Abstract
The p90 ribosomal S6 family of kinases (RSK) are potential drug targets, due to their involvement in cancer and other pathologies. There are currently only two known selective inhibitors of RSK, but the basis for selectivity is not known. One of these inhibitors is a naturally occurring kaempferol-α-L-diacetylrhamnoside, SL0101. Here, we report the crystal structure of the complex of the N-terminal kinase domain of the RSK2 isoform with SL0101 at 1.5 Å resolution. The refined atomic model reveals unprecedented structural reorganization of the protein moiety, as compared to the nucleotide-bound form. The entire N-lobe, the hinge region and the αD-helix undergo dramatic conformational changes resulting in a rearrangement of the nucleotide binding site with concomitant formation of a highly hydrophobic pocket spatially suited to accommodate SL0101. These unexpected results will be invaluable in further optimization of the SL0101 scaffold as a promising lead for a novel class of kinase inhibitors.
Protein phosphorylation is a key regulatory mechanism in all eukaryotic cells. The phosphorylation of either Ser/Thr or Tyr residues on target proteins is catalyzed in humans by 518 protein kinases, collectively known as the human kinome.1 Intense interest in the structure and function of protein kinases is primarily driven by their potential as drug targets, particularly in cancer therapy. Eleven drugs acting on protein kinases have already been approved by the FDA, and approximately 150 other small molecule inhibitors are at various stages of clinical development.2, 3
One of the families of Ser/Thr protein kinases recently recognized for their potential as drug targets are the p90 ribosomal S6 (RSK) kinases.4 The four known RSK isoforms (RSK1–4), as well as the related mitogen- and stress-activated kinases MSK1 and MSK2, display unique molecular architecture, whereby their polypeptide chains contain two distinct kinase domains in tandem, i.e. the regulatory C-terminal kinase domain (CTKD), and the physiologically active N-terminal kinase domain (NTKD).4–6 The activation of RSK kinases involves several phosphorylation steps, initiated by the docking of extracellular signal-regulated kinases (ERK1/2) at the C-terminus with subsequent phosphorylation and activation of CTKD, intramolecular phosphorylation of the interdomain linker by CTKD, the recruitment of phosphoinositide-dependent kinase 1 (PDK1) to this newly phosphorylated site, and finally PDK1-dependent phosphorylation and activation of the NTKD.4 Two of the most ubiquitously expressed isoforms, i.e. RSK1 and RSK2, are increasingly attracting attention because of their involvement in various cancers.7–15 Further, RSK1 has been implicated in ischaemia/reperfusioninduced injury.16 Hence, there is considerable interest in the development of RSK specific inhibitors, particularly those selective for the physiologically important NTKD.7, 8, 17
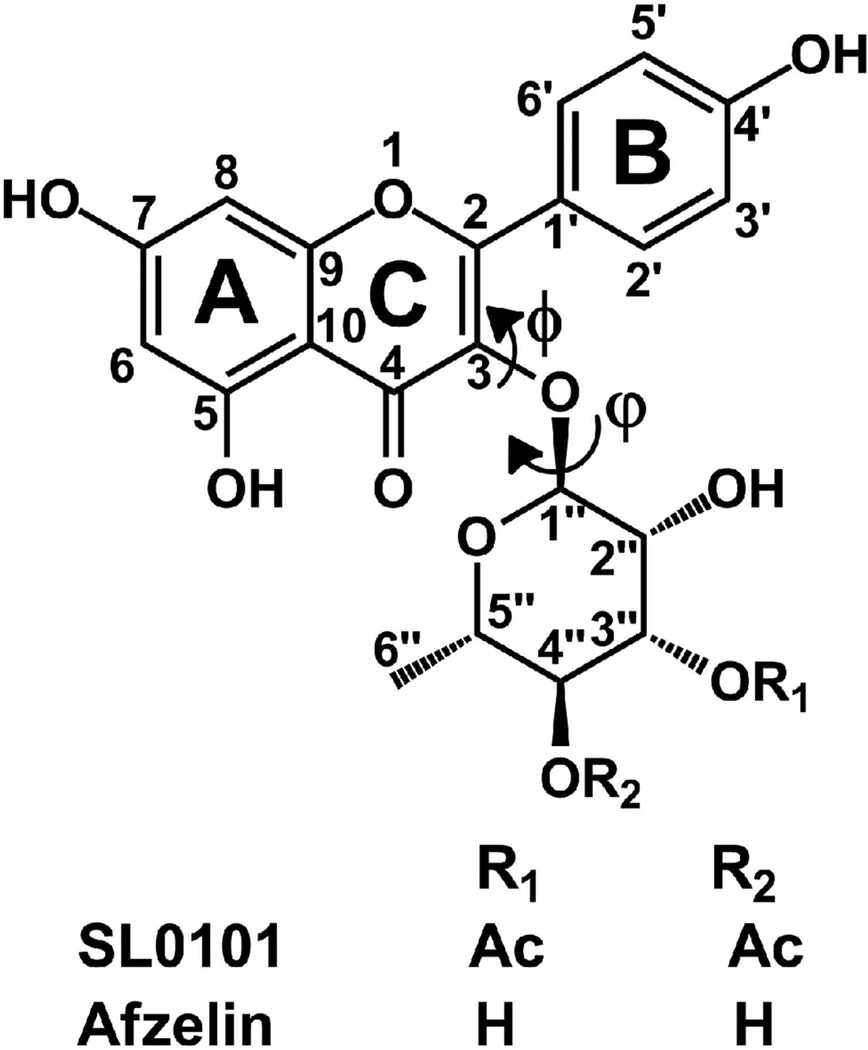
Only two inhibitors with relative specificity for the RSK family have been described to date: BI-D1870, derived from the pyrido[2,3-d]pyridimidine group of Src inhibitors18, 19, and a naturally occurring compound, SL0101, identified in extracts of a tropical plant Forsteronia refracta.9 The latter is a flavonol glycoside, or more specifically kaempferol-3-O-(3”,4”-di-O-acetyl-α-L-rhamnopyranoside (Figure 1). Flavonols, ubiquitous compounds synthesized by plants as antioxidants, constitute an important part of human diet, with up to ~50 mg ingested daily. Flavonols are currently under intense scrutiny as key factors responsible for the reduced risk of cancer and cardiovascular disease.20–22 Several flavonols, such as quercetin, kaempferol, fisetin and others, are either sold as food supplements or constitute vital ingredients of such supplements.
Figure 1.
The chemical structure of the SL0101 inhibitor (kaempferol-3-O-(3”,4”-di-O-acetyl-α-L-rhamnopyranoside) and its analogue afzelin.
Importantly, flavonols are now also recognized as inhibitors of kinases.20, 23, 24 For example, kaempferol inhibits myosin light chain kinase25 and phosphatidylinositol-3-kinase26, quercetin has been found to inhibit 16 kinases relevant to cancer cell growth23, myricetin inhibits Akt27 and phosphoinositide-3-kinase28, while fisetin inhibits Akt and the rapamycin kinase (mTOR).29 The inhibitory activity is due to the fact that the planar benzopyran moiety of the flavonol is able to compete with ATP, by mimicking the purine heterocycle. However, low affinities of such interactions and poor selectivity have traditionally rendered the flavonol scaffold as unattractive for further drug development.
Flavonols are typically synthesized in plants as either 7-O or 3-O glycosides, which in general limits their ability to inhibit kinases. However, at least some glycosides do show inhibitory activity. For example, luteolin-7-O-glucoside, has been shown to be a highly specific inhibitor of JNK3 kinase.30 We were therefore intrigued by the molecular basis of the specificity of SL0101 towards the RSK family.
Fortuitously, crystal structures of NTKDs from both RSK1 and RSK2 have been determined: the human RSK1NTKD, residues 33 – 353, has had its structure determined in complexes with AMP–PCP, staurosporine, and purvalnol A31, while the structure of mouse RSK2NTKD was determined with bound ATP surrogate – AMP-PNP.32 In general terms, both crystal structures show a typical molecular architecture of AGC kinases6, with distinct N- and C-terminal subdomains, or lobes. The N-terminal lobe (the N-lobe), contains a five-stranded, antiparallel β-sheet, with a distinctive, flexible P-loop (also referred to as the Gly-rich loop) between strands β1 and β2. The N-lobe is involved primarily in ATP/Mg2+-binding and is subject to regulatory phenomena. The larger, C-terminal lobe (C-lobe) contains a rigid, very stable core made up of six α-helices (αD-αI); this lobe contains the substrate binding site and most of the catalytic machinery.33 The ATP/Mg2+-binding site is located within a large cleft between the two lobes.
Unexpectedly, the crystal structure of the mRSK2NTKD in complex with AMP-PNP32 revealed some unusual features of the N-lobe. Specifically, the stretch normally folded in the N-lobe of canonical protein kinases into the αB-helix (residues 104 to 109), in mRSK2NTKD forms a β-strand (denoted the βB strand), which along with an N-terminal fragment extraneous to the canonical kinase domain (residues 48 to 52), and a segment immediately downstream of the DFG motif (residues 214 to 217) assemble into a three stranded β-sheet. Further, a portion of the αC-helix is disordered, which is expected to impair the catalytic function. A similar structure of the N-lobe, has been previously reported for the mitogen- and stress-activated protein kinase MSK134, and was interpreted as a novel autoinhibited conformation. This raised the possibility that the RSK-specific inhibitors achieve selectivity by binding to a unique, inactive conformation.
To address this issue, we solved the crystal structure of mouse RSK2 NTKD (mRSK2NTKD) with SL0101. As there has been evidence that the two acetyl groups on rhamnose enhance the binding35, we also solved the crystal structure of mRSK2NTKD with deacetylated SL0101 (kaempferol-3-O-α-L-rhamnopyranoside, or afzelin). The two complexes have virtually identical structures, except for the absence of the acetyl groups in afzelin. Surprisingly, we find that the inhibition of mRSK2NTKD by SL0101 or afzelin, is associated with dramatic, unprecedented structural rearrangements within the protein moiety, when compared to the AMP-PNP-bound form. This work provides novel and unexpected insights into the mechanism of kinase inhibition and constitutes vivid illustration of the dangers of in silico predictions of protein-inhibitor interactions, based on insufficient or inadequate structural information.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The N-terminal domain of murine RSK2 encompassing amino acids 47–346 (mRSK2NTKD) was cloned into pHisUni136 vector using BamHI and SalI restriction sites. Because BamHI site encodes amino acids Gly and Ser which are also found in positions 45 and 46 of mRSK2, identity of the cloned fragment to murine RSK2 starts with Gly45. Point mutants of RSK2 were generated as described elsewhere37 with the use of the Phusion™ polymerase. E.coli BL21(RIPL) cells were transformed with mRSK2NTKD expression construct and grown in Terrific Broth (TB) media in the presence of 100 µg/ml ampicilin until reaching OD600 of 4–4.5. Thereafter the temperature was lowered to 16 °C, protein expression was induced by the addition of IPTG to a final concentration of 0.3 mM and carried overnight. Cells were harvested by centrifugation and disrupted by high pressure homogenization in the buffer containing 50 mM Tris pH 8.0 and 500 mM NaCl (Buffer A). RSK2 was purified using His-Select nickel resin (Sigma), eluted with Buffer A containing 200 mM imidazole and digested with rTEV protease overnight with concomitant dialysis against Buffer A containing 5 mM 2-mercaptoethanol. Dialyzed sample was passed through the 1 mL His-Select column, purified by size exclusion on Sephadex 200 column and concentrated to 6–8 mg/mL. The obtained protein was mixed with SL0101 (20 mM stock solution in ethylene glycol) or afzelin (20 mM aqueous solution in 100 mM sodium acetate) using about 10% excess of ligands, dialyzed against the Buffer A containing 5 mM 2-mercaptoethanol and 5 mM EDTA and used for crystallization setups.
