Abstract
Small peptides and oligosaccharides are important antigens for the development of vaccines and the production of monoclonal antibodies. Because of their small size, peptides and oligosaccharides are non-immunogenic on their own and typically must be conjugated to a larger carrier protein to elicit an immune response. Selection of a suitable carrier protein, conjugation method, and hapten density is critical for generating an optimal immune response. In this study, we use a glycan array to evaluate the effects of hapten density on the spectrum of antibodies elicited to a tumor-associated carbohydrate antigen. We demonstrate that high hapten density produces a broader range of antibodies while low hapten density induces a narrower range of antibodies.
Keywords: carbohydrates, glycoconjugates, ligand density, vaccine, antibody
Introduction
Antibodies, soluble proteins produced by B cells, are a key element of the immune response to pathogens and vaccines. Antibodies function in vivo by binding their target antigen, which results in aggregation of the antigen, tagging of the antigen for elimination by effector cells of the immune system, and/or potentially blocking key steps in infection. The ability of antibodies to bind a target antigen with high affinity and selectivity has also made them indispensable research tools. As a result, strategies to induce an optimal antibody response are critical for vaccine development and reagent antibody production.
Many important and useful target antigens, such as peptides and oligosaccharides, are too small to elicit an immune response on their own. To overcome this problem, small molecules are typically conjugated to a large carrier protein prior to vaccination. Many features of immunogen design, such as the choice of carrier protein and the hapten density, can affect the magnitude of the ensuing immune response.[1–6] The breadth and selectivities of the induced antibody repertoire can also be crucial factors for vaccine efficacy and monoclonal antibody development; however, much less is known about the effects of immunogen design on the selectivity and spectrum of the induced antibodies, primarily due to the difficulties in measuring binding to a broad range of potential antigens. Selectivity has often been evaluated by measuring binding to cell lines and/or tissue samples, but the complex nature of these materials makes it difficult to draw specific conclusions regarding selectivity at a molecular level. Binding to structurally-defined antigens has also been used to measure selectivity. These studies are limited by the availability of pure antigens and the throughput of the assay used for evaluation. For certain classes of antigens that are difficult to obtain, such as carbohydrates, these studies have typically been limited to a very small number of antigens.
Antigen arrays contain many different antigens or fragments of antigens immobilized on a solid support in a spatially-defined arrangement.[7] These arrays provide a high-throughput approach to evaluate binding to many potential antigens in a single experiment. The array format is especially useful for studying recognition of carbohydrate antigens, since only tiny amounts of scarce materials are required for the array. Carbohydrate antigen arrays, or glycan arrays, have been used extensively to evaluate binding of antibodies, lectins, cells, and viruses to glycans.[8–11]
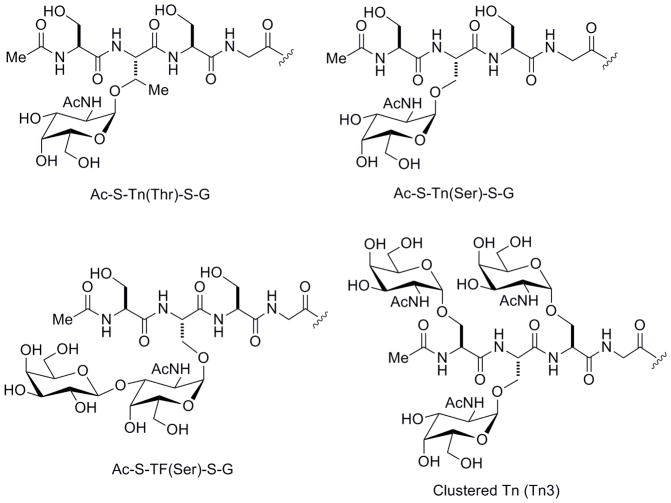
One especially important carbohydrate antigen is the tumor-associated Tn antigen. This antigen is composed of a GalNAc residue alpha linked to either a serine or threonine of a polypeptide. It is reported to be expressed in 70–90% of carcinomas of the breast, colon, prostate, and lung, making it an appealing target for therapeutic and diagnostic development.[12, 13] In particular, the Tn antigen has been studied extensively as a cancer vaccine antigen, and Tn-based vaccines have progressed into clinical trials for the treatment of breast and prostate cancer.[14–16] Nevertheless, clinical responses to Tn vaccines are not optimal, and a number of studies have been directed at improving immunogenicity.[17–25] While the focus of these efforts has been on increasing the magnitude of the antibody response (especially IgG), selectivity is also a key factor. The Tn antigen can be present in many forms and contexts such as GalNAc attachment to either serine [Tn(Ser)] or threonine [Tn(Thr)], single versus clustered units (2 or more Tn residues linked consecutively on a peptide chain), high versus low density of those units, and within a variety of peptide sequences [see Ac-S-Tn(Thr)-S-G, Ac-S-Tn(Ser)-S-G, and Tn3 in Figure 1]. Previous studies have shown that some antibodies can distinguish between different forms of the Tn antigen and/or can require a particular peptide sequence for binding.[24, 26–30] Other antibodies, however, can recognize multiple forms of the Tn antigen.[31–33] Broader selectivity may facilitate recognition of a larger percentage of tumors but may also lead to cross-reactivity with normal tissues and/or binding to related normal carbohydrates. Therefore, the selectivity and breadth of the antibody repertoire is relevant to vaccine research and antibody development.
