Abstract
The posttranslational methylation of N-terminal α-amino groups (α-N-methylation) is a ubiquitous reaction found in all domains of life. Although this modification usually occurs on protein substrates, recent studies have shown that it also takes place on ribosomally synthesized natural products. Here we report an investigation of the bacterial α-N-methyltransferase CypM involved in the biosynthesis of the peptide antibiotic cypemycin. We demonstrate that CypM has low substrate selectivity and methylates a variety of oligopeptides, cyclic peptides such as nisin and haloduracin, and the ε-amino group of lysine. Hence it may have potential for enzyme engineering and combinatorial biosynthesis. Bayesian phylogenetic inference of bacterial α-N-methyltransferases suggests that they have not evolved as a specific group based on the chemical transformations they catalyze, but that they have been acquired from various other methyltransferase classes during evolution.
Keywords: α-N-methyltransferase, peptide antibiotic, lantibiotic, catalytic promiscuity, evolution
1. Introduction
Posttranslational methylation of N-terminal α-amino groups (α-N-methylation) is a ubiquitous reaction found in both bacteria and eukarya [1–10]. These reactions occur usually on protein substrates that function as part of macromolecular complexes, including the ribosome, chromatin, respiratory chains, photosynthetic complexes, myofibrils and bacterial pili [4]. Although the exact role and the enzymology of α-N-methylation remains largely unknown, recent studies have revealed that the N-terminal methylation of Ran guanine nucleotide exchange factor RCC1 catalyzed by NRMT (N-terminal RCC1 methyltransferase) is critical for normal bipolar spindle formation and chromosome segregation in the mitotic phase of mammalian cells [7,8]. NRMT was shown to be promiscuous and is able to methylate protein substrates other than RCC1, such as SET (also known as TAF-I or PHAPII) and the retinoblastoma protein RB [8]. An ortholog of NRMT in Drosophila melanogaster, dNTMT, was shown to catalyze methylation of histone H2B [10]. For bacteria, methylation of the ribosomal protein L11 has been relatively well characterized. This reaction is catalyzed by PrmA, a versatile enzyme that methylates not only the N-terminal α-amino group but also the ε-amino group of Lys residues [11,12].
In addition to protein substrates, ribosomally synthesized peptides also serve as substrates for α-N-methylation. The recently-characterized thiazole/oxazole-modified peptide plantazolicin B, for example, is N-terminally methylated to produce plantazolicin A containing an N,N-dimethylarginine residue (Fig. 1) [13–15]. The N,N-dimethylalanine (Me2-Ala) residue in cypemycin and its structural variant grisemycin also results from α-N-methylation [16,17] (Fig. 1). Cypemycin is a posttranslationally modified peptide antibiotic produced by Streptomyces sp. OH-4156 with potent in vitro activity against mouse leukemia cells [18]. Historically, it has been grouped in the lantibiotic family because of its dehydrobutyrine (Dhb) residues and (Z)-2-aminovinyl-cysteine (Avi-Cys) moiety (Fig. 1), which are also found in some lantibiotics [19,20]. However, cypemycin does not contain lanthionine or methyllanthionine rings, and the Dhb and Avi-Cys residues are proposed to be introduced by a pathway distinct from that of lantibiotic biosynthesis [16]. CypM, an enzyme of the methyltransferase 11 family, is responsible for the synthesis of the N-terminal Me2-Ala residue, as deletion of cypM from the cypemycin biosynthetic gene cluster resulted in a demethylated cypemycin derivative, which could be methylated by CypM in vitro to produce cypemycin (Fig. 1) [16]. Interestingly, the antimicrobial activity of demethylated cypemycin was almost completely abolished. A similar result was found for plantazolicin, as plantazolicin B was devoid of activity against B. anthracis whereas plantazolicin A displayed potent activity [15]. These results demonstrated a new role of α-N-methylation in the biological activity of natural products.
Fig. 1.

