Abstract
The central nervous system can experience a number of stresses and neurological insults, which can have numerous adverse effects that ultimately lead to a reduction in neuronal population and function. Damaged axons can release excitatory molecules including potassium or glutamate into the extracellular matrix, which in turn, can produce further insult and injury to the supporting glial cells including astrocytes and oligodendrocytes 8, 16. If the insult persists, cells will undergo programmed cell death (apoptosis), which is regulated and activated by a number of well-established signal transduction cascades 14. Apoptosis and tissue necrosis can occur after traumatic brain injury, cerebral ischemia, and seizures. A classical example of apoptotic regulation is the family of cysteine-dependent aspartate-directed proteases, or caspases. Activated proteases including caspases have also been implicated in cell death in response to chronic neurodegenerative diseases including Alzheimer's, Huntington's, and Multiple Sclerosis 4, 14, 3, 11, 7.
In this protocol we describe the use of the NucView 488 caspase-3 substrate to measure the rate of caspase-3 mediated apoptosis in immortalized N19-oligodendrocyte (OLG) cell cultures 15, 5, following exposure to different extracellular stresses such as high concentrations of potassium or glutamate. The conditionally-immortalized N19-OLG cell line (representing the O2A progenitor) was obtained from Dr. Anthony Campagnoni (UCLA Semel Institute for Neuroscience) 15, 5, and has been previously used to study molecular mechanisms of myelin gene expression and signal transduction leading to OLG differentiation (e.g.6, 10). We have found this cell line to be robust with respect to transfection with exogenous myelin basic protein (MBP) constructs fused to either RFP or GFP (red or green fluorescent protein) 13, 12. Here, the N19-OLG cell cultures were treated with either 80 mM potassium chloride or 100 mM sodium glutamate to mimic axonal leakage into the extracellular matrix to induce apoptosis 9. We used a bi-functional caspase-3 substrate containing a DEVD (Asp-Glu-Val-Asp) caspase-3 recognition subunit and a DNA-binding dye 2. The substrate quickly enters the cytoplasm where it is cleaved by intracellular caspase-3. The dye, NucView 488 is released and enters the cell nucleus where it binds DNA and fluoresces green at 488 nm, signaling apoptosis. Use of the NucView 488 caspase-3 substrate allows for live-cell imaging in real-time 1, 10. In this video, we also describe the culturing and transfection of immortalized N19-OLG cells, as well as live-cell imaging techniques.
Keywords: Neuroscience, Issue 59, myelin basic protein, apoptosis, neuroprotection, caspase-3, live-cell imaging, glia, oligodendrocytes
Protocol
1. Thawing and Culturing Cells
Obtain stock of frozen immortalized N19-oligodendroglial cells from long term liquid nitrogen stores.
Immerse vial containing cells at 37 °C water bath until the cell suspension is completely thawed.
Add 7 mL of DMEM (Dulbecco's Modified Eagle Medium) with high-glucose supplemented with 10% FBS (Fetal Bovine Serum) and 1% penicillin/streptomycin to a 10 cm culture plate.
Add the cell-suspension drop-wise to the plate, and gently agitate and rock the Petri plate to disperse the cells evenly.
Culture cells at 34 °C/5% CO2 incubator.
After 4 hours, aspirate media from plates to remove any remaining DMSO (dimethyl sulphoxide) that had been used as a cryoprotectant, and replace with fresh media (7 mL DMEM high-glucose media supplemented with 10% FBS and 1% penicillin/streptomycin).
2. Passaging Cells
At 70-80% confluence (4-7 days growth), aspirate media from cells.
Add 1 mL of 0.25% trypsin to plate. Pipette trypsin to detach cells (about 5 minutes).
Make the appropriate dilutions for experimental conditions and passage an additional 10 cm plate for future experiments at a cell density no less that 0.1 x 106 cells / mL.
Note: You should passage your cells at least twice after thawing before using them for experiments.
3. Counting and Plating Cells
To plate cells for live-cell imaging, trypsinize as previously described.
Remove approximately 30 μL of cells and count using a haemocytometer.
Place an uncoated glass coverslip in a 6 well plate. Add cells to the well at a density of 0.1 x 106 cells/mL in 2 mL phenol-free DMEM high-glucose media, supplemented with 10% FBS and 1% penicillin/streptomycin, at 34 °C/5% CO2.
