Abstract
JNK (c-Jun N-terminal kinase) is part of a MAPK (mitogen-activated protein kinase) signalling cascade. Scaffold proteins simultaneously associate with various components of the MAPK signalling pathway and play a crucial role in signal transmission and MAPK regulation. WDR62 (WD repeat domain 62) is a JNK scaffold protein. Recessive mutations within WDR62 result in severe cerebral cortical malformation. In the present study we demonstrate the association of WDR62 with endogenous and overexpressed proteins of both JNK2 and the JNK2-activating kinase MKK7 (MAPK kinase 7). Association of WDR62 with JNK2 and MKK7 occurs via direct protein–protein interactions. We mapped the docking domain of WDR62 responsible for the association with JNK. WDR62 interacts with all JNK isoforms through a D domain motif located at the C-terminus. A WDR62 mutant lacking the putative JNK-binding domain fails to activate and recruit JNK to cellular granules. Furthermore, a synthetic peptide composed of the WDR62 docking domain inhibits JNK2 activity in vitro. WDR62 association with JNK2 requires both the JNK CD and ED domains, and the binding requisite is distinct from that of the previously described JNK2 association with JIP1 (JNK-interacting protein 1). Next, we characterized the association between WDR62 and MKK7. WDR62 associates directly with the MKK7β1 isoform independently of JNK binding, but fails to interact with MKK7α1. Furthermore, MKK7β1 recruits a protein phosphatase that dephosphorylates WDR62. Interestingly, a premature termination mutation in WDR62 that results in severe brain developmental defects does not abrogate WDR62 association with either JNK or MKK7. Therefore such mutations represent a loss of WDR62 function independent of JNK signalling.
Keywords: c-Jun N-terminal kinase (JNK), D domain, mitogen-activated protein kinase (MAPK), mitogen-activated protein kinase 7 (MKK7), scaffold, WD repeat domain 62 (WDR62)
Abbreviations: aa, amino acids; CIP, calf intestinal phosphatase; DAPI, 4′,6-diamidino-2-phenylindole; DTT, dithiothreitol; EGFP, enhanced green fluorescent protein; FBS, fetal bovine serum; GFP, green fluorescent protein; GST, glutathione transferase; HRP, horseradish peroxidase; JDP2, Jun dimerization protein 2; JNK, c-Jun N-terminal kinase; JIP, JNK-interacting protein; HA, haemagglutinin; HEK-293T cells, human embryonic kidney 293 cells expressing the large T-antigen of SV40 (simian virus 40); MAPK, mitogen-activated protein kinase; MAP2K, MAPK kinase; MAP3K, MAPK kinase kinase; MBP, maltose-binding protein; MKK, MAPK kinase; SAPK, stress-activated protein kinase; Tat, transactivator of transcription; WCE, whole-cell extraction; WDR62, WD repeat domain 62
INTRODUCTION
MAPKs (mitogen-activated protein kinases) regulate a variety of cellular processes in response to extracellular signals. MAPKs are activated through a protein kinase cascade in which a MAP3K (MAPK kinase kinase) activates an MKK (MAPK kinase, also known as MAP2K) that, in turn, activates a MAPK [1]. Three main modules exist in mammals: the ERKs (extracellular-signal-regulated kinases), SAPKs [stress-activated protein kinases, also known as JNKs (c-Jun N-terminal kinases)] and p38. In many instances, these MAPK modules have been shown to regulate distinctly different cellular responses in a cell-type- or signal-specific manner. During the last decade, the importance of scaffold proteins for providing signal specificity and fidelity has become evident [2]. Scaffold proteins act as multidomain-interacting surfaces that serve as a meeting platform for kinases and substrates to orchestrate specific transmission of signalling. By the interaction with two or more components of the cascade, scaffold proteins increase the efficiency of signalling by concentrating proteins locally and positioning kinases in close proximity to their substrates. The association of the signalling components with the scaffold protein allows signal channelling to a specific outcome. In addition, some of the scaffold proteins are able to allosterically activate the associated kinase or, alternatively, they may restrict the activation of the signalling pathway to a specific subcellular compartment [3]. Multiple scaffold proteins have now been described for the MAPK cascade and have been found to enhance signal transduction by promoting the assembly of multiprotein complexes [4–8].
Recently, we described the isolation of a novel JNK scaffold protein WDR62 (WD repeat domain 62) that associates with JNKs and the upstream MKK7 [9]. WDR62 is recruited to stress granules following the exposure of cells to arsenite [9]. WDR62 is suggested to control mRNA homoeostasis. Interestingly, recessive mutations in the WDR62 were found to be involved in severe brain malformations, including microcephaly and pachygyria [10]. The mutations found in patients span multiple regions of the WDR62 protein and include missense mutations as well as small deletions resulting in a frameshift followed by a stop codon. Thus biochemical characterization of WDR62 structure and function would be important for understanding the functional consequence of WDR62 mutations found in affected individuals. In the present paper we describe in detail the distinct association domains of WDR62 with JNK and MKK7 and their effect on JNK activity.
MATERIALS AND METHODS
Plasmids
The mammalian expression plasmid pCAN was used to express all Myc-tagged human WDR62 deletion mutants [9]. Two full-length WDR62 splice variants exist: CS2 and CS5; all deletion constructs were designated according to the amino acid position within CS5. The letters N and C represent aa (amino acids) 1 and 1523 respectively. The following constructs were used: Myc-N-1284 (aa 1–1284), Myc-1018-C (aa 1018–1523), Myc-1018-1284 (aa 1018–1284), Myc-1018-1345 (aa 1018–1345), Myc-NG 26-1 (aa 1018–1401; NG 26-1 contains a naturally occurring mutation that results in premature termination at amino acid 1401 in human patients with microcephaly), Myc-1212-C (aa 1212–1523) and Myc-1232-C (aa 1232–1523). Constructed plasmids were verified by DNA sequencing. Human WDR62 fragments fused to GST (glutathione transferase) were designed in the mammalian expression plasmid pcDNA3·AIRAP-GST-V3 [11] in which the AIRAP fragment was replaced with WDR62 fragments by HindIII/XhoI restriction.
Deletion mutants in Myc-WDR62 CS5 (encoding CS2, aa Δ1074–1078), Myc-WDR62ΔD, 1018-CΔD–GST, and Myc-1018-CΔD (aa Δ1294–1301) were generated using the QuikChange® site-directed mutagenesis kit (Stratagene) according to the instructions of the manufacturer. Mutations were verified by DNA sequencing.
HA (haemagglutinin)-tagged human MKK4, MKK7 and JNK2 were expressed using the pSRα mammalian expression plasmid. Myc-tagged human JNK3 was expressed using the pcDNA3-Myc6 mammalian expression plasmid. GFP (green fluorescent protein)-tagged human JNK (1–3) expression plasmids were kindly provided by Dr M. Courtney (Department of Neurobiology, A.I. Virtanen Institute, Kuopio, Finland). The bacterial PET28 expression plasmids expressing His-tagged human JNK2 and MKK7β1 were kindly provided by Dr D. Engelberg (Department of Biological Chemistry, The Hebrew University, Jerusalem, Israel). The mammalian expression plasmid pEBG was used to express GST-tagged human JNK2α2 and MKK7 (α1 and β1). The bacterial expression plasmid pMAL-p4X (NEB) was used to express MBP (maltose-binding protein) fused to human WDR62 1018-C fragment (pMBP-1018-C).
