Abstract
The field of human trophoblast research aids in understanding the complex environment established during placentation. Due to the nature of these studies, human in vivo experimentation is impossible. A combination of primary cultures, explant cultures and trophoblast cell lines1 support our understanding of invasion of the uterine wall2 and remodeling of uterine spiral arteries3,4 by extravillous trophoblast cells (EVTs), which is required for successful establishment of pregnancy. Despite the wealth of knowledge gleaned from such models, it is accepted that in vitro cell culture models using EVT-like cell lines display altered cellular properties when compared to their in vivo counterparts5,6. Cells cultured in the rotating cell culture system (RCCS) display morphological, phenotypic, and functional properties of EVT-like cell lines that more closely mimic differentiating in utero EVTs, with increased expression of genes mediating invasion (e.g. matrix metalloproteinases (MMPs)) and trophoblast differentiation7,8,9. The Saint Georges Hospital Placental cell Line-4 (SGHPL-4) (kindly donated by Dr. Guy Whitley and Dr. Judith Cartwright) is an EVT-like cell line that was used for testing in the RCCS.
The design of the RCCS culture vessel is based on the principle that organs and tissues function in a three-dimensional (3-D) environment. Due to the dynamic culture conditions in the vessel, including conditions of physiologically relevant shear, cells grown in three dimensions form aggregates based on natural cellular affinities and differentiate into organotypic tissue-like assemblies10,11,12 . The maintenance of a fluid orbit provides a low-shear, low-turbulence environment similar to conditions found in vivo. Sedimentation of the cultured cells is countered by adjusting the rotation speed of the RCCS to ensure a constant free-fall of cells. Gas exchange occurs through a permeable hydrophobic membrane located on the back of the bioreactor. Like their parental tissue in vivo, RCCS-grown cells are able to respond to chemical and molecular gradients in three dimensions (i.e. at their apical, basal, and lateral surfaces) because they are cultured on the surface of porous microcarrier beads. When grown as two-dimensional monolayers on impermeable surfaces like plastic, cells are deprived of this important communication at their basal surface. Consequently, the spatial constraints imposed by the environment profoundly affect how cells sense and decode signals from the surrounding microenvironment, thus implying an important role for the 3-D milieu13.
We have used the RCCS to engineer biologically meaningful 3-D models of various human epithelial tissues7,14,15,16. Indeed, many previous reports have demonstrated that cells cultured in the RCCS can assume physiologically relevant phenotypes that have not been possible with other models10,17-21. In summary, culture in the RCCS represents an easy, reproducible, high-throughput platform that provides large numbers of differentiated cells that are amenable to a variety of experimental manipulations. In the following protocol, using EVTs as an example, we clearly describe the steps required to three-dimensionally culture adherent cells in the RCCS.
Keywords: Bioengineering, Issue 59, Extravillous trophoblasts, cytotrophoblast, invasion, matrix metalloproteinase, 3-D cell culture, RCCS, ECM, microcarriers
Protocol
1. Collagen Bead Preparation
- Prior to loading EVTs for 3-D cell culture, one needs to prepare the Cytodex-3 microcarrier beads:
- Weigh out the appropriate amount of Cytodex-3 beads required for the experiment. This protocol is adapted for the 10ml RCCS vessel, in which 0.05g of beads are needed. For a 50ml RCCS vessel, scale accordingly. In a 50mL autoclavable conical tube, mix 250 mg Cytodex-3 beads with 12mL Dulbecco phosphate buffered solution (DPBS). This amount is sufficient for 5 RCCS vessels.
Ensure adequate volume is present in the conical tube, as the autoclave process will result in evaporation. Loosely cap the tube and autoclave for 10 min at 110°C.
Remove the conical tube at the completion of the autoclave cycle and allow the Cytodex-3 bead solution to cool.
After the conical tube has cooled to room temperature, use sterile technique to bring the total volume to 12.5mL using 1X DPBS.
Cap and store the prepared Cytodex-3 beads at room temperature. Allow the prepared beads to swell and cool to room temperature prior to use. The above preparation of Cytodex-3 beads will provide for five 10mL RCCS vessels. We have observed that extended storage of Cytodex-3 beads at room temperature will result in diminished performance, as the collagen will destabilize after approximately one month. Cytodex-3 beads range in size from 133-215 μm.
