Abstract
The present study establishes relationships between structure and reactivity for the pyrroloquinoline and phenanthroline quinones. The electrochemical reductions of 1,7- and 1,10-phenanthroline-5,6-quinones, like other quinones, are reversible and occur by 2e- transfer in a single step in aqueous solution and by two 1e(-)-transfer steps in aprotic media. The electron-withdrawing pyridine moieties both increase their potentials and stabilize their aprotic semiquinones. The electrochemistry of the cofactor methoxatin and its trimethylester derivative is similar to the phenanthroline quinones in aqueous solution. However, the electrochemical reductions of methoxatin and its triester in aprotic solutions are characterized by at least three potentials, each accounting for less than 1e-. This has been explained by the proposal of semiquinone complexing with itself and with quinone. Despite an electron-donating pyrrole moiety, methoxatin and its trimethylester have relatively high potentials in aprotic solution. This is presumably due to stabilization of radical anions by the aforementioned complexing or by delocalization with carboxylic acid and ester groups. The reduction potential of methoxatin, in both aqueous and aprotic solvent, suggests that oxidation of methanol should be a thermodynamically favorable process. No evidence for an electrochemically reduced state lower than the quinol was found for any of the compounds. Chemical reactivity is influenced by the orientation of the pyridine nitrogen. The two quinones with a pyridine nitrogen peri to a quinone carbonyl add and oxidize nucleophiles most readily.
Full text
PDF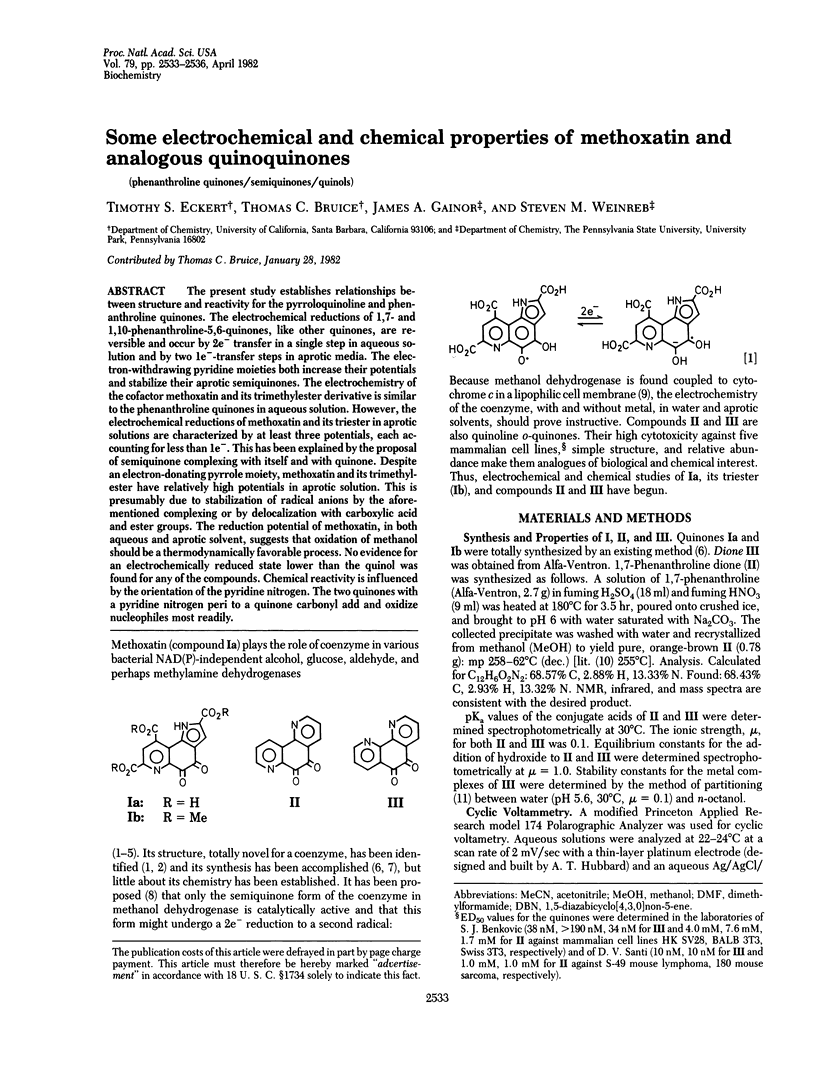

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alefounder P. R., Ferguson S. J. A periplasmic location for methanol dehydrogenase from Paracoccus denitrificans: implications for proton pumping by cytochrome aa3. Biochem Biophys Res Commun. 1981 Feb 12;98(3):778–784. doi: 10.1016/0006-291x(81)91179-7. [DOI] [PubMed] [Google Scholar]
- Ameyama M., Matsushita K., Ohno Y., Shinagawa E., Adachi O. Existence of a novel prosthetic group, PQQ, in membrane-bound, electron transport chain-linked, primary dehydrogenases of oxidative bacteria. FEBS Lett. 1981 Aug 3;130(2):179–183. doi: 10.1016/0014-5793(81)81114-3. [DOI] [PubMed] [Google Scholar]
- Duine J. A., Frank J., Jr The prosthetic group of methanol dehydrogenase. Purification and some of its properties. Biochem J. 1980 Apr 1;187(1):221–226. doi: 10.1042/bj1870221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duine J. A., Frank J., van Zeeland J. K. Glucose dehydrogenase from Acinetobacter calcoaceticus: a 'quinoprotein'. FEBS Lett. 1979 Dec 15;108(2):443–446. doi: 10.1016/0014-5793(79)80584-0. [DOI] [PubMed] [Google Scholar]
- Forrest H. S., Salisbury S. A., Sperl G. Crystallization of a derivative of a new coenzyme, methoxatin. Biochim Biophys Acta. 1981 Aug 17;676(2):226–229. doi: 10.1016/0304-4165(81)90191-4. [DOI] [PubMed] [Google Scholar]
- Mincey T., Bell J. A., Mildvan A. S., Abeles R. H. Mechanism of action of methoxatin-dependent alcohol dehydrogenase. Biochemistry. 1981 Dec 22;20(26):7502–7509. doi: 10.1021/bi00529a027. [DOI] [PubMed] [Google Scholar]
- Ribbons D. W., Harrison J. E., Wadzinski A. M. Metabolism of single carbon compounds. Annu Rev Microbiol. 1970;24:135–158. doi: 10.1146/annurev.mi.24.100170.001031. [DOI] [PubMed] [Google Scholar]
- Salisbury S. A., Forrest H. S., Cruse W. B., Kennard O. A novel coenzyme from bacterial primary alcohol dehydrogenases. Nature. 1979 Aug 30;280(5725):843–844. doi: 10.1038/280843a0. [DOI] [PubMed] [Google Scholar]
- Verwiel P. E., Frank J., Verwiel E. J. Characterization of the second prosthetic group in methanol dehydrogenase from hyphomicrobium X. Eur J Biochem. 1981 Aug;118(2):395–399. doi: 10.1111/j.1432-1033.1981.tb06415.x. [DOI] [PubMed] [Google Scholar]
- de Beer R., Duine J. A., Frank J., Large P. J. The prosthetic group of methylamine dehydrogenase from Pseudomonas AM1: evidence for a quinone structure. Biochim Biophys Acta. 1980 Apr 25;622(2):370–374. doi: 10.1016/0005-2795(80)90050-1. [DOI] [PubMed] [Google Scholar]



