamHI and SalI restriction sites of pWPT to replace eGFP. The cDNAs of mouse Sco2 and LC3 were purchased from Eurofins MWG Operon and obtained from K. Ryan, respectively, then cloned between the same restriction sites of pWPT.
Measurements of glucose, lactate, ATP and oxygen
Glucose consumption, lactate secretion and intracellular ATP levels were measured as described previously13,14. Briefly, cells were seeded onto 60-mm tissue-culture dishes, media were changed 6 h later and assays were carried out after a 48-h culture in normal medium (basal determinations) or in medium containing no or low glucose (glucose starvation), as specified. Glucose and lactate concentrations were measured in the culture media using the Glucose Assay Kit and the Lactate Assay Kit II (Biovision), respectively, according to the manufacturer’s instructions. Glucose consumption was extrapolated by subtracting the measured glucose concentrations in the media from the original 25 mM glucose concentration. Intracellular ATP levels were determined in cell lysates using the ATP Bioluminescent Somatic Cell Assay Kit (Sigma-Aldrich), according to the manufacturer’s instructions. Oxygen uptake was measured in 24-well plates using a Seahorse XF24 extracellular flux analyser9. Cells were seeded at 2–5 × 104 cells per well in 8.3 g l−1 DMEM base medium (pH 7.4) supplemented with 200 mM GlutaMax-1, 100 mM Na pyruvate, 32 mM NaCl and 40 μM phenol red, in the presence of either 25 mM (basal determinations) or 6 mM (glucose starvation) glucose, and oxygen consumption was measured continuously as described previously9. Mitochondrial oxygen consumption rates were calculated by subtracting the residual oxygen consumption in the cells after treatment with antimycin (1 μM) plus rotenone (1 μM). All values were finally normalized on a per-cell basis.
Soft-agar colony assays
Soft-agar colony formation was assessed using standard methods14. Briefly, early passage MEFs were infected with the viruses indicated and poured 6 days later at a density of 2 × 104 cells per dish together with 0.5%-agarose F12 medium onto a pre-solidified bottom layer of 0.5% agarose in the same medium, in 60-mm tissue-culture dishes. Colony formation was assessed 3 weeks later.
Cell death and macroautophagy analyses
Cell viability and HMGB1 extracellular release were assessed by trypan blue exclusion assays and western blotting, respectively, as reported previously34,37. HMGB1 assessment was carried out both in media and cellular fractions. For the analyses of macroautophagy, cells were infected with pWPT–eGFP–LC3, expressing an eGFP fusion protein of LC3, and eGFP–LC3 translocation was monitored as described previously38. Macroautophagic vesicles were quantified by counting 100 cells and scoring as positive those showing any signs of punctuate, rather than diffuse, eGFP–LC3 fluorescence, using an inverted Leica SP5 confocal microscope.
Cell-cycle analyses
Cell-cycle analyses were carried out using BrdU and 7-AAD labelling, as described previously39. Briefly, early passage or immortalized MEFs were seeded at a density of 1 × 106 cells per dish in 100-mm tissue-culture dishes and 24 h later either left untreated or subjected to glucose starvation (0 mM and 6 mM glucose, respectively) for a further 48 h. Cells were then incubated with BrdU (30 μM) for 3 h, trypsinized, fixed in 70% ethanol and permeabilized with 2N HCl and 0.5% Triton X-100, followed by neutralization in 0.1 M sodium borate. Samples were finally stained with FITC-labelled anti-BrdU antibody and 7-AAD, and fluorescence signals were acquired using a FACS Caliber (Beckman).