Antibodies
The following antibodies were used: anti-Myc monoclonal (clone 9E10, Abcam), anti-HA monoclonal (clone 12CA5, Abcam), anti-GFP polyclonal (Santa Cruz Biotechnology, catalogue number sc-8334), anti-GST rabbit polyclonal (made in our laboratory), anti-JNK polyclonal (Cell Signaling Technology, catalogue number 9252), anti-MKK7 polyclonal (Cell Signaling Technology, catalogue number 4172), anti-MBP polyclonal (Santa Cruz Biotechnology, catalogue number sc-808), anti-pJNK (Sigma–Aldrich, catalogue number J4644), anti-WDR62 monoclonal antibody (clone 3G8, Sigma–Aldrich, catalogue number W3269).
Fluorochrome-tagged secondary antibodies for immunofluorescence assays were obtained from Jackson ImmunoResearch Laboratories. All antibodies were prepared at a dilution of 1:500 or as otherwise stated.
Peptide synthesis
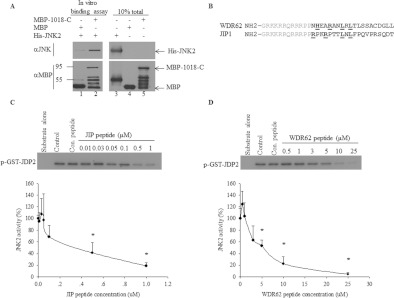
Peptides were synthesized by Synthetic Biomolecules. Peptide purification was performed by HPLC, achieving >85% purity. The peptides used were: JIP (JNK-interacting protein) (GRKKRRQRRRPPRPK-RPTTLNLFPQVPRSQDT); WDR62 (GRKKRRQRRRPPNHEARANLRLTLSSACDGLL); and influenza HA elution peptide (Sigma–Aldrich). Bold letters indicate Tat (transactivator of transcription) membrane-permeant sequences [12]. Underlined letters correspond to amino acids matching the D-box consensus sequence.
Cell culture and transient transfection
HEK-293T cells [human embryonic kidney 293 cells expressing the large T-antigen of SV40 (simian virus 40)] and HeLa cells were maintained in DMEM (Dulbecco's modified Eagle's medium) containing 10% (v/v) FBS (fetal bovine serum), 100 i.u./ml penicillin and 0.1 μg/ml streptomycin and grown at 37°C and 5% CO2.
HEK-293T cells were transfected with the appropriate expression plasmid using the calcium phosphate method [13]. The total amount of plasmid DNA was adjusted to 10–12 μg in a total volume of 1 ml. Cell culture medium was replaced with fresh medium 4–5 h post-transfection and cells were harvested 24 h post-transfection.
HeLa cells were transfected with jetPEI (Polyplus Transfection) according to the manufacturer's instructions.
For the immunofluorescence assay, cells were grown in six-well plates and transfection was performed in a total volume of 400 μl.
Western blot analysis
Cells were lysed in WCE (whole-cell extraction) buffer [25 mM Hepes, pH 7.7, 0.3 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.1% Triton X-100, 0.5 mM DTT (dithiothreitol), 20 mM 2-glycerophosphate, 0.1 mM Na3VO4, 100 μg/ml PMSF and protease inhibitor cocktail (Sigma–Aldrich, 1:100 dilution)]. The proteins were then separated by SDS/PAGE (10% gel), followed by transfer on to a nitrocellulose membrane. Blots were blocked in 5% (w/v) non-fat dried skimmed milk powder in PBS and washed in PBS. Proteins were detected using the corresponding HRP (horseradish peroxidase)-conjugated secondary antibodies obtained from Sigma–Aldrich (anti-rabbit A0545, anti-mouse A0168) and Jackson ImmunoResearch Laboratories (anti-mouse light chain 115-035-174, anti-rabbit light chain 211-032-171).
Immunoprecipitations
Protein extract (400–600 μg in WCE buffer) was pre-cleared with non-relevant antibodies. The lysate was then incubated with antibodies against Myc or WDR62 overnight at 4°C. Protein A–Sepharose beads (Sigma–Aldrich) were then added to the lysate and incubated for 1 h. Samples were washed four times with WCE buffer. The precipitated proteins were eluted using SDS/PAGE sample buffer, boiled and then processed for Western blot analysis. Phosphatase treatment was performed with CIP (calf intestinal phosphatase; Roche); enzyme (20 units) in 1× phosphatase buffer (supplied with the enzyme) was added to the immunoprecipitation complex and incubated at 37°C for 30 min.
GST pull-down assay
Glutathione–agarose beads (Sigma–Aldrich) were pre-blocked with 5% (w/v) BSA and were then incubated with 400–600 μg of the protein extract (in WCE buffer). Following four washes with WCE buffer, the precipitated proteins were eluted using freshly made glutathione elution buffer [50 mM Tris/HCl, pH 8.0, 20 mM L-glutathione (Sigma-Aldrich), 1 mM DTT and 1 mM PMSF]. Samples were boiled and then processed for Western blot analysis.
In vitro protein-binding assay
His–JNK2 and His–MKK7β were purified from bacteria using Ni-NTA (Ni2+-nitrilotriacetate)–agarose beads (Qiagen) according to the instructions of the manufacturer. Recombinant MBP–1018-C protein was purified using amylose resin (NEB) according to the instructions of the manufacturer. His-tagged protein (5 μg) was incubated with 10 μg of MBP-1018-C and 0.5 mg of BSA for 2 h at 37°C. Amylose resin was pre-blocked with 0.5 mg of BSA and was then incubated with the indicated pre-incubated protein complexes. Following five washes with column buffer (20 mM Tris/HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA and 1 mM DTT), the precipitated proteins were eluted using elution buffer [column buffer containing 1 mM DTT and 10 mM D-maltose (Sigma–Aldrich)]. Samples were boiled and then processed by Western blot analysis.
In vitro kinase assay
An in vitro kinase assay was performed using bacterially purified activated His–JNK2–FLAG [14,15] and purified GST–JDP2 (Jun dimerization protein 2) as substrate. First, the activated JNK2 was incubated for 30 min at 30°C with the indicated concentrations of synthetic peptides. The JNK substrate (GST–JDP2) and [γ-32P]ATP were then added to the reaction mixture and incubated for another 30 min at 30°C. The reaction was stopped by the addition of SDS/PAGE sample buffer. The samples were then boiled, and the phosphorylated proteins were resolved by SDS/PAGE (10% gel). The gel was dried and subjected to radiography. Phosphorylated JDP2 product was quantified using a FLA-2000 phosphorimager (Fujifilm). GST–JDP2 phosphorylation was determined using TotalLab software.
