Abstract
In humans, assembly of spliceosomal snRNPs (small nuclear ribonucleoproteins) begins in the cytoplasm where the multi-protein SMN (survival of motor neuron) complex mediates the formation of a seven-membered ring of Sm proteins on to a conserved site of the snRNA (small nuclear RNA). The SMN complex contains the SMN protein Gemin2 and several additional Gemins that participate in snRNP biosynthesis. SMN was first identified as the product of a gene found to be deleted or mutated in patients with the neurodegenerative disease SMA (spinal muscular atrophy), the leading genetic cause of infant mortality. In the present study, we report the solution structure of Gemin2 bound to the Gemin2-binding domain of SMN determined by NMR spectroscopy. This complex reveals the structure of Gemin2, how Gemin2 binds to SMN and the roles of conserved SMN residues near the binding interface. Surprisingly, several conserved SMN residues, including the sites of two SMA patient mutations, are not required for binding to Gemin2. Instead, they form a conserved SMN/Gemin2 surface that may be functionally important for snRNP assembly. The SMN–Gemin2 structure explains how Gemin2 is stabilized by SMN and establishes a framework for structure–function studies to investigate snRNP biogenesis as well as biological processes involving Gemin2 that do not involve snRNP assembly.
Keywords: Gemin2, survival of motor neuron (SMN), small nuclear ribonucleoprotein (snRNP) assembly, spinal muscular atrophy
Abbreviations: APS, Advanced Photon Source; CHESS, Cornell High Energy Synchrotron Source; DTT, dithiothreitol; GST, glutathione transferase; HSQC, heteronuclear single-quantum coherence; MTSL, S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate; NOE, nuclear Overhauser effect; NP40, Nonidet P40; PRE, paramagnetic relaxation enhancement; RDC, residual dipolar coupling; RMSD, root mean square deviation; SAXS, small-angle X-ray scattering; SMA, spinal muscular atrophy; SMN, survival of motor neuron; snRNA, small nuclear RNA; snRNP, small nuclear ribonucleoprotein
INTRODUCTION
The SMN (survival of motor neuron) protein is found in the cytoplasm and nucleus of cells, where it binds tightly to Gemin2 as part of a large multiprotein ‘SMN complex’ [1]. The SMN complex mediates the ordered assembly of spliceosomal snRNP (small nuclear ribonucleoprotein) particles that are responsible for removing introns from pre-mRNA [2–4]. During the early steps of snRNP biogenesis, seven Sm proteins are assembled to form a ring around the Sm site of uridine-rich snRNAs (U-snRNAs), resulting in a stable snRNP core. In human cells, assembly of the Sm ring on the snRNA occurs in the cytoplasm and is catalysed by the SMN complex. In addition to SMN and Gemin2, six additional Gemin proteins, plus the protein Unrip, contribute to the cytoplasmic phase of snRNP biogenesis [5]. However, in simpler organisms, snRNP assembly is carried out by a more modest complex that lacks some of the accessory factors found in human cells. This observation, together with demonstrations that the basic steps of snRNP assembly can be carried out in vitro with minimal recombinant complexes, suggests that the crucial conserved functions of the SMN complex are carried out by SMN and Gemin2 [6,7].
SMN was first identified as the product of a gene that was found to be deleted or mutated in patients with the disease SMA (spinal muscular atrophy), the leading genetic cause of infant mortality [8]. SMA is an autosomal recessive disorder characterized by degeneration of α-motor neurons in the spinal cord, leading to muscle weakness and poor muscle tone [9,10]. In the most severe cases, patients typically die within the first two years of life due to difficulties in breathing and poor airway protection [11].
Humans are unusual in having two copies of the SMN gene, located in a large inverted duplication on chromosome 5q. The telomeric SMN1 gene is altered in SMA patients, whereas the centromeric SMN2 gene is unchanged and is often present in multiple copies, where the copy number varies inversely with disease severity [8,12]. Most transcripts from the SMN2 gene are alternatively spliced, resulting in a truncated SMN that is rapidly degraded [13,14]. Thus SMA patients have sharply reduced amounts of functional SMN protein due to homozygous mutations in SMN1; their functional SMN is derived from low levels of expression from one or more copies of the SMN2 gene.
A strong body of evidence suggests that SMA is caused by defects in snRNP assembly that arise from low levels of SMN [15–18]. However, SMN appears to have additional functions in specific tissues, including muscle and neural cells [19,20]. The Gemin2 protein has also been implicated in biological processes independent of snRNP assembly [21,22]. An important goal for understanding the function of SMN and Gemin2 has been to establish structural models that can be used to probe the various biological activities of the proteins. Until recently, the only structural information available for either SMN or Gemin2 has been NMR and crystal structures of the central Tudor domain of SMN [23,24]. To address this gap in our understanding of this important system, we have determined the solution structure of SMN bound to Gemin2. The present study reveals the structures of Gemin2 and the Gemin2-binding domain of SMN, the nature of the conserved SMN–Gemin2 interface, and the roles of conserved SMN residues at or near the interface in Gemin2 binding. The structure establishes a framework for structure–function studies in snRNP assembly and other biological processes that involve SMN and Gemin2.
MATERIALS AND METHODS
Expression and purification of human SMN and Gemin2
Gemin2, Gemin295–280 and SMN26–51 were fused to a C-terminal Mxe intein (New England Biolabs) containing chitin-binding and hexa-histidine tags in pETDuet (Novagen) and expressed in BL21(DE3) cells. Cells were induced with 0.5 mM IPTG (isopropyl β-D-thiogalactopyranoside) at 15°C for 20 h in LB (Luria–Bertani) medium or for 24 h in Mops minimal medium with 15NH4Cl as the main nitrogen source and U-[12C/1H] or U-[13C/1H]glucose (Cambridge Isotope Laboratories) as the main carbon source. For leucine-selective 15N labelling, the Neidhardt et al. [35] protocol for amino acid-supplemented Mops was followed by the substitution of [15N]leucine (Cambridge Isotope Laboratories); fractional 13C labelling for stereo-specific methyl assignments was achieved with 10% U-[13C/1H]glucose.
Following purification on Ni-NTA (Ni2+-nitrilotriacetate) (Qiagen) and chitin (New England Biolabs) resins, Gemin2 and SMN26–51 were cleaved from the intein by addition of 50 mM 2-mercaptoethanol and overnight incubation at 20°C. The cleaved proteins were then purified by anion exchange using a Mono Q column (GE Healthcare) with a 0.1–0.4 M NaCl gradient at pH 8.0. SMN26–51 was further purified by gel filtration on a Superdex-75 column (GE Healthcare). To form the SMN–Gemin2 complex, SMN26–51 was combined in molar excess with Gemin295–280 and the complex was purified on Superdex-75. Purified proteins were concentrated and stored at −80°C in 50 mM sodium/potassium phosphate, pH 6.5, 50 mM NaCl, 50 mM DTT (dithiothreitol) and 10% (v/v) glycerol.