Figure 1.
Structures of selected glycopeptides
In this study, we used a glycan array to evaluate the effects of hapten density on the immune response to the tumor-associated Tn antigen. We demonstrate that the hapten density has a significant effect on the induced antibody repertoire.
Results and Discussion
Preparation of Tn-conjugates and induction of antibodies
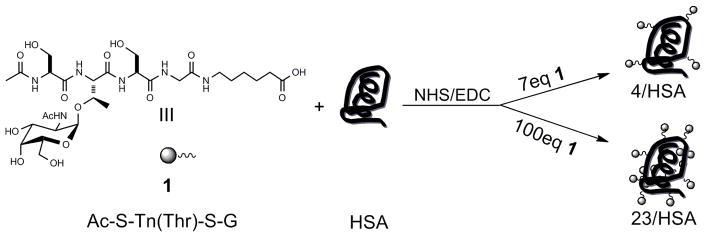
To evaluate the effects of hapten density, we prepared Tn-HSA conjugates with either high or low hapten density. Since neighboring amino acid residues can be important for recognition by Tn binding antibodies and lectins,[24, 26–30] a hapten containing a Tn(Thr) in the context of a short peptide was used. The selected peptide sequence, Ser-Tn(Thr)-Ser, is found in a number of mucin tandem repeat sequences, including those of Muc3b, Muc6, and Muc16, and within a region of glycophorin A that is bound by antibody MLS128.[26, 27, 34–36] Tn glycopeptide 1[32]was coupled to HSA via activation of the C-terminal carboxylic acid with 1-ethyl-3-(3-dimethylaminopropyl)-1-carbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS) followed by reaction with amine groups on HSA (see Scheme 1).[25, 37] At a molar ratio of 7:1 glycopeptide to HSA, we obtained an average of 4 haptens per HSA as judged by MALDI-MS. To get the high density conjugate, 100 equivalents of glycopeptide was used, resulting in an average of 23 haptens per HSA as judged by MALDI-MS. High and low density bovine serum albumin (BSA) conjugates were also prepared.
Scheme 1.
Preparation of the Tn-HSA conjugates.
Rabbits were vaccinated subcutaneously with either the low or high density HSA-conjugate mixed with complete Freund’s adjuvant (CFA) followed by boosts in incomplete Freund’s adjuvant (IFA) on days 21 and 42. Serum was collected prior to the first vaccination and on day 51.
We began by evaluating the magnitude of the antibody response to the hapten. Antibody titers were measured using standard ELISA with Ac-S-Tn(Thr)-S-G-BSA-coated plates. Vaccination produces antibodies to the hapten and the carrier protein. The use of a BSA-conjugate, rather than an HSA-conjugate, allows one to measure antibody responses specific to the hapten. The rabbits produced high titers of IgG antibodies along with modest IgM titers. Interestingly, hapten density did not have a significant effect on the magnitude of the response (Table 1). All rabbits had similar titers to the hapten.
Table 1.
