Abstract
Due to the high costs associated with purification of recombinant proteins the protocols need to be rationalized. For high-throughput efforts there is a demand for general methods that do not require target protein specific optimization1 . To achieve this, purification tags that genetically can be fused to the gene of interest are commonly used2 . The most widely used affinity handle is the hexa-histidine tag, which is suitable for purification under both native and denaturing conditions3 . The metabolic burden for producing the tag is low, but it does not provide as high specificity as competing affinity chromatography based strategies1,2.
Here, a bispecific purification tag with two different binding sites on a 46 amino acid, small protein domain has been developed. The albumin-binding domain is derived from Streptococcal protein G and has a strong inherent affinity to human serum albumin (HSA). Eleven surface-exposed amino acids, not involved in albumin-binding4 , were genetically randomized to produce a combinatorial library. The protein library with the novel randomly arranged binding surface (Figure 1) was expressed on phage particles to facilitate selection of binders by phage display technology. Through several rounds of biopanning against a dimeric Z-domain derived from Staphylococcal protein A5, a small, bispecific molecule with affinity for both HSA and the novel target was identified6 .
The novel protein domain, referred to as ABDz1, was evaluated as a purification tag for a selection of target proteins with different molecular weight, solubility and isoelectric point. Three target proteins were expressed in Escherishia coli with the novel tag fused to their N-termini and thereafter affinity purified. Initial purification on either a column with immobilized HSA or Z-domain resulted in relatively pure products. Two-step affinity purification with the bispecific tag resulted in substantial improvement of protein purity. Chromatographic media with the Z-domain immobilized, for example MabSelect SuRe, are readily available for purification of antibodies and HSA can easily be chemically coupled to media to provide the second matrix.
This method is especially advantageous when there is a high demand on purity of the recovered target protein. The bifunctionality of the tag allows two different chromatographic steps to be used while the metabolic burden on the expression host is limited due to the small size of the tag. It provides a competitive alternative to so called combinatorial tagging where multiple tags are used in combination1,7.
Keywords: Molecular Biology, Issue 59, Affinity chromatography, albumin-binding domain, human serum albumin, Z-domain
Protocol
1. Cloning of the target gene/ABDz1-tagged fusion construct
Prepare PCR fragments of the ABDz1 gene flanked by suitable restriction sites for N-terminal ligation of the target gene in the expression plasmid (a plasmid containing ABDz1 is freely available through a material transfer agreement).
Cleave purified expression vector and PCR fragments with chosen restriction enzymes in a suitable reaction buffer. Purify the products before ligation.
Ligate the restricted ABDz1 fragment into the expression vector containing the gene of interest and transform the ligation product to E. coli (in the original method the RR1ΔM15 strain was used8). Spread transformed cells on agar plates supplemented with suitable antibiotics for selection.
PCR screen a few colonies and sequence verify the resulting expression cassette by DNA sequencing.
Prepare plasmid from an overnight culture of a sequence verified colony and transform to the preferred expression strain (in the original method E. coli Rosetta (DE3), hosting a pRARE plasmid to improve the production of proteins encoded by human genes, were used).
2. Protein expression
Inoculate a single bacterial colony into 10 ml of tryptic soy broth supplemented with 5 g/l yeast extract and appropriate antibiotics. Incubate at 150 rpm at 37 °C overnight.
Inoculate 1 ml of overnight culture in 100 ml fresh medium and induce protein expression (originally 1 mM isopropyl-β-D-thiogalactoside when a lac operon was utilized) when the cells reach the logarithmic growth phase.
Incubate at 150 rpm at 25 °C overnight and harvest cells by centrifugation.
3. Orthogonal affinity purification
Resuspend the pellet in 25 ml running buffer (25 mM Tris-HCl, 200 mM NaCl, 1 mM EDTA, 0.05 % (w/v) Tween 20, pH 8.0) and disrupt the cells by sonication at 60 % amplitude and 1.0/1.0 pulses for 3 min. Centrifuge the sample and filter the target protein containing supernatant (0.45 μm) before further purification.
Equilibrate either a 1 ml HSA Sepharose column or an NHS-activated column, previously coupled with HSA according to the supplier's recommendations, with 10 column volumes (CV) of running buffer at 1 ml/min on a suitable protein purification system. Load the bacterial lysate at 0.5 ml/min and then reset the flow rate to 1 ml/min.
Wash the column with 5 CV of running buffer followed by 5 CV of washing buffer (5 mM NH4Ac, pH 5.5).
Elute the sample with elution buffer (0.5 M HAc, pH 2.5) at 1 ml/min and collect fractions, monitor absorption at 280 nm to select fractions from the eluted peak for further purification.