SMN peptide–Gemin2 binding
Purified SMN26–51 peptides and an unrelated 32-mer control peptide were labelled with amine-reactive fluorescein-5-isothiocyanate (Invitrogen) in 0.1 M sodium bicarbonate, pH 9.0, to favour labelling of the N-terminus. Binding reactions (100 μl) containing 15 nM FITC-labelled SMN and increasing concentrations of Gemin2 (0–1 μM) were analysed in PBS (10 mM Na2HPO4, 2 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4, and 2 mM DTT) using a Beacon 2000 fluorescence polarization instrument (Panvera). Data were fitted to a simple binding isotherm corrected for receptor depletion. Similar results were obtained by titrating labelled peptide fixed at a concentration of 5 nM, at the expense of weaker fluorescence intensities.
NMR spectroscopy and analysis
All NMR samples were prepared in Shigemi NMR tubes in buffer containing 50 mM sodium/potassium phosphate, pH 6.5, 50 mM NaCl, 50 mM DTT, 50 μM EDTA, 0.2 mM sodium azide and either 90% H2O/10% 2H2O or 99.99% 2H2O. Spectra were collected at 25°C at either 750 MHz or 500 MHz (1H) on Bruker AVANCE III NMR spectrometers equipped with cryo-probes. Proton chemical shifts were referenced to DSS in the buffer, whereas 13C and 15N chemical shifts were indirectly referenced. Data were processed using FELIX (Molecular Simulations) and analysed in SPARKY (Goddard, T. and Kneller, D., SPARKY 3, University of San Francisco, 2008). Specific experiments used to assign backbone and side chain resonances are described in the Supplementary methods at http://www.BiochemJ.org/bj/445/bj4450361add.htm.
NOE (nuclear Overhauser effect) distance restraints were derived, in H2O, from three-dimensional 13C-edited NOESY-HSQC (heteronuclear single-quantum coherence) and from four-dimensional HNCH-NOESY-HSQC experiments [37] and, in 2H2O, from a four-dimensional HCCH-NOESY-HMQC [38,39]. All mixing times were 110 ms. NOE-derived interproton distances were sorted into classes in each NOESY spectrum, corresponding to approximate distance ranges: 1.8–3.0 Å (1 Å=0.1 nm) (strong), 1.8–3.5 Å (medium-strong), 1.8–4.0 Å (medium) and 1.8–5.0 Å (weak). All interproton restraints derived from the aromatic-optimized 13C NOESY-HSQC were classified into the 1.8–5.0 Å distance range. A distance restraint involving one or two methyl groups was afforded an empirical correction of 0.5 Å per methyl group added to its upper distance boundary [40].
Backbone torsion angle restraints were derived from chemical shifts using the TALOS+ program [41]. Hydrogen bonds were entered on the basis of secondary structure as indicated by calculated backbone torsion angles for residues that demonstrated slowed hydrogen/deuterium exchange when dissolved in 2H2O buffer. For residual dipolar couplings, 15N-labelled protein was aligned in bacteriophage Pf1 (ASLA Biotech) to a final concentration of 12 mg/ml. 1DHN couplings were obtained from a two-dimensional (1H,15N)-HSQC IPAP (in-phase/antiphase) experiment [42]. SAXS (small-angle X-ray scattering) data were collected at CHESS (Cornell High Energy Synchrotron Source) beamline G1, and APS (Advanced Photon Source) BioCAT 18-ID and PRE (paramagnetic relaxation enhancement) distance restraints were derived from the attenuation of signal intensity of amide resonances in (1H,15N)-HSQCs collected in the presence of MTSL [S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate]-labelled (paramagnetic) SMN26–51-Gemin295–280 (C154S/C221S/C264S) and its diamagnetic analogue. Further details of SAXS and PRE restraints are described in the Supplementary methods at http://www.BiochemJ.org/bj/445/bj4450361add.htm.
Structure calculations
Structures were calculated using the simulated annealing molecular dynamics refinement protocol within the program XPLOR-NIH [43]. The minimized target function comprised NOE- and PRE-derived interproton distance restraints, torsion angle restraints, RDC (residual dipolar coupling) restraints, a SAXS restraint and a quartic van der Waals repulsion term for the non-bonded contacts. A total of 400 structures were generated, of which the 32 lowest-energy structures formed the ensemble. Structure figures were generated using PyMOL (http://www.pymol.org).
RESULTS AND DISCUSSION
Identification of a minimal SMN–Gemin2 complex
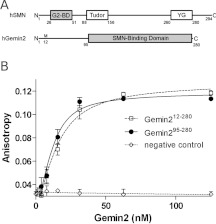
SMN and Gemin2 form the core of a large oligomeric complex that sediments similarly to ribosomes on sucrose and glycerol gradients [25]. To identify a minimal complex that contains all of the protein elements required for a stable SMN–Gemin2 interaction, we used bacterial two-hybrid and in vitro binding assays to identify the minimal domain boundaries (Figure 1A and Supplementary Figure S1 at http://www.BiochemJ.org/bj/445/bj4450361add.htm). We found that an SMN fragment including residues 1–62 (encoding exons 1–2b) binds as well to Gemin2 as fragments including residues 1–209 (exons 1–4) and full-length SMN, but we observed no binding for a fragment including residues 52–209 (exons 2b, 3 and 4). Further deletion of the 1–62 fragment led to a peptide corresponding to residues 26–51 of SMN that was still able to bind with high affinity to Gemin2 (Figure 1B). These results are consistent with some previous reports [1,26], but do not support a role for exon 2b in binding independently to Gemin2 [27].
Figure 1. A minimal SMN–Gemin2 complex.
(A) Domain structures of human (h) SMN and Gemin2. The conserved Gemin2-binding domain, Tudor domain and YG-box are shown for SMN. Minimal interaction domains are shaded. The numbering for Gemin2 follows that used in the original report [1]. (B) Binding of SMN26–51 to Gemin2 or to Gemin295–280 using fluorescence anisotropy of SMN labelled at the α-amino group with FITC. The average Kd values fit to binding curves from three independent experiments are 7.0±2.5 and 3.3±1.8 nM for Gemin2 and Gemin295–280 respectively. Error bars represent one S.D. from the mean for three independent experiments. No binding was observed to a labelled unrelated peptide (NC, negative control).
