Abstract
The functioning of the neuronal dendrite results from a variety of biological processes including mRNA transport to and protein translation in the dendrite. The complexity of the mRNA population in dendrites suggests that specific biological processes are modulated through the regulation of dendritic biology. There are various classes of mRNAs in dendrites whose translation modulates the ability of the dendrite to receive and integrate presynaptic information. Among these mRNAs are those encoding selective transcription factors that function in the neuronal soma and ionotropic glutamate receptors that function on the neuronal membrane. Conclusive evidence that these mRNAs can be translated is reviewed, and identification of the endogenous sites of translation in living dendrites is presented. These data, as well as those described in the other articles resulting from this colloquium, highlight the complexity of dendritic molecular biology and the exquisitely selective and sensitive modulatory role played by the dendrite in facilitating intracellular and intercellular communication.
Since the time of Cajal, it has been apparent that neurons have a striking morphological polarity. It was recognized with time that a neuron's morphological appearance relates to the function of these cells. Neurons typically have a cell soma from which multiple dendrites and a single axon protrude. Presynaptic cells communicate with postsynaptic cells through direct connections between the presynaptic axon and the postsynaptic dendrite. Presynaptic information is processed within the dendrites and transferred to the cell soma where additional signal integration occurs. The axon, in turn, transfers the postsynaptic cells' integrated response to the next postsyanptic neuron. Information processing in the dendrite is complex, involving both local dendritic and more global soma modulatory components.
Dendrites are the primary locus of postsynaptic connectivity and can comprise >90% of the postsynaptic surface of some neurons. In general the presynaptic axon forms synapses on dendritic spines of the postsynaptic membrane. The establishment of such postsynaptic specializations reflects how neurons elaborate discrete plasma membrane domains that differ focally in their regulatory properties. Such focal domains are critical in determining specificity of information flow in the neuronal circuitry of the central nervous system. In response to prolonged periods of synaptic plasticity, including long-term potentiation and long-term depression, subsets of these synapses undergo a series of enduring changes in spine shape and density as well as electrophysiological characteristics. These changes result, in part, from local regulation of the functioning of the postsynaptic density (a protein–mRNA complex present in the dendritic spine). In this paper, we review some of the work from our laboratory detailing aspects of neuronal dendrite functioning.
mRNA Complexity of Individual Neuronal Dendrites
It has been clear since the mid-1960s (1) that RNAs are localized in dendrites, with the first RNAs found being ribosomal RNA as visualized by electron microscopy. It took several more years for mRNAs to be conclusively shown to be localized to dendrites. For nearly a decade ribosomal RNA and a handful of mRNAs were the only ones known to exist in dendrites (2). In 1994, Miyashiro and colleagues (3) used mRNA amplification techniques to show that many more mRNAs existed in dendrites including the mRNAs for all of the ionotropic glutamate receptors and various mRNAs encoding proteins involved in modulating the translation of proteins. The repertoire of dendritically localized mRNAs was further expanded through the work of Crino and Eberwine (4) in which expression profiling and differential display showed the presence of more than 30 identified and many additional expressed sequence tag mRNAs in dendritic processes. These studies also established that there is molecular individuality in neuronal dendrites, because different processes can contain different mRNAs. These studies initially were controversial because some of the mRNAs, such as the ionotropic glutamate receptor subunit mRNAs, had been considered absent from dendrites based on the inability to detect the mRNAs by in situ hybridization. It is important to note that the earlier in situ hybridization papers suffered from a lack of technique sensitivity. With improved sensitivity many of these dendritically localized mRNAs, including members of the glutamate receptor family, have been shown to be present in dendrites by in situ hybridization methodologies.
