Abstract
Breast cancers can be classified into those that express the estrogen (ER) and progesterone (PR) receptors, those with ERBB2 (HER-2/Neu) amplification, and those without expression of ER, PR, or amplification of ERBB2 (referred to as triple-negative or basal-like breast cancer). In order to identify potential molecular targets in breast cancer, we performed a synthetic siRNA-mediated RNAi screen of the human tyrosine kinome. A primary RNAi screen conducted in the triple-negative/basal-like breast cancer cell line MDA-MB231 followed by secondary RNAi screens and further studies in this cell line and two additional triple-negative/basal-like breast cancer cell lines, BT20 and HCC1937, identified the G2/M checkpoint protein, WEE1, as a potential therapeutic target. Similar sensitivity to WEE1 inhibition was observed in cell lines from all subtypes of breast cancer. RNAi-mediated silencing or small compound inhibition of WEE1 in breast cancer cell lines resulted in an increase in γH2AX levels, arrest in the S-phase of the cell cycle, and a significant decrease in cell proliferation. WEE1-inhibited cells underwent apoptosis as demonstrated by positive Annexin V staining, increased sub-G1 DNA content, apoptotic morphology, caspase activation, and rescue by the pan-caspase inhibitor, Z-VAD-FMK. In contrast, the non-transformed mammary epithelial cell line, MCF10A, did not exhibit any of these downstream effects following WEE1 silencing or inhibition. These results identify WEE1 as a potential molecular target in breast cancer.
Keywords: RNAi screen, Breast cancer, Tyrosine kinase, WEE1, Apoptosis
Introduction
Breast cancer is a heterogeneous group of diseases. Approximately 60–70% of breast cancers express estrogen receptors (ER) and/or progesterone receptors (PR), and approximately 20–30% of breast cancers have amplified ERBB2 (HER-2) thus expressing high levels of the HER-2 protein [1, 2]. In approximately 15–20% of patients with breast cancer, the tumors do not express ER or PR and do not have amplification of ERBB2 [1]. These are often categorized as triple-negative breast cancer, and patients with these tumors have a poor prognosis [1]. Molecular classification by expression profiling of primary breast cancers and breast cancer cell lines has determined that the majority of these triple-negative tumors share expression profiles with basal epithelial cells of the breast duct [3–6] and hence are also referred to as basal-like tumors. Currently, the mainstay of treatment for these tumors is chemotherapy [1]. Thus, identification of novel, molecularly targeted therapies for triple-negative/basal-like breast cancer in particular or for breast cancer in general, would be of great benefit.
The tyrosine kinases (TKs) constitute a protein family of approximately 90 members that play an integral role in signal transduction of mammalian cells including critical cellular processes as diverse as proliferation, apoptosis, differentiation, and cell motility [7]. Thus, it is not surprising that deregulation of TKs activity has been observed in numerous types of malignancy [8]. A number of TKs have been validated as therapeutic targets in human malignancies including both receptor TKs (e.g., EGFR, ERBB2/HER-2/Neu, Kit, and VEGF receptors) and non-receptor TKs (e.g., BCR-ABL) [9]. We took advantage of the description of the complete human kinome [7] to apply a systematic functional genomics approach to reduce specifically the expression of each of the TKs using RNA interference (RNAi) in breast cancer cell lines and to investigate the consequences of the kinase loss of function on cell growth. Genome-wide application of RNAi-based screening has been used previously to identify genes that regulate processes such as apoptosis and cell cycle progression [10–12]. Thus, this methodology is particularly well suited in evaluating the role that each TK plays in the growth and survival of breast cancer cells and has the potential to identify novel molecular targets for patients with breast cancer.
Using synthetic siRNA-mediated RNAi screens of the human tyrosine kinome, we have identified the G2/M checkpoint kinase WEE1 as a potential molecular target for breast cancer. Further, we show that inhibition of WEE1 results in the accumulation of DNA damage, alteration in cell cycle regulation, and induction of apoptosis in breast cancer cells.
Materials and methods
Cell culture
The MDA-MB231 (MB231), HCC38, HCC1954, MB453, MB468, MCF7, MCF10A, SKBR3, T47D, and ZR75 cell lines were obtained from ATCC (Manassas, VA); BT20 and HCC1937 were obtained from Reinhard Ebner (Avalon Pharmaceuticals, Germantown, MD); NIH/3T3 cells were a gift from Dr. Micheal Birrer, Harvard Medical School, Boston, MA. MB231 cells were grown in RPMI 1640 supplemented with 5% fetal bovine serum (FBS), (R5), NIH/3T3 cells were grown in DMEM supplemented with 10% FBS, MCF10A cells were grown in DMEM F12 supplemented with 5% horse serum, 1.4 μM cortisone, 10 μg/ml insulin, 100 ng/ml cholera toxin, and 20 ng/ml Epidermal Growth Factor and all other cells were grown in RPMI 1640 supplemented with 10% FBS (R10). All growth media contained 100 units/ml of penicillin and 100 units/ml of streptomycin.
Gene-specific RNAi analysis
Gene-targeted silencing was performed as described in the Supplementary methods.
Lysate preparation and histone extraction
Cell lysates were made as described previously [13]. Protein concentration was determined by using the Bio-Rad colorimetric assay (Bio-Rad, Hercules, CA). To extract histones, the pellets obtained after clarification were solubilized in 0.2 N HCl overnight at 4°C, neutralized with 2.0 M NaOH, and the protein was measured [14].
Immunoblotting
Immunoblotting was performed as described earlier [13], and the antibodies are listed in Supplementary Table 1.
Inhibitors
WEE1 inhibitor II (6-Butyl-4-(2-chlorophenyl)-9-hydroxy-pyrrolo[3,4-c]carbazole-1,3-(2H,6H)-dione, 681641, Calbiochem, La Jolla, CA) and the pan-caspase inhibitor, Z-VAD-FMK (P416, Biomol International, Plymouth Meeting, PA) dissolved in DMSO, were used at 10 and 100 μM concentrations, respectively.
Trypan blue staining
Cells were incubated with Trypan blue stain (15250-061, Invitrogen, Carlsbad, CA) after transfection with siRNA or treatment with inhibitors. Cells that excluded the dye (viable) and cells that retained the dye (dead) were counted.
Annexin V staining
After treatment with WEE1 inhibitor or DMSO, cells were stained using Annexin V-FITC Apoptosis Detection kit II (51-6710AK, BD Pharmingen, San Diego, CA) as per the manufacturer’s protocol and were analyzed on a BD FACSCalibur (BD Biosciences, San Diego, CA) using FLOWJO software.