A minimal SMN-binding domain of Gemin2 was identified as residues 95–280 (Figure 1A and Supplementary Figure S1). Additional deletions of up to 15 residues from the N-terminus were tolerated, but secondary structure predictions indicated that these involved truncations of a long α-helix. These results are in broad agreement with previous findings based on a mammalian two-hybrid assay [26]. The Gemin295–280 core domain binds to SMN26–51 tightly, and with similar affinity compared with full-length Gemin2 (Figure 1B). Gemin2 and Gemin295–280 also bind to larger SMN constructs with similar efficiency in qualitative interaction assays (Supplementary Figure S1). The complex formed between SMN26–51 and Gemin295–280 (24 kDa; shaded regions in Figure 1A) proved to be highly soluble and monodisperse in solution, with no evidence of oligomerization on the basis of analytical ultracentrifugation and static light scattering analyses (Supplementary Figure S2 at http://www.BiochemJ.org/bj/445/bj4450361add.htm). This complex was refractory to crystallization, but was sufficiently soluble to be a candidate for structure solution by NMR methods.
Solution structure of the core SMN–Gemin2 complex
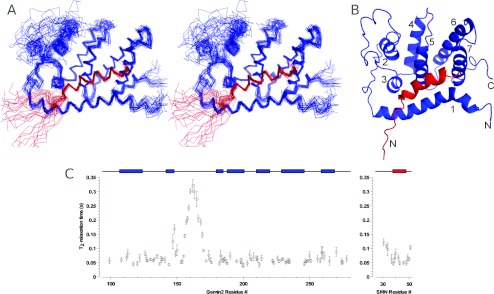
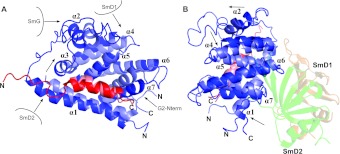
To determine the solution structure of the core SMN–Gemin2 complex using heteronuclear NMR methods, we expressed and purified 15N- and 13C-enriched SMN26–51 and Gemin295–280 and obtained comprehensive resonance assignments using triple resonance, total correlation spectroscopy and NOE-based strategies (see the Materials and methods section). Structural restraints were primarily interproton distances derived from three- and four-dimensional 15N- and 13C-edited NOE spectra and backbone dihedral restraints based on chemical shifts. Orientational restraints included in refinement were derived from residual dipolar couplings measured in a liquid crystalline medium containing bacteriophage Pf1, and a restraint on overall molecular shape was imposed from SAXS data (additional SAXS data are presented in Supplementary Figure S3 at http://www.BiochemJ.org/bj/445/bj4450361add.htm). Additional distance restraints were obtained from PREs, where Cys241 of Gemin2 was coupled to a paramagnetic nitroxide spin label. The structure was determined by simulated annealing molecular dynamics refinement using these restraints. Out of 400 trial structures, we selected the 32 with the lowest energy to define an ensemble that represents the structure of SMN26–51–Gemin295–280 (Figures 2A and 2B). Most of the structure is well defined (Table 1). The major exception is a large poorly conserved loop in Gemin2 (residues 150–175) that is disordered, with no long-range NOEs observed. This region of the protein is highly mobile, as indicated by the relatively long 15N T2 relaxation time constants (Figure 2C). In addition, the first seven residues in the SMN peptide (residues 26–32) are disordered, which is consistent with their lack of importance for Gemin2 binding as indicated by our interaction assays.
Figure 2. Structure of SMN bound to Gemin2.
(A) Stereo overlay of Cα-traces for the 32 lowest-energy structures. (B) Ribbon drawing of the lowest-energy structure. SMN26–51 is in red and Gemin295–280 is in blue. The Gemin2 helices are numbered. (C) Backbone amide 15N T2 relaxation times of Gemin2 and the bound SMN peptide. Obtained at 750 MHz (1H). Helices are depicted above the graph as coloured rectangles for reference. High T2 times are indicative of increased backbone flexibility.
Table 1. Summary of structural ensemble statistics.
| Parameter | Value |
|---|---|
| Experimental restraints | |
| NOE distance restraints | |
| All | 1849 |
| Intraresidue | 616 |
| Sequential (|i−j|=1) | 541 |
| Medium-range (1<|i−j|<5) | 453 |
| Long-range (|i−j|>5) | 239 |
| Interprotein | 50 |
| PRE distance restraints | 31 |
| Hydrogen bond restraints | 55 |
| Dihedral angle restraints | |
| Φ | 139 |
| Ψ | 134 |
| 15N-1H RDCs | 84 |
| RMSD from idealized covalent geometry | |
| Bonds (Å) | 0.0040±0.0001 |
| Angles (°) | 0.589±0.006 |
| Impropers (°) | 0.441±0.011 |
| RMSD from solution NMR restraints | |
| NOE distance restraints (Å) | 0.021±0.001 |
| PRE distance restraints (Å) | 0.042±0.009 |
| Dihedral restraints (°) | 0.787±0.069 |
| RDC restraints (Hz) | 0.116±0.014 |
| RMSD from SAXS data* | 0.740±0.050 |
| RDC quality factor | 0.311±0.012 |
| Precision of atomic co-ordinates | |
| Secondary structure† | |
| Backbone (N, Cα, C', O) (Å) | 0.596±0.096 |
| Heavy atoms (Å) | 1.056±0.081 |
| Ramachandran statistics | |
| Favoured (%) | 98.0 |
| Allowed (%) | 1.8 |
| Disallowed (%) | 0.2 |
*RMSD between observed and predicted solution X-ray scattering intensities.
†The secondary elements used were as follows: Gemin2 residues 107–124,142–148, 180–185, 188–201, 210–220, 229–246 and 259–269; SMN residues 38–48.
Gemin295–280 folds into a helical bundle containing seven α-helices (Figure 2B). The locations of helical secondary structure agree well with those predicted by a number of prediction algorithms. The Gemin2-binding domain of SMN also forms an α-helix and this helix is embedded in an elongated hydrophobic cavity formed by Gemin2 helices 1, 3, 5 and 7. A search of the Dali database [28] using either the lowest-energy Gemin295–280 structure or the SMN–Gemin2 complex yielded no strong matches. The highest scoring hits had RMSDs (root mean square deviations) >3 Å and typically identified the four-helix bundle formed by helices 4–7 of Gemin2, which is similar to that found in a number of helical subdomains. Thus the overall spatial arrangement of helices in Gemin2 and in the SMN–Gemin2 complex appears to be unique.