Indeed, using antisense RNA (aRNA) amplification of dendritic mRNA followed by differential display and microarray analysis, we estimate that ≈400 mRNAs can be localized to dendrites of rat hippocampal neurons in primary cell culture (Fig. 1) (3, 5, 6). In Fig. 1A, a cultured rat hippocampal neuron is shown. The processes marked HP 3–6 and HP 3–5 were independently harvested and the endogenous mRNA amplified with the aRNA procedure, and then differential display was performed on the dendritic aRNA. In Fig. 1B, a comparison between the two processes of the differential display pattern using two different differential display primer sets is shown. The same 5′ primer was used for each PCR while the 3′ primer was varied (lanes A and C). Bands that comigrated between the different dendrites correspond to mRNAs that are localized to both dendrites. Fig. 1C shows the display pattern generated when using nine different primer sets on the aRNA made from dendrite HP 3–5. The large number of bands highlights the complexity of the mRNA localized to dendrites. To put this number of 400 dendritically localized mRNAs into context, using the same methodologies we estimate that there are ≈10,000 different mRNAs expressed in the cell soma of these same hippocampal neurons. Consequently, ≈5% of the mRNAs complexity expressed in a neuron can be targeted to the dendritic domain.
Figure 1.
Differential display analysis of dendritic processes from a single hippocampal neuron in culture. (A) Isolated hippocampal cells free of overlapping processes from neighboring cells were identified in low-density cultures (5). Individual dendrites were harvested by transecting at the branch point and aspirated as described (3). (B) Comparisons of differential display products between dendrites with a single 5′ primer, OPA-5 (Operon Technologies, Alameda, CA), in combination with anchor primers A (T11AC) or C (T11GC) show the presence of common (closed arrowheads) and unique (open arrowheads) PCR products. (C) When a single 5′ primer (OPA-13) is used in combination with all nine anchor primers a large population of mRNAs are present within neuronal processes.
The mechanism by which mRNAs are targeted to dendrites is thought to involve the binding of RNA-binding proteins to cis-acting elements in the transported mRNAs. There are more than 500 RNA binding proteins estimated to be encoded by the cellular genome. The cis-acting elements are likely composed of a primary nucleic acid sequence and a secondary RNA structure such that a specific RNA-binding protein recognition binding site is generated. The specificity of these cis-acting elements and the fact that only a limited number of RNAs are targeted to dendrites suggest that different RNAs may have different rates of transport into the dendrite.
The regulated transport of mRNA into neuronal dendrites recently has become a topic of examination by a number of groups. One approach to determining the rates of mRNA transport into dendrites is to treat neuronal cells with various pharmacological agents that are known to modulate dendritic functioning followed by characterization of the mRNA populations that are present in the dendrite as a function of time after stimulation. Primary hippocampal neuronal cultures were stimulated for various time periods with a metabotrobic glutatmate receptor agonist DHPG [(RS)-3,5-dihydroxyphenylglycine] that is also a known modulator of dendritic protein translation (7). At increasing exposure times the dendrites were harvested, the mRNAs were amplified, and the amplified products used as a probe to screen macroarrays and microarrays containing thousands of different cDNAs. Data presented in Fig. 2 show that this approach can be used to monitor the movement of mRNAs into the dendrite. The arrows in Fig. 2 A–D point to a pair of identical cDNA clones (one cDNA spotted in the two positions) that hybridize to the aRNA probe with greater intensity as a function of time after metabotrobic glutamate receptor stimulation. The aRNA probe used in this study was a pooled probe made from multiple dendrites from multiple neurons to ensure that differences in hybridization intensity are not caused by biological differences between individual neuronal dendrites.
Figure 2.
Time course for movement of mRNAs into the neuronal dendrite. Intact cultured rat hippocampal neurons were treated with DHPG. mRNA was harvested from dendrites at times of 0, 24, 32, and 45 min posttreatment, and aRNA was amplified. This probe was used to screen various types of arrays including the macroarrays shown. The hybridization intensity of selected spots corresponding to different immobilized cDNAs increased as a function of time after DHPG treatment. These data indicate that the movement of mRNA into the dendrite can be regulated by DHPG.