N-Methylation of cypemycin and plantazolicin A catalyzed by CypM and PznL, respectively. The amino groups on which methylation occurs are highlighted by dashed boxes. Dhb, dehydrobutyrine; a-Ile, allo-isoleucine; Avi-Cys, S-[(Z)-2-aminovinyl]-cysteine; SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine. A shorthand notation for these structures is shown below each chemical structure in the red box.
Given the essentiality of α-N-methylation for the antibiotic activity and the catalytic versatility found for NRMT and PrmA, α-N-methyltransferases might possibly serve as a useful tool for structural diversification and functional optimization of peptide antibiotics. Modification of the N-terminus is an often-used strategy for synthetic peptides to decrease the susceptibility to aminopeptidases. Here we probed the catalytic specificities of CypM using various synthetic and natural peptides. Our data show that CypM has low substrate specificity and can methylate a series of structurally distinct substrates, suggesting that catalytic promiscuity might be a common property among α-N-methyltransferases and demonstrating a potentially useful tool for future combinatorial biosynthesis studies. Phylogenetic analysis using the Bayesian Markov chain Monte Carlo (MCMC) method [21] indicates that bacterial α-N-methyltransferase may not have evolved from a common ancestor, but were likely acquired several times from other ancient methyltransferases.
2. Materials and Methods
2.1. Chemicals, biochemicals, plasmids and strains
This information is provided in the Supplementary Methods.
2.2. Synthesis of oligopeptides
Peptides were synthesized using standard fluorenylmethyloxycarbonyl (Fmoc) based solid phase peptide synthesis (SPPS) techniques using a Rainin PS3 peptide synthesizer. Preloaded resin (either Wang or 2-chlorotrityl; 0.1 mmol) was first swollen in dimethylformamide (DMF) (3 × 5 mL × 10 min). Fmoc-amino acids (0.4 mmol, 4 equiv) were coupled using O-(1H-6-chlorobenzotriazol-1-yl)-N,N,N',N'-tetramethyluronium hexafluorophosphate (HCTU, 165 mg, 0.4 mmol, 4 equiv) as coupling reagent and 0.4 M N-methyl morpholine (NMM) as activating reagent (45 min). Fmoc deprotection was performed with piperidine (3 × 5 mL × 3 min; 20% in DMF). After completion of the coupling of the final amino acid, the Fmoc group was removed to generate the free amino group.
Peptides were cleaved from the resin by adding a solution of trifluoroacetic acid (TFA) (5 mL), triisopropylsilane (TIPS) (100 μL), and H2O (100 μL) to the resin (0.1 mmol), and stirring the solution for 2 h at room temperature. The solution was concentrated by purging with a nitrogen stream and peptides were precipitated with cold diethyl ether. The crude peptides were dissolved in 0.1% aqueous TFA, lyophilized, and purified by preparative RP-HPLC on a Waters Delta-PakTM C18 column (2.5 cm × 10.0 cm) employing a water-acetonitrile solvent system. HPLC fractions containing products as confirmed by (MALDI) mass spectrometry (MS) were collected and lyophilized.
2.3. In vitro CypM assays
Detailed procedures for overexpression and purification of C-terminally hexa His-tagged CypM are described in the Supplementary Methods. For in vitro methylation assays, peptide substrates (100 μM) were incubated with CypM-His6 (20 μM) in a reaction buffer containing 50 mM HEPES (pH 7.2), 0.5 mM S-adenosylmethionine (SAM), 1 mM DTT, 100 mM NaCl and 0.01 U of S-adenosylhomocysteine hydrolase (from rabbit erythrocytes, Sigma-Aldrich). The reactions were incubated at 37 °C for 5 h. Reactions were quenched by 5% TFA and the protein precipitate was removed by centrifugation. Samples were analyzed directly by liquid chromatography electrospray ionization tandem mass spectrometry (LC/ESI-MS/MS) as described in the Supplementary Methods. For MALDI-TOF MS analysis, samples were either purified by reverse phase (C4) solid phase extraction or desalted using ZipTipC18 before analysis.