Caspase-Glo 3/7 assay
Cells were plated in 100 μl at a concentration of 3,000 cells/well, in replicates of 6 wells, in a black-walled 96-well plate and allowed to adhere overnight. Z-VAD-FMK or DMSO was added for 2 h, and cells were then treated with WEE1 inhibitor or DMSO for 4, 8, or 24 h at 37°C. The plate was brought to room temperature and 100 μl of the Caspase-Glo 3/7 reagent (G8091, Promega, Madison, WI) was added to each well. The plate was incubated at room temperature for 45 min in the dark and read using a luminescence plate reader (VICTOR X, PerkinElmer Waltham, MA). Results are shown as mean ± SE of three independent experiments normalized to DMSO-treated cells.
Immunofluorescence
Cells grown in chamber slides were treated with WEE1 inhibitor or DMSO (control) for 4 or 24 h, washed with PBS and fixed in 3.7% paraformaldehyde for 15 min at room temperature. Cells were washed three times with PBS for 10 min, permeabilized with 0.2% Triton X-100 in PBS/ 1% BSA on ice for 5 min, washed in PBS/1% BSA, blocked at room temperature (PBS/3% BSA) for 1 h and incubated with anti-phosphohistone H2AX antibody at room temperature (1:500 in PBS/1% BSA/0.5% Tween 20) for 1 h. After washing with PBS/1% BSA/0.5% Tween 20 (3×, 10 min each), cells were incubated with secondary antibody (1:5,000 of anti-mouse Alexa-Fluor 488) in 3%BSA/PBS at room temperature for 1 h, washed (4×, 10 min each), mounted using Vectashield containing DAPI (H1200, Vector Laboratories, Burlingame, CA), and visualized under a confocal microscope (LSM 510, Carl Zeiss Microimaging, Germany).
Cell cycle analysis
Cells (1 × 106) were plated overnight, treated with WEE1 inhibitor as indicated, incubated with 10 μM BrdU (5002, Sigma–Aldrich, St Louis, MO) for 15 min, trypsinized, washed with PBS, and fixed in cold 70% ethanol. Staining was performed with anti-BrdU-FITC antibody (347583, BD Biosciences) and 5 μg/ml propidium iodide (PI) (P3566, Invitrogen). Analysis was performed on the BD FACSCalibur using FLOWJO software.
Statistics
Statistical comparison of mean values was performed using the paired Student’s t-tests. All P values are 2 tailed.
Results
Identification and validation of WEE1 as a target from the RNAi screen
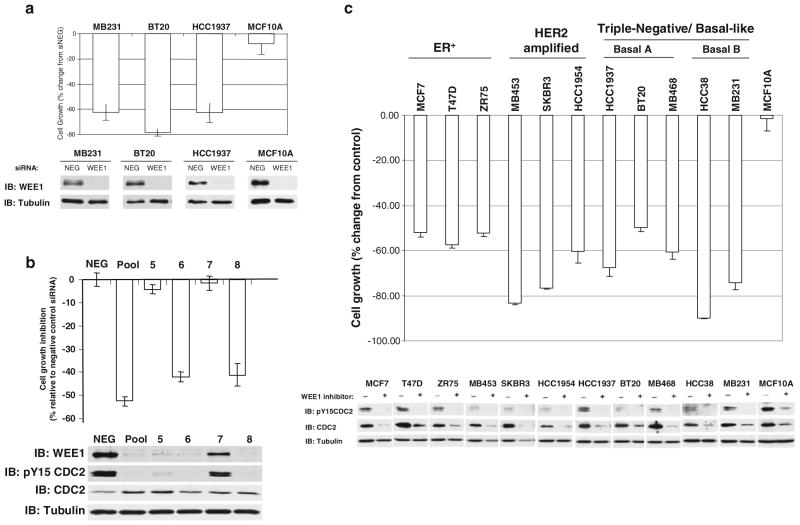
To identify TKs that may be therapeutic targets for treatment of breast cancer, we performed an RNAi-based functional genomic screen of the human tyrosine kinome in cell lines representative of triple-negative/basal-like breast cancer (see supplementary methods, supplementary results and supplementary Figs. 1–3). This screen identified the G2/M checkpoint protein WEE1 as a potential target kinase. The viability of three triple-negative/basal-like breast cancer cell lines MB231, BT20, and HCC1937 was inhibited by more than 60% upon silencing of WEE1 (WEE1 siRNA pool) 5 days post-transfection compared to a non-targeting control siRNA pool (Fig. 1a). Triple-negative/basal-like breast cancer cells are classified into two subtypes: Basal A (BaA) and Basal B (BaB) based on the hierarchical clustering of their transcriptional profiles [6]. The three cell lines used in these studies represented both BaA (BT20 and HCC1937) and BaB (MB231) subtypes of triple-negative/basal-like breast cancer. Importantly, silencing of WEE1 in the non-transformed basal-like breast epithelial cell line, MCF10A, had little or no effect on cell viability (Fig. 1a). To further confirm that the specific inhibition of WEE1 by the siRNA pool was responsible for the reduction in the viability of MB231 cells, each siRNA of the pool was tested individually (Fig. 1b). Of the four siRNAs, two: numbers 6 and 8, reduced WEE1 protein and cell viability to levels comparable to that seen following transfection of the pooled WEE1 siRNAs. MB231 cells transfected with the WEE1 siRNA pool and the individual WEE1 siRNAs numbers 6 and 8 also exhibited a decrease in the phosphorylation of CDC2 on tyrosine 15, the substrate for WEE1 [15]. By contrast, WEE1 siRNA number 7 had little effect on WEE1 protein levels or CDC2 tyrosine phosphorylation and did not significantly affect cell viability. Interestingly, WEE1 siRNA number 5 had an intermediate effect on WEE1 protein levels and CDC2 tyrosine phosphorylation but no significant effect on cell viability. This suggests that there may be a threshold level above which WEE1 function is adequate to maintain cell viability.