Inhibitors
SL0101 was synthesized as described elsewhere.38 Deacyl-SL0101 (afzelin) was obtained by incubating SL0101 solution with 5 molar equivalents of NaOH at room temperature for 1 hr followed by neutralization of solution with 3 molar equivalents of acetic acid.
Crystallization and Structure Determination
Crystals of mRSK2NTKD-SL0101 complex and isomorphous crystals of mRSK2NTKD-afzelin complex grew in 2–3 days at room temperature from vapor diffusion setups consisting of equal volumes (250 nL) of the complex solution and a reservoir buffer containing 0.1 M HEPES pH 7.5 and 30% of Jeffamine ED2003. Crystals were harvested in reservoir buffer and flash cooled in liquid nitrogen. Single wavelength (λ = 1.000 Å) X-ray diffraction data were collected at 100 K at Southeast Regional Collaborative Access Team (SER-CAT) 22-BM beamline at the Advanced Photon Source, Argonne National Laboratory. Data were indexed, integrated and scaled with HKL2000.39 R-free was monitored by setting aside 5% of reflections as test set. Initial phase estimates were obtained by automated molecular replacement with BALBES.40 Large part of the model was automatically built with ARP/wARP41 and further improved manually with COOT42. Restrained positional and isotropic atomic displacement parameters (ADP) refinement was performed with PHENIX.43 CIF dictionaries for SL0101 or afzelin were generated with eLBOW using structure of trifolin (kaempferol-3-O-β-galactopyranoside)44 and used to refine positions of ligands in unaccounted electron density. A Ramachandran plot calculated with PROCHECK45 indicated that 97.6% and 2.4% of all non-Gly and non-Pro residues lie in most favored and additional allowed regions. Data collection and refinement statistics are listed in Table 1. Figures were prepared using PYMOL (http://www.pymol.org/).
Table 1.
Data collection and refinement statisticsa
| RSK2-SL0101 | RSK2-Afzelin | |
|---|---|---|
| Data collection | ||
| Space group | C2 | C2 |
| Cell dimensions | ||
| a, b, c (Å) | 98.45, 40.70, 83.35 | 99.12, 40.86, 83.87 |
| β (°) | 114.54 | 114.65 |
| Resolution (Å) | 1.53 (1.56–1.53)b | 1.55 (1.58–1.55) |
| Number of unique reflections | 46201 (2182) | 43194 (1739) |
| Rsym | 0.062 (0.471) | 0.053 (0.355) |
| <I /σ(I)> | 24.0 (2.2) | 23.2 (2.0) |
| Completeness (%) | 99.5 (95.1) | 96.8 (76.9) |
| Redundancy | 4.5 (2.7) | 4.1 (2.2) |
| Refinement | ||
| Resolution (Å) | 1.53 | 1.55 |
| No. reflections | 44706 | 41260 |
| Rwork / Rfree | 0.19/0.22 | 0.20/0.23 |
| No. atoms | ||
| Protein (non-hydrogen) | 2364 | 2354 |
| Inhibitor (all atoms) | 60 | 50 |
| Water | 337 | 285 |
| B-factors (Å2) | ||
| Protein | 26.1 | 26.4 |
| Inhibitor | 26.0 | 22.9 |
| Water | 34.5 | 32.8 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.006 | 0.006 |
| Bond angles (°) | 0.97 | 0.94 |
Each dataset was collected from one native crystal
Values in parentheses are for highest-resolution shell.
Isothermal Titration Calorimetry
Isothermal titration calorimetry (ITC) was performed at 25 °C using a Microcal ITC-200 instrument (MicroCal, Northhampton, MA). The mRKS2NTKD samples were dialyzed against buffer A containing 5 mM β-mercaptoethanol prior to the experiment and all ligands were dissolved in the same buffer. Contents of the sample cell were stirred continuously at 700 rpm during the experiment. A typical titration of mRSK2NTKD involved 18–22 injections of SL0101 (0.5 mM, 2 µl, 4 minute intervals) or AMPPNP (0.9 mM, 2.4 µl, 3 minute intervals) into a sample cell containing 0.2 ml of NTKDRSK2 (60–150 µM). The baseline-corrected data were analyzed with Microcal Origin 5.0 software to determine the enthalpy change (ΔH), the association constant (Ka) and the stoichiometry of binding (n) by fitting to the association model for single set of identical sites.
Thermal Shift Assay
Melting temperatures for WT and F79A mutant of mRSK2NTKD were obtained by the thermal shift assay (TSA).46 The assay was performed using StepOnePlus™ Real-Time PCR instrument. Protein samples were diluted to 1.1 mg/ml in a buffer A containing 5 mM β-mercaptoethanol. The protein samples were mixed with 5× SYPRO Orange dye (Molecular Probes, S6650) with a ratio of 5:1 in a 20 µl reaction volume. Temperature range was 20° C to 95° C in 1° C steps. At each step, fluorescence was measured after excitation at 480 nm. Melting curves were calculated using the StepOnePlus™ software. The melting curve minima were calculated using derivative of the normalized fluorescence measured at 520 nm wavelength and represent the half-maximal fluorescence and the stability of the protein sample.
Enzyme Activity Assay
Wild type mRSK2NTKD or F79A point mutant were diluted to 1 µM with kinase buffer (20 mM Tris-HCl pH-7.5, 1 mM MgCl2, 25 mM KCl, 1 mM DTT and 100 µM [γ-32P]ATP) and incubated with 100 nM of PDK1 (Promega) at 25°C for 20 minutes. Kinase activity was assayed using myelin basic protein (MBP) as a substrate (2.75 µM) in the presence of varying amounts of SL0101. The reaction was initiated by the addition of activated kinase to the substrate and carried out for 60 minutes at 25° C with frequent mixing. The reaction was stopped by the addition of SDS-PAGE sample buffer. Samples were separated on 15% SDS-PAGE gel, stained with Coomassie Blue, dried onto Whatman paper together with aliquots of [γ-32P]ATP and exposed to Molecular Dynamics Phosphor Screen overnight. Storm 860 phospho-scanner, by Molecular Dynamics, was used to scan Phosphor Screen and the resulting images were processed with ImageQuant software to calculate amounts of PO43− incorporated into proteins.
RESULTS
Overview
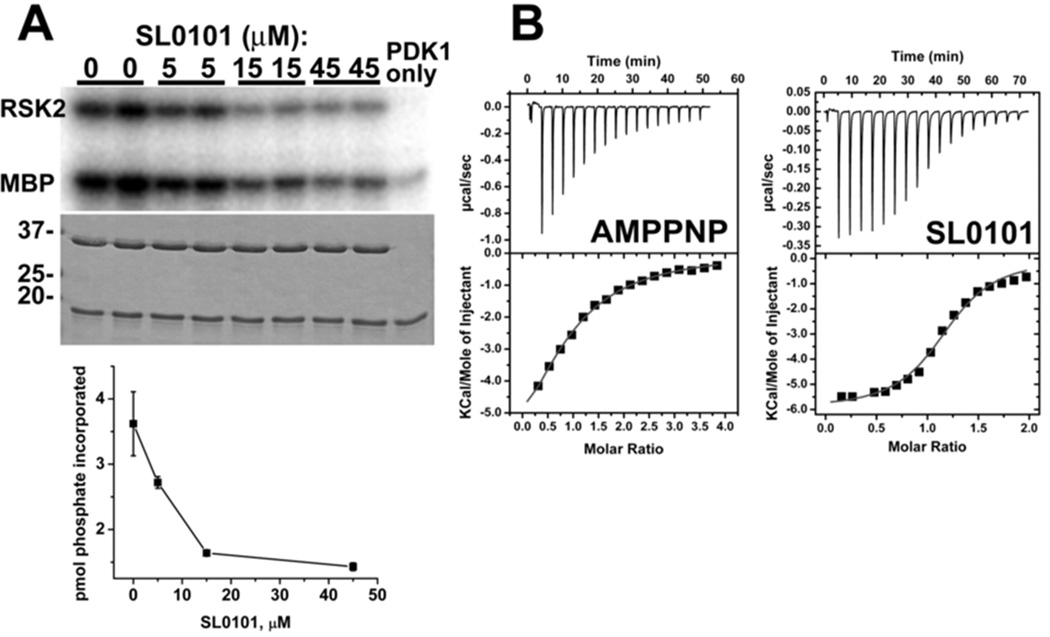
The mRSK2NTKD domain, encompassing residues 45–346 was expressed in E.coli and purified (see Experimental Procedures). This construct contains the canonical kinase domain and a short N-terminal extension which was found to be folded and to contain a β-strand incorporated into the atypical 3-stranded sheet in the complex of mRSK2NTKD with AMP-PNP.32 In agreement with the data reported for the mRSK2NTKD construct encompassing residues 1–373,47 our recombinant, isolated kinase domain has no measurable catalytic activity (data not shown). However, upon incubation with PDK1, which phosphorylates the activation (or T) loop on Ser 227,48 mRSK2NTKD shows detectable activity that is inhibited, as expected, by SL0101 (Figure 2A). Isothermal titration calorimetry (ITC) shows that even the inactive, unphosphorylated protein binds AMP-PNP and SL0101 with 50 µM and 2.9 µM dissociation constants (KD), respectively (Figure 2B). The latter value is in agreement with estimates obtained for the activated full-length, wild-type RSK2 kinase,9 and attests to the fact that the isolated N-terminal kinase domain of RSK2 is a good model for the action of SL0101 on the full-length protein. The crystal structures of the complexes of mRSK2NTKD with SL0101 and afzelin were refined at 1.53 Å, and 1.55 Å resolution, respectively (Table 1). Each complex was co-crystallized individually, but the corresponding crystals are isomorphous, with the protein moieties virtually identical within experimental error. Given this result, our description refers hereafter to the mRSK2NTKD/SL0101 complex.
Figure 2.
Functional properties of the recombinant mRSK2NTKD.
(A) Inhibition by SL0101 of the catalytic activity of the phosphorylated protein. The kinase assays were performed in the presence of indicated amounts of SL0101 using myelin basis protein (MBP) as a substrate. Incorporated phosphate was analyzed by autoradiography of gel samples and proteins were visualized by Coomassie Blue staining (shown beneath corresponding autoradiography images). Lower panel: kinase activity data (averaged from duplicate measurements) were expressed as picomoles of incorporated phosphate and plotted against SL0101 concentration. The decreasing amount of phosphorylated mRSK2NTKD following incubation with SL0101 suggests that autophosphorylation of the kinase is also subject to inhibition by SL0101.