Formal Molecular Proof of Protein Translation in Dendrites and Identification of in Vivo Translation Sites
The presence of ribosomes and mRNAs within dendrites suggests that the dendrites are translationally competent. Indeed various groups have shown by using immunohistochemistry that various protein components of the translational machinery are present within dendrites (8, 9). Additional data suggesting that translation can occur in dendrites was provided by monitoring radioactive amino acid incorporation into individual dendrites (10) (not an actual proof of translation) and through the use of synaptoneurosome preparations to show that radioactivity could be incorporated into a product whose synthesis was diminished by protein synthesis inhibitors (7, 11). Conclusive evidence that protein synthesis can occur in dendrites was provided by transfecting isolated dendrites with mRNA constructs that encode a protein fused to an epitope tag (c-myc) (4). This mRNA was lipid-encoated and applied to transected live dendrites by using the patch pipette as a delivery device. The reasoning behind this experiment was that the only way in which the myc epitope would be visualized is if the transfected mRNA was translated. Indeed, in the presence of brain-derived neurotrophic factor or neurotrophin-3 (to stimulate protein synthesis) the myc epitope was visible, thus providing direct evidence that proteins can be synthesized in dendrites. These data additionally provided support for stimulated protein synthesis in dendrites, which is compatible with the work in acute hippocampal slice preparations by Kang and Schuman (12).
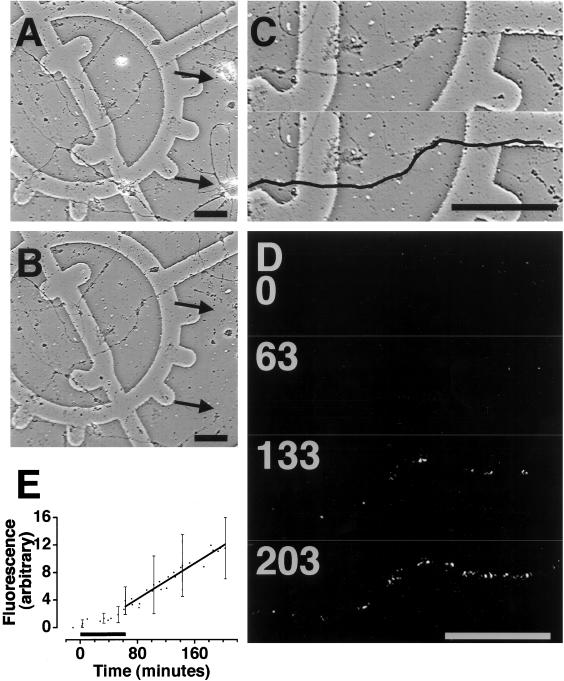
Protein synthetic machinery has been identified within dendrites and translation of exogenous mRNAs has clearly demonstrated that dendritic translation occurs independently of neuronal cell bodies. However, although dendritic ribosomes, mRNAs, and membraneous structures have been extensively characterized, the actual sites of protein synthesis in living dendrites have not been explored. To examine and characterize the endogenous dendritic translation sites, we used an assay that monitors protein synthesis in living dendrites whose cell bodies had been removed. These “isolated dendrites” were transfected with mRNA encoding green fluorescent protein (GFP) (Fig. 3 A–C) and green fluorescence that appeared upon translation of the GFP mRNA was recorded with a multiphoton laser-scanning microscope (Fig. 3 D and E). Over a time course of hours fluorescence was detected in some dendrites (Fig. 3 D and E). This “basal translation” was linear and nonsaturating over a time course longer than 3 h posttransfection (Fig. 3E). These studies highlight the “site-specific” nature of translation within dendrites because the translation sites do not appear to move or, in other words, they are immobile in the dendrite. Visualization and additional characterization of these in vivo translation sites will permit a careful dissection of the kinetics and biology of stimulated protein synthesis in dendrites.
Figure 3.
Transfection of isolated dendrites with GFP mRNA results in fluorescence. (A and B) Phase images of 4-day-old primary hippocampal neurons grown on grided coverslips. Arrows indicate positions of cell bodies removed with a micropipette. (C) Two identical enlarged images of one dendrite from B. Bold black line indicates the position of the dendrite. (D) Four fluorescence images of dendrite in B before (0) and after (63, 133, and 203 min) transfection with GFP mRNA. Images were contrast-stretched in National Institutes of Health image for display purposes. (Scale bars = 50 μm.) (E) Mean fluorescence in isolated dendrites over a time course of hours. Black bar indicates period of transfection with GFP mRNA. n = 3.