2.4. Phylogenetic analysis
The protein sequences in this study were obtained from the GenBank database; their accession numbers and the source organisms are listed in Supplementary Table S2. The sequences were aligned in ClustalX [22] using default parameters with iteration at each alignment step, and the alignments were manually fine-tuned afterwards. Bayesian inference was used to calculate posterior probability of clades utilizing the program MrBayes (version 3.1) [23]. Final analyses consisted of two sets of eight chains each (one cold and seven heated), run for about 3 million generations with trees saved and parameters sampled every 100 generations. Analyses were run to reach a convergence with standard deviation of split frequencies <0.005. Posterior probabilities were averaged over the final 75% of trees (25% burn in). The analysis utilized a mixed amino acid model with a proportion of sites designated invariant, and rate variation among sites modeled after a gamma distribution divided into eight categories, with all variable parameters estimated by the program based on BioNJ starting trees. The figures of the Bayesian phylograms were prepared by using TreeView [24].
3. Results and Discussion
3.1. CypM-catalyzed methylation of short oligopeptides
Cypemycin biosynthesis follows a common paradigm of ribosomal natural product maturation, involving posttranslational modifications of the C-terminal core region of a precursor peptide and subsequent proteolytic removal of an N-terminal leader sequence [25]. CypM methylates the α-amino group of an N-terminal Ala residue that is released by removal of the leader peptide. Because the leader peptide is typically indispensable for precursor peptide modifications [25], α-N-methylation of Ala1 probably occurs as the last step of cypemycin biosynthesis. Accordingly, the natural substrate of CypM would be a peptide containing the Dhb residues and a C-terminal Avi-Cys moiety (Fig. 1).
We synthesized a series of short oligopeptides resembling the N-terminal sequence of cypemycin, in which the Dhb residues were replaced with Ala, Ser or Thr. As shown in Table 1, in the presence of S-adenosylmethionine (SAM) these peptides (entries 1–5) were methylated by CypM (Table 1 and Supplementary Fig. S1–S5). In contrast to the reaction with native substrate in which only di-methylated product (cypemycin) was observed [15], mono- and di-methylated products were both identified in most cases, reflecting the relatively inefficient methylation of these short oligopeptides as compared to the native substrate. However, the tolerance of CypM to Thr- and Ser- containing peptide indicates the rather relaxed substrate specificity of the enzyme, as the N-terminus as well as the overall scaffold of cypemycin is highly hydrophobic (Fig. 1). Moreover, the methylation is not confined to N-terminal Ala. Peptides with N-terminal Gly, Ser, and Met residues were also modified by CypM (Table 1, entries 6–8), albeit with reduced efficiencies (Supplementary Fig. S6–S8). Collectively, these results indicate that CypM has a relaxed substrate specificity and can methylate a series of short oligopeptides that mimic the N-terminal sequence of cypemycin.
Table 1.
Methylation of short oligopeptides by CypM.
| Peptide | Sequence | Observed methylation |
|---|---|---|
| 1 | AAPAAPA | Mono-, Di- |
| 2 | AAPAAPS | Di- |
| 3 | ASPAAPA | Di- |
| 4 | ATPAAPA | Mono-, Di- |
| 5 | ATPATPA | Mono-, Di- |
| 6 | GAPAAPA | Di- |
| 7 | SAPAAPA | Di- |
| 8 | MAPAAPA | Di- |
| 9 | KAPAAPA | No reaction |
| 10 | AKPAAPA | No reaction |
| 11 | AAPAAPK | Di-, Tetra- |
| 12 | AAAATPT | No reaction |
| 13 | APKAAPA | No reaction |
The ability of PrmA to methylate not only the N-terminal α-amino group but also the ε-amino group of Lys led us to investigate whether CypM can also methylate the ε-amino group of Lys, which could be useful in histone research. We synthesized several oligopeptides similar to peptide 1 (AAPAAPA) but with Lys replacements in various positions (peptides 9–11). As shown in Table 1, peptides with Lys in the first or second position from the N-terminus (entries 9–10) were not modified by CypM, but a peptide with a Lys at the C-terminus (entry 11) was methylated, with both di- and tetra-methylated products detected in the reaction mixture (Supplementary Fig. S11). Detailed MS-MS analysis clearly showed that di-methylation occurred almost entirely on the ε-amino group of Lys instead of the N-terminus (Supplementary Fig. S12), suggesting that the ε-amino group of Lys might be preferred over the N-terminal amino group provided it is not near the N-terminus. We note that cypemycin does not contain any Lys residues.