(B) ITC profiles of the interaction of AMP-PNP and SL0101 with wild type mRSK2NTKD. The individual dissipated heats were plotted against the molar ratio of interacting partners (squares) to estimate thermodynamic parameters. The data were fitted with a “one set of sites model” shown as a solid red line on lower panels. The best fits resulted in Kd = 50 µM, n = 0.85, ΔH = −9.1 kcal/mol and ΔS = −10.9 cal/mol/deg (TΔS = −3.2 kcal/mol) for AMP-PNP (ΔG = −5.9 kcal/mol) ; and Kd = 2.9 µM, n = 1.16, ΔH = −5.9 kcal/mol and ΔS = 4.9. cal/mol/deg (TΔS = 1.4 kcal/mol) for SL0101(ΔG = − 7.3 kcal/mol). Only representative experiments are shown out of several, which all yielded reproducible results.
To solve the structure of the two mRSK2NTKD complexes we used the automated molecular replacement system BALBES.40 Using the template of the known structure of mRSK2NTKD with AMP-PNP,32 BALBES was able to locate correctly the C-lobe using MOLREP49, while the N-lobe was rebuilt by ARP/wARP41 with partial refinement with REFMAC550. The inhibitors were built manually (see Experimental Procedures). Crystallographic details are shown in Table 1.
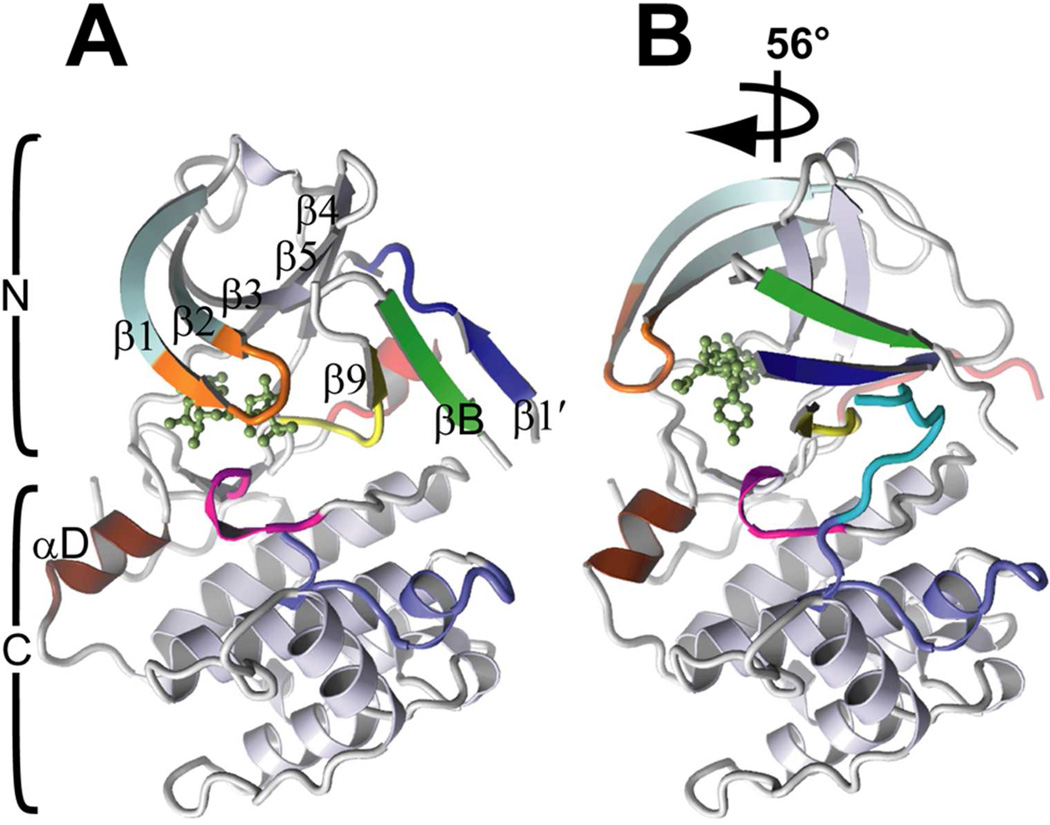
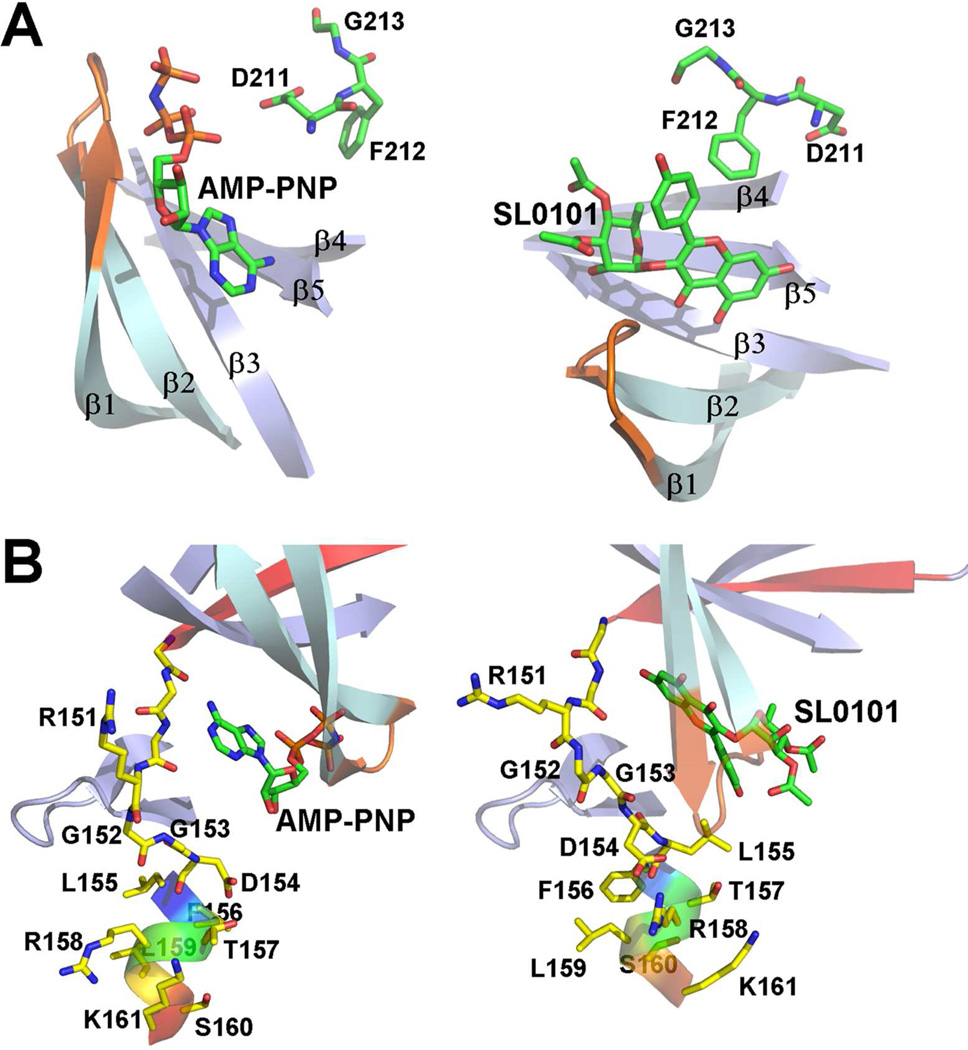
A cartoon view, comparing the mRSK2NTKD/SL0101 complex with the structure of mRSK2NTKD/AMP-PNP is shown in Figure 3. Most of the polypeptide chain is well ordered in the crystal structure of the complex with SL0101, with only two loops lacking interpretable electron density, i.e. residues 114–119 and 218–222, the latter being a part of the activation loop. The SL0101 molecule, as well as afzelin, are very well resolved in the electron density maps, and are located as expected in the cleft between the N- and C-lobes. The cores of the C-lobes in the SL0101 and AMP-PNP structures are highly similar, with an r.m.s. difference of 0.56 Å for main chain atoms. In contrast, the N-lobe undergoes a dramatic rearrangement in the SL0101 complex compared to the AMP-PNP bound structure, including changes in both the topology and architecture of the novel three-stranded β-sheet. A closer structural comparison reveals additional differences between the two complexes within the C-lobe. The DFG-motif, located upstream of the activation loop undergoes a structural reorganization, while the C-terminal portion of the activation loop, beginning with residue 223, becomes ordered and clearly visible in the electron density map. Finally, the αD-helix, which normally remains inert and not affected by the binding of ATP or inhibitors, significantly alters its conformation.
Figure 3.
A general comparison of the structure of RSK2NTKD in complex with (A) AMP-PNP and (B) SL0101. The two structures are superposed on the C-lobe. The N- and C-lobes are indicated by brackets. The arrow shows the relative rotation of the N-lobe in the SL0101 complex. All the β-strands in the core sheet and in the secondary, three-stranded-β-sheet are identified with symbols, the αD-helix is labeled and shown in red, the P-loop is orange, and the inhibitors are shown as ball-and-stick.
The overall effect of the structural differences observed within the protein moiety of the two complexes is an unprecedented rearrangement of the nucleotide binding site. Although SL0101 binds in the cleft between the N- and C-lobes, as expected for most kinase inhibitors, the nature of this cleft and the identities of residues that make it up are significantly different from the canonical ATP-binding site.
Next, we describe the details of the differences between mRSK2NTKD/SL0101 and mRSK2NTKD/AMP-PNP, followed by the description of the specific interactions of SL0101 with the protein, and experiments designed to probe the mechanism of selective inhibition.