Pool the fractions with the highest protein concentration, dilute two times in SuRe running buffer (20 mM phosphate, 150 mM NaCl, pH 7.2) and make sure the pH is around neutral. Add 1 M Tris-HCl pH 8 to increase pH if necessary.
Load the sample at 0.5 ml/min on a 1 ml HiTrap MabSelect SuRe column that has been equilibrated with 10 CV of SuRe running buffer at 1 ml/min. Reset the flow rate to 1 ml/min after loading.
Wash the column with 5 CV of SuRe running buffer (step 5) and elute the proteins with 0.2 M HAc, pH 2.7. For sensitive target proteins the eluate can be neutralized directly upon collection by addition of Tris-HCl.
If the chromatographic steps are reversed the eluate from the MabSelect SuRe column can be diluted in running buffer (step 3.1) and the pH increased to around 8 by addition of 1 M Tris-HCl. In general, narrower peaks are observed when the SuRe matrix is employed in the second step.
4. Evaluation of purity by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE)
Load the purified fractions on a reducing SDS-PAGE and, if desired, analyze the molecular weight of the purified product by mass spectrometry. It is important to fully reduce the sample since a free cysteine in ABDz1 may cause dimerization of the product under non-reducing conditions.
5. Representative Results:
As a proof of principle, orthogonal affinity purification facilitated by the ABDz1 tag was evaluated for three human target proteins representing different solubility classes, molecular weights and isoelectric points (Table 1). The ABDz1 gene was genetically fused to the target genes and the constructs were expressed in E. coli, Figure 2 shows a flow chart for all steps in the method consecutively. Following purification by the orthogonal protocol, samples collected at different points were analyzed by SDS-PAGE (Figure 3). The results clearly show the utility of a dual tag and two highly specific purification steps. Even though reasonable purity is achieved after the initial purification step as seen from the gels, the successive step yields a very pure product well suited for highly demanding applications.
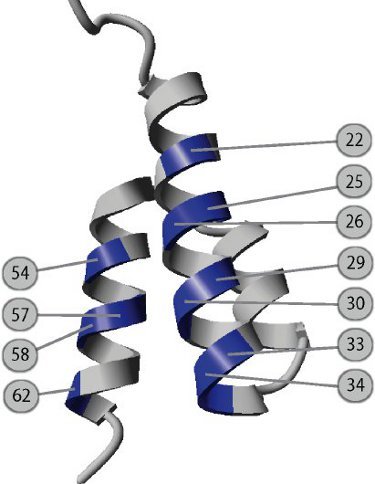 Figure 1. Design of the combinatorial library used for selection of the bispecific ABDz1 tag. The 46 amino acid albumin-binding domain folds into a stable three helix bundle and it contains a binding site for human serum albumin mainly located to the second helix. By genetically randomizing eleven surface exposed amino acids located in the first- and third helix, a novel binding surface was engineered. The eleven positions are indicated in the figure and numbered according to Kraulis et al.9 . Following biopanning against a dimer of the Z-domain of Staphylococcal protein A with the combinatorial library expressed on phage, the bispecific ABDz1 molecule was identified.
Figure 1. Design of the combinatorial library used for selection of the bispecific ABDz1 tag. The 46 amino acid albumin-binding domain folds into a stable three helix bundle and it contains a binding site for human serum albumin mainly located to the second helix. By genetically randomizing eleven surface exposed amino acids located in the first- and third helix, a novel binding surface was engineered. The eleven positions are indicated in the figure and numbered according to Kraulis et al.9 . Following biopanning against a dimer of the Z-domain of Staphylococcal protein A with the combinatorial library expressed on phage, the bispecific ABDz1 molecule was identified.
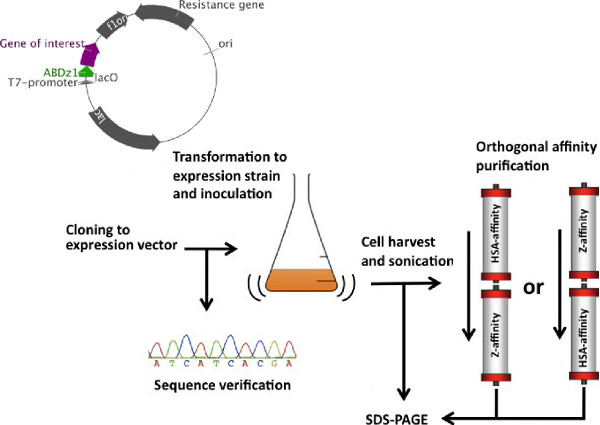 Figure 2. A simplified flow chart for the orthogonal affinity purification protocol. A PCR-fragment containing the ABDz1 sequence is ligated (N-terminally) with a gene of interest in a suitable expression vector. The ligation product is transformed to E. coli for sequence verification and plasmid preparation. Following transformation to an expression host, a large-scale culture for recombinant protein expression is set up from an initial overnight culture. Bacteria are harvested by centrifugation and lysed by sonication to produce bacterial lysate containing the target protein. After a filtration step to remove residual solid particles, the lysate is subjected to orthogonal affinity purification on a liquid handling system by undergoing two purification steps in succession. Eluted peaks from both purification steps are sampled and evaluated by SDS-PAGE to assess purity and compared to the lysate before purification.