Allow cells to grow overnight (16-20 h) at 34 °C/5% CO2 before transfection.
4. Transfection
Combine 100 μL serum-free media, 0.5-4 μg purified plasmid DNA, and 4 μL FuGENE HD (Roche). Vortex gently to mix.
Vortex again briefly and allow the DNA to complex for 5 min at room temperature, and then add FuGENE HD DNA mixture directly to the cultured cells. Tilt the plate gently to mix.
Culture the cells for an additional 48 h at 34 °C/5% CO2 prior to treatment or experimentation.
5. Preparing Cells for Live-Cell Imaging (LCI)
Turn on the LCI Chamlide live-cell instrument control box at least 1 h before you want to start your experiment. The control box also regulates the temperature and humidity within the LCI Chamlide, and regulates the flow of the premixed 5% CO2.
Note: It is advisable to turn on the control box up to 3 h before beginning experiments to ensure that the entire stage reaches 34 °C. This step will reduce focal drift, which is caused by fluxulation and thermal expansion of the metal stage as it warms, during imaging.
Working in a flow-hood, spray the Chamlide magnetic-type culture chamber, and tweezers, with 70% ethanol and allow them to dry for 5 minutes.
Remove cells from incubator and confirm that they are healthy. They should appear adherent and well-spread on the glass coverslip with numerous membrane processes. Cells that are stressed from transfection are not suitable for experimentation, and will have a reduced number of process extensions, and often have an irregularly-shaped nucleus.
Tilt the 6-well plate and remove the coverslip with tweezers. Quickly place the coverslip cell side up into the bottom plate of the culture chamber. Do not allow the coverslip to dry.
Attach the magnetic main body of the culture chamber and add 500 μL of media from the original 6-well plate onto the top of the coverslip. Re-using this media will decrease the amount of stress placed on the cells caused by environmental changes, and can also be useful for assessing extracellular secreted factors.
Place the glass cover on the culture chamber.
Use a KimWipe sprayed with 70% ethanol to remove any residual material from the bottom of the coverslip, which would interfere with microscopy.
Place the culture chamber in the 34 °C/5% CO2 incubator for 30 min. This step will ensure that the chamber itself warms to 34 °C to reduce shifting of the coverslip as the metal warms up.
6. Microscope Settings
Images were acquired here using a Leica DMIRE2 inverted microscope with a custom relay lens and emission filter wheel housing for cartridge loading of multiple wheels (Quorum Technologies Inc., Guelph, ON).
Brightfield - Lamp set to 2.5 V and exposure time to 240 ms.
Red Fluorescent Protein (RFP) - Gain set at 140 for 200 ms.
Green Fluorescent Protein (GFP) - Gain set at 140 for 500 ms.
Our microscope has a dimmable lamp rather than a spinning disk to control the amount of light available to cells. We run our experiments with the light at 90% of maximum lamp intensity.
7. Treatment of Cells with Apoptosis Inducers and NucView 488 Caspase-3 substrate
It is important to prepare large stocks of apoptosis inducers ahead of time and freeze them in aliquots, so that the concentrations will be consistent between experiments.
The NucView 488 substrate is light sensitive. Prepare aliquots of 15 μL to reduce freeze/thawing, and store tubes at -20 °C covered in aluminum foil. Work with the ambient room lighting as low as possible to reduce exposure of the NucView 488 substrate to light, prior to its use.
Retrieve the culture chamber quickly from the incubator so that the cells are not exposed to a reduction in temperature. Place the culture chamber onto the environmental chamber of the microscope, and use clamps to keep the culture chamber from moving. At this point you should also dim the room lights.
Turn on the 5% premixed CO2 tank with dual regulator.
Using the 10x objective, focus to find cells on the computer monitor using bright-field microscopy. Note: you would want to use the largest numerical aperture on the objective with the desired magnification to reduce the exposure time.
- Deliver inducers of apoptosis to the cells (in our case 80 mM potassium, or 100 mM glutamate), one of the following methods can be used. We will use delivery of 80 mM potassium as an example, using KCl dissolved as a 10x concentrate in our typical culture media.
- Media exchange by slow and local perfusion: - Use a peristaltic pump to exchange media in the culture chamber with new media containing 80 mM potassium. - One end of the tubing will deliver 10 mL of a stock of 80 mM potassium dissolved in phenol-free DMEM, and the other end of the tubing will remove media from the chamber. - You want at least a 10x exchange of media to ensure that the media left in the chamber has the correct concentration of potassium. - After media is exchanged, add 3 μL of NucView 488 substrate to the media in the chamber. Pipette to mix.