Fig. 1.
siRNA-mediated silencing or inhibition of WEE1 inhibits breast cancer cells. a Breast cancer cell lines MB231, BT20, and HCC1937 and the non-transformed breast cell line MCF10A were transfected with either a WEE1 or non-targeting (NEG) pool of siRNAs. Cell viability was measured by MTS assay 5 days post-transfection and is plotted as percent change from the NEG pool. The degree of WEE1 silencing was assessed by immunoblotting shown below the graphs. Tubulin protein levels were measured as a loading control. b MB231 cells were transfected with NEG pool, a pool of four WEE1 siRNAs or each individual WEE1 siRNA used within the pool (siWEE1 numbers 5, 6, 7, and 8). Cell viability is plotted as a change from the NEG pool control 72 h post-transfection. Immuno-blots of WEE1, phosphorylated CDC2 (pY15CDC2), total CDC2 and the loading control tubulin are shown. c Cells were incubated with 10 μM WEE1 inhibitor (+) or DMSO (−). Cell viability was assessed by MTS assay 3 days post-treatment, and results are plotted as a percent change relative to the DMSO control. WEE1 inhibition was assessed by phosphorylation of CDC2 (Tyr15). a–c All MTS values represent the mean of three experiments ± SE
Inhibition of WEE1 decreases the viability of breast cancer cell lines but not of non-transformed cells
Having identified WEE1 as a potential molecular target by the RNAi screen, we tested the effect of a specific inhibitor of WEE1 (WEE1 inhibitor II) on the survival of different breast cancer cells. WEE1 inhibitor II is a relatively specific small molecule inhibitor targeting the ATP-binding site of WEE1 [16]. Treatment of ER-positive breast cancer cell lines (MCF7, T47D, ZR75), HER2 amplified cell lines (MB453, SKBR3, HCC1954), and triple-negative/basal-like breast cancer cell lines [HCC1937, BT20, MB468 (BaA) or HCC38, MB231 (BaB)] with the WEE1 inhibitor resulted in decreased cell growth similar to WEE1 silencing (Fig. 1c). Importantly, the WEE1 inhibitor had no effect on the viability of the non-transformed MCF10A cells (Fig. 1c) or NIH3T3 fibroblasts (data not shown). Also, WEE1 inhibition led to a decrease in phosphorylation on Tyr15 of CDC2 (Fig. 1c), consistent with our data obtained following transfection of WEE1 siRNAs (Fig. 1b). This data suggests that the breast cancer cell lines of various subtypes (ER-positive, HER2 amplified and triple-negative/basal-like A or B) are sensitive to the downstream effects of WEE1 inhibition, and thus WEE1 could be a potential target in treatment of all subsets of breast cancer. Sensitivity to WEE1 loss has been linked previously to high levels of WEE1 expression [17]. However, we did not observe a difference in either expression or function of WEE1 that correlated with the sensitivity of the cancer cells to the loss of WEE1 function (Supplementary Fig. 4). Sequence analysis of WEE1 mRNA did not reveal any evidence of activating mutations nor is there evidence of gene amplification of the WEE1 locus in the cancer cell lines (data not shown).
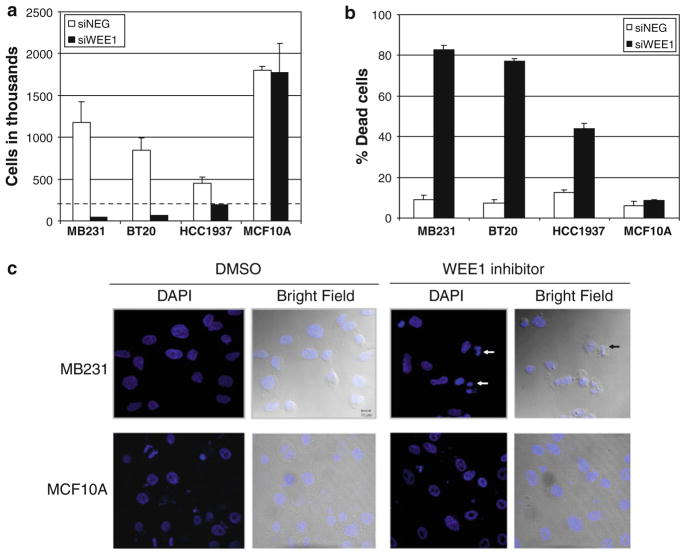
The MTS assay measures the metabolic activity of cells and as such is used as an estimate of cell viability. To more directly assess the effect of WEE1 loss of function on the number of viable and dead cells within a cell population, we stained the cells transfected with either WEE1 siRNA or control siRNA with Trypan blue. Silencing of WEE1 reduced the number of viable cells significantly in all three breast cancer cell lines tested (MB231, BT20, and HCC1937) but not in MCF10A cells (Fig. 2a). There was also a corresponding increase in the percentage of dead cells in the breast cancer cell lines but not in MCF10A cells upon WEE1 silencing (Fig. 2b). Also, the effect of WEE1 inhibition on non-transformed NIH/3T3 fibroblasts was examined (Supplementary Fig. 6a, b). WEE1 inhibition resulted in a modest decrease in the proliferation of the NIH/3T3 cells over the course of the experiment (2.6 doublings in DMSO-treated cells compared to 2 doublings in WEE1-inhibited cells) (Supplementary Fig. 6a). Notably, there was no increase in the percentage of dead cells upon WEE1 inhibition in NIH/ 3T3 cells (Supplementary Fig. 6b). Thus, this data indicate that the loss of WEE1 function induces death of breast cancer cell lines but not of non-transformed cell lines.
Fig. 2.
Silencing or inhibition of WEE1 in breast cancer cells results in cell death. a The number of viable and b the percentage dead cells were assessed 5 days post-transfection of the cell lines with either a NEG siRNA pool (white bars) or a WEE1 siRNA pool (black bars). The dotted line in a represents the number of cells plated at the start of the experiment. Data represent mean cell counts ± SE from three experiments. WEE1 silencing resulted in a statistically significant decrease in viable cells (P < 0.05) and an increase in percentage of dead cells (P < 0.005) in the breast cancer cells but not in the MCF10A cells. c Cells treated with WEE1 inhibitor or DMSO control for 4 h were fixed, stained with DAPI, and observed under a LSM 510 confocal microscope. DNA fragmentation (white arrows) and membrane blebbing (black arrow) were observed in inhibitor treated MB231 cells but not in the MCF10A cells
Inhibition of WEE1 in breast cancer cell lines induces apoptosis
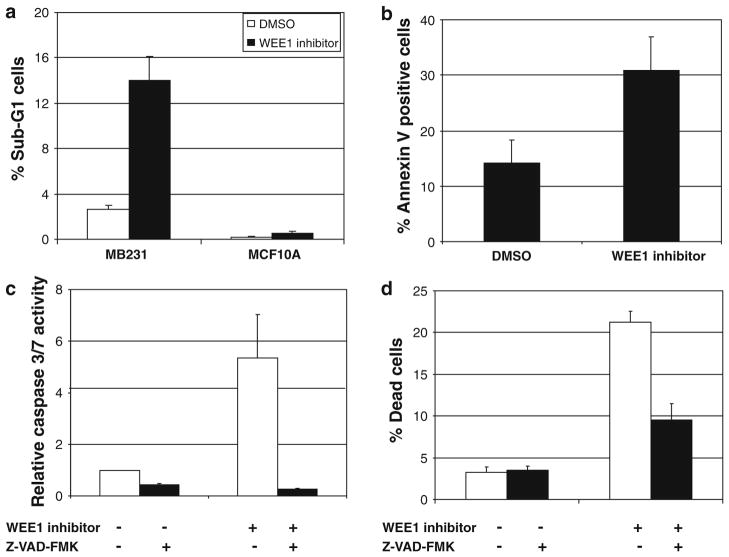
WEE1 inhibitor treated MB231 cells but not MCF10A cells exhibited membrane blebbing and fragmented nuclei consistent with apoptosis (Fig. 2c). To further determine if apoptosis underlies the cell death seen following loss of WEE1 function, we measured DNA fragmentation by FACS analysis of PI stained cells following WEE1 inhibition. In MB231 cells, we observed a fourfold increase in cells with sub-G1 DNA content, whereas no increase was observed in MCF10A cells (Fig. 3a). We also performed Annexin V staining of MB231 cells treated with the WEE1 inhibitor. There was a significant increase in the percentage of Annexin V-positive cells upon WEE1 inhibition (Fig. 3b). Also, fivefold increase in caspase-3/7 activity was observed upon WEE1 inhibitor treatment in MB231 cells that was blocked by the pan-caspase inhibitor, Z-VAD-FMK (Fig. 3c). Corresponding to the decrease in caspase activity, Z-VAD-FMK significantly reduced the percentage of dead cells as measured by Trypan blue staining (Fig. 3d). Together, these results indicate that WEE1 inhibition results in caspase-mediated apoptosis in breast cancer cells.