2. Media Preparation
Prepare 1L of RCCS optimized GTSF-2 media (adapted from 22), which consists of 40% MEM alpha, plus supplements and 60% L-15 Leibovitz's media (adapted from 22). First, prepare 400mL in total volume of MEM alpha supplemented with: 21.2mM sodium bicarbonate 0.06% Peptone 0.7mM Fructose 1.4mM Galactose 5.6mM Glucose 1% HEPES 1% L-glutamine 0.5% ITS 10% FBS
To prepare a stock solution of ITS, dissolve in 5mL sterile acidified H2O prepared by addition of glacial acetic acid (approx. 0.05mL). Swirl to dissolve, follow with 45mL sterile water.
Weigh out enough L-15 Leibovitz's powder for 600ml of medium by dissolving in tissue-culture grade H2O.
Bring the total volume of cell culture medium to 1L with L-15 Leibovitz's medium. Filter-sterilize and store at 4°C in the dark. Add 1% Penicillin-Streptomycin individually to each aliquot of medium used.
Many media formulations other than GTSF-2 media have been successfully tested in the RCCS. Individual laboratories must decide for themselves which medium is optimal for the experiment being performed.
3. Cells and Bead Incubation
Propagate the EVTs to ~80% confluency, trypsinize and count using accepted cell culture practices.
Suspend 1x106 EVTs in 4mL of warmed media.
Gently mix prepared Cytodex-3 beads. Using sterile techniques, remove 2.5mL of prepared beads using a wide tip, 10mL serological pipette and transfer to an unused 15mL conical tube. Note that there may be a small loss of beads as they attach to the pipette.
Allow the Cytodex-3 beads to settle on the bottom of the tube. After sedimentation, use a pipettor to remove the top layer of DPBS without disturbing the Cytodex-3 beads.
Mix the 1x106 prepared EVTs in media with the prepared Cytodex-3 beads.
Incubate the cell-bead mixture at room temperature for 30 min. Periodically mix gently. Incubate for a further 30 min at 37°C and 5% CO2. Periodically mix gently.
4. Loading the RWV
In a laminar flow cabinet, remove a 10mL RCCS from the packaging and place it in a sterile, 6-well culture plate for stability. Refer to Figure 1 for a labeled diagram of the RCCS.
Remove the large stopper from the port.
Bring the total volume of the incubating cell-bead mixture to 10mL with warmed media.
Load the cell/bead-mixture into the RCCS through the large port. The RCCS should be tilted at a 45° angle (large port up) when loading to aid in removal of potential air bubbles. Prevent build-up of positive pressure by filling the vessel slowly and steadily.
Replace the stopper in the large port.
Remove the pistons from the 3-mL syringes and place the empty syringes onto the small ports. Add 1-3mL of media to each syringe. Slowly open the two valves. Replace the syringe pistons on the syringes gently. Add media from one syringe until all bubbles are removed from the inside chamber.
Pick up the RCCS and gently tap the side while rotating it in front of you to check for bubbles. If there are any bubbles, you must get them out of the chamber, as air bubbles will interfere with formation of cell aggregates and introduce shear forces. Remove bubbles by rotating the RCCS until the bubbles are under the small port. Then gently push down on the syringe on the opposite side to force the bubbles into the port and out of the chamber.
Close one side port. Gently push down on the other syringe piston to introduce a small amount of positive pressure into the vessel, which prevents bubbles from forming. Close second valve.
Load the RCCS onto the rotor. Start the rotation at 19rpm in a 37°C CO2 incubator.
5. Changing the Media
Change the media every other day for the first three days, then every day thereafter.
Turn off the rotor and remove the RCCS. Pull the pistons up to create some suction, remove the syringes from each small port and place the RCCS on an angle so that the beads settle opposite of the large port.
After the beads have all settled, open one of the small valves and allow the media to flow out of the RCCS and into a waste container. Remove 2/3 of the media in this manner. Be sure to not disturb the beads and to not discard any aggregates.