Quantitative real-time polymerase-chain reaction
Total RNA was extracted with Trizol, and purified using the PureLink RNA mini-kit (Invitrogen). RNA (1 μg) was added as a template to reverse-transcriptase reactions carried out using the GeneAmp RNA PCR Kit (Applied Biosystems). Quantitative real-time PCRs (qRT-PCRs) were carried out with the resulting cDNAs in triplicate using SYBR Green PCR Master Mix (Applied Biosystems), the primers listed in Supplementary Table SII and an ABI 7900 real-time PCR machine. Experimental Ct values were normalized to hypoxanthine–guanine phosphoribosyltransferase (HPRT), and relative mRNA expression was calculated versus a reference sample.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was carried out as described previously40. Briefly, unstimulated, TNF -treated and glucose-starved immortalized RelA−/− and wild-type MEFs were treated with 1% formaldehyde for 10 min at room temperature to crosslink protein–DNA complexes; then reactions were quenched using 125 mM glycine. Chromatin extracts were sonicated with a Bioruptor sonicator (Diagenode), and protein–DNA complexes were immunoprecipitated using anti-RelA or rabbit control IgG antibodies. Quantitative PCR (qPCR) assays were then carried out using anti-RelA and IgG precipitates and the primers listed in Supplementary Table SII, encompassing the κB-containing regions of the p53 and IκBα promoters or control genomic regions (control 1–3), to assess RelA DNA-binding specificity. RelA-specific DNA binding was calculated relative to the background signal yielded by control IgG and expressed as fold change.
Mouse allografts
Early passage CT-26 wild-type cells expressing non-specific or RelA shRNAs were collected, washed twice with PBS, and resuspended in PBS at a concentration of 1 × 106 cells ml−1 before being inoculated subcutaneously into both flanks (non-specific shRNA cells on the right flank, and RelA-shRNA cells on the left flank; 1 × 105 cells per flank) of nude mice (6-week-old females; Charles River). Metformin was dissolved in PBS (50 mg ml−1) and administered intraperitoneally daily (250 mg × kg body weight) from day 3 after tumour cell injection. The control group received PBS, using the same administration schedule and route. Tumour volume (mm3) was measured twice a week at the indicated times and estimated from caliper measurements using the following formula: volume = A × B2/2 (with A being the larger diameter, and B the smaller diameter of the tumour). Experiments were carried out under the Home Office Authority (Cambridge, UK; PPL 70/6874).
Statistical analysis
Results are expressed as mean ± s.e.m. from an appropriate number of samples as indicated in the figure legends. Student’s t-test was used to determine statistical significance.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Pagano, F. Dazzi, P. Ashton-Rickardt, F. Marelli-Berg and G. Screaton for critical comments on the manuscript. We also thank K. Ryan (Beatson Institute for Cancer Research, Glasgow, UK) for the eGFP–LC3 plasmid; T. Lindsten and C. B. Thompson (University of Pennsylvania, Philadelphia, USA) for the immortalized Bax−/−/Bak−/− MEFs; D. Trono (Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland) for the pWPT lentiviral vector; C. Chevtzoff and D. G. Hardie for assistance with the use of the Seahorse machine; and K. R. Chng for assistance with the analyses of the p53 promoter. C.M. was supported in part by a fellowship from AIRC (Italy). S.C.L. is supported by a scholarship from A*STAR (Singapore). M.M. is supported by a fellowship from the Pasteur Institute, Cenci Bolognetti Foundation (Italy). This work was supported by NIH grants R01 CA084040 and R01 CA098583 and Cancer Research UK grant C26587/A8839 to G.F., and NIH grant R01 CA123067 to N.S.C.
Footnotes
Note: Supplementary Information is available on the Nature Cell Biology website
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints
References
- 1.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karin M. Nuclear factor-κB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 4.Leary SC. Redox regulation of SCO protein function: controlling copper at a mitochondrial crossroad. Antioxid. Redox Signal. 2010;13:1403–1416. doi: 10.1089/ars.2010.3116. [DOI] [PubMed] [Google Scholar]
- 5.Vousden KH, Ryan KM. p53 and metabolism. Nat. Rev. Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 6.Jones RG, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 7.Matoba S, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 8.Cuezva JM, et al. The tumor suppressor function of mitochondria: translation into the clinics. Biochim. Biophys. Acta. 2009;1792:1145–1158. doi: 10.1016/j.bbadis.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Wu M, et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am. J. Physiol. Cell Physiol. 2007;292:125–136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- 10.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 11.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 12.Kirch HC, et al. Expression of human p53 requires synergistic activation of transcription from the p53 promoter by AP-1, NF-κB and Myc/Max. Oncogene. 1999;18:2728–2738. doi: 10.1038/sj.onc.1202626. [DOI] [PubMed] [Google Scholar]