The conserved SMN–Gemin2 interface
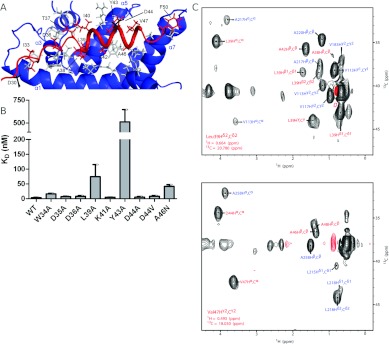
The definition of the SMN–Gemin2 interface is largely derived from 50 intermolecular NOEs (Figure 3). Eleven of the 18 SMN residues from Ile33 through Phe50 are positioned close to the Gemin2 core domain. The SMN helix is strongly amphipathic, with its hydrophobic face largely buried in a cavity formed by Gemin2 helices 1, 3, 5, and 7. In the lowest-energy structure, the SMN–Gemin2 interface buries a total of 1800 Å2 of solvent-accessible surface area, which is consistent with a high-affinity interaction. There are also several polar interactions flanking the hydrophobic SMN–Gemin2 interface, which we infer from side chain positions. The Gemin2 residues involved in binding to SMN are located throughout the sequence, explaining why more extensive deletions into the 95–280 construct result in loss of SMN-binding activity [29] and why a smaller SMN-binding domain has not been previously reported.
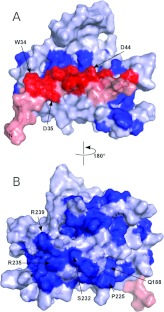
Figure 3. The conserved SMN–Gemin2 interface.
(A) Close-up of the interface. Gemin2 residues that make up the interface are grey. SMN residues involved in binding Gemin2 are red. The residues mutated in some SMA patients (Asp30 and Asp44) are black. Only selected SMN residues are labelled. (B) Binding of SMN26–51 mutants to Gemin295–280 using the fluorescence anisotropy assay described in Figure 1. Each binding titration was repeated 3–6 times and the average Kd value is shown. Error bars represent one S.D. from the mean. WT, wild-type. (C) Planes of a four-dimensional HCCH-NOESY-HMQC spectrum of the SMN26–51–Gemin295–280 complex, aliased one sweep width upfield and downfield in the 13C dimension, illustrating some of the intermolecular 1H-1H NOEs defining the interface of the complex. Red labels indicate 1H-1H NOEs to SMN and blue labels indicate 1H-1H NOEs to Gemin2. Obtained at 750 MHz (1H) with a mixing time of 110 ms.
Given the hydrophobic nature of the SMN–Gemin2 interface observed in the NMR structure, we examined the effects of ionic strength, temperature and non-denaturing detergents on the affinity of the interaction. In PBS, SMN26–51 binds to Gemin295–280 with Kd=3.3±1.8 nM (Figure 1B). As the salt concentration is increased to 300 mM and then 600 mM, the binding affinity remains similar, with Kd values of 5.2±1.8 and 2.4±1.5 nM respectively. Binding is relatively unaffected in PBS at 4°C or in PBS at 25°C containing 0.05% NP40 (Nonidet P40) (Kd=2.3±0.92 and 2.2±1.7 nM respectively). Importantly, however, the combination of 4°C and detergent substantially weakens the affinity, with Kd=180±27 nM. These observations, summarized in Table 2, are consistent with the primarily hydrophobic interface observed in the structure and shown in Figure 3(A).
Table 2. SMN26–51–Gemin295–280 dissociation constants.
Kd values reported are the average of three to six individual binding titrations. The Kd value for SMN D44V in NP40-containing buffer at 4°C could not be determined precisely, but a lower limit of 4 μM could be established.
| SMN26–51 | Gemin2 | Buffer* | Temperature (°C) | Kd (nM) |
|---|---|---|---|---|
| Wild-type | 12–280 | PBS | 25 | 7.0±2.5 |
| Wild-type | 95–280 | PBS | 25 | 3.3±1.8 |
| Wild-type | 95–280 | PBS, 300 mM NaCl | 25 | 5.2±1.8 |
| Wild-type | 95–280 | PBS, 600 mM NaCl | 25 | 2.4±1.5 |
| Wild-type | 95–280 | PBS | 4 | 2.3±0.92 |
| Wild-type | 95–280 | PBS+NP40 | 25 | 2.2±1.7 |
| Wild-type | 95–280 | PBS+NP40 | 4 | 180±27 |
| W34A | 95–280 | PBS | 25 | 16±2.2 |
| D35A | 95–280 | PBS | 25 | 6.8±2.1 |
| D36A | 95–280 | PBS | 25 | 8.3±2.5 |
| L39A | 95–280 | PBS | 25 | 74±41 |
| K41A | 95–280 | PBS | 25 | 4.8±1.0 |
| Y43A | 95–280 | PBS | 25 | 510±130 |
| D44A | 95–280 | PBS | 25 | 5.3±2.4 |
| D44V | 95–280 | PBS | 25 | 8.4±2.4 |
| D44V | 95–280 | PBS | 4 | 1.5±0.47 |
| D44V | 95–280 | PBS+NP40 | 25 | 1800±240 |
| D44V | 95–280 | PBS+NP40 | 4 | >4000 |
| A46N | 95–280 | PBS | 25 | 42±5.9 |
| p28, p31 | 95–280 | PBS | 25 | 4.2±1.3 |
*PBS: 10 mM Na2HPO4, 2 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4, and 2 mM DTT; NaCl concentration is modified where indicated.
SMN sequence alignments reveal that most of the residues involved in the interface with Gemin2 are conserved (Supplementary Figure S4 at http://www.BiochemJ.org/bj/445/bj4450361add.htm). Leu39, Ala42, Tyr43, Ala46 and Val47 are largely buried (Figure 3A), with little solvent-accessible surface in members of the low-energy structural ensemble. In addition, Asp36 is directed towards the Gemin2 interface, where it could interact with His120 and/or Trp124. To examine the role of conserved SMN residues in Gemin2 binding, we prepared a series of SMN26–51 alanine substitutions and tested them for binding to Gemin295–280 (Figure 3B). Binding affinity was reduced more than 100-fold for Y43A, 20-fold for L39A and 10-fold for A46N, all in accord with their apparent roles in Gemin2-binding. Asp44 is the site of a SMA patient mutation in SMN, where a valine residue is substituted in this position [30]. Although Asp44 is well positioned to make an electrostatic interaction with Arg213 of Gemin2, it is unclear whether this would make a significant energetic contribution to binding. We therefore tested the D44A and D44V mutants for interaction with Gemin2. As peptides, each bound as well as wild-type SMN, suggesting that the functional defect in patients with the D44V mutation is unlikely to be the loss of an Asp44 interaction with Gemin2.