Close the small valve and open the large port. Add media back into the RCCS through the large port. Replace the stopper in the large port.
Repeat steps 4.6 - 4.9.
As the aggregates increase in size, you need to increase the rotation speed to keep them in suspension at all times, ideally in a small circulatory pattern at optimal speed. Generally, increase the speed between 0.3-0.7 rpm after each feeding once the aggregates start growing visibly.
6. Collecting Propagated Cells
Remove the RCCS from the rotor. Remove the syringes from each small port and place the RCCS on an angle so that the beads may settle opposite of the large port.
After the beads have all settled, open one of the small valves and allow the media to flow out of the RCCS and into a waste container. Remove 1/3 of the media in this manner.
Close the small valve and open the large port. Gently swirl the vessel to disperse the aggregates back into solution. Empty liquid mixture into a sterile 50ml conical tube. Allow the collected aggregates to settle in the conical tube before using supernatant to thoroughly wash the culture vessel to maximize recovery of aggregates. The aggregates may be used for downstream assays immediately, such as flow cytometry, invasion assays, immunofluorescence and others.
7. Representative Results
An example of EVT-like cells (SGHPL-4 trophoblast cell line) grown in the RCCS on Cytodex-3 beads is shown in Figure 2. The EVT-like cell line displays projections extending away from the main cluster and attaching to neighboring clusters. Many of the beads are completely covered with propagating cells. Once removed from the RCCS and plated on an extracellular matrix, the EVT-like 3-D grown cells aggressively invade and/or migrate (Figure 3). RT-PCR data confirms the increased expression of MMPs seen in 3-D aggregates as opposed to traditional cell culture monolayers (Figure 4). Interestingly, genes not associated with invasion are also upregulated in the RCCS (Table 1) and may aid in delineating such areas as immune interactions with invading trophoblasts cells.
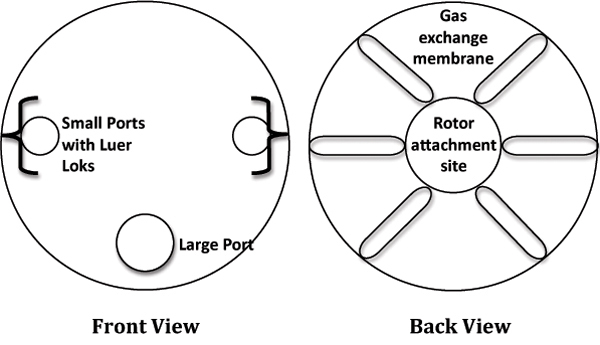 Figure 1. Cartoon depiction of the Rotating Cell Culture System (RCCS).
Figure 1. Cartoon depiction of the Rotating Cell Culture System (RCCS).
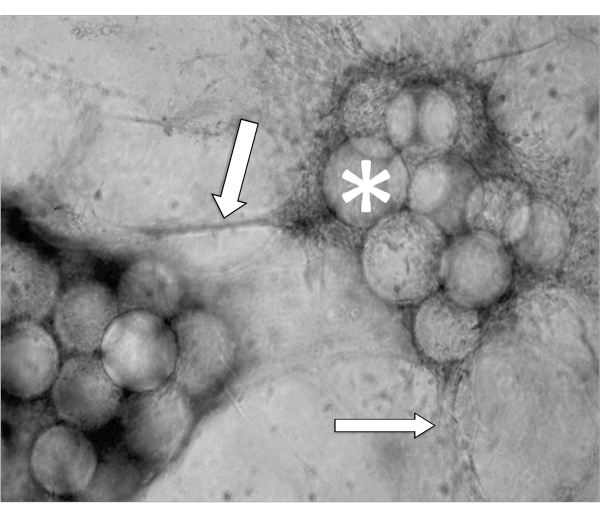 Figure 2. Phase contrast micrograph of a representative aggregate after 5 days growth in a RCCS. Arrows indicate invading cells and * denotes the Cytodex-3 microcarrier beads.
Figure 2. Phase contrast micrograph of a representative aggregate after 5 days growth in a RCCS. Arrows indicate invading cells and * denotes the Cytodex-3 microcarrier beads.