- Direct addition of 80 mM K+ and NucView 488 substrate to media in culture chamber: - Prepare a 10x stock solution of 80 mM potassium dissolved in phenol-free DMEM. - Add 3 μL of NucView 488 substrate to 50 μL of 10x 80 mM potassium. Pipette to mix. Add to 500 μL phenol-free DMEM in culture chamber. Note: this method is preferred if you are interested in maintaining or assessing growth or other secreted factors that may be present in the original media. We have found that the NucView 488 substrate is stable in cell culture for experiments lasting as long as 36 h.
8. Live-cell Imaging
Click on the red channel to display transfected cells.
Select and save around a dozen frames where there are several transfected cells, and save these "X-Y stage points". Depending on the amount of computer memory, you may be limited to the number of stage points that you will be able to acquire.
Have the microscope capture images (in bright-field, red channel, and green channel) at these saved stage points every 6 minutes.
Any green staining that is visible during the early stages of the experiments likely indicates cells that are already undergoing apoptosis, due usually to transfection and/or environmental stress. Apoptosis due to the experimental treatments will be detected at a later timepoint in the experiment.
9. Statistical Analysis
For each experiment, we program the microscope to acquire multiple X-Y points to gather a large data set efficiently. Each experiment is performed in duplicate or triplicate, and data are compiled from separate experiments performed on different days. From each data set, we analyze up to 15 fields of view and compare the ratio of caspase-negative cells to caspase-positive cells (total number of cells in field of view/total number of caspase-positive cells).
The recorded measurements from each data set are grouped into a larger sample set, and are then compared to one another using an ANOVA table (p = 0.05). We show the standard errors of the mean (SEM) of each experiment, and then compare the difference in means by performing a Tukey means comparison test (p = 0.05) to determine which treatments are significantly different from each other.
10. Representative Results
We have described an experiment to illustrate how the NucView 488 substrate can indicate an increased rate of apoptosis of N19-OLG cell cultures following a treatment with a high extracellular potassium concentration. The N19-cells were transfected with RFP, and were either treated with 3 μL NucView 488 substrate (control), or 3 μL NucView 488 substrate and 80 mM [K+] (treatment). Cells were monitored and images were acquired over a 12 h time course, which is adequate for studies involving neurological insults (Figure 1A). In control conditions, we did not observe significant amounts of apoptosis compared to the 80 mM [K+] treated cultures (hashed box), which showed approximately 45% cell death after 12 h (Figure 1B). The background green signal observed in the control conditions indicates the cells that are undergoing apoptosis without the addition of extracellular potassium. Virtually none of the cells in the control experiment exhibit apoptosis by the 12 h time point, although in other situations the experiments may be required to run longer. Images were acquired using a 10x objective. Bar = 100 μm.
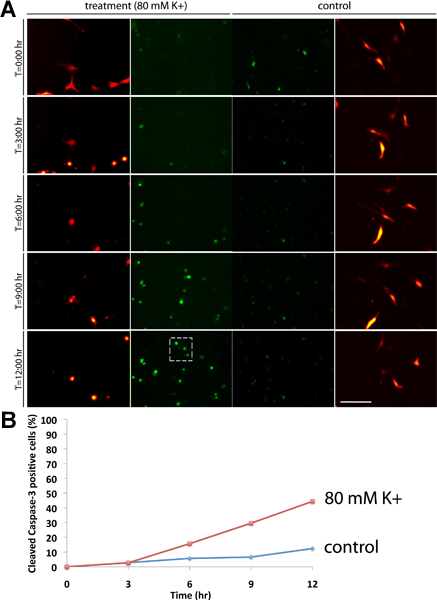 Figure 1. (A) A 12 h time course experiment of N19 OLG cultures 48 h post-transfection expressing RFP-MBP (red channel) along with 3 μL of NucView 488 substrate (green channel). Cultures were either treated with a final concentration of 80 mM [K+] (left panels), or no treatment as a control (right panels). Images were acquired at 6 min intervals, and significant activation of cleaved caspase-3 (green signal) can be observed in cultures treated with 80 mM [K+] within the cell nuclei (hashed box) compared to the control experiment. (B) Percentage of cleaved caspase-3 cells (calculated by dividing the total number of cells in the field of view by the total number of caspase-positive cells). In comparison to control conditions, we observed around 45% cell death following K+-treatment by 12 h.