Antibody IgG and IgM ELISA titers to high and low density Ac-S-Tn(Thr)-S-G-BSA conjugates
| IgG Titers | IgM Titers | |||
|---|---|---|---|---|
| 4/BSA | 24/BSA | 4/BSA | 24/BSA | |
| Rabbit 1[a] | 781,250 | 781,250 | 1,250 | 1,250 |
| Rabbit 2[a] | 312,500 | 312,500 | 1,250 | 1,250 |
| Rabbit 3[a] | 781,250 | 312,500 | 1,250 | 1,250 |
| Rabbit 4[a] | 781,250 | 781,250 | 3,125 | 1,250 |
Rabbits 1 and 2 were vaccinated with low density Tn conjugates; Rabbits 3 and 4 were vaccinated with high density Tn conjugates
We have previously found that monoclonal antibodies and certain subpopulations of naturally-occurring serum antibodies can distinguish different densities of the same antigen.[32] One of our primary questions was whether vaccination with conjugates of different density would induce antibodies specific for high or low density presentations of the hapten. As can be seen from Table 1, variations in hapten density on the immunogen did not lead to density preferences in the induced antibodies. For example, the rabbits vaccinated with low density Tn had equal titers to both low and high density Ac-S-Tn(Thr)-S-G-BSA (compare 4/BSA vs 24/BSA, Table 1).
Glycan array profiling of immune responses
We used carbohydrate microarray technology to evaluate the spectrum of antibodies induced by the low and high density conjugates. Carbohydrate microarrays, or glycan arrays, contain many different carbohydrate structures immobilized on a solid support in a spatially-controlled arrangement.[38–43] The miniaturized format allows for high-throughput analysis of binding while using only tiny amounts of each carbohydrate. Our group has developed a carbohydrate array composed of neoglycoproteins and natural glycoproteins.[28, 31, 44–47] For this study, an array with 170 components was used. It contained a variety of Tn peptides including serine and threonine variants, peptides with various amino acids on either side of the Tn antigen residue, and both single and clustered forms of Tn (see Figure 1). Besides variations in carbohydrate structure, the array contained conjugates of varying density as an added element of diversity. A full list of array components can be found in the supporting information (Table S1).
Serum was profiled on the array at a dilution of 1:2000 following the previously published procedure.[28] Overall, the induced antibodies displayed good selectivity for Tn-containing structures; however, some reactivity was observed for the corresponding TF peptide, Ac-S-TF(Ser)-S-G, and the related non-glycosylated peptide, Ac-S-S-S-G, in all rabbits. Binding was also observed to the parent peptide, Ac-S-T-S-G (IgG titers were as follows: rabbit1=256,000; rabbit2=12,800; rabbit3=128,000; rabbit4=32,000). Recognition of the TF peptide was dependent on the presence of the amino acid backbone, since binding was not observed to the TF disaccharide alone (see GA1 and GA1di; Figure 2). No binding was observed to other GalNAc terminal structures including blood group A [GalNAcα1-3(Fucα1-2)Gal], the A disaccharide (GalNAcα1-3Gal), or the Forssman disaccharide (GalNAcα1-3Gal).
Figure 2. Microarray profiles of induced IgG antibodies.

Colored boxes represent median fluorescence intensities after background correction and log2 transformation, with dark blue representing the highest intensity signals and white representing no signal. Rabbits 1 and 2 were vaccinated with low density Tn conjugate; Rabbits 3 and 4 were vaccinated with high density Tn conjugate. Selected array components are shown; full results and descriptions can be found in supporting information. The haptens used for vaccination are listed in purple.
Interestingly, variations in hapten density affected the spectrum of induced antibodies. Rabbits vaccinated with high density Tn (rabbit 3 and 4) produced antibodies that reacted with a broader variety of Tn containing peptides. Serum antibodies recognized both serine and threonine variants of the Tn antigen, all peptide sequence variants, and both single and clustered forms of the Tn antigen (see Figure 2). In addition, recognition of the monosaccharide, GalNAc-α, was also observed. Antibodies from rabbits vaccinated with low density Tn (rabbit 1 and 2) recognized fewer Tn variants and did not react with clustered Tn or GalNAc-α. The difference in binding to the clustered Tn antigen is especially important since this form of the antigen is reported to be highly tumor specific.[26, 27] Based on our results, the high density conjugate induced a broader spectrum of antibody reactivity than the low density conjugate. Full microarray profiling results are available in the supporting information (Table S2).