Cypemycin and many α-N-methylated proteins contain Pro residues within a few amino acids of the N-termini [4,8,16], which might be important for enzyme recognition. Indeed, a peptide in which Pro3 was replaced by Ala (entry 12) was not modified by CypM. CypM also did not methylate a peptide with an N-terminal APK motif (entry 13) that is the target of NRMT; CypM and NRMT share no sequence similarity.
3.2. Methylation of other peptide antibiotics
We next interrogated whether CypM could methylate structurally unrelated peptide antibiotics. Nisin (Fig. 2A) represents the best studied member of the lantibiotic family and has been widely used in the food industry because of its potent antimicrobial activity and unique mode of action [26,27]. There is interest in expanding its use to clinical applications in both human and animal health products [28]. As nisin possesses an unmodified Ile residue at the N-terminus, the α-N-methylation of this residue may potentially improve the stability and the pharmacological property of the molecule by preventing proteolytic degradation by aminopeptidases. Incubation of CypM with nisin resulted in addition of three methyl groups (Fig. 2B). Detailed MS-MS analysis revealed that one methylation occurred on the N-terminal Ile with the other two methylations on Lys12 (Supplementary Fig. S15). The MIC value of the methylated nisin A against Lactococcus Lactis HP (ATCC11602) and Bacillus subtillis 168 (ATCC6633) was determined to be 1.0 μM and 4 μM, 8-fold and 4-fold higher than that of nisin A, respectively, indicating that the methylated nisin is still a potent antibiotic. CypM may thus serve a methylating tool for diversification of peptide natural products.
Fig. 2.
(A) Structure of nisin and its methylated derivatives. The amino groups on which methylation occurred are highlighted by dashed boxes. Abu, 2-aminobutyric acid; Dha, dehydroalanine; Dhb, dehydrobutyrine. For lanthionine and methyllanthionine structures, the segments derived from Ser/Thr are in red and those derived from Cys are in blue. A shorthand notation for these structures used in the natural product drawings is shown below each chemical structure (B) MS spectra of nisin A and nisin A in vitro modified by CypM.
We also evaluated haloduracin as substrate for CypM. Haloduracin is a two-component lantibiotic containing two distinct peptides halduracin α (Halα) and β (Halβ) (Fig. 3A), which act synergistically to exhibit potent antibiotic activity [29,30]. Halα contains a Cys residue at its N-terminus, which links to Cys8 by a disulfide bond and forms a macrocyclic system, and Halβ possesses an N-terminal methyllanthionine ring (Fig. 3A). Intriguingly, both Halα and Halβ were methylated by CypM, and the mono-methylated product was found to be the major product in both cases (Fig. 3B). MS-MS spectrometric analysis indicated that, although both Halα and Halβ contain Lys residues, methylation occurred on the N-terminal amino group in both cases (Supplementary Fig. S16–17). Given the considerable structural difference between the native substrate of CypM (demethyl-cypemycin) and nisin and haloduracin, the catalytic promiscuity of CypM demonstrated herein is surprising. The factors that determine methylation of either α-N-amino or Lys ε-amino groups in different structural contexts, however, is currently unclear and requires structural studies.
Fig. 3.
(A) Structure of Halα, Halβ and their methylated derivatives. The amino groups on which methylation occurred are highlighted by dashed boxes. Structures are similarly represented as in Fig. 2. (B) MS spectra of Halα and Halβ modified by CypM.