- 13.Ide T, et al. GAMT, a p53-inducible modulator of apoptosis, is critical for the adaptive response to nutrient stress. Mol. Cell. 2009;36:379–392. doi: 10.1016/j.molcel.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Kawauchi K, Araki K, Tobiume K, Tanaka N. p53 regulates glucose metabolism through an IKK-NF-κB pathway and inhibits cell transformation. Nat. Cell Biol. 2008;10:611–618. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]
- 15.Meylan E, et al. Requirement for NF-κB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462:104–107. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64:2627–2633. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- 17.Kondoh H, et al. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65:177–185. [PubMed] [Google Scholar]
- 18.Bensaad K, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 19.Vousden KH. Alternative fuel—another role for p53 in the regulation of metabolism. Proc. Natl Acad. Sci. USA. 2010;107:7117–7118. doi: 10.1073/pnas.1002656107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Locasale JW, Cantley LC, Vander Heiden MG. Cancer’s insatiable appetite. Nat. Biotechnol. 2009;10:916–917. doi: 10.1038/nbt1009-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanson JL, Hawke NA, Kashatus D, Baldwin AS. The nuclear factor-κB subunits RelA/p65 and c-Rel potentiate but are not required for Ras-induced cellular transformation. Cancer Res. 2004;64:7248–7255. doi: 10.1158/0008-5472.CAN-03-3898. [DOI] [PubMed] [Google Scholar]
- 22.Luo JL, et al. Inhibition of NF-κB in cancer cells converts inflammation-induced tumour growth mediated by TNFα to TRAIL-mediated tumour regression. Cancer Cell. 2004;6:297–305. doi: 10.1016/j.ccr.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 23.Greten FR, et al. IKK links inflammation and tumourigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 24.Buzzai M, et al. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res. 2007;67:6745–6752. doi: 10.1158/0008-5472.CAN-06-4447. [DOI] [PubMed] [Google Scholar]
- 25.Tergaonkar V, Perkins ND. p53 and NF-κB crosstalk: IKKα tips the balance. Mol. Cell. 2007;26:158–159. doi: 10.1016/j.molcel.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 26.Sablina AA, et al. The antioxidant function of the p53 tumor suppressor. Nat. Med. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, et al. RelA/p65 functions to maintain cellular senescence by regulating genomic stability and DNA repair. EMBO Rep. 2009;10:1272–1278. doi: 10.1038/embor.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dajee M, et al. NF-κB blockade and oncogenic Ras trigger invasive human epidermal neoplasia. Nature. 2003;421:639–643. doi: 10.1038/nature01283. [DOI] [PubMed] [Google Scholar]
- 29.Gapuzan M-ER, et al. Immortalized fibroblasts from NF-κB RelA knockout mice show phenotypic heterogeneity and maintain increased sensitivity to tumor necrosis factor after transformation by v-Ras. Oncogene. 2005;24:6574–6583. doi: 10.1038/sj.onc.1208809. [DOI] [PubMed] [Google Scholar]
- 30.He G, et al. Hepatocyte IKKβ/NF-κB inhibits tumor promotion and progression by preventing oxidative stress-driven STAT3 activation. Cancer Cell. 2010;17:286–297. doi: 10.1016/j.ccr.2009.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bassères DS, Ebbs A, Levantini E, Baldwin AS. Requirement of the NF-κB subunit p65/RelA for K-Ras-induced lung tumorigenesis. Cancer Res. 2010;71:3537–3546. doi: 10.1158/0008-5472.CAN-09-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinberg F, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumourigenicity. Proc. Natl Acad. Sci. USA. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo JY, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumourigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pham CG, et al. Ferritin heavy chain upregulation by NF-κB inhibits TNFα-induced apoptosis by suppressing reactive oxygen species. Cell. 2004;119:529–542. doi: 10.1016/j.cell.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 35.Yang H, et al. TNF-α inhibits asbestos-induced cytotoxicity via a NF-κB-dependent pathway, a possible mechanism for asbestos-induced oncogenesis. Proc. Natl Acad. Sci. USA. 2006;103:10397–10402. doi: 10.1073/pnas.0604008103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dull T, et al. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zong WX, et al. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 2004;18:1272–1282. doi: 10.1101/gad.1199904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crighton D, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 39.Zinszner H, et al. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fullwood MJ, et al. An oestrogen-receptor-α-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.