Discovery of a Second Messenger System in Dendrites
The identification of multiple dendritically localized mRNAs and the evidence of local dendritic translation suggests that translation of mRNA in dendrites may produce proteins that serve various biological functions. Clearly, the mechanisms of signaling between presynaptic and postsynaptic neurons are quite complex. The production of action potentials was the first of these signaling mechanisms characterized. It has previously been shown that presynaptic activation of signal transduction cascades in the postsynaptic cell can converge on the postsynaptic nucleus to alter transcription factor activity, potentially resulting in activation or suppression of gene transcription. One example of how this can occur is shown in the intracellular convergence of signaling events that modify the activity of localized cAMP responsive element binding protein (CREB), resulting in an averaging of the cellular response to presynaptic stimulation. This averaging results in the loss of the spatially activated influences of presynaptic input on individual dendrites. Pertinent to this discussion is that upon analysis of amplified mRNA from isolated dendrites and growth cones of hippocampal neurons, select transcription factor mRNAs were shown to be present in this subcellular compartment (13). This discovery led to the hypothesis that synthesis of transcription factor proteins within dendrites would provide a novel and direct signaling pathway between the distal dendrite and the nucleus, resulting in modulation of gene expression important for neuronal functioning (13, 14). Among the transcription factor mRNAs shown to be present within developing dendrites was CREB (13). This initial mRNA discovery was expanded on to show that CREB is present in dendrites, that translation of CREB mRNA in isolated dendrites is feasible, and that CREB found in dendrites can interact with the cis-acting CRE DNA sequence using a novel in situ Southwestern assay (13). Further it was shown that CREB in dendrites is not transported to this site from the cell body because fluorescently tagged CREB microperfused into the soma did not move into the dendrites (13). In addition, CREB microperfused into dendrites was rapidly transported to the nucleus, its likely site of bioactivity. Lastly, using the isolated dendrite system it was shown that phosphorylation of Ser133 on CREB (which will greatly stimulate cAMP-dependent gene expression) can occur in isolated dendrites independently of the nucleus (13). These data suggest the existence of a novel regulatory pathway in which transcription factors, synthesized and posttranslationally modified in dendrites, directly move to the neuronal nucleus to alter gene expression bypassing the integration of signal transduction pathways that converge on the nucleus (13, 14).
Demonstration of Synthesis and Membrane Insertion of Integral Membrane Proteins in Isolated Dendrites
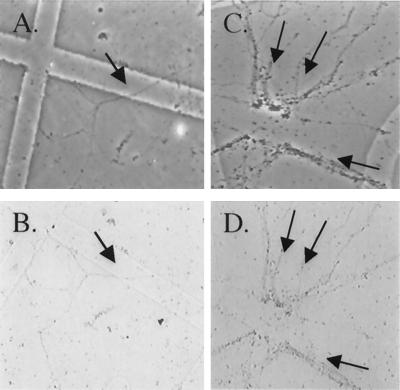
As mentioned previously in this article, our laboratory showed that glutamate receptor mRNAs are present in dendrites. This was an unexpected result because the proteins encoded by these mRNAs are integral membrane receptors and there is little electron microscopic evidence of classical rough endoplasmic reticulum or Golgi apparatus in the dendritic compartment. These are the subcellular structures necessary for synthesis and membrane insertion of integral membrane proteins into the cellular membrane. Immunohistochemical analysis at the light microscopy level has shown the presence of some protein components of the rough endoplasmic reticulum and Golgi in dendrites most notably at the base of the dendritic spine (8, 9, 15). The initial discovery of integral membrane protein encoding mRNAs in dendrites recently has been followed by showing that integral membrane proteins can indeed be synthesized in synaptic regions (Fig. 4) (16). This was accomplished by creating fusion constructs between glutamate receptor mRNA and a sequence encoding the c-myc epitope such that the translated glutamate receptor protein is extended with c-myc. Fig. 4 shows a GluR2-cMyc (c-myc engineered onto the C-terminal end of GluR2 protein) construct transfected into isolated dendrites. Transfected dendrites show low basal levels of GluR2-c-myc translation as evidenced by nearly undetectable GluR2-c-myc immunoreactivity (Fig. 4, A transmission image, B diaminobenzidine immunoreactivity). Upon DHPG stimulation GluR2-c-myc immunoreactivity is dramatically increased (Fig. 4 C and D).