3.3. Evolution of bacterial α-N-methyltransferases
A search of CypM homologs in the National Center for Biotechnology Information (NCBI) sequence database revealed that except for GrmM, involved in the biosynthesis of grisemycin (a cypemycin structural variant), proteins with homology to CypM share only modest sequence identities with CypM. These homologous proteins include the α-N-methyltransferase PznL for plantazolicin A biosynthesis (Fig. 1), LmbJ and CcbJ that are involved in the biosynthesis of lincomycin and celesticetin, respectively [31,32], and several proteins that are designated as ubiquinone/menaquinone methylase UbiE [33]. However, most proteins with sequence homology to CypM in the NCBI database are functionally uncharacterized.
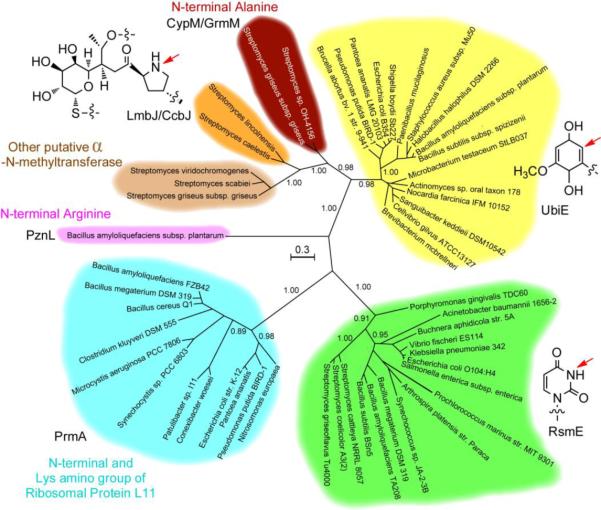
We used the Bayesian MCMC method to construct the phylogenetic inference of CypM and other bacterial methyltransferases [34]. Several UbiE enzymes identified from the ubiquinone/menaquinone biosynthetic gene clusters of different genera and three methyltransferases proposed to be involved in producing cypemycin-like natural products [16] were included in the analysis. Several RsmE proteins that methylate the imide nitrogen N3 of a unique uridine (U1489 for E. coli) of 16S rRNAs [35,36] were also included for comparative analysis. The Bayesian MCMC tree shown in Fig. 4 clearly indicates that bacterial α-N-methyltransferases did not evolve together, as CypM, PnzL and PrmA fall into different clades with strong posterior probability support. CypM and GrmM, LmbJ and CcbJ, and other methyltransferases for putative cypemycin-like natural product biosynthesis form a distinct clade. Given the close phylogenetic relationship and sequence similarities of CypM with UbiE enzymes, it is likely that CypM and other enzymes in the same clade have evolved from ancient UbiE enzymes despite the latter being C-methyltransferases [33]. PrmA [12,37,38] seems to have a closer relationship with RsmE than with CypM, suggesting that these two α-N-methyltransferases evolved from different ancestors. Although PznL is an α-N-methyltransferase involved in ribosomal natural product biosynthesis like CypM, it is phylogenetically distant from CypM and falls into a distinct clade (Fig. 4). The phylogenetic analysis demonstrates that α-N-methyltransferases did not evolve based on the chemical reaction of methylating N-terminal amino groups, but rather were acquired through convergent evolution from diverse precursors with different functions. It has previously been suggested that some plant O-methyltransferases might also have undergone convergent evolutionary processes [39].
Fig. 4.
Unrooted tree of selected bacterial methyltransferases generated by Bayesian MCMC method. Support for the major clades is indicated by posterior probability values. Substrates for each clade of enzymes are shown, and the red arrows indicate the methylation sites.
In conclusion, we demonstrate that CypM, an α-N-methyltransferase involved in cypemycin biosynthesis, has high catalytic flexibility and can act on a series of structurally distinct substrates, including the lantibiotic nisin. Combined with the studies of NRMT [8] and PrmA [12,37,38], it seems that catalytic promiscuity is a common property of α-N-methyltransferase enzymes, which appear to have evolved by distinct pathways. The flexibility of α-N-methytransferases may find utility in methylation of therapeutic peptides that are metabolized by aminopeptidases.