Fig. 3.
Inhibition of WEE1 in breast cancer cells results in caspase-mediated apoptosis. a The percentage of cells with sub-G1 DNA content was measured by PI staining after 24 h of WEE1 inhibition. The increase in the percentage of sub-G1 cells in MB231 treated with WEE1 inhibitor was statistically significant (P < 0.05). b The percentage of Annexin V-FITC positive MB231 cells seen following treatment with either DMSO or WEE1 inhibitor for 24 h was calculated as the number of cells that were Annexin V-positive and PI-negative relative to the total cells analyzed. WEE1 inhibition increased the percentage of apoptotic cells significantly (P < 0.005). c MB231 cells were pre-incubated with 100 μM of pan-caspase inhibitor Z-VAD-FMK and treated with WEE1 inhibitor or DMSO. Increase in caspase-3/7 activation measured after 24 h of WEE1 inhibition was significantly blocked by Z-VAD-FMK treatment (P < 0.005). d Cell counts with Trypan blue were performed in MB231 cells after pre-treatment with Z-VAD-FMK and 48 h of WEE1 inhibition. Pre-treatment with Z-VAD-FMK resulted in a decrease in the percentage of dead cells (P < 0.005). All graphs represent mean values ± SE of three independent experiments
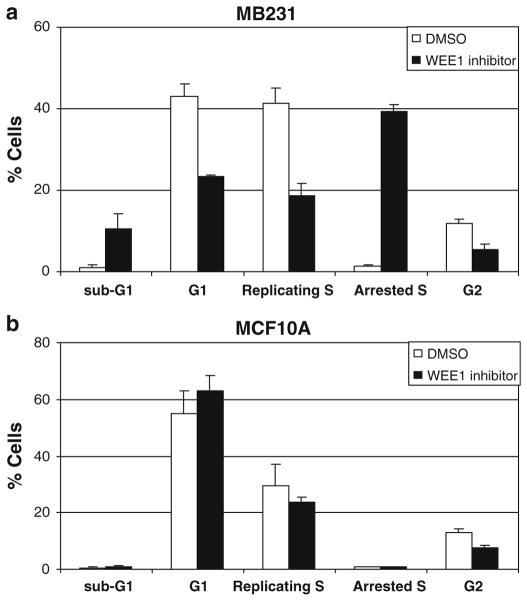
Inhibition of WEE1 causes S-phase arrest in breast cancer cell lines
As WEE1 is an important negative regulator of the G2/M checkpoint that controls mitotic entry of the cells [15], we used PI and BrdU labeling to study the effects of WEE1 inhibition on the cell cycle profile of the breast cancer cells and non-transformed MCF10A cells (Fig. 4, Supplementary Fig. 5). In MB231 cells, WEE1 inhibition at 24 h resulted in an increase in the fraction of cells arrested in S-phase and a concomitant decrease in the fraction of cells in the replicating S-phase (Fig. 4a and Supplementary Fig. 5a). Also, there was an increase in the fraction of cells with sub-G1 DNA content (~tenfold) and a decrease in the fraction of cells in the G1- and the G2-phase of the cell cycle (Fig. 4a). Similar results were observed in HCC1937 (Supplementary Fig. 5b). In contrast, WEE1 inhibition of the non-transformed MCF10A cells did not lead to S-phase arrest or increased cells with a sub-G1 DNA content (Fig. 4b and Supplementary Fig. 5c). In the MCF10A cells, a slight decrease in the G2-phase and the replicating S-phase cells and a slight increase in the G1-phase cells were seen, but these differences were not significantly different (Fig. 4b). Similar to MCF10A, in NIH/3T3 cells there was no significant increase in the percentage of cells arrested in S-phase or in cells with sub-G1 DNA content upon WEE1 inhibition (Supplementary Fig. 6c).
Fig. 4.