Figure 2. A simplified flow chart for the orthogonal affinity purification protocol. A PCR-fragment containing the ABDz1 sequence is ligated (N-terminally) with a gene of interest in a suitable expression vector. The ligation product is transformed to E. coli for sequence verification and plasmid preparation. Following transformation to an expression host, a large-scale culture for recombinant protein expression is set up from an initial overnight culture. Bacteria are harvested by centrifugation and lysed by sonication to produce bacterial lysate containing the target protein. After a filtration step to remove residual solid particles, the lysate is subjected to orthogonal affinity purification on a liquid handling system by undergoing two purification steps in succession. Eluted peaks from both purification steps are sampled and evaluated by SDS-PAGE to assess purity and compared to the lysate before purification.
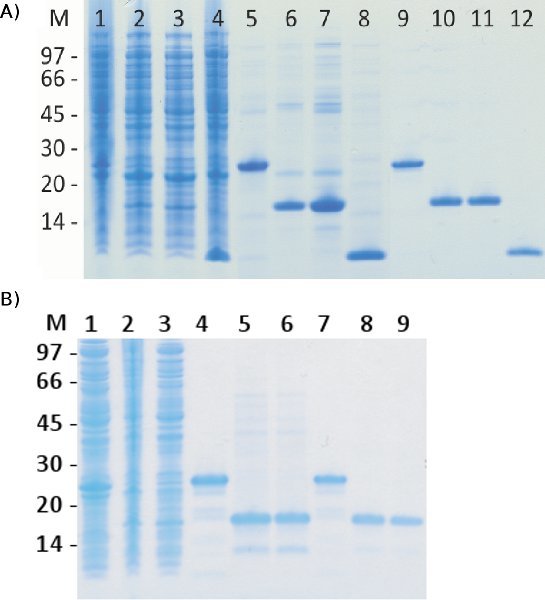 Figure 3. SDS-PAGE analysis of target proteins expressed and purified in fusion with ABDz1. Samples taken from the bacterial lysate of three proteins expressed in fusion to ABDz1, representing different properties, and the ABDz1 tag itself were collected together with samples from the corresponding peaks from purification on a HSA-column followed by a MabSelect SuRe-column (A). Lanes 1-4 represent samples from lysates before purification, lanes 5-8 from the HSA-purification (first step) and lanes 9-12 from the MabSelect SuRe purification (second step). Samples are loaded in the following order: ABDz1-141377, ABDz1-HT875, ABDz1-HT2375 and ABDz1. In addition, samples acquired from the same lysates purified in the reverse order on the same columns were analyzed (B). Lanes 1-3 represent samples from lysates before purification, lanes 4-6 from MabSelect SuRe-purification (first step) and lanes 7-9 from HSA-purification (second step). Samples were loaded in the same order as in (A) but the tag itself is not included. From those results it is clear that this two-step method yields highly purified proteins regardless of the order in which the steps are applied.
Figure 3. SDS-PAGE analysis of target proteins expressed and purified in fusion with ABDz1. Samples taken from the bacterial lysate of three proteins expressed in fusion to ABDz1, representing different properties, and the ABDz1 tag itself were collected together with samples from the corresponding peaks from purification on a HSA-column followed by a MabSelect SuRe-column (A). Lanes 1-4 represent samples from lysates before purification, lanes 5-8 from the HSA-purification (first step) and lanes 9-12 from the MabSelect SuRe purification (second step). Samples are loaded in the following order: ABDz1-141377, ABDz1-HT875, ABDz1-HT2375 and ABDz1. In addition, samples acquired from the same lysates purified in the reverse order on the same columns were analyzed (B). Lanes 1-3 represent samples from lysates before purification, lanes 4-6 from MabSelect SuRe-purification (first step) and lanes 7-9 from HSA-purification (second step). Samples were loaded in the same order as in (A) but the tag itself is not included. From those results it is clear that this two-step method yields highly purified proteins regardless of the order in which the steps are applied.
Molecular weight of ABDz1 tag alone is 6.3 kDa, isoelectric point 6.7.
Target protein represents a part of the Uniprot protein.
Solubility class of protein fragment with N-terminal His6ABP10 .