Supplementary Material
Highlights
CypM can methylate both α-N-terminal amino groups and lysine ε-amino groups
CypM methylates a variety of cyclic peptides such as nisin and haloduracin
α-N-methyltransferases did not evolve as a specific clade
Acknowledgements
We thank Prof. Mervyn Bibb (John Innes Centre, UK) for providing cosmid pIJ12404. This work was supported by the U.S. National Institutes of Health (GM58822 to W.A.v.d.D). Mass spectra were recorded in part on an instrument purchased with funds from grant S10RR027109-01 from the National Institutes of Health.
Abbreviations
- Dha
dehydroalanine
- Dhb
dehydrobutyrine
- Avi-Cys
S-[(Z)-2-aminovinyl]-cysteine
- SAM
S-adenosylmethionine
- SAH
S-adenosylhomocysteine
- NRMT
N-terminal RCC1 methyltransferase
- MCMC
Markov chain Monte Carlo
- SPPS
solid phase peptide synthesis
- DMF
dimethylformamide
- NMM
N-methyl morpholine
- MALDI
matrix assisted laser desorption ionization
- Abu
2-aminobutyric acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplementary data Supplementary data associated with this article can be found in the online version at xxxxxxxxxxx
References
- [1].Pettigrew GW, Smith GM. Novel N-terminal protein blocking group identified as dimethylproline. Nature. 1977;265:661–2. doi: 10.1038/265661a0. [DOI] [PubMed] [Google Scholar]
- [2].Henry GD, Dalgarno DC, Marcus G, Scott M, Levine BA, Trayer IP. The Occurrence of Alpha-N-Trimethylalanine as the N-Terminal Amino-Acid of Some Myosin Light-Chains. FEBS Lett. 1982;144:11–15. doi: 10.1016/0014-5793(82)80558-9. [DOI] [PubMed] [Google Scholar]
- [3].Martinage A, Briand G, Van Dorsselaer A, Turner CH, Sautiere P. Primary structure of histone H2B from gonads of the starfish Asterias rubens. Identification of an N-dimethylproline residue at the amino-terminal. Eur. J. Biochem. 1985;147:351–9. doi: 10.1111/j.1432-1033.1985.tb08757.x. [DOI] [PubMed] [Google Scholar]
- [4].Stock A, Clarke S, Clarke C, Stock J. N-terminal methylation of proteins: structure, function and specificity. FEBS Lett. 1987;220:8–14. doi: 10.1016/0014-5793(87)80866-9. [DOI] [PubMed] [Google Scholar]
- [5].Grimm R, Grimm M, Eckerskorn C, Pohlmeyer K, Rohl T, Soll J. Postimport methylation of the small subunit of ribulose-1,5-bisphosphate carboxylase in chloroplasts. FEBS Lett. 1997;408:350–4. doi: 10.1016/s0014-5793(97)00462-6. [DOI] [PubMed] [Google Scholar]
- [6].Sadaie M, Shinmyozu K, Nakayama J. A conserved SET domain methyltransferase, Set11, modifies ribosomal protein Rpl12 in fission yeast. J. Biol. Chem. 2008;283:7185–95. doi: 10.1074/jbc.M709429200. [DOI] [PubMed] [Google Scholar]
- [7].Chen T, Muratore TL, Schaner-Tooley CE, Shabanowitz J, Hunt DF, Macara IG. N-terminal alpha-methylation of RCC1 is necessary for stable chromatin association and normal mitosis. Nat. Cell. Biol. 2007;9:596–603. doi: 10.1038/ncb1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tooley CE, et al. NRMT is an alpha-N-methyltransferase that methylates RCC1 and retinoblastoma protein. Nature. 2010;466:1125–8. doi: 10.1038/nature09343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sekine F, Horiguchi K, Kashino Y, Shimizu Y, Yu LJ, Kobayashi M, Wang ZY. Gene sequencing and characterization of the light-harvesting complex 2 from thermophilic purple sulfur bacterium Thermochromatium tepidum. Photosynth. Res. 2012;111:9–18. doi: 10.1007/s11120-011-9658-9. [DOI] [PubMed] [Google Scholar]