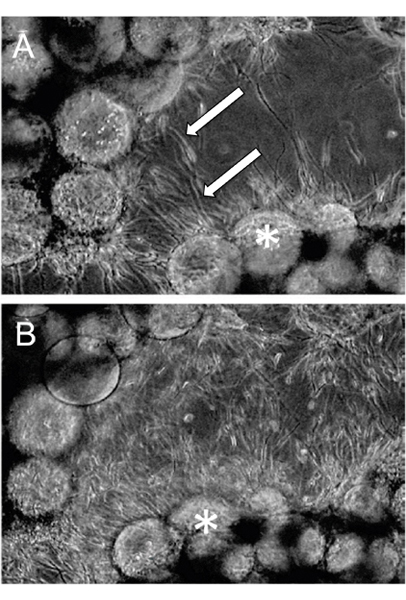 Figure 3. RCCS Grown aggregates were embedded in fibrin gels. Invasion through the fibrin gel was observed as early as (A) 24 hours post-fibrin embedding and (B) continued through 48 hours. Arrows indicate invading cells and * denotes the Cytodex-3 microcarrier beads (Adapted, with permission, from Ref. 7).
Figure 3. RCCS Grown aggregates were embedded in fibrin gels. Invasion through the fibrin gel was observed as early as (A) 24 hours post-fibrin embedding and (B) continued through 48 hours. Arrows indicate invading cells and * denotes the Cytodex-3 microcarrier beads (Adapted, with permission, from Ref. 7).
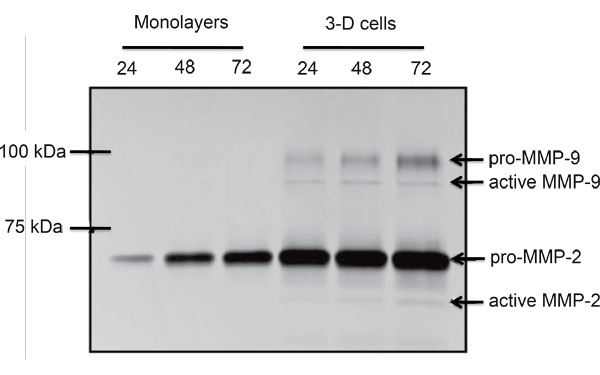 Figure 4. Comparative gelatin zymogram of monolayer and RCCS propagated SGHPL-4 EVT-like cells. Pro- and active forms of MMP-2 and MMP-9 were secreted by the RCCS grown trophoblast cells (Adapted, with permission, from Ref. 7).
Figure 4. Comparative gelatin zymogram of monolayer and RCCS propagated SGHPL-4 EVT-like cells. Pro- and active forms of MMP-2 and MMP-9 were secreted by the RCCS grown trophoblast cells (Adapted, with permission, from Ref. 7).
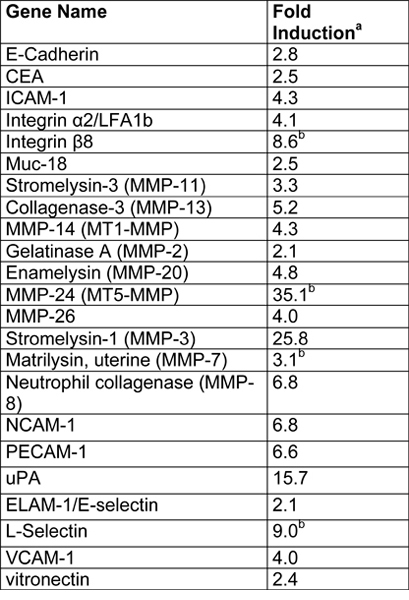 Table 1. Summary of microarray results from SGHPL-4 cells grown in a RCCS (Adapted, with permission, from Ref. 7).
Table 1. Summary of microarray results from SGHPL-4 cells grown in a RCCS (Adapted, with permission, from Ref. 7).
Fold induction in the 3-D aggregates was calculated as follows: normalized 3-D gene value/normalized monlayer gene value.
Indicates genes that were below the limit of detection in monolayers.