Inhibition of WEE1 induces S-phase arrest in breast cancer cells. a MB231 cells b and MCF10A cells were treated with WEE1 inhibitor or DMSO, labeled with BrdU and PI and analyzed by flow cytometry as described in the “Materials and methods”. WEE1 inhibition significantly increased the percentage of cells arrested in S-phase for MB231 (P < 0.005) but not for MCF10A. Data represent mean values ± SE of three experiments
Inhibition of WEE1 in cancer cell lines results in DNA damage
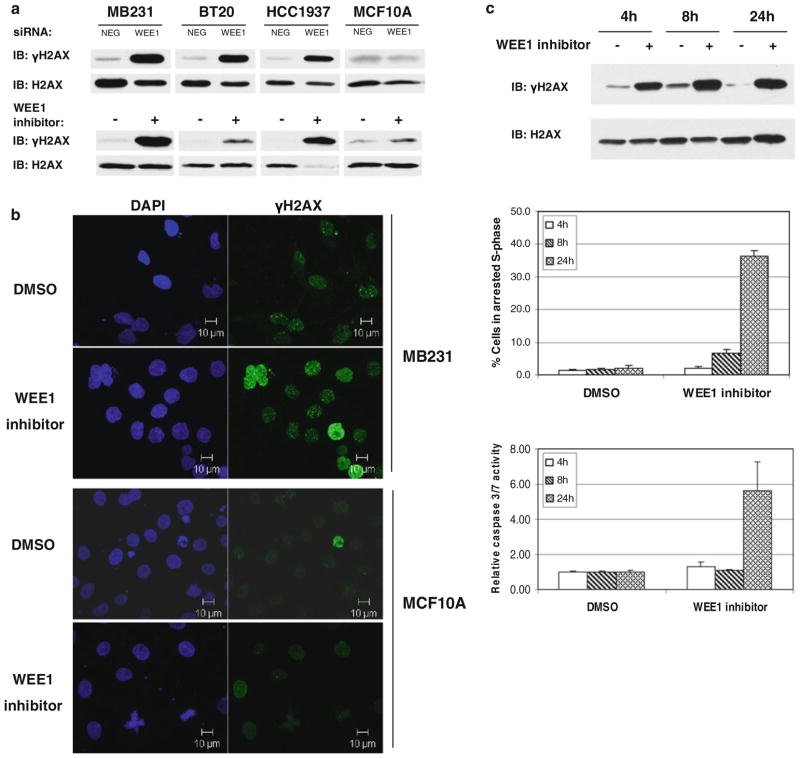
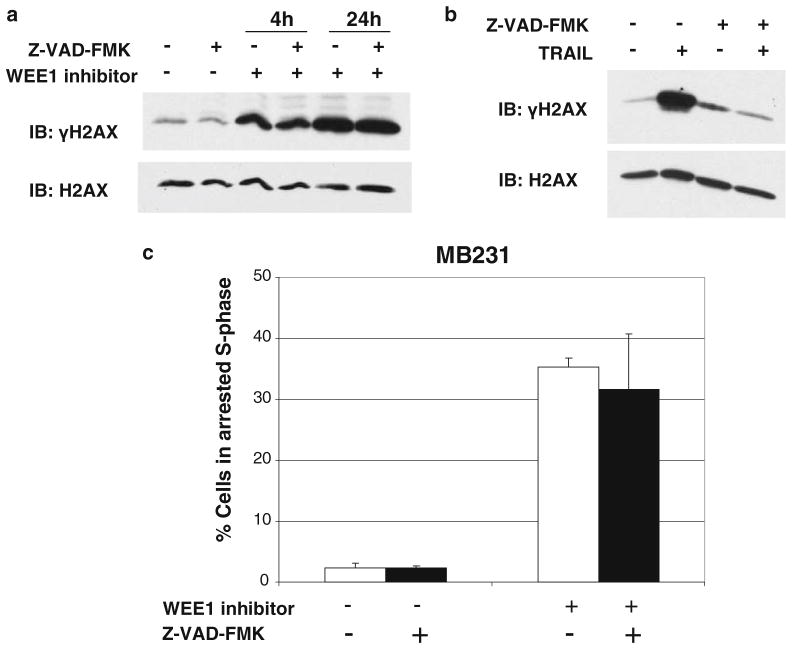
WEE1 is activated downstream of DNA damage sensed by the ATM and ATR kinases [18]. Thus, we looked for evidence of DNA damage in cells that had lost WEE1 function using phosphorylation of histone H2AX on Ser139 (γH2AX) as a marker of DNA double-strand breaks [19]. An increase in γH2AX levels was observed in the breast cancer cell lines, but not in the non-transformed MCF10A cells upon siRNA-mediated silencing of WEE1 or treatment with WEE1 inhibitor (Fig. 5a). In addition, WEE1 inhibition did not increase γH2AX levels in the non-transformed NIH/3T3 cells (Supplementary Fig. 6d). Also, evident was an increase in γH2AX foci in MB231 cells (Fig. 5b) after 4 h of treatment with the WEE1 inhibitor. In contrast, no increase in γH2AX foci was observed in MCF10A cells (Fig. 5b). To check if DNA damage preceded cell cycle arrest and caspase activation, γH2AX levels, the percent of cells arrested in the S-phase, and caspase activation were measured in MB231 cells at different time points after WEE1 inhibition. An increase in γH2AX was evident as early as 4 h after the addition of the WEE1 inhibitor (Fig. 5c upper panel). WEE1 inhibition induced a moderate increase in the percentage of arrested S-phase cells by 8 h that increased by 24 h (Fig. 5c middle panel). Interestingly, caspase activation was observed only at 24 h (Fig. 5c lower panel). The caspase inhibitor Z-VAD-FMK blocks caspase activation and the death of MB231 cells following WEE1 inhibition (Fig. 3d). However, Z-VAD-FMK did not prevent an increase in γH2AX levels following WEE1 inhibition (Fig. 6a). In contrast, the induction of DNA damage (as measured by increased γH2AX) by TNF-related apoptosis-inducing ligand (TRAIL) is reversed in the presence of Z-VAD-FMK (Fig. 6b). This suggests that WEE1 inhibition causes DNA damage followed by caspase activation, whereas TRAIL triggers caspase activation that is upstream of DNA damage. Similarly, the fraction of cells undergoing S-phase arrest upon WEE1 inhibition was not significantly affected by caspase inhibition (Fig. 6c). Thus, at least in the context of breast cancer cells, WEE1 inhibition results in DNA damage, followed by S-phase arrest, caspase activation, and caspase-mediated apoptotic cell death.
Fig. 5.
WEE1 inhibition induces DNA damage in breast cancer cells. a γH2AX was measured in MB231, BT20, HCC1937, and MCF10A after silencing of WEE1 for 48 h or inhibition of WEE1 for 24 h by immunoblotting. Total H2AX was used as loading control. b Cells treated with WEE1 inhibitor or DMSO for 4 h were fixed and immunostained for γH2AX, counterstained with DAPI and imaged on an LSM 510 confocal microscope. c MB231 cells treated with (+) or without (−) WEE1 inhibitor for 4, 8, or 24 h were analyzed for γH2AX levels by immunoblotting (upper panel), for S-phase arrested cells by cell cycle analysis (middle panel), and for caspase activity by Caspase-Glo assay (lower panel). The increase in the percentage of S-phase arrested cells was significant at 8 h (P < 0.05) and 24 h (P < 0.005) of WEE1 inhibition compared to untreated cells. Values represent mean ± SE of three experiments
Fig. 6.