To gain more detailed information about the repertoire of antibodies induced by the low and high density conjugates, we carried out a series of inhibition studies on the array. The objective was to determine if the HSA conjugates induced a single population of polyspecific antibodies or multiple subpopulations of antibodies that bind a single glycan or a small subset of antigens. Briefly, various carbohydrate BSA-conjugates were pre-incubated with the serum at a dilution of 1:2000 and then profiled on the array as before. Both Ac-S-Tn(Thr)-S-G-4/BSA and Ac-S-Tn(Thr)-S-G-24/BSA were found to completely inhibit all signals on the array. In contrast, soluble Tn3-27/BSA and Ac-S-S-S-G-BSA each inhibited binding to the equivalent antigen on the array but displayed minimal or no inhibition of the other antigens (see Figure 3). The inhibition data indicates that vaccination induced multiple subpopulations of antibodies with distinct specificities. For example, the subset of antibodies that bound the clustered Tn antigen had little or no reactivity with AcS-S-S-G, while the subpopulation of antibodies that bound Ac-S-S-S-G had little or no reactivity with clustered Tn.
Figure 3. Inhibition of antibody binding.
Sera (1:2000) for rabbits 1–4 were pre-incubated with BSA conjugates (20 μg/mL; listed at the top of each graph) and then profiled on the array. Bars represent the percentage of inhibition. Antigens on the array are listed on the x-axis. “*” indicates no inhibition was possible.
Analysis of whole cell binding
To determine if the observed differences in specificity measured on the array correlate with recognition of the Tn antigen on cell surfaces, we evaluated binding to Jurkat cells using flow cytometry. Binding of serum antibodies was evaluated prior to vaccination and on day 51 for each rabbit. Rabbits 1, 2, and 3 had only modest increases in binding to Jurkat cells relative to pre-vaccination sera. In contrast, rabbit 4 had a substantial increase in whole cell binding. Since serum from rabbit 4 recognized the most glycans on the array but did not have a bigger antibody response to the hapten, the results suggest that a broader antibody response improves whole cell binding; however, additional studies will be needed to confirm this conclusion.
Conclusion
In this study, we demonstrate that hapten density can significantly affect the spectrum of antibodies induced by a conjugate. At low density, a narrower distribution is produced whereas at high density a broader spectrum of antibodies is produced. Therefore, modulation of hapten density provides a simple strategy to control the selectivity and breadth of the induced antibody repertoire. This can be useful for a variety of objectives, such as monoclonal antibody production and vaccine development, but the optimal hapten density will depend on the particular goals. Although this study focused on a single tumor-associated carbohydrate antigen, alterations of hapten density may also prove useful with other antigens. Finally, the study illustrates the utility of carbohydrate antigen arrays for providing a more comprehensive evaluation of immune responses.
Experimental Section
Chemicals and Reagents
Unless indicated, all chemicals and organic solvents were purchased from Sigma-Aldrich (St. Louis, MO) and used without further purification. Goat anti-rabbit IgG and IgM-alkaline phosphatase conjugates and TRITC-labeled goat anti-rabbit IgG were obtained from Southern Biotech (Birmingham, Al).
Synthesis of high and low density Tn immunogen
Glycopeptides were synthesized via solid phase peptide synthesis. Fmoc-protected Tn was obtained from Sussex Research (Ottawa, Canada). Conjugation of Tn acid 1 with HSA was carried out using NHS/EDC activation of the carboxylic acid. Briefly, acetyl-Ser-Tn(Thr)-Ser-Gly-aminohexanoic acid (Ac-S-Tn(Thr)-S-G, 150 mM in H2O), NHS (300mM in DMF), and EDC (300 mM in 50/50 H2O/ DMF) were combined in a 2:1:1 ratio and allowed to react for 1 h at r.t. to pre-form the NHS ester in situ. The NHS ester was then added to a solution of BSA (4 mg/mL in in 10 mM sodium borate, 90 mM NaCl, pH 8.0) pre-cooled to 4 °C. The reaction was allowed to stand at 4 °C for 5 min, warmed to r.t., and then allowed to stand for 2 h. Conjugates were dialyzed (SpectraPor7, MWCO 10,000, Spectrum Laboratories, Inc. Rancho Dominguez, CA) against 5 mM aqueous NaCl and then analyzed by MALDI-MS to determine the average number of haptens per HSA. HSA conjugates were sterile filtered through a 0.2μm filter. BSA conjugates were prepared using the same procedure.