Discussion
The culture technique presented here provides investigators with highly invasive EVT-like cells. It has now been recognized that a loss of differentiation occurs in monolayers due to inhibition of cellular responses to chemical and molecular cues in three dimensions (apical, basal, and lateral cell surfaces)10,13. This technique reflects characteristics noted in utero on invading EVT cells. As the procedure mimics conventional monolayer tissue culture time kinetics, but provides cells with differential expression, comparative assays may be employed. Cell clusters may be harvested for use in experiments at any time. They may be used as clusters or collagen degradation may be employed to isolate cells as single cell suspension. The Cytodex-3 microcarrier beads may be substituted with scaffolds or microcarrier beads coated with other extracellular matrices (ECMs) that may be more conducive to individual laboratories' specific interests. Alternatively, there are many other 3-D culture platforms that may be more conducive to specific cell type culture conditions, like cell culture inserts and spinner flasks10,23. It should be noted that culture kinetics could be different under these circumstances and should be established individually. Culture kinetics may be easily tested for by removing samples from the RCCS at regular intervals and analyzing for expression of distinctive differentiation markers, proliferation, and viability15. Optimal proliferation and differentiation kinetics may be ascertained by comparing results over time and against conventional monolayer tissue culture techniques. Using the SGHPL-4 cells as an example, aggregates were removed daily and stained with Ki67 (to determine proliferation) and caspase-3 (to determine apoptosis), Markers specific to the individual laboratories' interests may be readily exchanged. The technique described herein can be applied to the study of spiral artery remodeling, shallow implantation, and maternal immune responses to invading trophoblasts cells.
As has been published previously, this 3-D cell culture model can also be used for a variety of other cell lines17,14-21. In every case, the utility of the model has to be tested by a variety of known differentiation markers. Like any in vitro cell culture model, 3-D cell cultures are inherently reductionistic. No single culture model will provide all of the mechanistic details, but when combined with published results from other models, 3-D cultures become an additional valuable alternative model for improving our understanding of disease-related mechanisms, which may hold enormous potential for the development of novel products and procedures in the diagnosis, prevention, and treatment of infectious diseases.
Disclosures
We have nothing to disclose.
Acknowledgments
This work was supported by the US National Institutes of Health grant NIH/NICHD #HD051998 (to C.A.M.).
References
- Knofler M. Critical growth factors and signalling pathways controlling human trophoblast invasion. Int. J. Dev. Biol. 2010;54:269–269. doi: 10.1387/ijdb.082769mk. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright JE. Remodelling at the maternal-fetal interface: relevance to human pregnancy disorders. Reproduction. 2010;140:803–803. doi: 10.1530/REP-10-0294. [DOI] [PubMed] [Google Scholar]
- Harris LK. IFPA Gabor Than Award lecture: Transformation of the spiral arteries in human pregnancy: key events in the remodelling timeline. Placenta. 2011;32(2):S154–S154. doi: 10.1016/j.placenta.2010.11.018. [DOI] [PubMed] [Google Scholar]
- Whitley GS, Cartwright JE. Trophoblast-mediated spiral artery remodelling: a role for apoptosis. J. Anat. 2009;215:21–21. doi: 10.1111/j.1469-7580.2008.01039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apps R. Genome-wide expression profile of first trimester villous and extravillous human trophoblast cells. Placenta. 2011;32:33–33. doi: 10.1016/j.placenta.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilban M. Trophoblast invasion: assessment of cellular models using gene expression signatures. Placenta. 2010;31:989–989. doi: 10.1016/j.placenta.2010.08.011. [DOI] [PubMed] [Google Scholar]