Immunofluorescence
HeLa cells were grown on glass coverslips. At 24 h after transfection, cells were fixed with 4% (v/v) formaldehyde for 10 min. After washing with PBS, the cells were permeabilized with 0.1% Triton X-100 for 5 min and incubated in blocking solution (5% FBS in PBS) for 30 min. The cells were then incubated with anti-Myc antibody (9E10), diluted 1:250 in PBS containing 1% FBS, for 1 h. The cells were washed three times with PBS and incubated with a Rhodamine Red-X-conjugated anti-mouse antibody (Jackson ImmunoResearch Laboratories, catalogue number 715-295-150), diluted 1:250 in PBS containing 2% BSA, 2% FBS and 0.1% Tween 20, for 1 h. The cells were washed twice with PBS and processed for nuclear staining using DAPI (4′,6-diamidino-2-phenylindole; Sigma–Aldrich) at a final concentration of 1 μg/μl in PBS. The stained cells were then washed twice with PBS and mounted in Fluoromount-G (Southern Biotechnology).
Confocal microscopy
Fluorescence microscopy was performed using the Zeiss LSM 510 Meta inverted confocal microscope equipped with a ×63/1.4 NA (numerical aperture) oil objective, multiline argon laser (488, 514 nm), DPSS (diode-pumped solid-state) laser (561 nm) and a UV diode laser (405 nm). Each image was acquired from a single 1-μm-thick Z-stack using 510 LSM software (Zeiss).
Statistical analysis
Statistical analysis was performed using Student's unpaired t tests with one-tailed distribution.
RESULTS
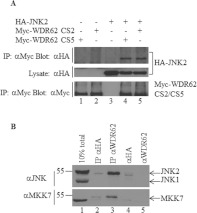
Previously, we demonstrated the biochemical association between overexpressed JNK1 and JNK2 with a C-terminal fragment of WDR62 [9]. To characterize this interaction further, we first verified that full-length WDR62 interacts with JNK2. Two full-length WDR62 splice variants exist: CS2 and CS5. The CS2 transcript lacks amino acids 1074–1078, the functional consequence of which is unknown [9]. HEK-293T cells were transfected with Myc-tagged WDR62 CS2 or CS5 together with HA-tagged JNK2. The WDR62 splice variants were immunoprecipitated from cell lysates using anti-Myc antibodies. Co-precipitated JNK2 was detected by Western blotting with anti-HA antibodies. As shown in Figure 1(A), JNK2 was efficiently co-precipitated with WDR62 CS2 and CS5 in a specific manner, indicating that JNK associates with both splice variants of full-length WDR62. To demonstrate the interaction between endogenous WDR62 and JNK2, we immunoprecipitated endogenous WDR62 from HEK-293T cells using the anti-WDR62 3G8 monoclonal antibody. Western blot analysis with the anti-JNK antibody revealed the presence of endogenous JNK2 in the WDR62 precipitate. Endogenous MKK7 was also detected in the WDR62 precipitate by blotting with the anti-MKK7 antibody (Figure 1B).
Figure 1. WDR62 association with JNK2 and MKK7.
(A) WDR62 CS2 and CS5 splice variants associate with JNK2. HEK-293T cells were transfected with Myc-WDR62 CS2, Myc-WDR62 CS5 and HA–JNK2 in various combinations as indicated. Cell lysates were subjected to immunoprecipitation (IP) with anti-Myc antibodies followed by Western blotting with either anti-HA or anti-Myc antibodies (top panel and bottom panel respectively). The expression level of HA–JNK2 was determined by blotting total cell lysate with anti-HA antibody (middle panel). (B) Co-immunoprecipitation of endogenous WDR62 with JNK and MKK7. Endogenous WDR62 was immunoprecipitated from HEK-293T cells using anti-WDR62 3G8 antibodies. Monoclonal anti-HA antibodies were used as a negative control (lane 2). Eluted proteins were analysed by Western blotting with either anti-JNK (upper panel) or anti-MKK7 (lower panel) antibodies. Cell lysate (10% of total) was used to assess the expression level of JNK1/2 and MKK7 in the cells (lane 1). Anti-(rabbit light chain) antibody linked to HRP was used as a secondary antibody. Protein molecular mass markers (in kDa) are indicated on the left-hand side. The anti-HA and anti-WDR62 monoclonal antibodies were loaded directly into lanes 4 and 5 respectively to demonstrate lack of cross-reactivity of the anti-rabbit secondary antibody with the mouse IgG heavy chain.
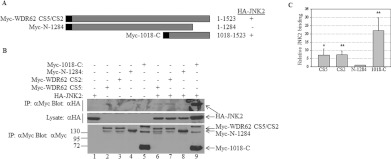
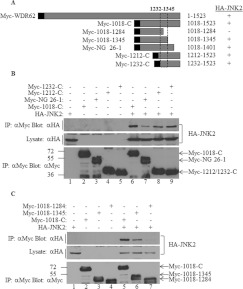
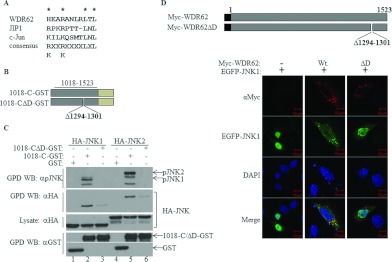
Next, we sought to map the JNK2-interaction domain within WDR62. A plasmid construct encoding a C-terminally truncated mutant of WDR62 was generated for use in the HA–JNK2 co-immunoprecipitation experiments. The encoded Myc-tagged WDR62 fragment Myc-N-1284 corresponds to amino acids 1–1284 (Figure 2A). The corresponding full-length WDR62 construct and WDR62 C-terminal fragment Myc-1018-C were used as positive controls. As shown in Figure 2(B), deletion of the last 239 amino acids from the C-terminus of WDR62 reduced the amount of precipitated JNK2 7-fold as compared with CS2 and CS5 full-length proteins (Figures 2B and 2C). In contrast, the WDR62 fragment with the N-terminal truncation (Myc-1018-C) resulted in a 3-fold increase in precipitated HA–JNK2 as compared with CS2 and CS5 WDR62 full-length isoforms (Figure 2C). This result suggests that a domain within the last 239 amino acids of WDR62 is necessary for the association with JNK2. To further map the JNK2-interaction domain, we generated a series of constructs encoding Myc-tagged Cterminal fragments of WDR62 (Figure 3A). These included C-terminal fragments starting at amino acids 1212 and 1232, as well as an internal fragment spanning amino acids 1018–1401. The latter fragment (NG 26-1) represents a naturally occurring mutation that results in premature termination at amino acid 1401 in human patients with microcephaly. Patients that harbour this mutation display a phenotype as severe as patients with upstream disruption of the WDR62 gene [10]. The WDR62 C-terminal fragment Myc-1018-C was used as a positive control. The two C-terminal fragments Myc-1212-C and Myc-1232-C associated with HA–JNK2 to the same extent as the control (Figure 3B, lanes 8 and 9), suggesting that the last 291 amino acids are both necessary and sufficient for WDR62 association with JNK2. The internal fragment, Myc-1018-1401, was still able to interact with HA–JNK2, although to a lesser extent (Figure 3B, lane 7). Thus we extended the deletion of the C-terminal domain by removing the last 178 and 239 amino acids, corresponding to constructs Myc-1018-1345 and Myc-1018-1284 respectively (Figure 3A). Association of HA–JNK2 with the former fragment was similar to that of the control. However, deletion of the next 60 amino acids completely abrogated the association (Figure 3C). Thus the domain responsible for JNK association resides within 113 amino acids located at position 1232–1345. JNK2 binding is typically mediated via a D domain found in many JNK substrates and JNK-associating proteins [16]. The D domain is composed of a core consensus sequence of K/RXXK/RXXXXLXL. We identified a domain within WDR62 that shares a high degree of homology with the consensus D domain sequence, located between residues 1294 and 1301 and composed of the amino acid sequence HEARANLRLTL (Figure 4A). To explore the possibility that this sequence represents a bona fide D domain with a role in JNK2 binding, we deleted it from wild-type WDR62-1018-C fused to GST using site-directed mutagenesis (Figure 4B). HEK-293T cells were transfected with the empty GST plasmid, 1018-C–GST or 1018-CΔD–GST together with either HA–JNK1 or HA–JNK2 (Figure 4C). At 24 h following transfection, cell lysates were used in GST pull-down assays. Eluted proteins were subjected to Western blot analysis with anti-pJNK and anti-HA antibodies. GST alone was unable to precipitate JNK proteins (Figure 4C, top two panels, lanes 1 and 4). The wild-type 1018-C–GST efficiently precipitated phosphorylated Myc–JNK1 and Myc–JNK2 isoforms (Figure 4C, top panel, lanes 2 and 5). In contrast, 1018-CΔD–GST displayed a relatively weak interaction with either HA–JNK1 or HA–JNK2 (Figure 4C, second panel from the top, lanes 3 and 6 respectively). Interestingly, no pJNK1/2 was detected in proteins precipitated with the WDR62 fragment lacking the D domain (Figure 4C, top panel, lanes 3 and 6 respectively), demonstrating the importance of the D domain in JNK association. Similar results were obtained with JNK3 (Supplementary Figure S1 at http://www.BiochemJ.org/bj/439/bj4390381add.htm).