- [10].Villar-Garea A, Forne I, Vetter I, Kremmer E, Thomae A, Imhof A. Developmental regulation of N-terminal H2B methylation in Drosophila melanogaster. Nucleic Acids Res. 2012;40:1536–49. doi: 10.1093/nar/gkr935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dognin MJ, Wittmann-Liebold B. Purification and primary structure determination of the N-terminal blocked protein, L11, from Escherichia coli ribosomes. Eur. J. Biochem. 1980;112:131–51. doi: 10.1111/j.1432-1033.1980.tb04995.x. [DOI] [PubMed] [Google Scholar]
- [12].Cameron DM, Gregory ST, Thompson J, Suh MJ, Limbach PA, Dahlberg AE. Thermus thermophilus L11 methyltransferase, PrmA, is dispensable for growth and preferentially modifies free ribosomal protein L11 prior to ribosome assembly. J. Bacteriol. 2004;186:5819–25. doi: 10.1128/JB.186.17.5819-5825.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Scholz R, Molohon KJ, Nachtigall J, Vater J, Markley AL, Sussmuth RD, Mitchell DA, Borriss R. Plantazolicin, a novel microcin B17/streptolysin S-like natural product from Bacillus amyloliquefaciens FZB42. J. Bacteriol. 2011;193:215–24. doi: 10.1128/JB.00784-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kalyon B, Helaly SE, Scholz R, Nachtigall J, Vater J, Borriss R, Sussmuth RD. Plantazolicin A and B: structure elucidation of ribosomally synthesized thiazole/oxazole peptides from Bacillus amyloliquefaciens FZB42. Org. Lett. 2011;13:2996–9. doi: 10.1021/ol200809m. [DOI] [PubMed] [Google Scholar]
- [15].Molohon KJ, Melby JO, Lee J, Evans BS, Dunbar KL, Bumpus SB, Kelleher NL, Mitchell DA. Structure Determination and Interception of Biosynthetic Intermediates for the Plantazolicin Class of Highly Discriminating Antibiotics. ACS Chem. Biol. 2011;6:1307–13. doi: 10.1021/cb200339d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Claesen J, Bibb M. Genome mining and genetic analysis of cypemycin biosynthesis reveal an unusual class of posttranslationally modified peptides. Proc. Natl. Acad. Sci. U. S. A. 2010;107:16297–302. doi: 10.1073/pnas.1008608107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Claesen J, Bibb MJ. Biosynthesis and regulation of grisemycin, a new member of the linaridin family of ribosomally synthesized peptides produced by Streptomyces griseus IFO 13350. J. Bacteriol. 2011;193:2510–6. doi: 10.1128/JB.00171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Komiyama K, et al. A new antibiotic, cypemycin. Taxonomy, fermentation, isolation and biological characteristics. J. Antibiot. 1993;46:1666–71. doi: 10.7164/antibiotics.46.1666. [DOI] [PubMed] [Google Scholar]
- [19].Knerr PJ, van der Donk WA. Discovery, biosynthesis, and engineering of lantipeptides. Annu. Rev. Biochem. 2012;81:479–505. doi: 10.1146/annurev-biochem-060110-113521. [DOI] [PubMed] [Google Scholar]
- [20].Sit CS, Yoganathan S, Vederas JC. Biosynthesis of aminovinyl-cysteine-containing peptides and its application in the production of potential drug candidates. Acc. Chem. Res. 2011;44:261–8. doi: 10.1021/ar1001395. [DOI] [PubMed] [Google Scholar]
- [21].Mau B, Newton MA, Larget B. Bayesian phylogenetic inference via Markov chain Monte Carlo methods. Biometrics. 1999;55:1–12. doi: 10.1111/j.0006-341x.1999.00001.x. [DOI] [PubMed] [Google Scholar]