- LaMarca HL. Three-dimensional growth of extravillous cytotrophoblasts promotes differentiation and invasion. Placenta. 2005;26:709–709. doi: 10.1016/j.placenta.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Jovanovic M, Stefanoska I, Radojcic L, Vicovac L. Interleukin-8 (CXCL8) stimulates trophoblast cell migration and invasion by increasing levels of matrix metalloproteinase (MMP)2 and MMP9 and integrins alpha5 and beta1. Reproduction. 2010;139:789–789. doi: 10.1530/REP-09-0341. [DOI] [PubMed] [Google Scholar]
- Husslein H. Expression, regulation and functional characterization of matrix metalloproteinase-3 of human trophoblast. Placenta. 2009;30:284–284. doi: 10.1016/j.placenta.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrila J. Organotypic 3D cell culture models: using the rotating wall vessel to study host-pathogen interactions. Nat. Rev. Microbiol. 2010;8:791–791. doi: 10.1038/nrmicro2423. [DOI] [PubMed] [Google Scholar]
- Hammond TG, Hammond JM. Optimized suspension culture: the rotating-wall vessel. Am. J. Physiol. Renal. Physiol. 2001;281:12–12. doi: 10.1152/ajprenal.2001.281.1.F12. [DOI] [PubMed] [Google Scholar]
- Unsworth BR, Lelkes PI. Growing tissues in microgravity. Nat. Med. 1998;4:901–901. doi: 10.1038/nm0898-901. [DOI] [PubMed] [Google Scholar]
- Schmeichel KL, Bissell MJ. Modeling tissue-specific signaling and organ function in three dimensions. J. Cell. Sci. 2003;116:2377–2377. doi: 10.1242/jcs.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentrup Hönerzu, K Three-dimensional organotypic models of human colonic epithelium to study the early stages of enteric salmonellosis. Microbes. Infect. 2006;8:1813–1813. doi: 10.1016/j.micinf.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Carterson AJ. A549 lung epithelial cells grown as three-dimensional aggregates: alternative tissue culture model for Pseudomonas aeruginosa pathogenesis. Infect. Immun. 2005;73:1129–1129. doi: 10.1128/IAI.73.2.1129-1140.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers TA. Closing the phenotypic gap between transformed neuronal cell lines in culture and untransformed neurons. J. Neurosci. Methods. 2008;174:31–31. doi: 10.1016/j.jneumeth.2008.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelm BE. Development and characterization of a three-dimensional organotypic human vaginal epithelial cell model. Biol. Reprod. 2010;82:617–617. doi: 10.1095/biolreprod.109.080408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub TM. In vitro cell culture infectivity assay for human noroviruses. Emerg. Infect. Dis. 2007;13:396–396. doi: 10.3201/eid1303.060549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson CA. Three-dimensional tissue assemblies: novel models for the study of Salmonella enterica serovar Typhimurium pathogenesis. Infect. Immun. 2001;69:7106–7106. doi: 10.1128/IAI.69.11.7106-7120.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho HM, Teel LD, Goping G, O'Brien AD. A three-dimensional tissue culture model for the study of attach and efface lesion formation by enteropathogenic and enterohaemorrhagic Escherichia coli. Cell. Microbiol. 2005;7:1771–1771. doi: 10.1111/j.1462-5822.2004.00594.x. [DOI] [PubMed] [Google Scholar]
- Sainz B, TenCate V, Uprichard SL. Three-dimensional Huh7 cell culture system for the study of Hepatitis C virus infection. Virol. J. 2009;6:103–103. doi: 10.1186/1743-422X-6-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelkes PI, Ramos E, Nikolaychik VV, Wankowski DM, Unsworth BR, Goodwin TJ. GTSF-2: a new, versatile cell culture medium for diverse normal and transformed mammalian cells. In Vitro Cell. Dev. Biol. Anim. 1997;33:344–344. doi: 10.1007/s11626-997-0004-7. [DOI] [PubMed] [Google Scholar]
- Lelkes PI, Ramos E, Nikolaychik VV, Wankowski DM, Unsworth BR, Goodwin TJ. GTSF-2: a new, versatile cell culture medium for diverse normal and transformed mammalian cells. In Vitro Cell. Dev. Biol. Anim. 1997;33:344–344. doi: 10.1007/s11626-997-0004-7. [DOI] [PubMed] [Google Scholar]
- GE Healthcare. Microcarrier Cell Culture - Principles and Methods. Handbooks. 2005. Available from: http://www.gelifesciences.com/aptrix/upp01077.nsf/Content/service_and_support~documents_and_downloads~handbooks.