Figure 2. WDR62 C-terminal domain is necessary for JNK association.
(A) Schematic representation of the WDR62 deletion constructs used in the experiments. Amino acid positions are numbered. The black square represents the position of the Myc epitope tag. Summary of the binding of the various WDR62 truncations to HA–JNK2 is indicated by + and −. (B) HEK-293T cells were transfected with full-length Myc-WDR62 CS2 and CS5 isoforms, the WDR62 C-terminus fragment (Myc-1018-C) or the C-terminal truncation mutant alone (Myc-N-1284), or with HA–JNK2 as indicated. Cell lysates were subjected to immunoprecipitation (IP) with anti-Myc antibodies followed by Western blotting with either anti-HA or anti-Myc antibodies (top panel and bottom panel respectively). The expression level of HA–JNK2 was determined by blotting the total cell lysate with anti-HA antibody (middle panel). The migration of the relevant proteins is indicated by arrows. Molecular mass markers (in kDa) are indicated on the left-hand side of the bottom panel. (C) Quantification of JNK2 binding as described in (B). JNK2 association with Myc-N-1284 is considered as 1 and the extent of JNK2 binding of the other WDR62 truncations is calculated relatively. Results shown are means±S.D. for three independent experiments. *P<0.05, **P<0.01.
Figure 3. JNK association is mapped to a 113-amino-acid region within WDR62 at position 1232–1345.
(A) Schematic representation of the WDR62 deletion constructs used in the experiments. The black square represents the position of the Myc epitope tag. Summary of the binding of the various WDR62 fragments to HA–JNK2 is indicated by + and −. The broken box indicates the minimal region that preserves JNK binding. (B and C) HEK-293T cells were transfected with a series of plasmids encoding various Myc-tagged WDR62 C-terminal fragments together with HA–JNK2 as indicated. Cell lysates were subjected to immunoprecipitation (IP) with anti-Myc antibodies followed by Western blotting with either anti-HA or anti-Myc antibodies (top panel and bottom panel respectively). Myc-1018-C was used as a positive control for the HA–JNK2 interaction. The expression level of HA–JNK2 was determined by blotting the total cell lysate with anti-HA antibody (middle panel). The migration of the relevant proteins is indicated by arrows. Molecular mass markers (in kDa) are indicated on the left-hand side.
Figure 4. A putative D domain within WDR62 is responsible for WDR62–JNK association.
(A) Alignment of the amino acid sequences of the D domains derived from WDR62, JIP1 and c-Jun. The consensus D domain sequence identified in numerous proteins is shown. (B) Schematic representation of the WDR62 deletion constructs used in the experiments. Amino acid positions are numbered. The light green rectangle represents the position of the GST tag. (C) HEK-293T cells were transfected with plasmids encoding GST, 1018-C–GST or 1018-CΔD–GST together with HA–JNK1/2 as indicated. GST-containing complexes were isolated from cell lysates with glutathione–agarose beads, washed extensively and eluted with reduced glutathione. The protein complexes were subjected to Western blotting (WB) with anti-pJNK (top panel), anti-HA (second panel from the top) or anti-GST (bottom panel) antibodies. The expression level of HA–JNK1/2 was determined by blotting the total cell lysate with anti-HA antibody (middle panel). The migration of the relevant proteins is indicated by arrows. (D) Top panel: schematic representation of the full-length WDR62 wild-type and deletion mutant constructs used in the HeLa cell immunofluorescence staining experiments. Amino acid positions are numbered. The black square represents the position of the Myc epitope tag. Bottom panels: HeLa cells were co-transfected with an EGFP–JNK1 expression plasmid together with plasmids encoding either wild-type WDR62 (Wt) or the D domain deletion mutant (ΔD). Cells were fixed and stained with anti-Myc antibody (red). EGFP–JNK1 is shown in green. Co-localization appears in yellow in the merged image. Nuclei were stained with DAPI (blue). Representative confocal microscopy images are shown. Scale bar, 10 μm. GPD, GST pull-down.
Previously, we showed that overexpression of full-length WDR62 in HEK-293T cells causes JNK to be recruited to discrete cellular granules [9]. Thus we examined whether this localization is dependent on the association of JNK with WDR62 via its consensus D domain. We co-transfected HeLa cells with either wild-type WDR62 (Myc-WDR62-wt) or a WDR62 mutant lacking the putative D domain (Myc-WDR62ΔD), together with EGFP (enhanced green fluorescent protein)–JNK1. Transfected cells were fixed and stained with anti-Myc antibodies. Both wild-type WDR62 and the WDR62ΔD deletion mutant localized to cellular granules (Figure 4D, top panels). As expected, EGFP–JNK1 was efficiently recruited to these granules in the presence of wild-type WDR62 (Figure 4D, middle panels). However, it failed to be recruited to cellular granules in the presence of the WDR62ΔD mutant (Figure 4D, right-hand panels). Similar results were obtained with EGFP–JNK2 and EGFP–JNK3 (Supplementary Figures S2A and S2B at http://www.BiochemJ.org/bj/439/bj4390381add.htm). Collectively, we propose that the WDR62–JNK interaction is mediated through a conserved D domain located within WDR62 at amino acid position 1294–1301. This domain is necessary for the association, activation and recruitment of JNK to the cellular granules.