Figure 1. (A) A 12 h time course experiment of N19 OLG cultures 48 h post-transfection expressing RFP-MBP (red channel) along with 3 μL of NucView 488 substrate (green channel). Cultures were either treated with a final concentration of 80 mM [K+] (left panels), or no treatment as a control (right panels). Images were acquired at 6 min intervals, and significant activation of cleaved caspase-3 (green signal) can be observed in cultures treated with 80 mM [K+] within the cell nuclei (hashed box) compared to the control experiment. (B) Percentage of cleaved caspase-3 cells (calculated by dividing the total number of cells in the field of view by the total number of caspase-positive cells). In comparison to control conditions, we observed around 45% cell death following K+-treatment by 12 h.
sVideo 1. A 12 h time course experiment of N19-OLG cultures 48 h post-transfection expressing RFP-MBP (red channel) with 3 μL of NucView 488 substrate (green channel), along with bright-field images and a three-way merged image, following treatment with 80 mM [K+]. Please click here to see/download this video file.
sVideo 2. A 12 h time course experiment of N19-OLG cultures 48 h post-transfection expressing RFP-MBP (red channel) with 3 μL of NucView 488 substrate (green channel), along with bright-field images and a three-way merged image. No treatment was applied to the cultures and N19-OLGs can be seen migrating through the microscope field in contrast to cell cultures treated with 80 mM [K+] (compare with sVideo 1). Please click here to see/download this video file.
Discussion
Although this is not the only fluorogenic product available for apoptosis detection, there are several significant advantages to using the NucView 488 substrate. One of the main benefits is the ability to follow apoptosis in live cells in real time, whereas most alternative products either require cell lysis or have poor cell permeability. Other benefits include high sensitivity for caspase-3 recognition, high cell permeability, low cytotoxicity, and no interference with the progression of apoptosis. The substrate also has low background fluorescence until it is cleaved and enters the nucleus, which eliminates background fluorescence. The caspase-3 recognition sequence contains 3 negative charges and the DNA-binding dye has one positive charge 2. The DEVD-NucView 488 molecule thus has a net negative charge, which prevents the activation and binding of the dye to DNA in cells where caspase is not active.
Maintaining consistency between experiments is required to be able to draw meaningful comparisons between replicates. One of the main parameters to keep consistent is cell density, as it affects the rate of transfection. Obtaining high transfection rates in immortalized N19-OLG cell cultures is more difficult than with other common cell lines such as HeLa or HEK293, and is highly dependent on density, in our experience 12, 13. Our best transfection efficiencies for these cells have been obtained with Fugene HD (Roche) and are normally about 15%, but can reach over 30%, depending on the construct. Careful cell counting will aid in obtaining consistent transfection rates. It is also important to expose cells from different treatments to the same degree of environmental stress, particularly when measuring rates of apoptosis. Specifically, the length of time over which the cells are exposed to transfection reagents, or the amount of light exposure, are important environmental variables to consider, and should remain constant across experiments. For our applications, we have found that collecting a large number of fields-of-view provides a sufficiently large sample size to make statistically-significant comparisons [ibid].
Disclosures
The authors have no conflicts of interest to disclose.
Acknowledgments
This laboratory has been supported by the Canadian Institutes of Health Research, the Natural Sciences and Engineering Research Council of Canada, and the Multiple Sclerosis Society of Canada (MSSC). GSTS was the recipient of a Doctoral Studentship from the MSSC. We are grateful to Dr. Joan Boggs (Hospital for Sick Children, Toronto) for many helpful discussions and comments on this manuscript. We are grateful to Biotium for their generous gift of additional NucView 488 caspase-3.