The Conformational Rearrangement of the N-lobe
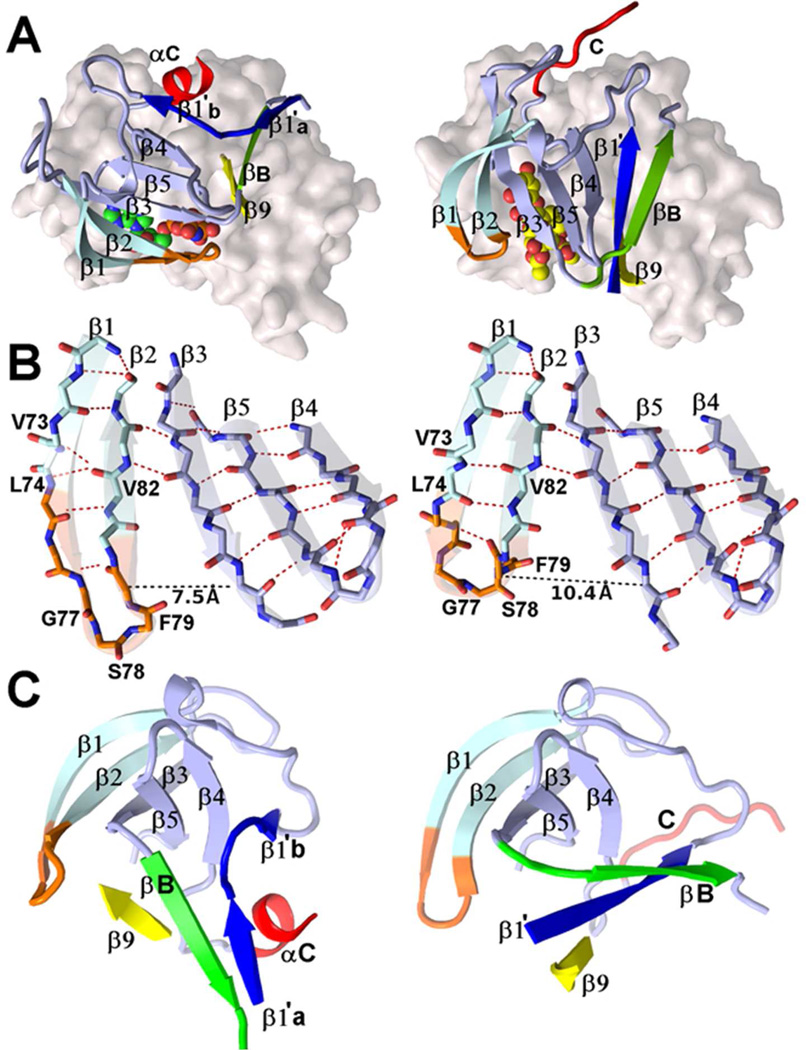
A particularly intriguing feature of the structure of the complex of mRSK2NTKD with SL0101 is the reorganization of the N-lobe compared to the AMP-PNP-bound structure (Figure 4A). The conformational changes within the N-lobe involve several distinct features. First, the main five-stranded β-sheet of the N-lobe undergoes a rotation of ~56° around an axis roughly perpendicular to the central β3-strand, pivoting around the N-terminal portion of the hinge region between the lobes. The β-sheet does not move as a rigid body: while strands β3 through β5 move in unison, the tip of the P-loop (strands β1 and β2) separates from strand β3, breaking the core sheet’s structural integrity. This is largely made possible by dissipation of the β-bulge in the β1 strand at the Leu74 position. It has been noted before that the β-bulges introduce flexibility to the otherwise rigid conformation of the β-sheets and, as a result, prevent aggregation of the proteins rich in β-sheets51. It is worth noting that most kinases have a β-bulge in this position. As a consequence of the disappearance of the β-bulge, the β1 strand straightens and pulls the β2 strand away from the rest of the β-sheet. The P-loop undergoes changes that shift the register of the main chain hydrogen bonds, and introduce a three-residue 310-helical structure in place of a classical hairpin (Figure 4B). The distance between the Cα of Phe79 (P-loop tip) and Cα of Lys100 (end of strand β3) increases from 7.5 Å in the AMP-PNP complex to 10.4 Å in the SL0101 complex. As a result, SL0101 is no longer sequestered under the P-loop, as is the case with ATP and canonical ATP-competitors, but instead becomes lodged between strands β2 and β3.
Figure 4.
A comparison of the secondary and tertiary structures of the N-lobe of the NTKDRSK2 in complexes with AMPPNP (left images) and SL0101 (right).
(A) The tertiary structures on the N-lobe (cartoon style) when the molecules are overlapped on the C-lobe (light grey surface). All core β-strands are labeled numerically 1 to 5. The navy blue, green, and yellow strands constitute the unique three stranded β-sheet of the RSK family: the N-terminal fragment extraneous to the canonical kinase domain (navy blue, residues 48 to 56, denoted β1’a and β1’b in the AMP-PNP complex, and β1’ in the SL0101 complex; green, residues 104–109, denoted βB, because the stretch typically forms a helical structure, αB, in protein kinases; yellow, residues 213–217, a segment immediately downstream of the DFG motif). A portion of the αC-helix, residues 120 – 126, shown in red, is observed in the AMP-PNP complex, while the same fragment has an extended structure in the SL0101 complex. The orange color denotes residues in the P- loop between strands β1 and β2. The two inhibitors are shown as van der Waals spheres.
(B) Differences in the secondary structures of the P-loops (orange), and H-bond networks connecting strands β1 and β2 (AMP-PNP complex on the left, SL0101 on the right).
(C) The N-lobes of the complexes with AMP-PNP (left) and SL0101 (right) are superposed on strands β1–β5 (the core β-sheet) to visualize the differences the secondary and tertiary structures of the RSK-specific, three-stranded β-sheet. The colors follow the scheme of (A).
As is the case in the AMP-PNP complex, the N-lobe contains an additional three-stranded β-sheet, which incorporates the fragment that normally folds into the αB-helix. A portion of a segment immediately following αB-helix and normally folded into a loop and the crucial αC-helix (residues 114–123) becomes disordered while the rest adopts an extended, non-helical conformation. Glu118, which is the residue that should engage in a conserved salt-bridge with Lys100, as anticipated from sequence comparisons, is disordered, but its approximate location is about ~30 Å away from Lys100, making it impossible for a salt-bridge to form. The three-stranded β-sheet is no longer in the same position as in the AMP-PNP complex, and there is a striking change in the topology. In the AMP-PNP complex, the sheet is antiparallel with the β1’ (the N-terminal extension) (↑) βB (↓), β9(↑) topology. In the SL0101 complex, the βB strand swings to the outside and positions itself on the solvent exposed side, while the β1’ strand reverses direction. As a consequence, the β-sheet is now mixed with a ↓↓↑ topology, for βB, β1’,β9 respectively (Figure 4C).
The DFG-motif
The invariant DFG motif (Asp211, Phe212, Gly213 in RSK2) is located immediately below the N-lobe and adjacent to the ATP-binding site. This motif toggles between the catalytically active (DFG-in) and catalytically inactive (DFG-out) states of kinases.52 In the typical active conformation (DFG-in), the aspartate points towards the triphosphate of ATP, whereas the side chain of the phenylalanine points in the opposite direction. In the absence of ATP, or in the complexes with inhibitors that bind to inactive conformations, the Phe side chain moves into the ATP-binding pocket, while the Asp flips out (DFG-out), as seen in the structure of the Abl kinase inhibited by Imatinib.53 In the SL0101 complex, the inhibitor leaves sufficient room to accommodate the benzene ring of Phe212 next to it, and consequently the catalytic Asp211 swings away from the active site, so that the entire DFG-motif assumes an out conformation (Figure 5A).
Figure 5.
Detailed comparison of the structures of mRSK2NTKD in complexes with AMP-PNP (left) and SL0101 (right).
(A) The differences in the conformation of the DFG-motif (Asp211, Phe212, G213) in the two complexes.
(B) The differences in the conformation of the hinge region and the αD helix.
The hinge loop and the αD-helix
The rotation of the N-lobe repositions the β4-strand, which consequently ‘pulls’ on the hinge loop, which plays a key role in the coordination of the purine moiety of ATP, and also provides anchoring H-bonds for most inhibitors. The portion of the hinge that includes Arg151, Gly152, Gly153, and Asp154, slides past the adjacent β6-strand, shifting by one the registry of H-bonds, compared to the AMP-PNP complex (Figure 5B). In response to this ‘pulling’ force exerted by the hinge oligopeptide, the αD-helix unwinds by one amino acid at its N-terminus. This causes Leu155, which normally packs against the αE-helix, to shift ~ 8 Å compared to the AMP-PNP structure, and move into the immediate proximity of the B-ring of SL0101. The αD-helix appears to rotate ~100 ° around its longitudinal axis, with a translation of 1.5 Å, resulting in a screw motion shifting the amino acid register by exactly one residue, leaving the helix short by one amino acid at its C-terminus. To accommodate this change, Glu162 is pulled into the αD helix from its position in the αD-αE loop.
While this comparison of the two crystal structures gives the appearance of a rotation of the αD-helix in the mRSK2NTKD/SL0101, it is quite clear that because of steric considerations this is not physically possible. Instead the entire fragment most likely unfolds transiently and refolds spontaneously into the new conformation. To the best of our knowledge, no similar rearrangement of the αD-helix has ever been reported for any kinase-inhibitor crystal structure.
The activation loop
In the AMP-PNP complex of mRSKNTKD, there is no electron density corresponding to residues 220–230 within the activation loop (including Ser227, which is the target for phosphorylation by PDK1). Unexpectedly, in the SL0101 complex the disorder is limited only to residues 218–222, while the stretch between Ala223 and Gly230 is clearly visible in the electron density map and shows low displacement (B) parameters. This ordering may be enhanced in part by the fact that residues Ala223 to Ser227 are involved in a crystal contact with the αD-αE loop and the C-terminus of an adjacent molecule. The electron density for the Ser227 side chain is very well resolved, and shows no indication of phosphorylation (Figure S1). Nevertheless, the activation loop assumes a conformation that bears significant resemblance to a number of reported phosphorylated kinases.54 Notably, the side chain of Ser227 is sequestered in a pocket flanked by several positively charged residues, i.e. Arg192, Arg242 and Arg110. Arg192, located downstream of the αE-helix and preceding the catalytic loop, is a highly conserved amino acid equivalent to Arg165 in PKA, which is instrumental in the binding of the pS/T motif of the activation loop55. Arg 242, on the other hand, is found in a loop upstream of the αF-helix, in a location that is also known to harbor a positively charged residue in a number of kinases, assisting in the binding of the pS/T motif.
In conclusion, the activation loop appears to assume a conformation characteristic of the active form of the kinase, even in the absence of phosphorylation of Ser227.