To determine if full-length SMN mutants have similar binding properties, we purified full-length versions of the same mutants shown in Figure 3(B) and tested their ability to bind to GST (glutathione transferase)–Gemin2 (Supplementary Figure S5 at http://www.BiochemJ.org/bj/445/bj4450361add.htm). The results with full-length SMN and Gemin2 proteins were qualitatively similar to those observed with the shorter constructs, with two exceptions. Full-length SMN D36A and D44V display modest Gemin2-binding defects that were not observed in the peptide-binding assay. Because the binding and wash buffers used in the GST pull-down assays contained detergent (routinely used to reduce non-specific binding), we questioned whether some of the mutants might be particularly sensitive to the presence of detergents. To address this possibility, we repeated peptide-binding experiments for SMN D44V in the presence and absence of 0.05% NP40, both at 25°C and at 4°C. Binding of SMN26–51 D44V to Gemin2 was reduced by a factor of ~500 at 25°C in the presence of detergent, but not at 4°C in the absence of detergent (Table 2). When the two effects were combined, only weak binding could be observed over the range of our titrations, indicating a Kd value greater than 4 μM. Thus the combined effects of low temperature and detergent observed for wild-type SMN26–51 are amplified for the D44V mutant. In the absence of detergent, full-length SMN D44V binds to GST–Gemin2 with three-fold higher efficiency (Supplementary Figure S5), qualitatively similar to wild-type SMN.
Conclusions from previous studies on the SMA patient mutation D44V have been mixed. In their original report of this mutation, Sun et al. [30] showed that binding to Gemin2 is unaffected. However, Ogawa et al. [26] have reported that binding of Gemin2 to a truncated SMN containing exons 1–5 is diminished with the D44V mutant, both in mammalian two-hybrid and in vitro pull-down assays. Most recently, Zhang et al. [29] have reported complete abrogation of the SMN D44V–Gemin2 interaction, using GST pull-down experiments carried out at 4°C in the presence of detergent. Interestingly, SMN has been shown to be cleaved by calpain in vivo at a site that is located in the C-terminal half of the protein [19]. The SMN D44A mutant is less efficiently cleaved by calpain, and this effect is dependent on SMN oligomerization [31]. This suggests that Asp44, and perhaps other conserved SMN residues in this region that are not strictly required for Gemin2 binding, may be involved in higher-order interactions within the oligomeric SMN complex [26]. Indeed, the idea that the region encoded by exon 4 may interact with the Gemin2-binding domain has already been proposed [27].
A second SMA patient mutation, D30N, is located upstream of the Gemin2-binding domain of SMN. Asp30 is present in the peptide used for our NMR and binding studies, but this region is unstructured. As expected, the SMN D30N mutant bound efficiently to Gemin2 (Supplementary Figure S5), in agreement with a previous report [30]. This region of SMN also contains known phosphorylation sites at Ser28 and Ser31 [32]. Although these residues play no obvious role in binding to Gemin2, we wanted to determine whether their phosphorylation would affect binding. We observed no change in the affinity of Gemin295–280 binding to SMN26–51 peptide containing phosphoserine at positions 28 and 31 (Table 2 and Supplementary Figure S5), in agreement with previous results showing that there was no effect in the context of full-length Gemin2 [32].
Surprisingly, several of the most strongly conserved residues in the Gemin2-binding domain of SMN play no obvious role in binding to Gemin2. The most conspicuous of these are the invariant Trp34 and Asp35, as well as strongly conserved Lys41 and Asp44. In accordance with the lack of SMN–Gemin2 NOEs involving these residues, their alanine substitutions each bind with high affinity to Gemin2. Of these, only the W34A substitution shows a modest (three-fold) reduction in binding affinity compared with the wild-type construct (Figure 3B and Table 2).
Integral role of SMN in stabilizing Gemin2 structure
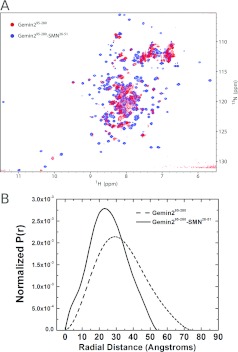
The arrangement of helices that form the SMN-binding pocket suggests that SMN may play a role in stabilizing the fold of Gemin2. Whereas helices 2–7 of Gemin2 could in principle form a globular helical bundle on their own, a substantial fraction of the interactions made by helix 1 involve SMN. It therefore seems unlikely that the Gemin2 structure observed when bound to SMN would be stable in the absence of SMN. Indeed, the HSQC spectrum of Gemin295–280 shows substantial line broadening and a loss of peak dispersion, indicating a poorly defined structure when SMN is not bound (Figure 4A). Solution scattering analysis of Gemin295–280 indicates an elongation of that domain in the absence of SMN, with an increase in radius of gyration from 19 to 25 Å and an increase in the maximum dimension from 54 to 75 Å (Figure 4B and Supplementary Figure S3). These changes could be interpreted as an unfolding of the N-terminal 30–40 residues of the Gemin2 core domain in the absence of stabilizing interactions involving SMN. A destabilization of Gemin2 in the absence of SMN could explain in part the decrease in cellular Gemin2 observed when SMN levels are experimentally reduced [33,34].
Figure 4. Properties of SMN-bound compared with unbound Gemin2.
(A) Overlay of (1H,15N)-HSQC spectra of Gemin295–280 (red) and Gemin295–280 bound to SMN26–51 (blue). (B) Comparison of the P(r) shape functions for Gemin295–280 (broken line) and Gemin295–280–SMN26–51 (continuous line). Geometric parameters derived from SAXS measurements are summarized in Supplementary Figure S3 at http://www.BiochemJ.org/bj/445/bj4450361add.htm.
Conserved surfaces of the SMN–Gemin2 complex
Many of the highly conserved residues in Gemin2 have obvious roles in interhelical packing and formation of the SMN-binding pocket. Some, however, are on or near the surface of the protein, suggesting that they may be involved in forming functional interaction surfaces. Similarly, only a subset of the highly conserved residues in SMN26–51 plays critical roles in binding to Gemin2. The others, including Trp34, Asp35, Lys41 and Asp44, contribute to the formation of a composite SMN–Gemin2 surface that we suggest is involved in some aspect of SMN complex structural organization and/or snRNP assembly (Figure 5).
Figure 5. Conserved surfaces in the SMN–Gemin2 complex.
(A) View of the SMN–Gemin2 surface in the same orientation as shown in Figure 2(B). (B) View of the opposite side of the complex. Conserved residues (boldface type in Supplementary Figure S4 at http://www.BiochemJ.org/bj/445/bj4450361add.htm) are drawn in red for SMN and dark blue for Gemin2. Other residues are in pink for SMN and pale blue for Gemin2.