We next examined whether WDR62 and JNK interact directly. To this end, the WDR62 C-terminal fragment (1018-C) was fused to MBP for bacterial expression and subsequent purification. An in vitro binding assay was performed using MBP-1018-C and His-tagged JNK2 purified proteins from bacteria. The proteins were allowed to interact for 2 h at 37°C and were subsequently washed extensively. Protein complexes were precipitated by amylose resin and the presence of JNK2 was assessed by Western blotting with anti-JNK antibodies. Indeed, MBP-1018-C, but not the MBP control, efficiently precipitated His–JNK2 (Figure 5A). Thus we can conclude that WDR62–JNK2 association occurs via a direct protein–protein interaction and is not mediated by a third protein.
Figure 5. WDR62–JNK association is direct and the WDR62 D domain inhibits JNK activity in vitro.
(A) Bacterially purified MBP, MBP-1018-C and His–JNK2 proteins were mixed as indicated for 2 h at 37°C. Subsequently, protein complexes were attached to amylose resin, washed extensively and then eluted using maltose. Western blot analysis was performed with either anti-JNK (upper panel) or anti-MBP (lower panel) antibodies. (B) Amino acid sequence alignment of WDR62 and JIP1 peptides. The amino acids representing the consensus D domain are underlined. Grey amino acids indicate HIV-Tat sequence that directs cellular import in cells. (C and D) In vitro kinase assay with activated JNK2 pre-incubated with the indicated concentration of JIP (C) and WDR62 (D) peptides. Control HA elution peptide (Con. peptide) was used at 25 μM (lanes 3). Kinase assay was performed in the presence of [γ-32P]ATP and purified GST–JDP2 (5 μg) as substrate. Lanes 1 contained substrate alone without JNK2, and lanes 2 contained JNK2 and substrate without competing peptide. Phosphorylated GST–JDP2 was resolved by SDS/PAGE and detected by autoradiography (upper panels). The extent of GST–JDP2 phosphorylation in the absence of competing peptide is considered as 100% and the extent of JDP2 phosphorylation in the presence of the peptides was calculated relatively (lower panels). Results shown are means±S.E.M. for three independent experiments. *P<0.05.
Thus far we have shown that the putative WDR62 D domain is necessary for the association with JNK2. We next examined whether the D domain can inhibit JNK activity in vitro by competing with a known substrate. A synthetic peptide composed of the Tat membrane-permeant sequence linked to 20 amino acids of the WDR62 putative D domain was prepared. The corresponding well-characterized JIP peptide [17] was used as a positive control (Figure 5B) and the HA elution peptide was used as a non-specific negative control. The peptides were pre-incubated at different concentrations with active JNK2. JNK activity was then measured in the presence of [γ-32P]ATP and bacterially purified GST–JDP2 substrate. JNK activity was evaluated by measuring the extent of 32P incorporation into GST–JDP2 (Figures 5C and 5D). The IC50 for the WDR62 and JIP D domain peptides was found to be approximately 5 and 0.3 μM respectively (Figures 5C and 5D). Collectively, we can conclude that the 20-amino-acid-long peptide derived from the WDR62 putative D domain is sufficient for direct association with JNK2 and is able to inhibit JNK activity in vitro. Nevertheless, the JIP D domain displayed a stronger association with JNK as compared with the WDR62 D domain.
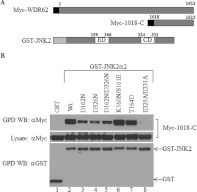
Previous studies have demonstrated that the CD and ED domains of MAPKs are important for docking to their associated proteins [18,19]. To reveal whether the ED and CD domains of JNK participate in its association with WDR62, we tested the ability of multiple JNK ED and CD mutants to bind to WDR62 (Figure 6A). All of the JNK2 mutants used were fused to GST [20]. HEK-293T cells were co-transfected with expression plasmids encoding either GST or various GST–JNK2 mutants together with the Myc-1018-C expression plasmid. Cell lysates were subjected to a GST pull-down assay and the presence of Myc-1018-C in the precipitated protein complex was detected by Western blot analysis. This analysis revealed that the GST–JNK2 mutants D162N or D326N, residing within the ED and CD domains respectively, had reduced association with Myc-1018-C (Figure 6B, lanes 3–5). These residues were found to be conserved in the docking domains of other MAPKs as well, but were not required for the association of JNK2 with the JIP1 scaffold protein [18–20]. The K160N/S161E and T164D mutations within the ED domain did not affect JNK2 association with WDR62 (Figure 6B, lanes 6 and 7). In contrast, double mutations within the JNK2 CD binding domain (E329A and E331A) resulted in complete loss of association with WDR62 (Figure 6B, lane 8). Collectively, the data suggest that WDR62–JNK association is mediated through the putative D domain within WDR62, and that amino acids within both the CD and ED domains of JNK2 mediate this association.
Figure 6. WDR62 association with wild-type GST–JNK2 and various ED/CD domain mutants.
(A) Schematic representation of the Myc-epitope-tagged wild-type WDR62-C construct (Myc-1018-C) and wild-type GST–JNK2 construct used in the experiments. The numbers above the JNK2 construct refer to the amino acid positions of the ED and CD domains. The light grey rectangle represents the position of the GST tag, and the black square represents the position of the Myc epitope tag. (B) HEK-293T cells were transfected with plasmids encoding Myc-1018-C together with GST, GST–JNK2 wild-type (Wt.) or GST–JNK2 mutants as indicated. GST-containing complexes were isolated from cell lysates with glutathione–agarose beads, washed extensively and eluted with reduced glutathione. The protein complexes were subjected to Western blotting (WB) with either anti-Myc or anti-GST antibodies (upper and lower panels respectively). The expression level of Myc-1018-C was determined by blotting the total cell lysate with anti-Myc antibody (middle panel). The migration of the relevant proteins is indicated by arrows. GPD, GST pull-down.