The binding mode of the SL0101 inhibitor and the molecular basis for specificity
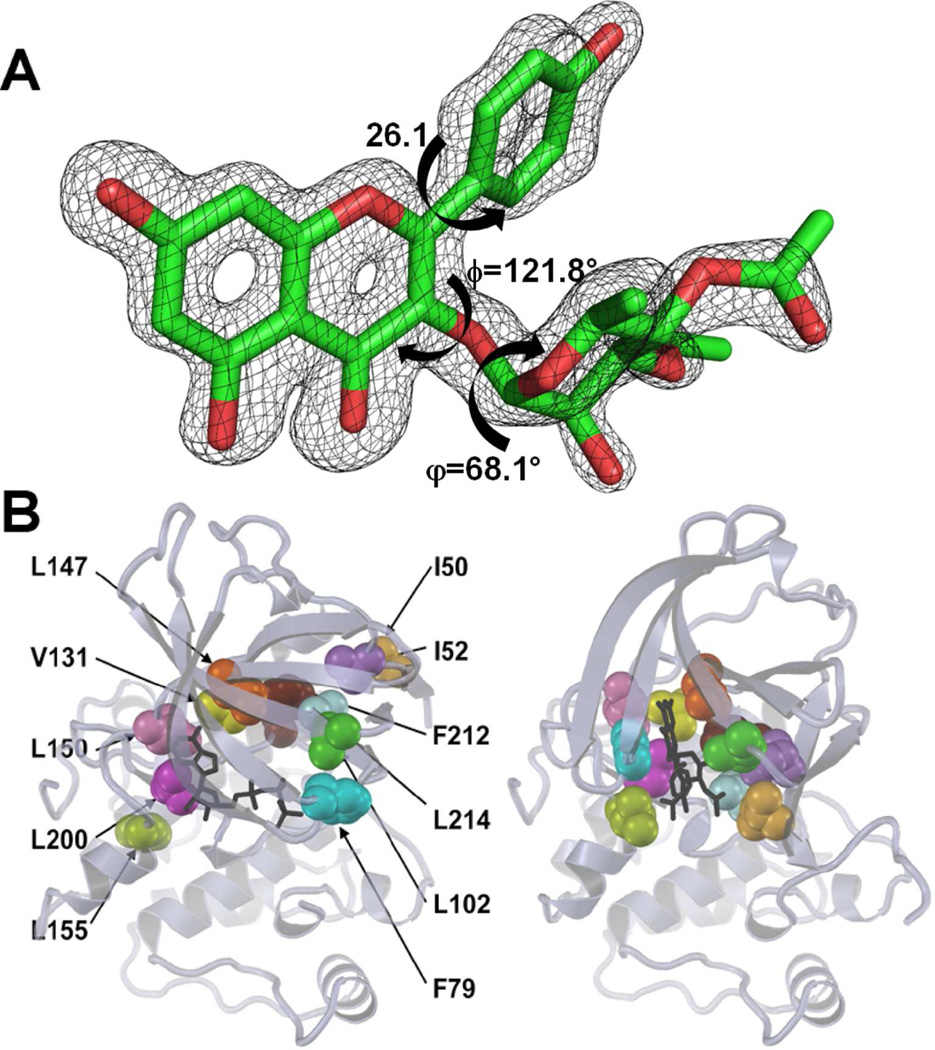
The unusually high resolution of the diffraction data (only 45 structures out of 1724 identified in the PDB have been determined at a resolution of 1.5 Å or better), allows for a detailed analysis of the stereochemistry of SL0101, and the mode of its binding to mRSK2NTKD. The electron density corresponding to the inhibitor is very well defined and the low atomic displacement (B) parameters are consistent with full occupancy (Figure 6A). The benzopyran moiety (rings A and C) is, as expected, planar within experimental error. However, unlike the situation in the free, non-glycosylated flavonols, where the B-ring is coplanar with benzopyran,56 in the present structure, the B-ring is twisted from the plane of benzopyran by 26.1 °, to avoid a clash with O6 (the glycosidic oxygen). The rhamnose ring adopts an expected 1C4 chair conformation, as observed in free α-L-rhamnose57 and in α-L-rhamnopyranosides.58 However, the stereochemistry of the glycosidic bond, defined by the two dihedral angles ϕ and φ, appears to be strained, away from a low energy conformation. The ϕ (C4-C3-O6-C1”) angle is 121.8 °, while the φ angle (C3-O6-C1”-H1”) is 68.1 °, resulting in a compact structure, in which the rhamnopyranoside ring packs against the phenolic B ring.
Figure 6.
The structure of the SL0101 inhibitor bound to mRSK2NTKD.
(A) Difference electron density (omit map) corresponding to the SL0101 inhibitor in the crystal structure. The SL0101 atoms were removed from the model and the remainder of the structure was refined to convergence. The map is contoured at 3σ.
(B) The reorganization of the eleven hydrophobic residues (labeled in the AMP-PNP complex), which form the SL0101 pocket. AMP-PNP and SL0101 are shown in black, each of the eleven residues is colored individually, and the color scheme in both pictures is identical.
Although, in general terms, SL0101 binds within the ATP site, its interactions with the protein differ from typical ATP mimetics, owing to the conformational changes in the protein that altered the stereochemistry of the ATP-binding site. The overall result of these changes within the protein moiety is a reorganization of the rather shallow cleft between the N- and C-domains, into a solvent occluded hydrophobic pocket. Eleven hydrophobic amino acids line this pocket, and all are in van der Waals contact with SL0101: Ile50, Ile52 (N-terminal β-strand), Phe79 from the P-loop, Leu102, Val131 and Leu147 from the N-lobe, Leu150 from the hinge region, Leu155 from the αD-helix, Leu200, and Phe212 (from the DFG loop) and Leu214. By contrast, in the complex with AMP-PNP, only four of these residues, i.e. Val131, Leu147, Leu150 and Leu200 are in direct contact with the adenine nucleoside. A cross section of this novel pocket reveals a surface that is remarkably complementary to the shape of SL0101 (Figure 6B; Figure S2).
We hypothesized that the formation of the binding pocket by the ensemble of eleven hydrophobic residues may result in increased stability of the complex, compared to the nucleotide-free and nucleotide-bound forms. Using the thermal shift assay (see Experimental Procedures) we discovered that binding of SL0101 increases the melting temperature (TM) of mRSKNTKD by ~5.1 °C, while AMP-PNP only by ~3.6 °C (Figure S3A).
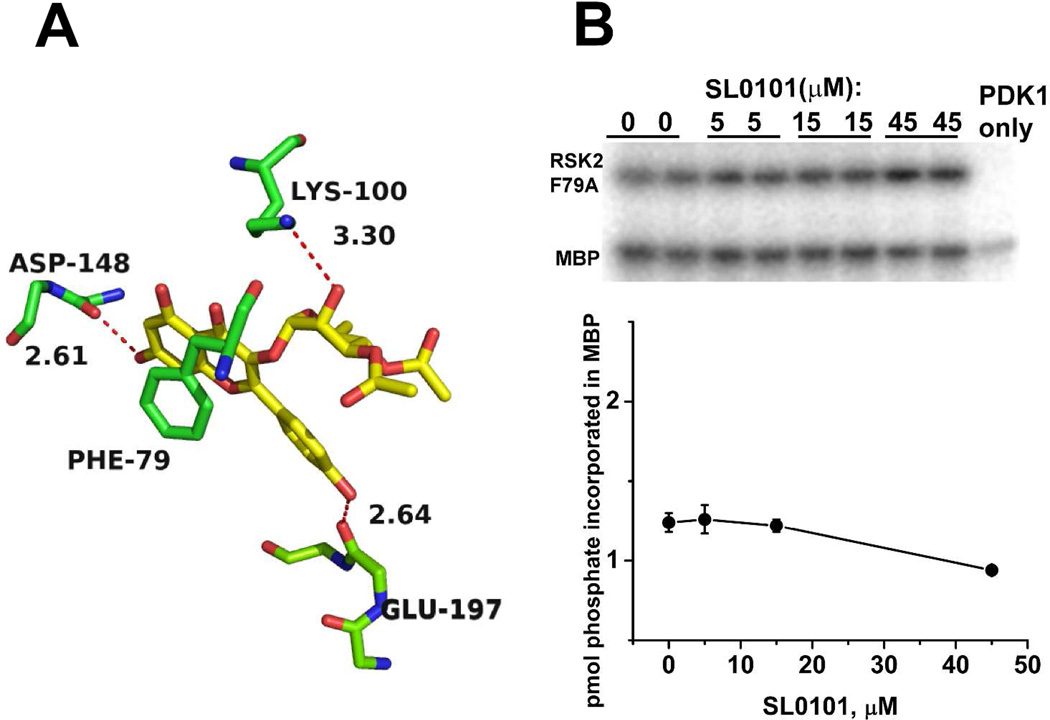
Aside from the non-specific hydrophobic pocket, there are only few specific interactions between the inhibitor and the protein moiety (Figure 7A). The 7-hydroxyl of the benzopyran forms an H-bond with the backbone carbonyl of Asp148 in the hinge region, while outside of the benzopyran core there are only two more H-bonds between SL0101 and the protein: the 4’-hydroxyl group (B ring) is a donor in an H-bond with the backbone carbonyl of Glu197, and the 2”-hydroxyl of the rhamnose moiety forms an H-bond with the side chain ε-amino group of Lys100.
Figure 7.
The role of Phe79 in RSK2 susceptibility to inhibition by SL0101.
(A) Details of select interactions of the SL0101 inhibitor with the protein. Phe79 from the P loop is seen in a π-stacking interaction with the C-ring. Hydrogen bonds with the main chain carbonyl groups of Asp148 and Glu197, and with the side chain of Lys100 are shown as red dotted lines and distances are shown.
(B) Catalytic activity of F79A in the presence of SL0101. Upper panel: analysis of gel samples by autoradiography performed simultaneously with wild-type RSK2NTKD as shown in Fig. 2A. Lower panel: kinase activity data averaged from duplicate measurements.
An intriguing feature of the binding mode of SL0101 by mRSK2NTKD is the unusual stereochemistry of the P-loop, which swings over the inhibitor so that the side chain of Phe79 forms an intimate π-stacking interaction with the C ring of the benzopyran of SL0101 (Figure 7A). Phe79, highly conserved as an aromatic residue, Phe or Tyr, occupies the position at the tip of the P-loop, and it is established that this residue serves to shield the active site from the solvent, although the phenyl ring never interacts with the nucleotide’s purine heterocycle. We therefore wondered how important this unusual interaction is for the RSK2 susceptibility to inhibition by SL0101. Using ITC as a binding assay, we found that the F79A mutant cannot bind SL0101, whereas it retains some affinity for ADP and AMP-PNP (Figure S4). The thermal denaturation temperature (TM) of the mutant is identical to that of the wild-type protein, but is not affected by the addition of SL0101 (Figure S3B). Furthermore, when phosphorylated by PDK1, the F79A mutant shows detectable catalytic activity of the wild-type mRSK2NTKD, but is no longer sensitive to inhibition by SL0101 (Figure 7B).
While the π-stacking interaction with Phe79 explains at least part of the mechanism of binding of SL0101, it does not explain the selectivity of the inhibitor. With the exception of Ile50 and Ile52, which are located in the N-terminal extension unique to the RSK family, all residues involved in the new inhibitor pocket sequestering SL0101 are well conserved among protein kinases, and only five interact with the adenine nucleotide (Phe79, Val131, Leu147, Leu150, and Leu200). We therefore wondered if the N-terminal extension was critical for the selective binding of SL0101. To that end, we generated two variants of the mRSK2NTKD, i.e. I50A and I52A, and carried out ITC assays to evaluate their ability to bind either AMP-PNP or SL0101. Interestingly, we found that neither variant was able to bind either the nucleotide analogue AMP-PNP or the inhibitor (Figure S4). Further studies will be required to evaluate the role on the N-terminal strand in nucleotide binding and catalytic activity.
The structure of afzelin in complex with mRSK2NTKD is virtually identical to that of SL0101, with the only difference being the absence of the acetyl groups (Figure S5). Interestingly, the previously reported IC50 values are 4.37 µM and 0.37 µM, respectively, suggest that two acetyl groups are of functional importance.35 However, given our structural analysis, the decrease in potency of afzelin as a RSK inhibitor must be associated with the kinetics of afzelin binding, or alternatively its binding to the full-length kinase may differ slightly from the binding to the isolated NTKD.
Crystal contacts and molecular packing
Given the magnitude of the differences observed between the complexes with AMP-PNP and SL0101, we asked if the molecular packing in the crystals of the latter might in any way be responsible for the unusual conformation. We found that the two major crystal contacts which bury ~960 Å2 and ~640 Å2 of solvent accessible surface, involve primarily amino acids from the C-lobe. There is nothing unusual in the packing that might account for a distortion of the structure due to packing forces (for a more detailed description see Figure S6 and its legend in the supporting information).