Chari et al. [6] have shown that a complex composed of Gemin2 bound to the N-terminal half of SMN binds to a 6S complex composed of the Sm proteins D1, D2, E, F and G, and the Sm chaperone pICln. In the context of a larger SMN complex, pICln is displaced, leading to assembly of the snRNP core from the bound Sm pentamer, SmD3–SmB, and snRNA. While the present paper was under review, Zhang et al. [29] reported the crystal structure of a SMN26–62–Gemin2–SmD1,D2,F,E,G complex, representing a minimal version of the complex identified by Chari et al. [6]. Surprisingly, the structure reveals that the Sm pentamer is bound by Gemin2, and neither SMN nor the SMN–Gemin2 interface is involved. In the context of the present study, some of the conserved surface elements shown in Figure 5(B) can now be identified as the binding site for SmD1–SmD2 in the Sm pentamer. However, the majority of the conserved surfaces in the core SMN–Gemin2 complex are not accounted for by the Sm pentamer interaction.
Changes in the SMN–Gemin2 core upon binding the Sm pentamer
The crystal structure of SMN26–62–Gemin2 bound to a Sm pentamer [29] reveals that the globular core domain of Gemin2 (residues 89–280) is responsible for interacting with SmD1 and SmD2, whereas the extended N-terminal region (residues 25–88) wraps around the Sm pentamer and engages SmD2, SmF, SmE and SmG. The present study provides an opportunity to investigate structural changes that may occur in the SMN–Gemin2 core domain upon forming these interactions. A superposition of the crystal and NMR structures of SMN–Gemin2 is shown in two orientations in Figure 6. The lobe of Gemin2 that forms the SmD1–SmD2 binding surface, formed by the turn between α5 and α6 and the C-terminal end of α7, is most similar between the two structures (Figure 6B). This implies that Gemin2 is pre-configured for interaction with the Sm pentamer when bound to SMN.
Figure 6. Superposition of the SMN–Gemin2 complex (lowest-energy structure) with the SMN–Gemin2-Sm pentamer structure (PDB code 3S6N).
The RMSD for Cα atoms in the Gemin2 helices is 1.98 Å. (A) View of the SMN-binding surface of Gemin2. The displacement of α2 and divergence of α1, α3, α4, and α7 are indicated by arrows. Crystal lattice contacts involving Sm proteins in different biological units are indicated schematically. The Sm pentamer is omitted for clarity. (B) Orthogonal view showing the interface with SmD1–SmD2 and the stronger overlap of NMR and crystal structures in the binding region.
In contrast, regions of Gemin2 not involved in Sm protein binding show substantial differences between the two structures, giving rise to an overall RMSD of 1.98 Å for Cα atoms in the seven Gemin2 helices. The short α2 helix, for example, is rotated by ~120° and translated by ~6 Å in the crystal structure, violating several long-range NOEs observed in solution. The positions of α1, α3, α4 and α7 are also displaced in the crystal structure relative to the low-energy ensemble of NMR structures. For α7, the helices overlap well at the C-terminus, near the interface with SmD1–SmD2, but diverge as the distance from the interface increases. Since the two structures overlap well in the region where SmD1 and SmD2 are bound, it seems unlikely that discrepancies in helical positions and orientations are due to a reorganization that occurs upon binding to Sm proteins. Examination of crystal packing and the N-terminus of the Gemin2 core domain provides more plausible explanations.
The α2 helix, which is flanked by disordered loops in the crystal structure, contacts an adjacent SmG molecule which shifts its position. The α4 helix makes close contact with a neighbouring SmD1 molecule in the crystal, an interaction that also appears to influence the position of the helix. Similarly, the N-terminus of the α1 helix is buttressed by an adjacent SmD2 molecule, resulting in shifts in not only α1 but in the positions of α3 and the SMN helix as well. Each of these lattice contacts, shown schematically in Figure 6, involve symmetry-related Sm proteins that are not part of the biological unit identified by the authors, and would therefore not be present in solution. In each case, the helix positions adopted by low-energy members of the NMR ensemble would not be possible in the context of the crystal structure owing to these crystal lattice contacts.
A shift in the orientation of α7 instead appears to be a consequence of the presence of additional residues at the N-terminus of the Gemin2 core domain. In the crystal structure, these residues serve as the connector to the N-terminal segment that wraps around the Sm pentamer. As a consequence of this structural motif, Gemin2 residues 90–98 cap one end of the core domain and shift α7 towards the centre of the domain (Figure 6). In this case, the crystal structure represents a more likely position for this helix when Gemin2 is bound to the Sm pentamer. In the absence of Sm proteins, it is unclear whether a similar position would be adopted by Gemin2. Residues 95–98 are present in the construct used for the NMR structure, but a conformation similar to that observed in the crystal structure is not energetically favoured.
An analysis of NOE measurements in the context of the crystal structure model reveals 215 violations where interproton distances exceed the expected range by more than 0.6 Å. These violations involve 80 Gemin2 residues and 11 SMN residues and include 10 intermolecular Gemin2–SMN NOEs. Only a small number of violations (~20) involve residues that interact directly with SmD1 and SmD2 and might therefore be expected as a consequence of side chain remodelling during the formation of the Gemin2–Sm interface. We attribute the majority of differences between the solution and crystal structures to artefacts introduced by formation of the crystal lattice and not to structural differences between the Sm-bound and unbound states. A smaller number of violations are due to the presence of additional structure at the N-terminus of the Gemin2 core domain in the crystal structure, as described above and shown in Figure 6.
Expanding our understanding of Gemin2
The present study both complements the crystal structure model of Gemin2 bound to a Sm pentamer and expands our understanding of Gemin2 and of the Gemin2–SMN interface. Zhang et al. [29] revealed that Gemin2 is primarily responsible for tethering the pentamer of Sm proteins in preparation for snRNP assembly and the crystal structure of the Gemin2–SMN–Sm5 complex provides a molecular understanding of how this occurs. The Sm proteins and the region of Gemin2 involved in binding SmD1 and SmD2 are very well defined in the structure, with an average B-factor of 38 Å2 for the Sm pentamer. However, the remainder of the Gemin2 core domain and the Gemin2–SMN interface are less well defined, with an average B-factor of 68 Å2 and 20 out of 186 residues missing from the model due to disorder (excluding the 150–175 loop that is also poorly defined in the NMR ensemble). Given the high average B-factors, the numerous lattice contacts that would prevent the adoption of a solution-like structure in the crystal and the large number of NOE violations, we would argue that some aspects of the NMR ensemble better describe the structure of the Gemin2 core bound to SMN and that the two structural models are highly complementary with respect to the information that they provide.