Figure 4.
Stimulation of glutamate receptor mRNA translation in isolated dendrites. (A and C) The phase-contrast images of transected dendrites that correspond to dendrites that have been transfected with GluR2-c-myc shown in B and D, respectively. The dendrites in D have been treated with DHPG to stimulate protein synthesis, whereas those in B are DHPG naïve. Arrows point to individual dendrites that are visible in the paired panels. See ref. 16 for more information.
The ability of the dendrite to translate an integral membrane protein, however, does not mean that the protein is inserted into the dendritic membrane. To examine this, we used the most current information concerning the topology of the glutamate receptor that shows that the N terminus of the protein is localized on external side of the cell membrane. Consequently, if a dendritically localized receptor is inserted into the membrane then the N terminus would be on the extracellular surface of the dendrite. Experimentally, the 5′ end of GluR2 was engineered to contain the c-myc sequence so that translated c-myc-GluR2 would exhibit c-myc on the exterior of expressing cells. When this mRNA was transfected into isolated living dendrites and the dendrites were stimulated by various neurotransmitters, c-myc-glutamate receptor immunoreactivity was clearly visible within the dendrites (16). Further, when the dendritic membrane was not permeabilized the c-myc-glutamate receptor fusion construct was visualized on the cell surface while Map2, a highly abundant dendritic protein, was absent from the membrane but present in the cytoplasm (16). These data show that integral membrane proteins can be synthesized in dendrites and that these proteins can be inserted into the membrane, suggesting that a functional rough endoplasmic reticulum and Golgi exist in dendrites (16). These results suggest a potential mechanism for the modulation of the receptor repertoire under specific synapses through alterations in glutamate receptor subunit representation locally in response to synaptic stimulation. Not only is it likely that receptor subunit composition is altered, but the posttranslational modifications and association with accessory proteins also may differ for dendritically synthesized receptors as compared with cytoplasmically synthesized receptors. There are clear implications of these data in furthering our understanding of the Hebbian synapse.
Summary
The localization of a subset of the cellular mRNAs (≈5%) to dendrites and the formal proof of local dendritic protein synthesis suggests that the dendrite serves a specialized role in regulating neuronal functioning. It is easy to envisage a dendrite as a passive entity capturing presynaptic information and passively transferring this information to the cell soma. To act in this manner the dendrite would not need to perform protein synthesis, it would just need to act as a cable, or wire, to propagate the information. The fact that stimulated protein synthesis occurs in dendrites, and data suggesting that this is important for various physiological properties, including long-term potentiation and long-term depression, suggests that incoming presynaptic information is either modified or modulated in postsynaptic dendrite. Alternatively, the postsynaptic dendrite may be modified in response to presynaptic input such that it responds differently to additional presynaptic input. As evidenced by data presented in this manuscript, and the work of many others, it is clear that both types of change occur. For example, a change in glutamate receptor repertoire at the synapse could alter the dendrite's responsiveness to presynaptic glutamate challenge. As another example, if a presynaptic signal causes CREB to be synthesized and specifically phosphorylated in the dendrite with its subsequent movement to the nucleus, specific alterations in gene expression could be induced related to the type and intensity of presynaptic input (17). It is currently unclear which is the dominating regulatory feature of postsynaptic responsiveness to presynaptic input. The dendrite responds to presynaptic input through the generation of a combinatorial set of coordinated responses that are integrated into a coherent signal for propagation to the next postsynaptic neuron. Questions that arise from these considerations include: How many signal transduction pathways are modulated in the dendrite in response to a specific type of presynaptic input? Is the percentage of synapses that are modified by local protein synthesis as important as the proximal or distal position of the modified synapses on the dendrite?