DISCUSSION
Kinases present unique challenges as drug targets because their tertiary architecture—with the highly conserved cleft adapted universally for ATP binding—makes it difficult to design inhibitors with adequate selectivity and specificity. However, kinases are also among the most structurally dynamic enzymes, sampling a wide range of conformations as they bind ATP/Mg2+, and interact with the substrate and/or regulatory proteins.59, 60 This structural malleability, inherent in the bilobal architecture of the core fold of a kinase catalytic domain, can be exploited for design of drugs that recognize unique, typically inactive conformations that differ from the canonical structures of the active domain. These are the so-called type II inhibitors, in contrast to type I molecules that bind within the ATP site without any concomitant conformational rearrangements.3 This is why understanding of conformational plasticity and dynamics of protein kinases is of special importance.
A wide spectrum of specific motions and conformational rearrangements has been described for kinases.59, 60 Among the AGC family of Ser/Thr kinases, the breathing open-and-close motion is associated with the relative movements of the two lobes: the open conformation is intrinsic to the nucleotide free form, while the closed conformation corresponds to a more compact molecule in which the two lobes sequester ATP/ Mg2+, to form a binary complex.59, 61–64 Nucleotide binding is often associated with the disorder-order transition of the P-loop, as it folds over the triphosphate of ATP and positions itself to assist in the phosphate transfer.65 The canonical kinase activation mechanism by phosphorylation of a Thr/Ser residue within the activation loop, involves a conformational rearrangement of the activation loop which vacates the catalytic site and positions itself at a different site on the protein’s surface, stabilized by interactions of the pS/T moiety with positively charged residues. Another flexible part of the molecule is the αC-helix, which adopts different conformations in the apo vs activated states.47 Finally, the DFG motif assumes different conformations in complexes with inhibitors, generally classified as either the in or the out conformation.
The breadth of the conformational flexibility of Ser/Thr kinase domains is evident from the vast number (1724 as of June 14, 2012) of structures of complexes with inhibitors deposited in the Protein Data Bank. Against this background, the mRSK2NTKD/SL0101 structure reported in this paper stands out as quite different: a comparison of the atomic coordinates with all PDB-deposited kinase structures using DALI66 shows that it differs on average by an r.m.s. of 4.0 Å (using Cα atoms only), with no structure showing structural similarity below an r.m.s. of 3.2 Å. This difference arises from the unusual relative rotation of the N- and C-lobes, and from the atypical three-stranded β-sheet in the N-lobe. By contrast, the complex with AMP-PNP differs from other kinase-inhibitor complexes by an r.m.s. of ~2.0 Å.
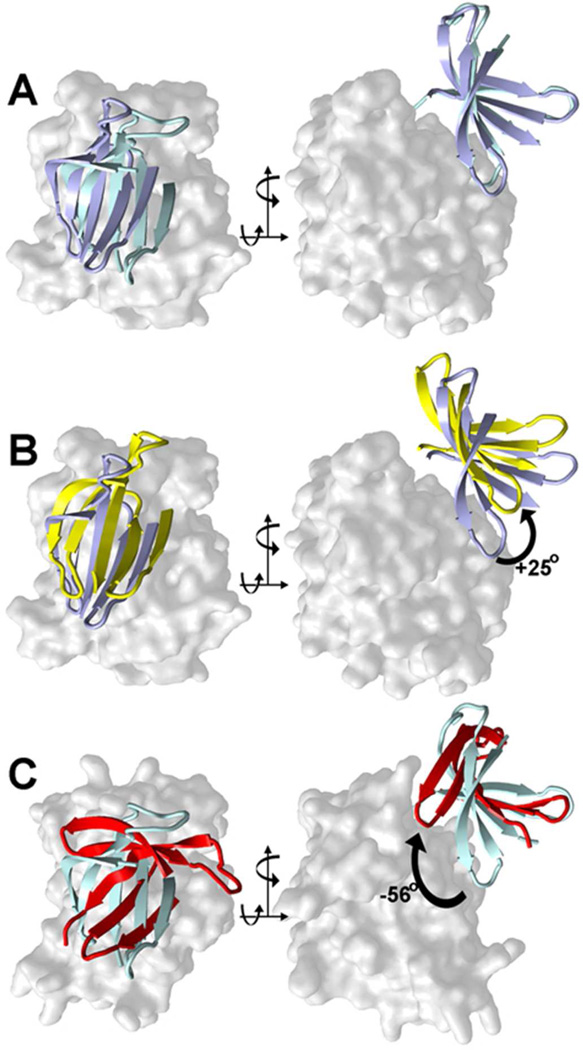
The unexpected structure of the mRSK2NTKD/SL0101 complex is of importance as it extends our understanding of kinase conformational malleability and kinase-inhibitor interactions. In contrast to the well-characterized canonical closed conformation typical for both type I and type II inhibition, the structure of mRSK2NTKD with bound SL0101 reveals a unique twisting motion of the N-lobe, with structural rearrangements of the core β-sheet and a dramatic change in the topology and structure of the auxiliary three-stranded β-sheet (Figure 8; Video S1).
Figure 8.
A comparison of the canonical molecular motions described for the N-lobe core β-sheet in PKA between the open and closed conformations, with the motion in RSK2 on going from the AMP-PNP32 to the SL0101-bound state, described in this paper. The proteins are superposed on the C-lobe, which is shown as solid surface; the core five-stranded β-sheet on the N-lobe is shown as a cartoon.
(A) A comparison between the AMP-PNP-bound structures of PKA (blue) and RSK2 (cyan);
(B) A comparison of PKA structures with AMP-PNP (blue) and in nucleotide-free state (yellow)64;
(C) A comparison of mRSK2NTKD with AMP-PNP (blue) and with SL0101 (red)
It is intriguing how the structure of the mRSK2NTKD/SL0101 complex is formed. It is important to realize that it is not clear at this point if the crystal structure of the mRSK2NTKD/AMP-PNP complex is actually representative of the conformation to which SL0101 would initially bind. There is also some controversy, if this structure corresponds to an unusual active form or perhaps to an autoinhibited form.32 Regardless of the details, the adenine nucleotide must dissociate prior to the binding of SL0101, and the structure of the nucleotide-free form of mRSK2NTKD is not known, and consequently the structure of the encounter complex is not easily predicted. One possibility is that SL0101 binds to mRSK2NTKD through conformational selection. This theory, which originated 50 years ago with the publication of the basis of cooperative transition by Monod, Wyman and Changeux,67 implies that proteins adapts a range of conformations in solution in the absence of the ligand, which then binds to the conformations with the highest affinity. The theory has gained wide acceptance and there is ample experimental evidence that indeed in many cases this is how ligand-protein interactions proceeds.68, 69 Conformational selection implies that the structure of the protein moiety in the mRSK2NTKD/SL0101 complex must be close to that in the encounter complex. However, it is difficult to envisage how the eleven hydrophobic amino acids might spontaneously coalesce to form a restructured binding site, prior to the binding of SL0101. Furthermore, the rearranged SL0101 binding site results in a tight pocket (Figure S2), which is likely to severely limit kon.
The other possibility, intuitively more probable, is an induced fit mechanism, as initially formulated by Koshland, Nemethy and Filmer.70, 71 This theory proposes that a gradual structural rearrangement occurs in the protein in response to ligand binding. Given the structural arguments listed above, it is tempting to hypothesize, that SL0101 binds to mRSK2NTKD when the latter is in a conformation broadly reminiscent of a canonical open conformation, probably with the kaempferol moiety mimicking the adenine of ATP, as seen in crystal structures of kinases with free flavonols.72 Following its formation, the encounter complex begins to undergo a conformational change, driven by the eleven residues coalescing to tighten the hydrophobic pocket around the SL0101 molecule. Such two-step mechanism is also consistent with the observation that kaempferol alone is able to bind to and inhibit RSK2, albeit with significantly lower IC50 of 15 µM.35 However, in spite of substantial efforts, we failed to obtain any crystals of the mRSK2NTKD/kaempferol complex under conditions that reproducibly yield crystals of the SL0101 complex, suggesting that the former exists in a conformation different from the one described here. However, if our speculation is correct, the conformational changes following the formation of the encounter complex occur on a very fast time scale, as we were unable to detect any slower events by routine means (unpublished data).
Given the complexity of the structural consequences of the binding of SL0101 to RSK, it is very likely that the actual binding mechanism proceeds by yet another route that incorporates aspects of both conformational selection and induced-fit. For example, NTKD might be unstable and partly unfolded in the absence of AMP-PNP, and SL0101 might bind to a unigue conformation in which the C-lobe and only fragments of the N-lobe are folded. Once such an encounter complex is formed, the remainder of the N-lobe might fold sequestering SL0101 in a novel pocket.
Another question concerns the molecular roots of susceptibility of the RSK family of kinases to SL0101. Interestingly, of the eleven hydrophobic amino acids that make up the novel pocket which sequesters the inhibitor, the only RSK-specific residues are Ile50 and Ile52, located in the N-terminal extension extraneous to the canonical domain. Indeed, we showed that if any of these are mutated to Ala, the protein is no longer capable of binding either AMP-PNP or SL0101. This strongly suggests that the N-terminus is essential for the integrity of the NTKD, and also confers of the RSK kinase susceptibility to SL0101.
Aside from the N-terminal extension, the P-loop plays an important role in the binding of SL0101. Phe79, which is involved in the π-stacking with the C-ring of the benzopyran, is a largely invariant aromatic residue located in the hairpin of the P-loop. This residue is essential for the expression of full activity in kinases, because it shields the triphosphate of ATP and the substrate phosphorylation site from solvent. Because of the inherent flexibility of the P-loop, the hydrophobic residue has been occasionally found to form π-π interactions with some aromatic moieties of inhibitors.73, 74 However, the P-loop never swings back far enough to make such interaction possible with an aromatic group that actually occupies the adenine site, and in that sense the interaction of Phe79 with benzopyran of SL0101 is unique. Interestingly the F79A mutant becomes resistant to SL0101, even though it retains a significant portion of catalytic activity.
Given its relative specificity as a RSK inhibitor, SL0101 has already been established as a powerful chemical tool in cell biology. For example, it has been used to demonstrate that the RSK2 kinase controls cell survival through its ability to regulate the formation of RNA granules during stress.75 The inhibitor was used to show that RSK regulates cell fate in the human breast ductal network76 and that it phosphorylates Y-box protein-1 on Ser102 in basal-like breast cancer cells77. When used in combination with a PKCζ pseudosubstrate, SL0101 completely abrogated ANG-II induced, RSK2 mediated cell proliferation.78 The compound inhibited glucose uptake in 3T3-L1 adipocytes, which occurs, at least in part, through a RSK-dependent phosphorylation of the Na+/H+ exchanger NHE1.79 The same signaling pathway may be exploited for enhanced cardiac protection against ischemia/reperfusion.80
Inhibitors of RSK kinases based on the SL0101 chemical scaffold may be ultimately useful as pharmacological agents for the treatment of cancer, diabetes and cardiomyocyte re-perfusion injury. Although the SL0101 template has been used as a starting point for in silico design of RSK inhibitors, in the absence of a crystal structure it was erroneously concluded that the binding mode was similar to that of free, unglycosylated flavonols.35, 81 The knowledge of the crystal structure reported in this paper will be of significant help in these efforts. In particular, the unique flexibility of the N-lobe and the P-loop in RSK2 revealed by the present structure may be exploited for the design of highly selective inhibitors that target the RSK-specific conformations.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Susan. S. Taylor, UCSD, Zbigniew Dauter, NCI, and Guy G. Dodson, University of York, for valuable discussions and comments on the manuscript.