Caspase inhibition does not rescue cancer cells from DNA damage or S-phase arrest induced by WEE1 inhibition. MB231 cells were pre-treated with 100 μM Z-VAD-FMK followed by a WEE1 inhibitor for 4 or 24 h or b TRAIL for 1 h and analyzed for γH2AX levels. H2AX is shown as a loading control. c The percentage of cells arrested in S-phase for MB231 cells treated as in a for 24 h, was determined as described in Fig. 4
Discussion
We performed a functional genomic screen of the tyrosine kinome to identify potential molecular targets in breast cancer cells and identified WEE1 as a promising target. TKs play an important role in cancer development and progression, and mutated TKs aid in the progression of human malignancies and cancer [8]. RNAi-mediated silencing of TKs has identified several potential molecular targets in cancer [12, 20]. To identify TK targets for the potential treatment of breast cancer, we used a systematic approach beginning with a primary screen of all 89 TKs in one representative triple-negative/basal-like (BaB) breast cancer cell line, MB231. Secondary screens and corroboration of the phenotypic effects of RNAi-mediated loss of function using independent siRNAs in other triple-negative/basal-like breast cancer cell lines followed. Of the 89 TKs targeted in the primary screen, the silencing of most of them reduced the viability of MB231 cell to some degree (Supplementary Fig. 1). This is not surprising as TKs play essential roles in regulating many fundamental processes such as growth and apoptosis [10, 11]. Several kinases were initially identified through our screening efforts as potential targets. However, a reduction in cell viability was not observed with an independent pool of siRNAs corresponding to these TKs. This phenotypic disparity could be a result of the difference in the degree of silencing mediated against each target TK gene by the two different pools and/or the differences in the combination of on- and off-target effects of each gene-specific siRNA pool. However, for WEE1, we saw consistency between the phenotypic effects mediated by both the pools of siRNAs targeting this TK and individual WEE1 siRNAs. Our follow-up screens and experimentation led us to focus on the G2/M checkpoint protein WEE1 (Supplementary Figs. 2–3).
WEE1 is a key element in the DNA damage response pathway. Upon DNA damage, the cellular sensors ATM and ATR are activated, leading to phosphorylation of the checkpoint kinases CHK1 and CHK2. CHK1 in turn phosphorylates WEE1 and activates it [21]. Activated WEE1 causes an inhibitory phosphorylation of CDC2/ CDK1 on Tyr15, which then prevents cells from entering mitosis allowing time for DNA repair prior to entering mitosis [15]. In our experiments, loss or inhibition of WEE1 in eleven breast cancer cell lines, representing ER-positive, HER2 amplified, and triple-negative/basal-like subtypes of breast cancer, resulted in cell death (Fig. 1). A recent functional genomic screen of several cancer cell lines identified WEE1 as a potential molecular target in HeLa cells and Cal51, a triple-negative breast cancer cell line with luminal features [17, 22]. Reports from several groups have suggested that inhibition of WEE1 can enhance the effects of radiation- or chemotherapy-induced DNA damage in colon cancer cells, lung cancer cells, and melanoma cells [23–26]. Thus, WEE1 may be a molecular target in a broad range of cancers, including breast cancers.
Loss or inhibition of WEE1 in breast cancer cell lines resulted in DNA damage, S-phase arrest, and the induction of caspase-mediated apoptosis (Figs. 2, 3, 4, 5 and 6). The mechanism that accounts for the caspase activation and subsequent cell death is not clear; although, the time course experiments demonstrate that the appearance of DNA damage (at 4 h) preceded cell cycle arrest (seen at 8 h) and caspase activation (seen at 24 h) (Fig. 5c). Caspase inhibition prevented cell death but not the accumulation of DNA damage or cell cycle arrest (Figs. 3, 6). This is consistent with a model whereby DNA damage results in S-phase arrest, which in turn activates caspases. Alternatively, the DNA damage itself may directly induce both caspase activation and cell cycle arrest. Interestingly, loss or inhibition of the upstream checkpoint kinases, CHK1 or CHK2, have been found to cause DNA damage, cell cycle arrest, and cell death in cancer cells [27, 28]. A recent publication describes that the loss of CHK1 triggers caspase-2 mediated apoptosis in TP53-deficient cells [29]. Breast cancers are documented to have mutations or deletions of TP53 [6, 30]. Of the cell lines we tested for sensitivity to WEE1 inhibition, MB231, T47D, HCC38, MB468, SKBR3, HCC1954, HCC1937 contain either a mutant copy of TP53 or are null for TP53. MCF7, MB453, BT20, and ZR75 have wild-type TP53 [6, 31]. Interestingly, the caspase-2 inhibitor Z-VDVAD-FMK blocked caspase-3/7 activation and cell death following WEE1 inhibition in MB231 cells similar to the inhibition by the pan-caspase inhibitor Z-VAD-FMK (data not shown). However, siRNA-mediated silencing of caspase-2 did not protect these cells from apoptosis induced by WEE1 inhibition (unpublished observation). Thus, it is possible that the degree of silencing of caspase-2 was insufficient to prevent apoptosis. However, while Z-VDVAD-FMK has been described as a caspase-2-specific inhibitor, recent work suggests that it may inhibit multiple caspases similar to Z-VAD-FMK [32–34]. Further investigation will be required to identify the mechanism by which caspase activation is triggered upon the loss or inhibition of WEE1.
This study and published data suggest that WEE1 inhibition has selective toxicity to cancer cells compared to normal cells [26]. In fact, WEE1 inhibitors are in phase I clinical trials in patients with cancer [35]. The basis for the selective toxicity of WEE1 loss or inhibition in breast cancer cell lines compared to non-transformed cells is not completely clear. In the functional genomic screen that identified WEE1 as a target in the Cal51 breast cancer cells, the authors found a relationship between high levels of WEE1 expression and sensitivity to WEE1 inhibition [17]. However, we found that the non-transformed breast basal epithelial cell line MCF10A is insensitive to WEE1 inhibition while expressing similar levels of WEE1 and had similar levels of WEE1 activity (measured by the levels of phosphorylated CDC2) as the cancer cells (Supplementary Fig. 4). This suggests that the cancer cells are more dependent on the function of WEE1. The loss or inhibition of WEE1 in the cancer cells, but not in non-transformed cells (MCF10A or NIH/3T3), results in the dramatic accumulation of DNA double-strand breaks as measured by the accumulation of γH2AX (Fig. 5, Supplementary Fig. 6d). Phosphorylation of H2AX (γH2AX) is triggered by DNA double-strand breaks and mediates the formation of clusters of proteins called DNA damage response foci at the site of damage [19, 36]. One possible explanation for the selectivity of WEE1 inhibition for cancer cells is that the cancer cells have a pre-existing defect in DNA repair pathways and/or ongoing DNA damage. It has been postulated that many breast cancers are deficient in the homologous DNA repair pathway [30]. Thus, the loss of the G2/M checkpoint that results from the loss or inhibition of WEE1 may allow DNA damage to accumulate that would otherwise be repaired at this checkpoint. We saw no evidence of DNA damage in the non-transformed MCF10A cell line upon the loss of WEE1 function suggesting that the cancer cells are more prone to DNA double-strand breaks (Fig. 5). In addition, it appears that the cancer cells have more γH2AX foci than the non-transformed MCF10A cells prior to the inhibition of WEE1 function (Fig. 5b). Consistent with this observation, recent studies have shown that untreated cancer cells have a relatively high number of γH2AX foci indicative of ongoing DNA double-strand breaks compared to normal cells [37]. Thus, the loss of the G2/M checkpoint may be particularly deleterious to cancer cells.