References
- Antczak C, Takagi T, Ramirez CN, Radu C, Djaballah H. Live-cell imaging of caspase activation for high-content screening. J. Biomol. Screen. 2009;14:956–969. doi: 10.1177/1087057109343207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen H, Mao F, Aronchik I, Fuentes RJ, Firestone GL. DEVD-NucView488: a novel class of enzyme substrates for real-time detection of caspase-3 activity in live cells. FASEB. J. 2008;22:2243–2252. doi: 10.1096/fj.07-099234. [DOI] [PubMed] [Google Scholar]
- de Calignon A, Fox LM, Pitstick R, Carlson GA, Bacskai BJ, Spires-Jones TL, Hyman BT. Caspase activation precedes and leads to tangles. Nature. 2010;464:1201–1204. doi: 10.1038/nature08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldadah BA, Faden AI. Caspase pathways, neuronal apoptosis, and CNS injury. J. Neurotrauma. 2000;17:811–829. doi: 10.1089/neu.2000.17.811. [DOI] [PubMed] [Google Scholar]
- Foster LM, Phan T, Verity AN, Bredesen D, Campagnoni AT. Generation and analysis of normal and shiverer temperature-sensitive immortalized cell lines exhibiting phenotypic characteristics of oligodendrocytes at several stages of differentiation. Dev. Neurosci. 1993;15:100–109. doi: 10.1159/000111322. [DOI] [PubMed] [Google Scholar]
- Fulton D, Paez PM, Fisher R, Handley V, Colwell CS, Campagnoni AT. Regulation of L-type Ca(++) currents and process morphology in white matter oligodendrocyte precursor cells by golli-myelin proteins. Glia. 2010;58:1292–1303. doi: 10.1002/glia.21008. [DOI] [PubMed] [Google Scholar]
- Hisahara S, Okano H, Miura M. Caspase-mediated oligodendrocyte cell death in the pathogenesis of autoimmune demyelination. Neurosci. Res. 2003;46:387–397. doi: 10.1016/s0168-0102(03)00127-5. [DOI] [PubMed] [Google Scholar]
- Lau A, Tymianski M. Glutamate receptors, neurotoxicity and neurodegeneration. Pflugers. Arch. 2010;460:525–542. doi: 10.1007/s00424-010-0809-1. [DOI] [PubMed] [Google Scholar]
- Lawrence MS, Ho DY, Sun GH, Steinberg GK, Sapolsky RM. Overexpression of Bcl-2 with herpes simplex virus vectors protects CNS neurons against neurological insults in vitro and in vivo. J. Neurosci. 1996;16:486–496. doi: 10.1523/JNEUROSCI.16-02-00486.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez PM, Spreuer V, Handley V, Feng JM, Campagnoni C, Campagnoni AT. Increased expression of golli myelin basic proteins enhances calcium influx into oligodendroglial cells. J. Neurosci. 2007;27:12690–12699. doi: 10.1523/JNEUROSCI.2381-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Mejia RO, Friedlander RM. Caspases in Huntington's disease. Neuroscientist. 2001;7:480–489. doi: 10.1177/107385840100700604. [DOI] [PubMed] [Google Scholar]
- Smith GST, De Avila M, Paez P, Spreuer V, Wills MKB, Jones N, Boggs JM, Harauz G. Proline substitutions and threonine pseudo-phosphorylation of the SH3-ligand of 18.5 kDa myelin basic protein decrease affinity for the Fyn-SH3-domain and alter process development and protein localization in oligodendrocytes. J. Neurosci. Res. 2011. Forthcoming. [DOI] [PMC free article] [PubMed]
- Smith GST, Paez PM, Spreuer V, Campagnoni CW, Boggs JM, Campagnoni AT, Harauz G. Classical 18.5-and 21.5-kDa isoforms of myelin basic protein inhibit calcium influx into oligodendroglial cells, in contrast to golli isoforms. J. Neurosci. Res. 2011;89:467–480. doi: 10.1002/jnr.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer JE, Azbill RD, Knapp PE. Activation of the caspase-3 apoptotic cascade in traumatic spinal cord injury. Nat. Med. 1999;5:943–946. doi: 10.1038/11387. [DOI] [PubMed] [Google Scholar]
- Verity AN, Bredesen D, Vonderscher C, Handley VW, Campagnoni AT. Expression of myelin protein genes and other myelin components in an oligodendrocytic cell line conditionally immortalized with a temperature-sensitive retrovirus. J. Neurochem. 1993;60:577–587. doi: 10.1111/j.1471-4159.1993.tb03188.x. [DOI] [PubMed] [Google Scholar]
- Yu SP. Regulation and critical role of potassium homeostasis in apoptosis. Prog. Neurobiol. 2003;70:363–386. doi: 10.1016/s0301-0082(03)00090-x. [DOI] [PubMed] [Google Scholar]