This research was supported by the University of Virginia Cancer Center through the NCI Cancer Center Support Grant, P30 CA44579 and in part by National Institutes of Health (R01 GM086457 to Zygmunt S. Derewenda and Avril V. Somlyo). The use of Advanced Photon Source (APS) was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under contract No. W-31-109-Eng-38. Supporting institutions of SER-CAT may be found at http://www.ser-cat.org/members.html.
ABBREVIATIONS
- RSK
p90 ribosomal S6 kinase
- MSK
mitogen- and stress-stimulated kinase
- CTKD
C-terminal kinase domain
- NTKD
N-terminal kinase domain
- ERK
extracellular signal-regulated kinase
- PDK1
phosphoinositide-dependent kinase 1
- ATP
adenosine triphosphate
- AMP-PCP
5’-adenylyl (β,γ-methylene)diphosphonate
- AMP-PNP
adenylyl-imidodiphosphate
- ITC
isothermal titration calorimetry
- TSA
thermal shift assay
- r.m.s.
root-mean-square
Footnotes
ASSOCIATED CONTENT
Supporting Information
Six supplementary figures (Figures S1–S6) with references and a movie (Video S1) illustrating structural rearrangements in mRSK2NTKD on going from the AMP-PNP bound state to the SL0101-bound complex. This material is available free of charge via the Internet at http://pubs.acs.org.
Accession Codes
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession code 3UBD, for the SL0101 complex, and 4EL9 for the afzelin complex.
The authors declare no competing financial interests/
REFERENCES
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Fabbro D, Cowan-Jacob SW, Mobitz H, Martiny-Baron G. Targeting cancer with small-molecular-weight kinase inhibitors. Methods Mol. Biol. 2012;795:1–34. doi: 10.1007/978-1-61779-337-0_1. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anjum R, Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat. Rev. Mol. Cell Biol. 2008;9:747–758. doi: 10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- 5.Jones SW, Erikson E, Blenis J, Maller JL, Erikson RL. A Xenopus ribosomal protein S6 kinase has two apparent kinase domains that are each similar to distinct protein kinases. Proc. Natl. Acad. Sci. U S A. 1988;85:3377–3381. doi: 10.1073/pnas.85.10.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 7.Stratford AL, Dunn SE. The promise and challenges of targeting RSK for the treatment of cancer. Expert Opin. Ther. Targets. 2011;15:1–4. doi: 10.1517/14728222.2011.537656. [DOI] [PubMed] [Google Scholar]
- 8.Romeo Y, Roux PP. Paving the way for targeting RSK in cancer. Expert Opin. Ther. Targets. 2011;15:5–9. doi: 10.1517/14728222.2010.531014. [DOI] [PubMed] [Google Scholar]
- 9.Smith JA, Poteet-Smith CE, Xu Y, Errington TM, Hecht SM, Lannigan DA. Identification of the first specific inhibitor of p90 ribosomal S6 kinase (RSK) reveals an unexpected role for RSK in cancer cell proliferation. Cancer Res. 2005;65:1027–1034. [PubMed] [Google Scholar]
- 10.Clark DE, Errington TM, Smith JA, Frierson HF, Jr, Weber MJ, Lannigan DA. The serine/threonine protein kinase, p90 ribosomal S6 kinase, is an important regulator of prostate cancer cell proliferation. Cancer Res. 2005;65:3108–3116. doi: 10.1158/0008-5472.CAN-04-3151. [DOI] [PubMed] [Google Scholar]
- 11.Kang S, Elf S, Lythgoe K, Hitosugi T, Taunton J, Zhou W, Xiong L, Wang D, Muller S, Fan S, Sun SY, Marcus AI, Gu TL, Polakiewicz RD, Chen ZG, Khuri FR, Shin DM, Chen J. p90 ribosomal S6 kinase 2 promotes invasion and metastasis of human head and neck squamous cell carcinoma cells. J. Clin. Invest. 2010;120:1165–1177. doi: 10.1172/JCI40582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuadrado A, Nebreda AR. New insights into RSK activation and hematopoietic cancer. Cancer Cell. 2007;12:187–189. doi: 10.1016/j.ccr.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 13.Kang S, Dong S, Gu TL, Guo A, Cohen MS, Lonial S, Khoury HJ, Fabbro D, Gilliland DG, Bergsagel PL, Taunton J, Polakiewicz RD, Chen J. FGFR3 activates RSK2 to mediate hematopoietic transformation through tyrosine phosphorylation of RSK2 and activation of the MEK/ERK pathway. Cancer Cell. 2007;12:201–214. doi: 10.1016/j.ccr.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang S, Elf S, Dong S, Hitosugi T, Lythgoe K, Guo A, Ruan H, Lonial S, Khoury HJ, Williams IR, Lee BH, Roesel JL, Karsenty G, Hanauer A, Taunton J, Boggon TJ, Gu TL, Chen J. Fibroblast growth factor receptor 3 associates with and tyrosine phosphorylates p90 RSK2, leading to RSK2 activation that mediates hematopoietic transformation. Mol. Cell. Biol. 2009;29:2105–2117. doi: 10.1128/MCB.00998-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mirmohammadsadegh A, Mota R, Gustrau A, Hassan M, Nambiar S, Marini A, Bojar H, Tannapfel A, Hengge UR. ERK1/2 is highly phosphorylated in melanoma metastases and protects melanoma cells from cisplatin-mediated apoptosis. J. Invest. Dermatol. 2007;127:2207–2215. doi: 10.1038/sj.jid.5700870. [DOI] [PubMed] [Google Scholar]
- 16.Berk BC, Maekawa N, Abe J, Itoh S, Ding B, Sharma VK, Sheu SS, Blaxall BC. Inhibiting p90 ribosomal S6 kinase prevents Na+-H+ exchanger-mediated cardiac ischemia-reperfusion injury. Circulation. 2006;113:2516–2523. doi: 10.1161/CIRCULATIONAHA.105.563486. [DOI] [PubMed] [Google Scholar]
- 17.Lu W, Liu X, Cao X, Xue M, Liu K, Zhao Z, Shen X, Jiang H, Xu Y, Huang J, Li H. SHAFTS: a hybrid approach for 3D molecular similarity calculation. 2. Prospective case study in the discovery of diverse p90 ribosomal S6 protein kinase 2 inhibitors to suppress cell migration. J. Med. Chem. 2011;54:3564–3574. doi: 10.1021/jm200139j. [DOI] [PubMed] [Google Scholar]
- 18.Alessi DR, Sapkota GP, Cummings L, Newell FS, Armstrong C, Bain J, Frodin M, Grauert M, Hoffmann M, Schnapp G, Steegmaier M, Cohen P. BI-D1870 is a specific inhibitor of the p90 RSK (ribosomal S6 kinase) isoforms in vitro and in vivo. Biochem. J. 2007;401:29–38. doi: 10.1042/BJ20061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calderon-Montano JM, Burgos-Moron E, Perez-Guerrero C, Lopez-Lazaro M. A review on the dietary flavonoid kaempferol. Mini Rev. Med. Chem. 2011;11:298–344. doi: 10.2174/138955711795305335. [DOI] [PubMed] [Google Scholar]
- 21.Neuhouser ML. Dietary flavonoids and cancer risk: evidence from human population studies. Nutr. Cancer. 2004;50:1–7. doi: 10.1207/s15327914nc5001_1. [DOI] [PubMed] [Google Scholar]
- 22.Maron DJ. Flavonoids for reduction of atherosclerotic risk. Curr. Atheroscler. Rep. 2004;6:73–78. doi: 10.1007/s11883-004-0119-1. [DOI] [PubMed] [Google Scholar]
- 23.Boly R, Gras T, Lamkami T, Guissou P, Serteyn D, Kiss R, Dubois J. Quercetin inhibits a large panel of kinases implicated in cancer cell biology. Int. J. Oncol. 2011;38:833–842. doi: 10.3892/ijo.2010.890. [DOI] [PubMed] [Google Scholar]
- 24.Hou DX, Kumamoto T. Flavonoids as protein kinase inhibitors for cancer chemoprevention: direct binding and molecular modeling. Antioxid. Redox. Signal. 2010;13:691–719. doi: 10.1089/ars.2009.2816. [DOI] [PubMed] [Google Scholar]
- 25.Rogers JC, Williams DL., Jr Kaempferol inhibits myosin light chain kinase. Biochem. Biophys. Res. Commun. 1989;164:419–425. doi: 10.1016/0006-291x(89)91736-1. [DOI] [PubMed] [Google Scholar]
- 26.Lee KM, Lee DE, Seo SK, Hwang MK, Heo YS, Lee KW, Lee HJ. Phosphatidylinositol 3-kinase, a novel target molecule for the inhibitory effects of kaempferol on neoplastic cell transformation. Carcinogenesis. 2010;31:1338–1343. doi: 10.1093/carcin/bgq102. [DOI] [PubMed] [Google Scholar]
- 27.Kim W, Yang HJ, Youn H, Yun YJ, Seong KM, Youn B. Myricetin inhibits Akt survival signaling and induces Bad-mediated apoptosis in a low dose ultraviolet (UV)-B-irradiated HaCaT human immortalized keratinocytes. J. Radiat. Res. (Tokyo) 2010;51:285–296. doi: 10.1269/jrr.09141. [DOI] [PubMed] [Google Scholar]
- 28.Walker EH, Pacold ME, Perisic O, Stephens L, Hawkins PT, Wymann MP, Williams RL. Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Mol. Cell. 2000;6:909–919. doi: 10.1016/s1097-2765(05)00089-4. [DOI] [PubMed] [Google Scholar]
- 29.Suh Y, Afaq F, Khan N, Johnson JJ, Khusro FH, Mukhtar H. Fisetin induces autophagic cell death through suppression of mTOR signaling pathway in prostate cancer cells. Carcinogenesis. 2010;31:1424–1433. doi: 10.1093/carcin/bgq115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goettert M, Schattel V, Koch P, Merfort I, Laufer S. Biological evaluation and structural determinants of p38alpha mitogen-activated-protein kinase and c-Jun-N-terminal kinase 3 inhibition by flavonoids. Chembiochem. 2010;11:2579–2588. doi: 10.1002/cbic.201000487. [DOI] [PubMed] [Google Scholar]
- 31.Ikuta M, Kornienko M, Byrne N, Reid JC, Mizuarai S, Kotani H, Munshi SK. Crystal structures of the N-terminal kinase domain of human RSK1 bound to three different ligands: Implications for the design of RSK1 specific inhibitors. Protein Science. 2007;16:2626–2635. doi: 10.1110/ps.073123707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malakhova M, Kurinov I, Liu KD, Zheng D, D'Angelo I, Shim JH, Steinman V, Bode AM, Dong ZG. Structural Diversity of the Active N-Terminal Kinase Domain of p90 Ribosomal S6 Kinase 2. PLoS One. 2009;4:e8044. doi: 10.1371/journal.pone.0008044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madhusudan, Akamine P, Xuong NH, Taylor SS. Crystal structure of a transition state mimic of the catalytic subunit of cAMP-dependent protein kinase. Nat. Struct. Biol. 2002;9:273–277. doi: 10.1038/nsb780. [DOI] [PubMed] [Google Scholar]
- 34.Smith KJ, Carter PS, Bridges A, Horrocks P, Lewis C, Pettman G, Clarke A, Brown M, Hughes J, Wilkinson M, Bax B, Reith A. The structure of MSK1 reveals a novel autoinhibitory conformation for a dual kinase protein. Structure. 2004;12:1067–1077. doi: 10.1016/j.str.2004.02.040. [DOI] [PubMed] [Google Scholar]