Another striking difference we observed was the arrest of the cancer cells in S-phase upon the loss or inhibition of WEE1, while the non-transformed cells showed a slight increase in G1 but no S-phase arrest (Fig. 4, Supplementary Figs. 5 and 6c). Eukaryotic cells have multiple checkpoints in the cell cycle that allow the faithful replication and transmission of DNA to daughter cells upon cell division [18]. Breast cancer cells have been shown to lack an active G1 checkpoint [30]. Interestingly, embryonic stem (ES) cells lack a functional G1 check point, and DNA damage induces S-phase arrest and apoptosis in them [38]. Restoration of the G1 check point in ES cells prevents radiation-induced apoptosis [38]. Our data are consistent with these observations, as the cancer cells, in the absence of a functional G1 checkpoint, arrest in S-phase when WEE1 function is lost. Consistent with the idea that transformed cells lose the G1 check point, a transformed variant of MCF10A cells exhibited increased γH2Ax levels, an increased percentage of cells arrested in S-phase, and an increased percentage of dead cells when treated with the WEE1 inhibitor (unpublished data). Whether it is the lack of DNA damage or the ability to repair DNA at the intact G1 checkpoint that allows the non-transformed cells to survive the loss of the G2/M checkpoint will require further study.
Our work suggests that WEE1 may be a useful target in breast cancer, including triple-negative/basal-like breast cancer. WEE1 inhibitors are currently undergoing clinical testing in phase I studies [35]. Recent work suggesting that triple-negative/basal-like breast cancers are defective in double-stranded DNA repair has led to clinical trials combining DNA-damaging agents such as platinum compounds with DNA repair inhibitors such as PARP inhibitors [39–41]. Interestingly, WEE1 serves as a critical component of the response to double-stranded DNA breaks by activating the G2/M checkpoint and allowing the cell to repair damaged DNA [18]. This suggests that the combination of DNA-damaging agents with WEE1 inhibition may be particularly effective in the triple-negative/basal-like breast cancer cells. Reports from several groups have suggested that inhibition of WEE1 can enhance the effects of radiation- or chemotherapy-induced DNA damage in colon cancer cells, lung cancer cells, and melanoma cells [23–26]. Our data, together with these observations, suggest that further study of WEE1 inhibition alone and particularly in combination with DNA-damaging agents in breast cancers is warranted.
Supplementary Material
Acknowledgments
We thank Marion Nau for critical reading of this manuscript. Financial Support This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10549-009-0571-2) contains supplementary material, which is available to authorized users.
Contributor Information
Lyndsay M. Murrow, Laboratory of Cellular and Molecular Biology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, 37 Convent Drive, Bethesda, MD 20892-4256, USA
Sireesha V. Garimella, Laboratory of Cellular and Molecular Biology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, 37 Convent Drive, Bethesda, MD 20892-4256, USA
Tamara L. Jones, Genetics Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, 37 Convent Drive, Bethesda, MD 20892, USA
Natasha J. Caplen, Genetics Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, 37 Convent Drive, Bethesda, MD 20892, USA
Stanley Lipkowitz, Email: lipkowis@mail.nih.gov, Laboratory of Cellular and Molecular Biology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, 37 Convent Drive, Bethesda, MD 20892-4256, USA.
References
- 1.Brenton JD, Carey LA, Ahmed AA, Caldas C. Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J Clin Oncol. 2005;23(29):7350–7360. doi: 10.1200/JCO.2005.03.3845. [DOI] [PubMed] [Google Scholar]
- 2.Irvin WJ, Jr, Carey LA. What is triple-negative breast cancer? Eur J Cancer. 2008;44(18):2799–2805. doi: 10.1016/j.ejca.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 3.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertucci F, Finetti P, Cervera N, Esterni B, Hermitte F, Viens P, Birnbaum D. How basal are triple-negative breast cancers? Int J Cancer. 2008;123(1):236–240. doi: 10.1002/ijc.23518. [DOI] [PubMed] [Google Scholar]
- 5.Charafe-Jauffret E, Ginestier C, Monville F, et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 2006;25(15):2273–2284. doi: 10.1038/sj.onc.1209254. [DOI] [PubMed] [Google Scholar]
- 6.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 8.Hunter T. A thousand and one protein kinases. Cell. 1987;50(6):823–829. doi: 10.1016/0092-8674(87)90509-5. [DOI] [PubMed] [Google Scholar]
- 9.Krause DS, Van Etten RA. Tyrosine kinases as targets for cancer therapy. N Engl J Med. 2005;353(2):172–187. doi: 10.1056/NEJMra044389. [DOI] [PubMed] [Google Scholar]
- 10.MacKeigan JP, Murphy LO, Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat Cell Biol. 2005;7(6):591–600. doi: 10.1038/ncb1258. [DOI] [PubMed] [Google Scholar]
- 11.Bettencourt-Dias M, Giet R, Sinka R, et al. Genome-wide survey of protein kinases required for cell cycle progression. Nature. 2004;432(7020):980–987. doi: 10.1038/nature03160. [DOI] [PubMed] [Google Scholar]
- 12.Giroux V, Iovanna J, Dagorn JC. Probing the human kinome for kinases involved in pancreatic cancer cell survival and gemcitabine resistance. FASEB J. 2006;20(12):1982–1991. doi: 10.1096/fj.06-6239com. [DOI] [PubMed] [Google Scholar]
- 13.Rahman M, Davis SR, Pumphrey JG, Bao J, Nau MM, Meltzer PS, Lipkowitz S. TRAIL induces apoptosis in triple-negative breast cancer cells with a mesenchymal phenotype. Breast Cancer Res Treat. 2009;113(2):217–230. doi: 10.1007/s10549-008-9924-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.http://www.stanford.edu/group/gozani/Histone%20extraction%20protocol.pdf [cited]