The complexities of the biological processes and information processing that occur within the dendrite suggest that the dendrite should be viewed as the primary site for filtering and modulating presynaptic input into the neuron. As highlighted in this manuscript and others from the National Academy of Sciences colloquium, data collected during the last decade have provided extensive information concerning the molecular composition of neuronal dendrites. With this foundation, the challenge for the coming decade will be to place this information into its functional context both in vitro and in vivo (14). Such information undoubtedly will be useful in many ways including (i) helping to develop drugs targeted to dendrite function and (ii) providing information that should be useful in the generation of better neural network algorithms.
Acknowledgments
We thank Margie Moronski for preparing cultured rat hippocampal neurons for these studies. Jim Sanzo's technical assistance with the multiphoton microscopy is greatly appreciated. This work was funded by National Institutes of Health Grants AG9900 and MH58561 to J.E.
Abbreviations
- aRNA
antisense RNA
- DHPG
(RS)-3,5-dihydroxyphenylglycine
- GFP
green fluorescent protein
- CREB
cAMP responsive element binding protein
Footnotes
This paper was presented at the National Academy of Sciences colloquium, “Molecular Kinesis in Cellular Function and Plasticity,” held December 7–9, 2000, at the Arnold and Mabel Beckman Center in Irvine, CA.
References
- 1.Bodian D. Proc Natl Acad Sci USA. 1965;53:418–425. doi: 10.1073/pnas.53.2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garner C, Tucker R, Matus A. Nature (London) 1988;336:374–377. doi: 10.1038/336674a0. [DOI] [PubMed] [Google Scholar]
- 3.Miyashiro K, Dichter M, Eberwine J. Proc Natl Acad Sci USA. 1994;91:10800–10804. doi: 10.1073/pnas.91.23.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crino P, Eberwine J. Neuron. 1996;17:1173–1187. doi: 10.1016/s0896-6273(00)80248-2. [DOI] [PubMed] [Google Scholar]
- 5.Buchhalter J, Dichter M. Brain Res Bull. 1991;26:333–338. doi: 10.1016/0361-9230(91)90003-3. [DOI] [PubMed] [Google Scholar]
- 6.Weiler I J, Irwin S, Klintsova A, Spencer C, Brazelton A, Miyashiro K, Comery T, Patel B, Eberwine J, Greenough W. Proc Natl Acad Sci USA. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiler I J, Greenough W. Proc Natl Acad Sci USA. 1993;90:7168–7171. doi: 10.1073/pnas.90.15.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tiedge H, Brosius J. J Neurosci. 1996;16:7171–7181. doi: 10.1523/JNEUROSCI.16-22-07171.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardiol A, Racca C, Triller A. J Neurosci. 1999;19:168–179. doi: 10.1523/JNEUROSCI.19-01-00168.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis L, Banker G, Stewart O. Nature (London) 1987;330:477–480. doi: 10.1038/330477a0. [DOI] [PubMed] [Google Scholar]
- 11.Weiler I J, Wang X, Greenough W. Prog Brain Res. 1994;100:189–194. doi: 10.1016/s0079-6123(08)60785-2. [DOI] [PubMed] [Google Scholar]
- 12.Kang H, Schuman E. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- 13.Crino P, Khodakhah K, Becker K, Ginsberg S, Hemby S, Eberwine J. Proc Natl Acad Sci USA. 1998;95:2313–2318. doi: 10.1073/pnas.95.5.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eberwine J. In: Dendrites. Stuart G, Spruston N, Hausser M, editors. Oxford: Oxford Univ. Press; 1990. pp. 68–84. [Google Scholar]
- 15.Torre E, Steward O. J Neurosci. 1996;16:5967–5978. doi: 10.1523/JNEUROSCI.16-19-05967.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estee Kacharmina J, Job C, Crino P, Eberwine J. Proc Natl Acad Sci USA. 2000;97:11545–11550. doi: 10.1073/pnas.97.21.11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eberwine J, Job C, Estee Kacharmina J, Miyashiro K, Therianos S. In: Cell Polarity and Subcellular RNA Localization. Richter D, editor. New York: Springer; 2001. pp. 57–68. [Google Scholar]