Understanding the Gemin2–SMN interaction has been the major focus of the present study. These results, summarized in Figure 3, Supplementary Figure S5 and Table 2, complement and greatly expand the work of Zhang et al. [29]. Our panel of alanine substitutions within the Gemin2-binding domain of SMN is readily explained by both the NMR and crystal structure models and provides a quantitative basis for understanding the roles of individual residues in binding to Gemin2. A particularly interesting subset of these residues includes those that are highly conserved yet contribute little energetically towards interacting with Gemin2. We suggest that these amino acids contribute to the formation of a conserved, solvent-accessible surface on the SMN–Gemin2 complex that is likely to be involved in higher-order interactions within the SMN complex. Finally, comparisons of SMN-bound versus unbound Gemin2 using solution scattering data and HSQC spectra indicate that SMN plays an integral role in stabilizing the folded three-dimensional structure of Gemin2.
One aspect of the present study that contrasts with the work of Zhang et al. [29] is the functional consequence of the type III SMA patient mutation D44V. We found that a SMN peptide containing this substitution bound to Gemin2 with similar affinity as the wild-type peptide, and full-length SMN D44V binding to Gemin2 was within a factor of two relative to wild-type SMN in qualitative assays carried out in the absence of detergents. Zhang et al. [29] reported that Gemin2 binding is abrogated for this substitution on the basis of GST–Gemin2 pull-down of in vitro-translated SMN compared with SMN D44V. On the basis of the structure of the SMN–Gemin2 interface, one rationale for a role of Asp44 interacting with Gemin2 is a potential electrostatic interaction with Arg213 that would be disrupted with the D44V substitution. Our binding studies using SMN peptides indicate no disruption of the SMN–Gemin2 interaction at high salt concentrations (600 mM NaCl), arguing against an essential electrostatic component of the interaction. We also investigated the effects of temperature and non-ionic detergent and found that although neither strongly affects the SMN–Gemin2 interaction alone, the combination of 4°C and 0.05% NP40 reduces the affinity of the wild-type SMN–Gemin2 interaction by a factor of ~60 and reduces the SMN D44V–Gemin2 interaction by at least 1000-fold. These conditions were used in the pull-down assay described by Zhang et al. [29], perhaps explaining the complete loss of interaction observed, rather than a modest effect that might be more consistent with the mild SMA phenotype of the D44V variant.
Other cellular functions for Gemin2
Gemin2's involvement in snRNP assembly as part of the SMN complex is well documented. However, previous studies have also implicated Gemin2 in other biological processes, including HIV infection and homologous recombination. During HIV-1 infection of cells, Gemin2 functions with the integrase protein to stimulate viral cDNA synthesis by facilitating reverse transcriptase loading on to viral RNA. This idea is supported by a direct interaction between Gemin2 and HIV-1 integrase, as well as strong negative effects in the efficiency of HIV infection when Gemin2 expression is reduced [21]. Similarly, an interaction between Rad51 and Gemin2 has been reported in a study showing that Gemin2 enhances Rad51's activity in homologous pairing and strand exchange [22]. It is not clear in these studies how HIV integrase and Rad51 bind to Gemin2 and which functional surfaces of the protein might be exploited in carrying out the respective biological activities. However, it should now be straightforward to identify the regions of interest using the structural framework for Gemin2 reported in the present paper.
Online data
AUTHOR CONTRIBUTION
Kathryn Sarachan, Kathleen Valentine, John Gledhill, and Veronica Moorman performed NMR experiments and analysed data. Kathryn Sarachan and Kushol Gupta performed SAXS and sedimentation experiments and analysed data. Kathryn Sarachan and Gregory Van Duyne performed binding experiments and analysed data. Matthew Bernens performed protein interaction experiments. Cecilia Tommos assisted with NMR data analysis. Joshua Wand and Gregory Van Duyne designed experiments and directed the research. Kathryn Sarachan and Gregory Van Duyne wrote the paper.
ACKNOWLEDGEMENTS
We thank Renee Martin, Nisha Ninan and Tina Glisovic for helpful discussions, Robert Sharp for technical assistance, and Richard Gillilan and Liang Guo for assistance with SAXS data collection. Gianluigi Veglia kindly shared XPLOR-NIH scripts.
FUNDING
G.D.V. is an Investigator of the Howard Hughes Medical Institute. This work was supported by the NIH (National Institutes of Health) [grant numbers DK39806 (to A.J.W.) and GM079190 (to C.T.)]. CHESS is supported by the NSF (National Science Foundation) and the NIH/NIGMS (National Institute of General Medical Sciences) [grant number DMR-0936384], and the MacCHESS (Macromolecular Diffraction Facility at CHESS) resource is supported by the NIH/NCRR (National Center for Research Resources) [grant number RR-01646]. Use of the APS, an Office of Science User Facility operated for the U.S. DOE (Department of Energy) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE [contract number DE-AC02-06CH11357]. BioCAT is a research centre supported by the NIH [grant number RR-08630].