- 15.McGowan CH, Russell P. Human Wee1 kinase inhibits cell division by phosphorylating p34cdc2 exclusively on Tyr15. EMBO J. 1993;12(1):75–85. doi: 10.1002/j.1460-2075.1993.tb05633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmer BD, Thompson AM, Booth RJ, et al. 4-Phenyl-pyrrolo[3, 4-c]carbazole-1, 3(2H, 6H)-dione inhibitors of the checkpoint kinase Wee1. Structure–activity relationships for chromophore modification and phenyl ring substitution. J Med Chem. 2006;49(16):4896–4911. doi: 10.1021/jm0512591. [DOI] [PubMed] [Google Scholar]
- 17.Iorns E, Lord CJ, Grigoriadis A, et al. Integrated functional, gene expression and genomic analysis for the identification of cancer targets. PLoS ONE. 2009;4(4):e5120. doi: 10.1371/journal.pone.0005120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niida H, Nakanishi M. DNA damage checkpoints in mammals. Mutagenesis. 2006;21(1):3–9. doi: 10.1093/mutage/gei063. [DOI] [PubMed] [Google Scholar]
- 19.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146(5):905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyner JW, Walters DK, Willis SG, et al. RNAi screening of the tyrosine kinome identifies therapeutic targets in acute myeloid leukemia. Blood. 2008;111(4):2238–2245. doi: 10.1182/blood-2007-06-097253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Connell MJ, Raleigh JM, Verkade HM, Nurse P. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 1997;16(3):545–554. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuli C, Shao N, Rao R, et al. BRCA1a has antitumor activity in TN breast, ovarian and prostate cancers. Oncogene. 2007;26(41):6031–6037. doi: 10.1038/sj.onc.1210420. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Li J, Booher RN, Kraker A, Lawrence T, Leopold WR, Sun Y. Radiosensitization of p53 mutant cells by PD0166285, a novel G(2) checkpoint abrogator. Cancer Res. 2001;61 (22):8211–8217. [PubMed] [Google Scholar]
- 24.Li J, Wang Y, Sun Y, Lawrence TS. Wild-type TP53 inhibits G(2)-phase checkpoint abrogation and radiosensitization induced by PD0166285, a WEE1 kinase inhibitor. Radiat Res. 2002;157(3):322–330. doi: 10.1667/0033-7587(2002)157[0322:wttigp]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Decker SJ, Sebolt-Leopold J. Knockdown of Chk1, Wee1 and Myt1 by RNA interference abrogates G2 checkpoint and induces apoptosis. Cancer Biol Ther. 2004;3(3):305–313. doi: 10.4161/cbt.3.3.697. [DOI] [PubMed] [Google Scholar]
- 26.Hashimoto O, Shinkawa M, Torimura T, Nakamura T, Selvendiran K, Sakamoto M, Koga H, Ueno T, Sata M. Cell cycle regulation by the Wee1 inhibitor PD0166285, pyrido [2, 3-d] pyrimidine, in the B16 mouse melanoma cell line. BMC Cancer. 2006;6:292. doi: 10.1186/1471-2407-6-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Syljuasen RG, Sorensen CS, Hansen LT, et al. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25(9):3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niida H, Tsuge S, Katsuno Y, Konishi A, Takeda N, Nakanishi M. Depletion of Chk1 leads to premature activation of Cdc2-cyclin B and mitotic catastrophe. J Biol Chem. 2005;280(47):39246–39252. doi: 10.1074/jbc.M505009200. [DOI] [PubMed] [Google Scholar]
- 29.Sidi S, Sanda T, Kennedy RD, et al. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell. 2008;133(5):864–877. doi: 10.1016/j.cell.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer. 2004;4(10):814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 31.Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28(6):622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 32.Chauvier D, Lecoeur H, Langonne A, Borgne-Sanchez A, Mariani J, Martinou JC, Rebouillat D, Jacotot E. Upstream control of apoptosis by caspase-2 in serum-deprived primary neurons. Apoptosis. 2005;10(6):1243–1259. doi: 10.1007/s10495-005-1681-x. [DOI] [PubMed] [Google Scholar]
- 33.Gregoli PA, Bondurant MC. Function of caspases in regulating apoptosis caused by erythropoietin deprivation in erythroid progenitors. J Cell Physiol. 1999;178(2):133–143. doi: 10.1002/(SICI)1097-4652(199902)178:2<133::AID-JCP2>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 34.Pereira NA, Song Z. Some commonly used caspase substrates and inhibitors lack the specificity required to monitor individual caspase activity. Biochem Biophys Res Commun. 2008;377 (3):873–877. doi: 10.1016/j.bbrc.2008.10.101. [DOI] [PubMed] [Google Scholar]
- 35.Schellens JH, Leijen S, Shaprio GI, et al. A phase I and pharmacological study of MK-1775, a Wee1 tyrosine kinase inhibitor, in both monotherapy and in combination with gemcitabine, cisplatin, or carboplatin in patients with advanced solid tumors. J Clin Oncol. 2009;27(15s):148s. [Google Scholar]
- 36.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. GammaH2AX and cancer. Nat Rev Cancer. 2008;8(12):957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sedelnikova OA, Bonner WM. GammaH2AX in cancer cells: a potential biomarker for cancer diagnostics, prediction and recurrence. Cell Cycle. 2006;5(24):2909–2913. doi: 10.4161/cc.5.24.3569. [DOI] [PubMed] [Google Scholar]
- 38.Hong Y, Cervantes RB, Tichy E, Tischfield JA, Stambrook PJ. Protecting genomic integrity in somatic cells and embryonic stem cells. Mutat Res. 2007;614(1–2):48–55. doi: 10.1016/j.mrfmmm.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 39.Opar A. Novel anticancer strategy targets DNA repair. Nat Rev Drug Discov. 2009;8(6):437–438. doi: 10.1038/nrd2916. [DOI] [PubMed] [Google Scholar]
- 40.O’Shaughnessy J, Osborne C, Pippen J, et al. Efficacy of BSI-201, a poly(ADP-ribose) polymerase-1 (PARP1) inhibitor, in combination with gemcitabine/carboplatin (G/C) in patients with metastatic triple-negative breast cancer (TNBC): results of a randomised phase II trial. J Clin Oncol. 2009;27(15s):6s. [Google Scholar]
- 41.Ashwell S, Zabludoff S. DNA damage detection and repair pathways—recent advances with inhibitors of checkpoint kinases in cancer therapy. Clin Cancer Res. 2008;14(13):4032–4037. doi: 10.1158/1078-0432.CCR-07-5138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.