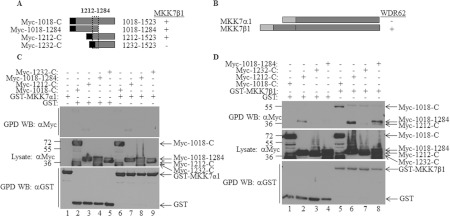
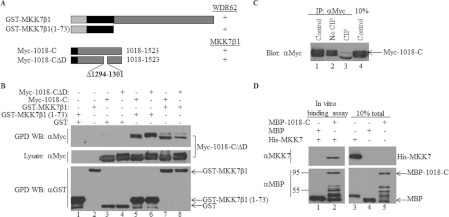
Next, we mapped the domain within WDR62 that interacts with the JNK-activating kinase MKK7. Various WDR62 C-terminal fragments (Myc-1018-C, Myc-1018-1284, Myc-1212-C and Myc-1232-C; Figure 7A) were used in a GST pull-down assay with either GST–MKK7α1 or GST–MKK7β1 isoforms (Figure 7B). Whereas MKK7α1 displayed no association with any of the WDR62 fragments (Figure 7C), MKK7β1 efficiently associated with WDR62 C-terminal fragments Myc-1018-C, Myc-1018-1284 and Myc-1212-C, but not Myc-1232-C (Figure 7D). These results suggest that a minimal binding domain of 72 residues between amino acids 1212 and 1284 is necessary for binding to MKK7β1. Interestingly, MKK7β1 interacted with the WDR62 fragment corresponding to amino acids 1018–1284 that was unable to bind JNK (compare Figure 7D, lane 8 with Figure 3C, lane 7). A similar pattern of binding was obtained with MKK4, although the MKK4–WDR62 interaction appeared to be weaker than the MKK7–WDR62 interaction (Supplementary Figure S3 at http://www.BiochemJ.org/bj/439/bj4390381add.htm). Collectively, the results suggest that distinct WDR62 domains are responsible for JNK and MKK7β1 association. To further examine this possibility, we tested the ability of the Myc-WDR62ΔD mutant lacking the JNK association domain to bind to GST–MKK7β1. GST pull-down assays were performed with cell lysates derived from cells co-transfected with GST–MKK7β1 together with either wild-type Myc-1018-C or Myc-1018-CΔD (Figure 8A). Both the wild-type WDR62 and WDR62ΔD mutant were efficiently precipitated with GST–MKK7β1 (Figure 8B, lanes 7 and 8) demonstrating that the WDR62–MKK7 interaction is independent of JNK. MKK7α1 and MKK7β1 isoforms are highly homologous, but MKK7α1 differs structurally from MKK7β1, having an additional 73-amino-acid N-terminal domain [21]. Since WDR62 displays association with MKK7β1 but not MKK7α1, we hypothesized that the domain within MKK7β1 responsible for interaction with WDR62 resides between amino acids 1 and 73. To examine this possibility, a MKK7β1 peptide corresponding to amino acids 1–73 fused to GST [20] was used in GST pull-down assays with a Myc-1018-C fragment encoding either the wild-type or the D domain mutant. Indeed, both WDR62 protein fragments displayed efficient binding to the MKK7β1 1–73 protein fragment (Figure 8B, lanes 5 and 6). Interestingly, WDR62-1018-C that was precipitated by full-length MKK7β1 migrated faster on the SDS/PAGE gel as compared with that precipitated by MKK7β1 1–73, raising the possibility that WDR62 undergoes dephosphorylation in the presence of full-length MKKβ1. Indeed, treatment of immunopurified Myc-1018-C with CIP resulted in a protein species with a faster migration (Figure 8C, lane 3). This analysis suggests that an as yet unidentified protein phosphatase is recruited to the protein complex by full-length MKK7β1. To examine whether MKK7β1 association with WDR62 occurs via a direct protein–protein interaction, bacterially purified MBP-1018-C and His–MKK7β1 proteins were incubated for 2 h at 37°C followed by precipitation using amylose resin. His–MKK7β1 was efficiently precipitated with MBP-1018-C, but not with the MBP control (Figure 8D). Collectively, results shown in Figures 7 and 8 demonstrate that MKK7β1, but not MKKα1, associates directly with WDR62 through a 73-amino-acid sequence located at the N-terminus of MKK7β1 and a 72-amino-acid sequence within WDR62. The MKK7β1–WDR62 association occurs via a direct protein–protein interaction in a JNK-independent manner and may involve dephosphorylation of WDR62.
Figure 7. WDR62 associates with MKK7β1, but not with MKK7α1.
(A and B) Schematic representation of the Myc-tagged WDR62 deletion constructs (A) and GST-fused MKK7 constructs (B) used in the experiments. The black square and light grey rectangle represent the position of the Myc epitope tag and GST fusion respectively. Summary of the binding of the various WDR62 fragments to MKK7 is indicated by + and −. The broken box represents the minimal region that preserves binding to MKK7β1. (C and D) HEK-293T cells were transfected with plasmids encoding the various WDR62 fragments (shown in A) together with GST–MKK7α1 (C) or GST–MKK7β1 (D). An empty GST plasmid was used as a negative control. GST-containing complexes were isolated from cell lysates with glutathione–agarose beads, washed extensively and eluted with reduced glutathione. The protein complexes were subjected to Western blotting (WB) with either anti-Myc or anti-GST antibodies (top and bottom panels respectively). The expression level of the WDR62 fragments was determined by blotting the total cell lysate with anti-Myc antibody (middle panel). The migration of the relevant proteins is indicated by arrows. Molecular mass markers (in kDa) are indicated on the left-hand side. GPD, GST pull-down.
Figure 8. MKK7β1 N-terminal extension associates directly with WDR62 independently of JNK.
(A) Schematic representation of the GST-fused MKK7β1 constructs and Myc-1018-C constructs used in the experiments. The black rectangle represents the MKK7β1 N-terminal spliced fragment encoding amino acids 1–73. The black square represents the position of the Myc epitope tag. The binding of the various WDR62 fragments to MKK7 is indicated by +/−. (B) HEK-293T cells were transfected with plasmids encoding Myc-tagged wild-type or ΔD mutant WDR62 fragments together with the full-length or N-terminal fragment of MKK7β1 fused to GST as indicated. An empty GST plasmid was used as a negative control. GST-containing complexes were isolated from cell lysates with glutathione–agarose beads, washed extensively and eluted with reduced glutathione. The protein complexes were subjected to Western blotting (WB) with either anti-Myc or anti-GST antibodies (upper and lower panels respectively). The expression level of the WDR62 fragments was determined by blotting the total cell lysate with anti-Myc antibody (middle panel). The migration of the relevant proteins is indicated by arrows. (C) HEK-293T cells were transfected with the Myc-1018-C expression plasmid. The WDR62 fragment was immunoprecipitated (IP) from the cell lysate with anti-Myc antibody and either eluted directly in sample buffer (lane 1) or resuspended in CIP buffer in the absence (lane 2) or presence (lane 3) of CIP. The samples were incubated at 37°C for 30 min. Eluted proteins were subjected to SDS/PAGE and Western blotting with anti-Myc antibody. The expression level of the WDR62 fragment was determined by blotting the total cell lysate with anti-Myc antibody (lane 4). (D) Bacterially purified MBP, MBP-1018-C and His–MKK7 proteins were mixed as indicated for 2 h at 37°C. Subsequently, proteins were attached to amylose resin, washed extensively and then eluted using maltose. The protein complexes were subjected to Western blot analysis with either anti-MKK7 (upper panel) or anti-MBP (lower panel) antibodies. The migration of the relevant proteins is indicated by arrows. Molecular mass markers (in kDa) are indicated on the left-hand side. GPD, GST pull-down.
DISCUSSION
In the present study we identified and characterized the docking domains of a novel JNK-binding protein, WDR62. Mutations in WDR62 are found to cause deleterious phenotypes such as microcephaly and pachygyria [10,22,23].