- 35.Smith JA, Maloney DJ, Hecht SM, Lannigan DA. Structural basis for the activity of the RSK-specific inhibitor, SL0101. Bioorg. Med. Chem. 2007;15:5018–5034. doi: 10.1016/j.bmc.2007.03.087. [DOI] [PubMed] [Google Scholar]
- 36.Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of "Parallel" expression vectors. Protein Expr. Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 37.Klock HE, Koesema EJ, Knuth MW, Lesley SA. Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins. 2008;71:982–994. doi: 10.1002/prot.21786. [DOI] [PubMed] [Google Scholar]
- 38.Maloney DJ, Hecht SM. Synthesis of a potent and selective inhibitor of p90 Rsk. Org. Lett. 2005;7:1097–1099. doi: 10.1021/ol0500463. [DOI] [PubMed] [Google Scholar]
- 39.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol.y A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 40.Long F, Vagin AA, Young P, Murshudov GN. BALBES: a molecular-replacement pipeline. Acta Cryst. D. 2008;64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cohen SX, Ben Jelloul M, Long F, Vagin A, Knipscheer P, Lebbink J, Sixma TK, Lamzin VS, Murshudov GN, Perrakis A. ARP/wARP and molecular replacement: the next generation. Acta Cryst D. 2008;64:49–60. doi: 10.1107/S0907444907047580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.da Silva I, Diaz JG, Gonzalez-Platas J. Structure determination of monohydrated trifolin (kaempferol 3-O-beta-D-galactopyranoside) from laboratory powder diffraction data. J Pharm Sci. 2011;100:1588–1593. doi: 10.1002/jps.22379. [DOI] [PubMed] [Google Scholar]
- 45.Laskowski RA, McArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:282–291. [Google Scholar]
- 46.Malawski GA, Hillig RC, Monteclaro F, Eberspaecher U, Schmitz AA, Crusius K, Huber M, Egner U, Donner P, Muller-Tiemann B. Identifying protein construct variants with increased crystallization propensity--a case study. Protein Sci. 2006;15:2718–2728. doi: 10.1110/ps.062491906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frodin M, Antal TL, Dummler BA, Jensen CJ, Deak M, Gammeltoft S, Biondi RM. A phosphoserine/threonine-binding pocket in AGC kinases and PDK1 mediates activation by hydrophobic motif phosphorylation. Embo J. 2002;21:5396–5407. doi: 10.1093/emboj/cdf551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jensen CJ, Buch MB, Krag TO, Hemmings BA, Gammeltoft S, Frodin M. 90-kDa ribosomal S6 kinase is phosphorylated and activated by 3-phosphoinositide-dependent protein kinase-1. J. Biol. Chem. 1999;274:27168–27176. doi: 10.1074/jbc.274.38.27168. [DOI] [PubMed] [Google Scholar]
- 49.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Cryst D. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 50.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Cryst D. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chan AW, Hutchinson EG, Harris D, Thornton JM. Identification, classification, and analysis of beta-bulges in proteins. Protein Sci. 1993;2:1574–1590. doi: 10.1002/pro.5560021004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y, Gray NS. Rational design of inhibitors that bind to inactive kinase conformations. Nat. Chem. Biol. 2006;2:358–364. doi: 10.1038/nchembio799. [DOI] [PubMed] [Google Scholar]
- 53.Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 54.Ko TP, Jeng WY, Liu CI, Lai MD, Wu CL, Chang WJ, Shr HL, Lu TJ, Wang AH. Structures of human MST3 kinase in complex with adenine, ADP and Mn2+ Acta Cryst D. 2010;66:145–154. doi: 10.1107/S0907444909047507. [DOI] [PubMed] [Google Scholar]
- 55.Knighton DR, Zheng JH, Ten Eyck LF, Ashford VA, Xuong NH, Taylor SS, Sowadski JM. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 56.Domagala S, Munshi P, Ahmed M, Guillot B, Jelsch C. Structural analysis and multipole modelling of quercetin monohydrate—a quantitative and comparative study. Acta Cryst. B. 2011;67:63–78. doi: 10.1107/S0108768110041996. [DOI] [PubMed] [Google Scholar]
- 57.Takagi S, Jeffrey GA. Neutron-Diffraction Refinement of Crystal-Structure of Alpha-L-Rhamnose Monohydrate. Acta Cryst. B. 1978;34:2551–2555. [Google Scholar]
- 58.Eriksson L, Stenutz R, Widmalm G. Methyl 2-O-beta-D-glucopyranosyl-alpha-L-rhamnopyranoside. Acta Cryst. C. 2002;58:O328–O329. doi: 10.1107/s0108270102005905. [DOI] [PubMed] [Google Scholar]
- 59.Taylor SS, Kornev AP. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem. Sci. 2011;36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 61.Karlsson R, Zheng J, Xuong N, Taylor SS, Sowadski JM. Structure of the mammalian catalytic subunit of cAMP-dependent protein kinase and an inhibitor peptide displays an open conformation. Acta Cryst D. 1993;49:381–388. doi: 10.1107/S0907444993002306. [DOI] [PubMed] [Google Scholar]
- 62.Karlsson R, Madhusudan, Taylor SS, Sowadski JM. Intermolecular contacts in various crystal forms related to the open and closed conformational states of the catalytic subunit of cAMP-dependent protein kinase. Acta Cryst D. 1994;50:657–662. doi: 10.1107/S0907444994001939. [DOI] [PubMed] [Google Scholar]
- 63.Masterson LR, Shi L, Metcalfe E, Gao J, Taylor SS, Veglia G. Dynamically committed, uncommitted, and quenched states encoded in protein kinase A revealed by NMR spectroscopy. Proc. Natl. Acad. Sci. U S A. 2011;108:6969–6974. doi: 10.1073/pnas.1102701108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Steichen JM, Kuchinskas M, Keshwani MM, Yang J, Adams JA, Taylor SS. Structural basis for the regulation of protein kinase A by activation loop phosphorylation. J. Biol. Chem. 2012;287:14672–14680. doi: 10.1074/jbc.M111.335091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hemmer W, McGlone M, Tsigelny I, Taylor SS. Role of the glycine triad in the ATP-binding site of cAMP-dependent protein kinase. J. Biol. Chem. 1997;272:16946–16954. doi: 10.1074/jbc.272.27.16946. [DOI] [PubMed] [Google Scholar]
- 66.Holm L, Kaariainen S, Wilton C, Plewczynski D. Using Dali for structural comparison of proteins. Curr. Protoc. Bioinformatics. 2006;Chapter 5(Unit 5):5. doi: 10.1002/0471250953.bi0505s14. [DOI] [PubMed] [Google Scholar]
- 67.Monod J, Wyman J, Changeux JP. On the Nature of Allosteric Transitions: A Plausible Model. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 68.Changeux JP. Allostery and the Monod-Wyman-Changeux Model After 50 Years. Annu Rev Biophys. 2012;41:103–133. doi: 10.1146/annurev-biophys-050511-102222. [DOI] [PubMed] [Google Scholar]
- 69.Changeux JP, Edelstein S. Conformational selection or induced fit? 50 years of debate resolved. F1000 Biol. Rep. 2011;3:19. doi: 10.3410/B3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koshland DE, Jr, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 71.Copeland RA. Conformational adaptation in drug-target interactions and residence time. Future Med. Chem. 2011;3:1491–1501. doi: 10.4155/fmc.11.112. [DOI] [PubMed] [Google Scholar]
- 72.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 73.Zhao B, Smallwood A, Yang J, Koretke K, Nurse K, Calamari A, Kirkpatrick RB, Lai Z. Modulation of kinase-inhibitor interactions by auxiliary protein binding: crystallography studies on Aurora A interactions with VX-680 and with TPX2. Protein Sci. 2008;17:1791–1797. doi: 10.1110/ps.036590.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Natarajan SR, Heller ST, Nam K, Singh SB, Scapin G, Patel S, Thompson JE, Fitzgerald CE, O'Keefe SJ. p38 MAP kinase inhibitors. Part 6: 2-arylpyridazin-3-ones as templates for inhibitor design. Bioorg. Med. Chem. Lett. 2006;16:5809–5813. doi: 10.1016/j.bmcl.2006.08.074. [DOI] [PubMed] [Google Scholar]
- 75.Eisinger-Mathason TS, Andrade J, Groehler AL, Clark DE, Muratore-Schroeder TL, Pasic L, Smith JA, Shabanowitz J, Hunt DF, Macara IG, Lannigan DA. Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. Mol. Cell. 2008;31:722–736. doi: 10.1016/j.molcel.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pasic L, Eisinger-Mathason TS, Velayudhan BT, Moskaluk CA, Brenin DR, Macara IG, Lannigan DA. Sustained activation of the HER1-ERK1/2-RSK signaling pathway controls myoepithelial cell fate in human mammary tissue. Genes Dev. 2011;25:1641–1653. doi: 10.1101/gad.2025611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stratford AL, Fry CJ, Desilets C, Davies AH, Cho YY, Li Y, Dong Z, Berquin IM, Roux PP, Dunn SE. Y-box binding protein-1 serine 102 is a downstream target of p90 ribosomal S6 kinase in basal-like breast cancer cells. Breast Cancer Res. 2008;10:R99. doi: 10.1186/bcr2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Godeny MD, Sayeski PP. ERK1/2 regulates ANG II-dependent cell proliferation via cytoplasmic activation of RSK2 and nuclear activation of elk1. Am. J. Physiol. Cell Physiol. 2006;291:C1308–C1317. doi: 10.1152/ajpcell.00618.2005. [DOI] [PubMed] [Google Scholar]
- 79.Chen S, Mackintosh C. Differential regulation of NHE1 phosphorylation and glucose uptake by inhibitors of the ERK pathway and p90RSK in 3T3-L1 adipocytes. Cell Signal. 2009;21:1984–1993. doi: 10.1016/j.cellsig.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 80.Avkiran M, Cook AR, Cuello F. Targeting Na+/H+ exchanger regulation for cardiac protection: a RSKy approach? Curr. Opin. Pharmacol. 2008;8:133–140. doi: 10.1016/j.coph.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 81.Nguyen TL, Gussio R, Smith JA, Lannigan DA, Hecht SM, Scudiero DA, Shoemaker RH, Zaharevitz DW. Homology model of RSK2 N-terminal kinase domain, structure-based identification of novel RSK2 inhibitors, and preliminary common pharmacophore. Bioorg. Med. Chem. 2006;14:6097–6105. doi: 10.1016/j.bmc.2006.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.