Serum antibody production
The rabbits were vaccinated at ProSci Inc. (Poway, CA). After initial pre-immune serum collection, New Zealand white rabbits were immunized subcutaneously with a 1:1 mixture of either low or high density HSA conjugate (200 μg; 1mg/mL in PBS) and complete Freund’s adjuvant (CFA). The rabbits were boosted on days 21 and 42 with a 1:1 mixture of antigen (100 μg) and Incomplete Freund’s adjuvant (IFA). Serum was collected on day 51.
Enzyme-Linked ImmunoSorbent Assay (ELISA)
384-well microtiter plates (Nunc, Roskilde, Denmark) were coated with 25 μL of Ac-S-Tn(Thr)-S-G-4/BSA (10 μg/mL) or Ac-S-Tn(Thr)-S-G-24/BSA (10 μg/mL) at 4 °C overnight followed by 100 μL of 3% BSA/PBS blocking buffer for 2 h at r.t. Serial dilutions of rabbit serum (35 μL diluted in 1% BSA/PBS) were incubated for 2 h at r.t. followed by 1 h incubation with 40 μL of goat anti-rabbit IgG (1:1000) or IgM (1:1000) alkaline phosphatase conjugate. Next, 50 μL of 4-methylumbelliferyl phosphate substrate solution [10mM in Tris (10mM Tris-HCl, 90mM NaCl, pH 9.0)] was added to each well and signals were monitored over 30 min using a FLx800 microplate fluorescence reader (Bio-Tek instruments Inc., Winooski, VT). The titer was defined as the largest dilution that produced a signal 3 times higher than the background after 30 min and was based on triplicate experiments.
Carbohydrate microarray assay
The carbohydrate microarrays were prepared as previously described with only minor modifications.[32, 48] Briefly, samples (125 μg/mL) were printed in duplicate on SuperEpoxy 2 Protein glass slides (TeleChem International, Inc., Sunnyvale, CA) using a Biorobotics MicroGrid II microarrayer (Genomic Solutions, Ann Arbor, MI) fitted with Stealth pins (Telechem International; #SMP3 which produce ~100 μm spots). 16 complete arrays were printed on each slide. The list of 170 different glycoprotein or glycoconjugate samples can be found in the supporting information (Table S1). Slides were kept at −20°C till use.
Serum antibody binding was evaluated using minor modifications of the previously reported protocol.[28] Slides were fitted with a 16-well chamber (Grace Bio-Labs, Inc., Bend, OR) to produce 16 subarrays. After blocking with 3%BSA/PBS (200 μL, per subarray well) at 4 °C overnight. Rabbit serum at a dilution of 1:2000 (diluted in 1%BSA/1%HSA/PBS 0.05% tween 20 buffer) were pre-incubated 1 h to remove non-specific antibodies generated by HSA before adding to the array wells for 2 h incubation at r.t. After washing with PBS 0.05% tween 20 buffer, bound antibody was detected with 100 μL of TRITC-conjugated goat anti-rabbit IgG (diluted 1:1000 in 1% BSA/PBS) for 2 h. Slides were washed with PBS 0.05% tween 20 buffer, dried by centrifugation at 900 × g in a 50 mL conical tube, and then scanned using a GenePix Scanner 4000B (Molecular Devices Corporation, Union City, CA) at pmt 480 for emission wavelength 532 nm. Genepix Pro 6.0 software was used for data processing. Background corrected median fluorescence intensities for each component were obtained. The average of the two spots for each array component was used as the final value. The data was logarithmic transformed (log2) and a heatmap (Figure 2) was generated using JcolorGrid[49] based on duplicate experiments. Data inflection point was 10 and data increment was 0.25.
For the inbihition assay, rabbit serum (diluted at 1:2000) was incubated with 20 μg/mL Ac-S-Tn(Thr)-S-G-4/BSA, Ac-S-Tn(Thr)-S-G-24/BSA, clustered Tn (Tn3)-27/BSA or Ac-S-S-S-G-BSA at r.t. for 1 h prior to adding into the well followed by the routine array assay procedure as before. Percentage of inhibition was determined by the average of duplicate experiments. Tabulated data can be found in the supporting information (Table S3).