JNK proteins play a major role in multiple stress responses and regulate diverse cellular functions. JNKs are part of the MAPK cascade mediated through association with MAP2Ks (MKK4/7) and MAP3Ks. JNK also associates with MAPK phosphatases as well as with various cellular substrates [24–26]. In addition, JNK proteins associate with scaffold proteins that also mediate binding to additional pathway components. Scaffolds facilitate and direct signalling to various cellular compartments [3]. WDR62 is a novel scaffold protein identified on the basis of its ability to associate with JNK in a yeast two-hybrid screen [9,27]. WDR62 was found to be recruited to cellular granules that are formed following cellular exposure to arsenite. The recruitment of WDR62 to stress granules and JNK to processing bodies has been suggested to play a role in mRNA homoeostasis [9]. Mutations in WDR62 have been found to cause deleterious phenotypes such as microcephaly and pachygyria [10,22,23]. Multiple mutations have been identified, including missense mutations, mutations affecting splicing, and mutations resulting in truncated WDR62 proteins as a result of premature termination. Patients harbouring any of these mutations display similar degrees of microcephaly and brain developmental defects [10,22,23]. WDR62 was suggested to be localized to centrosomes during mitosis and thus may play a role in neurite proliferation [22,23]. Revealing WDR62 functional domains might help explain the biological role of WDR62 in neurological development.
In the present study, we show for the first time that endogenous WDR62 is associated with JNK2 and MKK7 (Figure 1). We hypothesize that the lack of interaction between WDR62 and JNK1 is simply due to its lower level of expression in HEK-293T cells.
We mapped the domains within WDR62 responsible for binding to JNK and its activating kinase MKK7 (Figures 2, 3, 7 and 8). We tested a WDR62 mutant with premature termination, a mutation that occurred in a human patient, for its ability to associate with JNK and MKK7. This mutant, lacking 122 amino acids from the C-terminus of the protein, is able to associate with both JNK and MKK7 (Figures 3 and 7), and thus microcephaly and the developmental phenotype are probably not due to the loss of either JNK or MKK7 association. Interestingly, WDR62 displays the strongest association with the neuronal-enriched JNK3 isoform (Figure 4C). In view of the results of the present study, we hypothesize that the mutations that cause microcephaly affect WDR62 interactions with a different protein partner yet to be identified.
The WDR62 domain found to be both necessary and sufficient for JNK association resides between amino acids 1232 and 1345 (Figure 3). Within this domain, we identified a highly conserved D domain at position 1294–1301. WDR62 lacking the putative D domain fails to associate with JNK (Figure 4C), activate JNK (Figure 4C) and recruit JNK to cellular granules (Figure 4D). In addition, a small peptide containing the putative D domain of WDR62 is able to inhibit JNK activity in a similar manner, although with reduced potency (IC50=5 μM, as compared with 0.3 μM for the JIP1 inhibitory activity; Figure 5) [17]. This is in agreement with the previously calculated JIP dissociation constant of 0.4 μM [28] and IC50 of 0.56 μM [29]. The WDR62 D domain has a unique histidine residue in place of a lysine or arginine and also a threonine residue within the LXL domain (Figure 4A). Whether these perturbations are responsible for the lower binding ability of this peptide remains to be elucidated. Alternatively, additional amino acids that are not included in the WDR62-competing peptide may contribute to JNK association. The WDR62ΔD mutant preserves its binding to MKK7β1 (Figure 8), indicating that the protein is folded correctly and that the association of WDR62 with MKK7β1 is JNK-independent. WDR62 associates with MKK7β1, but not MKK7α1, via the MKK7β1 N-terminal extension composed of 73 amino acids and 72 amino acids within WDR62 (Figure 7). This is in agreement with previous results that show that MKK7α1 has much weaker binding to JNK and DLK (Delta-like protein) as compared with MKK7β1 [20,21]. However, this is in contrast with JIP1 scaffold protein, which interacts with both MKK7α and MKK7β [20]. This indicates a different selective association between WDR62 and JIP1 scaffolds in the JNK pathway. The MKK7β1 fragment 1–73 is composed of two low-affinity D domains [30]. The association between the two JNK scaffold proteins JIP/WDR62 and MKK7β1 competes with the direct binding of JNKs to their upstream kinase. This allows proper organization of the JNK tier with the corresponding scaffold, and certainly contributes to signalling selectivity and specificity. A striking difference in WDR62 gel migration is observed when co-expressing MKK7β1 full-length protein. Indeed, we observed that WDR62 is constitutively phosphorylated in the cell and the recruitment of full-length MKK7β1 results in dephosphorylation of WDR62 (Figure 8C). MKK7 has been shown to interact with the dual-specificity phosphatase SKRP1 (SAPK-pathway-regulating phosphatase 1) through a domain that resides between residues 128 and 214 within MKK7 [31]. This is consistent with the dephosphorylation of the WDR62-C fragment only in the presence of full-length MKK7β1, but not with the N-terminal 1–73 fragment. It would be of interest to identify the WDR62-specific phosphatase and the biological role of WDR62 phosphorylation and dephosphorylation; however, this is beyond the scope of the present study.
The basic residues of numerous D domain-containing proteins make electrostatic contacts with two closely spaced acidic residues on the MAPK. Surprisingly, mutation of the acidic residues Asp162 and Asp326 within the JNK2 ED and CD domains respectively resulted in no loss of JIP1–JNK interaction [20]. In contrast, WDR62–JNK2 interaction was highly affected by mutations of these residues, suggesting a conserved MAPK association between the WDR62 D domain and the JNK2 ED and CD domains. Interestingly, glutamic acid substitutions to alanine residues within the CD domain (E329A/E331A) completely abrogate JNK2 binding to both JIP family members [20] and WDR62 (Figure 6).
Overall, these results indicate involvement of the MKK7–JNK2–WDR62 module in a distinct signalling pathway leading to the formation of stress granules through interaction with distinct domains that serve as a platform for the association of both positive and negative regulators of the pathway.
Online data
AUTHOR CONTRIBUTION
Ksenya Cohen-Katsenelson and Tanya Wasserman carried out the experiments, performed data analysis and helped with Figure preparation. Samer Khateb helped with the in vitro binding analysis. Alan Whitmarsh provided DNA constructs and helped with experimental design, data interpretation and manuscript editing prior to submission. Ami Aronheim designed the study, interpreted data and wrote the manuscript. All authors read and approved the final version of the manuscript.
ACKNOWLEDGEMENTS
We thank Ms Aviva Cohen for excellent technical assistance, Dr Ariel Stanhill (Department of Biochemistry, Technion, Haifa, Israel) for pEBG expression plasmids, Dr David Engelberg for the His–JNK2 and His–MKK7β1 expression plasmids, and Dr Michael Courtney for kindly providing EGFP–JNK expression plasmids.
FUNDING
This work was supported by the Israeli Science Foundation [grant number 641/07 (to A.A.)]; and by the Wellcome Trust [grant number 088232/Z/09/Z (to A.J.W.)].