References
- 1.Liu Q., Fischer U., Wang F., Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 2.Meister G., Bühler D., Pillai R., Lottspeich F., Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 2001;3:945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- 3.Pellizzoni L., Yong J., Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- 4.Will C. L., Lührmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 5.Kolb S. J., Battle D. J., Dreyfuss G. Molecular functions of the SMN complex. J. Child Neurol. 2007;22:990–994. doi: 10.1177/0883073807305666. [DOI] [PubMed] [Google Scholar]
- 6.Chari A., Golas M. M., Klingenhäger M., Neuenkirchen N., Sander B., Englbrecht C., Sickmann A., Stark H., Fischer U. An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell. 2008;135:497–509. doi: 10.1016/j.cell.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 7.Kroiss M., Schultz J., Wiesner J., Chari A., Sickmann A., Fischer U. Evolution of an RNP assembly system: a minimal SMN complex facilitates formation of UsnRNPs in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10045–10050. doi: 10.1073/pnas.0802287105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lefebvre S., Bürglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 9.Crawford T. O., Pardo C. A. The neurobiology of childhood spinal muscular atrophy. Neurobiol. Dis. 1996;3:97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- 10.Lunn M. R., Wang C. H. Spinal muscular atrophy. Lancet. 2008;371:2120–2133. doi: 10.1016/S0140-6736(08)60921-6. [DOI] [PubMed] [Google Scholar]
- 11.Markowitz J. A., Tinkle M. B., Fischbeck K. H. Spinal muscular atrophy in the neonate. J. Obstet. Gynecol. Neonatal. Nurs. 2004;33:12–20. doi: 10.1177/0884217503261125. [DOI] [PubMed] [Google Scholar]
- 12.Parsons D. W., McAndrew P. E., Iannaccone S. T., Mendell J. R., Burghes A. H., Prior T. W. Intragenic telSMN mutations: frequency, distribution, evidence of a founder effect, and modification of the spinal muscular atrophy phenotype by cenSMN copy number. Am. J. Hum. Genet. 1998;63:1712–1723. doi: 10.1086/302160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorson C. L., Hahnen E., Androphy E. J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. U.S.A. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monani U. R., Lorson C. L., Parsons D. W., Prior T. W., Androphy E. J., Burghes A. H., McPherson J. D. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 15.Burghes A. H. M., Beattie C. E. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat. Rev. Neurosci. 2009;10:597–609. doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chari A., Paknia E., Fischer U. The role of RNP biogenesis in spinal muscular atrophy. Curr. Opin. Cell Biol. 2009;21:387–393. doi: 10.1016/j.ceb.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 17.Gabanella F., Butchbach M. E. R., Saieva L., Carissimi C., Burghes A. H. M., Pellizzoni L. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS ONE. 2007;2:e921. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker M. P., Rajendra T. K., Saieva L., Fuentes J. L., Pellizzoni L., Matera A. G. SMN complex localizes to the sarcomeric Z-disc and is a proteolytic target of calpain. Hum. Mol. Genet. 2008;17:3399–3410. doi: 10.1093/hmg/ddn234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossoll W., Jablonka S., Andreassi C., Kröning A., Karle K., Monani U. R., Sendtner M. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of β-actin mRNA in growth cones of motoneurons. J. Cell Biol. 2003;163:801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishitsuji H., Hayashi T., Takahashi T., Miyano M., Kannagi M., Masuda T. Augmentation of reverse transcription by integrase through an interaction with host factor, SIP1/Gemin2 Is critical for HIV-1 infection. PLoS ONE. 2009;4:e7825. doi: 10.1371/journal.pone.0007825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takizawa Y., Qing Y., Takaku M., Ishida T., Morozumi Y., Tsujita T., Kogame T., Hirota K., Takahashi M., Shibata T., et al. GEMIN2 promotes accumulation of RAD51 at double-strand breaks in homologous recombination. Nucleic Acids Res. 2010;38:5059–5074. doi: 10.1093/nar/gkq271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selenko P., Sprangers R., Stier G., Bühler D., Fischer U., Sattler M. SMN tudor domain structure and its interaction with the Sm proteins. Nat. Struct. Biol. 2001;8:27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- 24.Sprangers R., Groves M. R., Sinning I., Sattler M. High-resolution X-ray and NMR structures of the SMN Tudor domain: conformational variation in the binding site for symmetrically dimethylated arginine residues. J. Mol. Biol. 2003;327:507–520. doi: 10.1016/s0022-2836(03)00148-7. [DOI] [PubMed] [Google Scholar]
- 25.Paushkin S., Gubitz A. K., Massenet S., Dreyfuss G. The SMN complex, an assemblyosome of ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:305–312. doi: 10.1016/s0955-0674(02)00332-0. [DOI] [PubMed] [Google Scholar]
- 26.Ogawa C., Usui K., Aoki M., Ito F., Itoh M., Kai C., Kanamori-Katayama M., Hayashizaki Y., Suzuki H. Gemin2 plays an important role in stabilizing the survival of motor neuron complex. J. Biol. Chem. 2007;282:11122–11134. doi: 10.1074/jbc.M609297200. [DOI] [PubMed] [Google Scholar]
- 27.Young P. J., Man N. T., Lorson C. L., Le T. T., Androphy E. J., Burghes A. H., Morris G. E. The exon 2b region of the spinal muscular atrophy protein, SMN, is involved in self-association and SIP1 binding. Hum. Mol. Genet. 2000;9:2869–2877. doi: 10.1093/hmg/9.19.2869. [DOI] [PubMed] [Google Scholar]
- 28.Holm L., Sander C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 29.Zhang R., So B. R., Li P., Yong J., Glisovic T., Wan L., Dreyfuss G. Structure of a key intermediate of the SMN complex reveals Gemin2's crucial function in snRNP assembly. Cell. 2011;146:384–395. doi: 10.1016/j.cell.2011.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Y., Grimmler M., Schwarzer V., Schoenen F., Fischer U., Wirth B. Molecular and functional analysis of intragenic SMN1 mutations in patients with spinal muscular atrophy. Hum. Mutat. 2005;25:64–71. doi: 10.1002/humu.20111. [DOI] [PubMed] [Google Scholar]
- 31.Fuentes J. L., Strayer M. S., Matera A. G. Molecular determinants of survival motor neuron (SMN) protein cleavage by the calcium-activated protease, calpain. PLoS ONE. 2010;5:e15769. doi: 10.1371/journal.pone.0015769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grimmler M., Bauer L., Nousiainen M., Körner R., Meister G., Fischer U. Phosphorylation regulates the activity of the SMN complex during assembly of spliceosomal U snRNPs. EMBO Rep. 2005;6:70–76. doi: 10.1038/sj.embor.7400301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jablonka S., Holtmann B., Meister G., Bandilla M., Rossoll W., Fischer U., Sendtner M. Gene targeting of Gemin2 in mice reveals a correlation between defects in the biogenesis of U snRNPs and motoneuron cell death. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10126–10131. doi: 10.1073/pnas.152318699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shpargel K. B., Matera A. G. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc. Natl. Acad. Sci. U.S.A. 2005;102:17372–17377. doi: 10.1073/pnas.0508947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neidhardt F., Bloch P., Smith D. Culture medium for enterobacteria. J. Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reference deleted.
- 37.Diercks T., Coles M., Kessler H. An efficient strategy for assignment of cross-peaks in 3D heteronuclear NOESY experiments. J. Biomol. NMR. 1999;15:177–180. doi: 10.1023/A:1008367912535. [DOI] [PubMed] [Google Scholar]
- 38.Ross A., Czisch M., Cieslar C., Holak T. Efficient methods for obtaining phase-sensitive, gradient-enhanced HMQC data. J. Biol. NMR. 1993;3:215–224. [Google Scholar]
- 39.Vuister G., Clore G., Gronenborn A., Powers R., Garrett D., Tschudin R., Bax A. Increased resolution and improved spectral quality in four-dimensional 13C/13C-separated HMQC-NOESY-HMQC spectra using pulsed field gradients. J. Mag. Res. B. 1993;101:210–213. [Google Scholar]
- 40.Clore G. M., Gronenborn A. M., Nilges M., Ryan C. A. Three-dimensional structure of potato carboxypeptidase inhibitor in solution. A study using nuclear magnetic resonance, distance geometry, and restrained molecular dynamics. Biochemistry. 1987;26:8012–8023. doi: 10.1021/bi00398a069. [DOI] [PubMed] [Google Scholar]
- 41.Shen Y., Delaglio F., Cornilescu G., Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ottiger M., Delaglio F., Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J. Magn. Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 43.Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.