Whole cell binding assay
Jurkat cells were obtained from ATCC (Rockville, MD) and cultured under standard conditions. To test serum binding, cells were counted using a hemacytometer. Cells were collected by centrifugation at 1100 rpm for 6 min, washed with complete media, and resuspended in SFM containing 0.1% BSA. 1 – 6 × 105 cells were aliquoted into individual eppendorf tubes. Serum samples were diluted 1/20 into the cell suspension, mixed, and incubated 30–60 min at 4 °C with rotation. Cells were collected at 4000 rpm for 2 minutes and washed twice with cold PBS. Donkey anti-rabbit AlexaFluor-488 was added to the suspended cells, incubated for 20 minutes at 4 °C with rotation. Cells were washed twice with cold PBS, once with PBS/EDTA, and resuspended in PBS/BSA. Cells were analyzed for fluorescence by Flow Cytometry.
Supplementary Material
Figure 4. Serum binding to Jurkat cells.

Rabbit sera before and after vaccination was evaluated for binding to Jurkat cells in a fluorescence-based assay using anti-rabbit Alexafluor-488 secondary antibodies. Bars represent the mean fluorescence per event (± SD). Binding of rabbit sera (rabbits 1–4), autofluorescence (cells only), and non-specific binding of the secondary antibody (2°) were measured. PolyTn, an affinity purified polyclonal antibody to Tn, was used as a positive control.
Acknowledgments
We thank Jack Simpson (Protein Chemistry Laboratory, SAIC/NCI-Frederick) for MALDI-MS analysis of HSA and BSA conjugates. We thank ProSci, Inc. for vaccinations, care, and handling of the rabbits. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Supporting information for this article is available on the WWW under http://www.chembiochem.org or from the author.
References
- 1.Desaymard C, Howard JG. Eur J Immunol. 1975;5:541–545. doi: 10.1002/eji.1830050807. [DOI] [PubMed] [Google Scholar]
- 2.Bachmann MF, Rohrer UH, Kundig TM, Burki K, Hengartner H, Zinkernagel RM. Science. 1993;262:1448–1451. doi: 10.1126/science.8248784. [DOI] [PubMed] [Google Scholar]
- 3.Jegerlehner A, Storni T, Lipowsky G, Schmid M, Pumpens P, Bachmann MF. Eur J Immunol. 2002;32:3305–3314. doi: 10.1002/1521-4141(200211)32:11<3305::AID-IMMU3305>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 4.Liu W, Peng Z, Liu Z, Lu Y, Ding J, Chen YH. Vaccine. 2004;23:366–371. doi: 10.1016/j.vaccine.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 5.Pedersen MK, Sorensen NS, Heegaard PMH, Beyer NH, Bruun L. J Immunol Methods. 2006;311:198–206. doi: 10.1016/j.jim.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Puffer EB, Pontrello JK, Hollenbeck JJ, Kink JA, Kiessling LL. ACS Chem Biol. 2007;2:252–262. doi: 10.1021/cb600489g. [DOI] [PubMed] [Google Scholar]
- 7.Robinson WH. Curr Opin Chem Biol. 2006;10:67–72. doi: 10.1016/j.cbpa.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 8.Wu CY, Liang PH, Wong CH. Org Biomol Chem. 2009;7:2247–2254. doi: 10.1039/b902510n. [DOI] [PubMed] [Google Scholar]
- 9.Song EH, Pohl NLB. Curr Opin Chem Biol. 2009;13:626–632. doi: 10.1016/j.cbpa.2009.09.021. [DOI] [PubMed] [Google Scholar]
- 10.Oyelaran O, Gildersleeve JC. Curr Opin Chem Biol. 2009;13:406–13. doi: 10.1016/j.cbpa.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Palma AS, Feizi T. Biol Chem. 2009;390:647–656. doi: 10.1515/BC.2009.071. [DOI] [PubMed] [Google Scholar]