References
- 1.Shaul Y. D., Seger R. The MEK/ERK cascade: from signaling specificity to diverse functions. Biochim. Biophys. Acta. 2007;1773:1213–1226. doi: 10.1016/j.bbamcr.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Weston C. R., Lambright D. G., Davis R. J. MAP kinase signaling specificity. Science. 2002;296:2345–2347. doi: 10.1126/science.1073344. [DOI] [PubMed] [Google Scholar]
- 3.Dard N., Peter M. Scaffold proteins in MAP kinase signaling: more than simple passive activating platforms. BioEssays. 2006;28:146–156. doi: 10.1002/bies.20351. [DOI] [PubMed] [Google Scholar]
- 4.Tibbles L. A., Woodgett J. R. The stress-activated protein kinase pathways. Cell. Mol. Life Sci. 1999;55:1230–1254. doi: 10.1007/s000180050369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis R. J. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 6.Chang L., Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 7.Holmberg C., Katz S., Lerdrup M., Herdegen T., Jaattela M., Aronheim A., Kallunki T. A novel specific role for IκB kinase complex-associated protein in cytosolic stress signaling. J. Biol. Chem. 2002;277:31918–31928. doi: 10.1074/jbc.M200719200. [DOI] [PubMed] [Google Scholar]
- 8.Rubinfeld H., Seger R. The ERK cascade: a prototype of MAPK signaling. Mol. Biotechnol. 2005;31:151–174. doi: 10.1385/MB:31:2:151. [DOI] [PubMed] [Google Scholar]
- 9.Wasserman T., Katsenelson K., Daniliuc S., Hasin T., Choder M., Aronheim A. A novel c-Jun N-terminal kinase (JNK)-binding protein WDR62 is recruited to stress granules and mediates a nonclassical JNK activation. Mol. Biol. Cell. 2010;21:117–130. doi: 10.1091/mbc.E09-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bilguvar K., Ozturk A. K., Louvi A., Kwan K. Y., Choi M., Tatli B., Yalnizoglu D., Tuysuz B., Caglayan A. O., Gokben S., et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467:207–210. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stanhill A., Haynes C. M., Zhang Y., Min G., Steele M. C., Kalinina J., Martinez E., Pickart C. M., Kong X. P., Ron D. An arsenite-inducible 19S regulatory particle-associated protein adapts proteasomes to proteotoxicity. Mol. Cell. 2006;23:875–885. doi: 10.1016/j.molcel.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 12.Schwarze S. R., Ho A., Vocero-Akbani A., Dowdy S. F. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 13.Batard P., Jordan M., Wurm F. Transfer of high copy number plasmid into mammalian cells by calcium phosphate transfection. Gene. 2001;270:61–68. doi: 10.1016/s0378-1119(01)00467-x. [DOI] [PubMed] [Google Scholar]
- 14.Katz S., Aronheim A. Differential targeting of the stress mitogen-activated protein kinases to the c-Jun dimerization protein 2. Biochem. J. 2002;368:939–945. doi: 10.1042/BJ20021127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katz S., Heinrich R., Aronheim A. The AP-1 repressor, JDP2, is a bona fide substrate for the c-Jun N-terminal kinase. FEBS Lett. 2001;506:196–200. doi: 10.1016/s0014-5793(01)02907-6. [DOI] [PubMed] [Google Scholar]
- 16.Bogoyevitch M. A., Kobe B. Uses for JNK: the many and varied substrates of the c-Jun N-terminal kinases. Microbiol. Mol. Biol. Rev. 2006;70:1061–1095. doi: 10.1128/MMBR.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonny C., Oberson A., Negri S., Sauser C., Schorderet D. F. Cell-permeable peptide inhibitors of JNK: novel blockers of β-cell death. Diabetes. 2001;50:77–82. doi: 10.2337/diabetes.50.1.77. [DOI] [PubMed] [Google Scholar]
- 18.Tanoue T., Adachi M., Moriguchi T., Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2000;2:110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- 19.Tanoue T., Maeda R., Adachi M., Nishida E. Identification of a docking groove on ERK and p38 MAP kinases that regulates the specificity of docking interactions. EMBO J. 2001;20:466–479. doi: 10.1093/emboj/20.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mooney L. M., Whitmarsh A. J. Docking interactions in the c-Jun N-terminal kinase pathway. J. Biol. Chem. 2004;279:11843–11852. doi: 10.1074/jbc.M311841200. [DOI] [PubMed] [Google Scholar]
- 21.Tournier C., Whitmarsh A. J., Cavanagh J., Barrett T., Davis R. J. The MKK7 gene encodes a group of c-Jun NH2-terminal kinase kinases. Mol. Cell. Biol. 1999;19:1569–1581. doi: 10.1128/mcb.19.2.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu T. W., Mochida G. H., Tischfield D. J., Sgaier S. K., Flores-Sarnat L., Sergi C. M., Topcu M., McDonald M. T., Barry B. J., Felie J. M., et al. Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat. Genet. 2010;42:1015–1020. doi: 10.1038/ng.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholas A. K., Khurshid M., Desir J., Carvalho O. P., Cox J. J., Thornton G., Kausar R., Ansar M., Ahmad W., Verloes A., et al. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat. Genet. 2010;42:1010–1014. doi: 10.1038/ng.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bardwell A. J., Frankson E., Bardwell L. Selectivity of docking sites in MAPK kinases. J. Biol. Chem. 2009;284:13165–13173. doi: 10.1074/jbc.M900080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bardwell L. Mechanisms of MAPK signalling specificity. Biochem. Soc. Trans. 2006;34:837–841. doi: 10.1042/BST0340837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enslen H., Davis R. J. Regulation of MAP kinases by docking domains. Biol. Cell. 2001;93:5–14. doi: 10.1016/s0248-4900(01)01156-x. [DOI] [PubMed] [Google Scholar]
- 27.Broder Y. C., Katz S., Aronheim A. The Ras recruitment system, a novel approach to the study of protein–protein interactions. Curr. Biol. 1998;8:1121–1124. doi: 10.1016/s0960-9822(98)70467-1. [DOI] [PubMed] [Google Scholar]
- 28.Heo Y. S., Kim S. K., Seo C. I., Kim Y. K., Sung B. J., Lee H. S., Lee J. I., Park S. Y., Kim J. H., Hwang K. Y., et al. Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125. EMBO J. 2004;23:2185–2195. doi: 10.1038/sj.emboj.7600212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barr R. K., Kendrick T. S., Bogoyevitch M. A. Identification of the critical features of a small peptide inhibitor of JNK activity. J. Biol. Chem. 2002;277:10987–10997. doi: 10.1074/jbc.M107565200. [DOI] [PubMed] [Google Scholar]
- 30.Ho D. T., Bardwell A. J., Grewal S., Iverson C., Bardwell L. Interacting JNK-docking sites in MKK7 promote binding and activation of JNK mitogen-activated protein kinases. J. Biol. Chem. 2006;281:13169–13179. doi: 10.1074/jbc.M601010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zama T., Aoki R., Kamimoto T., Inoue K., Ikeda Y., Hagiwara M. A novel dual specificity phosphatase SKRP1 interacts with the MAPK kinase MKK7 and inactivates the JNK MAPK pathway. Implication for the precise regulation of the particular MAPK pathway. J. Biol. Chem. 2002;277:23909–23918. doi: 10.1074/jbc.M200837200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.