- [22].Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- [24].Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12:357–8. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- [25].Oman TJ, van der Donk WA. Follow the leader: the use of leader peptides to guide natural product biosynthesis. Nat. Chem. Biol. 2010;6:9–18. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Delves-Broughton J, Blackburn P, Evans RJ, Hugenholtz J. Applications of the bacteriocin, nisin. Antonie van Leeuwenhoek. 1996;69:193–202. doi: 10.1007/BF00399424. [DOI] [PubMed] [Google Scholar]
- [27].Lubelski J, Rink R, Khusainov R, Moll GN, Kuipers OP. Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell. Mol. Life Sci. 2008;65:455–476. doi: 10.1007/s00018-007-7171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].van Kraaij C, de Vos WM, Siezen RJ, Kuipers OP. Lantibiotics: biosynthesis, mode of action and applications. Nat. Prod. Rep. 1999;16:575–87. doi: 10.1039/a804531c. [DOI] [PubMed] [Google Scholar]
- [29].McClerren AL, Cooper LE, Quan C, Thomas PM, Kelleher NL, van der Donk WA. Discovery and in vitro biosynthesis of haloduracin, a two-component lantibiotic. Proc. Natl. Acad. Sci. U. S. A. 2006;103:17243–8. doi: 10.1073/pnas.0606088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lawton EM, Cotter PD, Hill C, Ross RP. Identification of a novel two-peptide lantibiotic, Haloduracin, produced by the alkaliphile Bacillus halodurans C-125. FEMS Microbiol. Lett. 2007;267:64–71. doi: 10.1111/j.1574-6968.2006.00539.x. [DOI] [PubMed] [Google Scholar]
- [31].Spizek J, Rezanka T. Lincomycin, cultivation of producing strains and biosynthesis. Appl. Microbiol. Biotechnol. 2004;63:510–9. doi: 10.1007/s00253-003-1431-3. [DOI] [PubMed] [Google Scholar]
- [32].Cermak L, Novotna J, Sagova-Mareckova M, Kopecky J, Najmanova L, Janata J. Hybridization analysis and mapping of the celesticetin gene cluster revealed genes shared with lincomycin biosynthesis. Folia Microbiol. (Praha) 2007;52:457–62. doi: 10.1007/BF02932104. [DOI] [PubMed] [Google Scholar]
- [33].Nowicka B, Kruk J. Occurrence, biosynthesis and function of isoprenoid quinones. Biochim. Biophys. Acta. 2010;1797:1587–605. doi: 10.1016/j.bbabio.2010.06.007. [DOI] [PubMed] [Google Scholar]
- [34].Hall BG. Comparison of the accuracies of several phylogenetic methods using protein and DNA sequences. Mol. Biol. Evol. 2005;22:792–802. doi: 10.1093/molbev/msi066. [DOI] [PubMed] [Google Scholar]
- [35].Basturea GN, Rudd KE, Deutscher MP. Identification and characterization of RsmE, the founding member of a new RNA base methyltransferase family. RNA. 2006;12:426–34. doi: 10.1261/rna.2283106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Basturea GN, Deutscher MP. Substrate specificity and properties of the Escherichia coli 16S rRNA methyltransferase, RsmE. RNA. 2007;13:1969–76. doi: 10.1261/rna.700507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Demirci H, Gregory ST, Dahlberg AE, Jogl G. Recognition of ribosomal protein L11 by the protein trimethyltransferase PrmA. EMBO J. 2007;26:567–77. doi: 10.1038/sj.emboj.7601508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Demirci H, Gregory ST, Dahlberg AE, Jogl G. Multiple-site trimethylation of ribosomal protein L11 by the PrmA methyltransferase. Structure. 2008;16:1059–66. doi: 10.1016/j.str.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lam KC, Ibrahim RK, Behdad B, Dayanandan S. Structure, function, and evolution of plant O-methyltransferases. Genome. 2007;50:1001–13. doi: 10.1139/g07-077. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.