- 12.Springer GF. Science. 1984;224:1198–1206. doi: 10.1126/science.6729450. [DOI] [PubMed] [Google Scholar]
- 13.Springer GF. J Mol Med. 1997;75:594–602. doi: 10.1007/s001090050144. [DOI] [PubMed] [Google Scholar]
- 14.Springer GF, Desai PR, Spencer BD, Tegtmeyer H, Carlstedt SC, Scanlan EF. Cancer Detect Prev. 1995;19:374–380. [PubMed] [Google Scholar]
- 15.Slovin SF, Ragupathi G, Musselli C, Olkiewicz K, Verbel D, Kuduk SD, Schwarz JB, Sames D, Danishefsky SJ, Livingston PO, Scher HI. J Clin Oncol. 2003;21:4292–4298. doi: 10.1200/JCO.2003.04.112. [DOI] [PubMed] [Google Scholar]
- 16.Slovin SF, Ragupathi G, Fernandez C, Diani M, Jefferson MP, Wilton A, Kelly WK, Morris M, Solit D, Clausen H, Livingston P, Scher HI. Cancer Immunol Immunother. 2007;56:1921–30. doi: 10.1007/s00262-007-0335-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cipolla L, Rescigno M, Leone A, Peri F, La Ferla B, Nicotra F. Bioorg Med Chem. 2002;10:1639–1646. doi: 10.1016/s0968-0896(01)00433-3. [DOI] [PubMed] [Google Scholar]
- 18.Vichier-Guerre S, Lo-Man R, BenMohamed L, Deriaud E, Kovats S, Leclerc C, Bay S. J Peptide Res. 2003;62:117–124. doi: 10.1034/j.1399-3011.2003.00074.x. [DOI] [PubMed] [Google Scholar]
- 19.Keding SJ, Danishefsky SJ. Proc Natl Acad Sci USA. 2004;101:11937–11942. doi: 10.1073/pnas.0401894101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo-Man R, Vichier-Guerre S, Perrsaut R, Deriaud E, Huteau V, BenMohamed L, Diop OM, Livingston PO, Bay S, Leclerc C. Cancer Res. 2004;64:4987–4994. doi: 10.1158/0008-5472.CAN-04-0252. [DOI] [PubMed] [Google Scholar]
- 21.Dziadek S, Hobel A, Schmitt E, Kunz H. Angew Chem Int Ed Engl. 2005;44:7630–7635. doi: 10.1002/anie.200501594. [DOI] [PubMed] [Google Scholar]
- 22.Kagan E, Ragupathi G, Yi SS, Reis CA, Gildersleeve J, Kahne D, Clausen H, Danishefsky SJ, Livingston PO. Cancer Immunol Immunother. 2005;54:424–430. doi: 10.1007/s00262-004-0584-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ingale S, Wolfert MA, Gaekwad J, Buskas T, Boons GJ. Nat Chem Biol. 2007;3:663–667. doi: 10.1038/nchembio.2007.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sorensen AL, Reis CA, Tarp MA, Mandel U, Ramachandran K, Sankaranarayanan V, Schwientek T, Graham R, Taylor-Papadimitriou J, Hollingsworth MA, Burchell J, Clausen H. Glycobiology. 2006;16:96–107. doi: 10.1093/glycob/cwj044. [DOI] [PubMed] [Google Scholar]
- 25.Kunz H, Birnbach S, Wernig P. Carbohydr Res. 1990;202:207–223. doi: 10.1016/0008-6215(90)84081-5. [DOI] [PubMed] [Google Scholar]
- 26.Nakada H, Numata Y, Inoue M, Tanaka N, Kitagawa H, Funakoshi I, Fukui S, Yamashina I. J Biol Chem. 1991;266:12402–12405. [PubMed] [Google Scholar]
- 27.Nakada H, Inoue M, Numata Y, Tanaka N, Funakoshi I, Fukui S, Mellors A, Yamashina I. Proc Natl Acad Sci. 1993;90:2495–2499. doi: 10.1073/pnas.90.6.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manimala JC, Roach TA, Li Z, Gildersleeve JC. Glycobiology. 2007;17:17C–23C. doi: 10.1093/glycob/cwm047. [DOI] [PubMed] [Google Scholar]