Table 1. Target proteins and fusion products with ABDz1 taga evaluated in a proof of principle study.
Three unique human proteins with different molecular weight, solubility and isoelectric point were chosen for expression and purification by the orthogonal affinity approach.
Discussion
The orthogonal affinity purification protocol presented here enables efficient purification of a wide range of target proteins. By combining the inherent binding site of the albumin-binding domain with a novel binding surface, a small bispecific protein tag was developed. It is very straightforward to utilize the tag since it can simply be cloned and expressed in fusion to any protein of interest in any preferred expression vector. Standard equipment available in most laboratories can be employed for the two-step protein purification. Since the function of the ABDz1 tag depends on correct folding of the domain to efficiently expose its two binding surfaces, the method is limited to proteins expressed in soluble form and not suited for purifications under denaturing conditions. Reducing agents should also be avoided since they interfere with the binding of ABDz1 to the Z-domain on the MabSelect SuRe matrix.
Orthogonal affinity purification provides a useful alternative to traditional methods in order to satisfy the requirements of highly purified proteins in many demanding applications. At the same time, this method eliminates the need for combinatorial tagging when different purification strategies are used in succession. For applications requiring a native target protein, a protease cleavage site may be introduced to render enzymatic removal of the ABDz1 tag possible11 . Furthermore, the ABDz1 tag is the first bispecific small and folded protein domain described to date. It is unique since it provides a purification strategy that depends on two target specific affinity interactions. In the future it would be interesting to expand this concept by developing novel bispecific tags that carry new binding surfaces that are compatible with readily available and inexpensive chromatographic resins. For example, by replacing the amino acids involved in albumin binding with basic residues, a tag compatible with ion exchange chromatography may be achievable. A similar concept has been proven effective by charge engineering of the Z-domain to generate tags known as Zacid12 and Zbasic13 compatible with anion- and cation exchange, respectively.
Disclosures
No conflicts of interest declared.
Acknowledgments
This project was funded by the Knut and Alice Wallenberg Foundation and the Swedish Research Council.
References
- Waugh DS. Making the most of affinity tags. Trends Biotechnol. 2005;23:316–320. doi: 10.1016/j.tibtech.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Hedhammar M. Protein engineering strategies for selective protein purification. CET. 2005;28:1315–1325. [Google Scholar]
- Porath J. Metal chelate affinity chromatography, a new approach to protein fractionation. Nature. 1975;258:206–213. doi: 10.1038/258598a0. [DOI] [PubMed] [Google Scholar]
- Linhult M. Mutational analysis of the interaction between albumin-binding domain from streptococcal protein G and human serum albumin. Protein Sci. 2002;11:206–213. doi: 10.1110/ps.02802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson B. A synthetic IgG-binding domain based on staphylococcal protein. A. Protein Eng. 1987;1:107–113. doi: 10.1093/protein/1.2.107. [DOI] [PubMed] [Google Scholar]
- Alm T. A small bispecific protein selected for orthogonal affinity purification. Biotechnol. J. 2010;5:605–617. doi: 10.1002/biot.201000041. [DOI] [PubMed] [Google Scholar]
- Nilsson J. Multiple affinity domains for the detection, purification and immobilization of recombinant proteins. J. Mol. Recognit. 1996;9:585–594. doi: 10.1002/(sici)1099-1352(199634/12)9:5/6<585::aid-jmr306>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Rüther U. pUR 250 allows rapid chemical sequencing of both DNA strands of its inserts. Nucleic Acids Res. 1982;10:5765–5772. doi: 10.1093/nar/10.19.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis PJ. The serum albumin-binding domain of streptococcal protein G is a three-helical bundle: a heteronuclear NMR study. FEBS Lett. 1996;378:190–194. doi: 10.1016/0014-5793(95)01452-7. [DOI] [PubMed] [Google Scholar]
- Hedhammar M. Novel flow cytometry-based method for analysis of expression levels in E coli giving information about precipitated and soluble protein. J. Biotechnol. 2005;119:133–146. doi: 10.1016/j.jbiotec.2005.03.024. [DOI] [PubMed] [Google Scholar]
- Carter P. Site-specific proteolysis of fusion proteins. ACS Sympos. Ser. 1990;427:181–193. [Google Scholar]
- Hedhammar M. Negatively charged purification tags for selective anion-exchange recovery. PEDS. 2004;17:779–786. doi: 10.1093/protein/gzh092. [DOI] [PubMed] [Google Scholar]
- Gräslund T. Charge engineering of a protein domain to allow efficient ion-exchange recovery. Protein Eng. 2000;13:703–709. doi: 10.1093/protein/13.10.703. [DOI] [PubMed] [Google Scholar]