- 29.Tarp MA, Sorensen AL, Mandel U, Paulsen H, Burchell J, Taylor-Papadimitriou J, Clausen H. Glycobiology. 2007;17:197–209. doi: 10.1093/glycob/cwl061. [DOI] [PubMed] [Google Scholar]
- 30.Oppezzo P, Osinaga E, Tello D, Bay S, Cantacuzene D, Irigoin F, Ferreira A, Roseto A, Cayota A, Alzari P, Pritsch O. Hybridoma. 2000;19:229–239. doi: 10.1089/02724570050109620. [DOI] [PubMed] [Google Scholar]
- 31.Li Q, Anver MR, Butcher DO, Gildersleeve JC. Mol Cancer Res. 2009;8:971–9. doi: 10.1158/1535-7163.MCT-08-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oyelaran OO, Li Q, Farnsworth DF, Gildersleeve JC. J Proteome Res. 2009;8:3529–38. doi: 10.1021/pr9002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Iwasaki H, Wang H, Kudo T, Kalka TB, Hennet T, Kubota T, Cheng L, Inaba N, Gotoh M, Togayachi A, Guo J, Hisatomi H, Nakajima K, Nishihara S, Nakamura M, Marth JD, Narimatsu H. J Biol Chem. 2003;278:573–584. doi: 10.1074/jbc.M203094200. [DOI] [PubMed] [Google Scholar]
- 34.Yin BWT, Lloyd KO. J Biol Chem. 2001;276:27371–27375. doi: 10.1074/jbc.M103554200. [DOI] [PubMed] [Google Scholar]
- 35.Pratt WS, Crawley S, Hicks J, Ho J, Nash M, Kim YS, Gum JR, Swallow DM. Biochem Biophys Res Comm. 2000;275:916–923. doi: 10.1006/bbrc.2000.3406. [DOI] [PubMed] [Google Scholar]
- 36.Parry S, Sutton-Smith M, Heal P, Leir SH, Palmai-Pallag T, Morris HR, Hollingsworth MA, Dell A, Harris A. Biochim Biophys Acta. 2005;1722:77–83. doi: 10.1016/j.bbagen.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 37.Krantz MJ, Weiner JW, Liu HH. Biochemistry. 1976;15:3963–3968. doi: 10.1021/bi00663a009. [DOI] [PubMed] [Google Scholar]
- 38.Paulson JC, Blixt O, Collins BE. Nat Chem Biol. 2006;2:238–248. doi: 10.1038/nchembio785. [DOI] [PubMed] [Google Scholar]
- 39.Oyelaran O, Gildersleeve JC. Expert Rev Vaccines. 2007;6:957–69. doi: 10.1586/14760584.6.6.957. [DOI] [PubMed] [Google Scholar]
- 40.Liang PH, Wu CY, Greenberg WA, Wong CH. Curr Opin Chem Biol. 2008;12:86–92. doi: 10.1016/j.cbpa.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laurent N, Voglmeir J, Flitsch SL. Chem Comm. 2008:4400–12. doi: 10.1039/b806983m. [DOI] [PubMed] [Google Scholar]
- 42.Park S, Lee M-R, Shin I. Chem Comm. 2008:4389–99. doi: 10.1039/b806699j. [DOI] [PubMed] [Google Scholar]
- 43.Horlacher T, Seeberger PH. Chem Soc Rev. 2008;37:1414. doi: 10.1039/b708016f. [DOI] [PubMed] [Google Scholar]
- 44.Manimala J, Li Z, Jain A, VedBrat S, Gildersleeve JC. ChemBioChem. 2005;6:2229–2241. doi: 10.1002/cbic.200500165. [DOI] [PubMed] [Google Scholar]
- 45.Manimala JC, Roach TA, Li ZT, Gildersleeve JC. Angew Chem Int Ed. 2006;45:3607–3610. doi: 10.1002/anie.200600591. [DOI] [PubMed] [Google Scholar]
- 46.Gildersleeve JC, Oyelaran O, Simpson JT, Allred B. Bioconjugate Chem. 2008;19:1485–90. doi: 10.1021/bc800153t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsu KL, Gildersleeve JC, Mahal LK. Mol BioSyst. 2008;4:654–662. doi: 10.1039/b800725j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oyelaran O, McShane LM, Dodd L, Gildersleeve JC. J Proteome Res. 2009;8:4301–10. doi: 10.1021/pr900515y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joachimiak MP, Weisman JL, May BCH. BMC Bioinformatics. 2006;7 doi: 10.1186/1471-2105-7-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.